Abstract
Herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) resides in the virus envelope in an oligomeric form and plays an essential role in virus entry into susceptible host cells. The oligomerizing domain is a movable element consisting of amino acids 626 to 653 in the gB external domain. This domain contains a single cysteine residue at position 633 (Cys-633) that is predicted to form an intramolecular disulfide bridge with Cys-596. In this study, we examined gB oligomerization, processing, and incorporation into mature virus during infection by two mutant viruses in which either the gB Cys-633 [KgB(C633S)] or both Cys-633 and Cys-596 [KgB(C596S/C633S)] residues were mutated to serine. The result of immunofluorescence studies and analyses of released virus particles showed that the mutant gB molecules were not transported to the cell surface or incorporated into mature virus envelopes and thus infectious virus was not produced. Immunoprecipitation studies revealed that the mutant gB molecules were in an oligomeric configuration and that these mutants produced hetero-oligomers with a truncated form of gB consisting of residues 1 to 43 and 595 to 904, the latter containing the oligomerization domain. Pulse-chase experiments in combination with endoglycosidase H treatment determined that the mutant molecules were improperly processed, having been retained in the endoplasmic reticulum (ER). Coimmunoprecipitation experiments revealed that the cysteine mutations resulted in gB misfolding and retention by the molecular chaperones calnexin, calreticulin, and Grp78 in the ER. The altered conformation of the gB mutant glycoproteins was directly detected by a reduction in monoclonal antibody recognition of two previously defined distinct antigenic sites located within residues 381 to 441 and 595 to 737. The misfolded molecules were not transported to the cell surface as hetero-oligomers with wild-type gB, suggesting that the conformational change could not be corrected by intermolecular interactions with the wild-type molecule. Together, these experiments confirmed that a disulfide bridge involving Cys-633 and Cys-596 is not essential for oligomerization but rather is required for proper folding and maintenance of a gB domain essential to complete posttranslational modification, transport, and incorporation into mature virus particles.
Herpes simplex virus type 1 (HSV-1) encodes at least 11 glycoproteins, 4 of which, glycoprotein B (gB), gD, gH, and gL, are essential for virus attachment and entry in cell culture (7, 9, 17, 29, 35). gB is among the most highly conserved herpesvirus structural components, suggesting a common and essential role in the life cycle of the Herpesviridae (1, 40). gB is expressed as an early gene product that persists following viral DNA synthesis, indicating that it is a member of the γ1 temporal class of genes (for a review, see reference 44). Molecular genetic studies of the functional domains of gB have revealed that it contains structures involved in controlling or defining the rate of virus entry, oligomer formation, temperature-dependent stability, nuclear membrane localization, antigenic sites, glycosylation, membrane anchorage, syncytial plaque formation, and binding to the heparan sulfate receptor (4, 10, 11, 13, 15, 19, 24, 26, 36, 38, 43). Despite these detailed studies, unanswered questions remain regarding gB function, processing, and incorporation into mature virions. For example, the molecular events leading to the incorporation of gB into mature virus envelopes and the mechanism through which gB cooperates with other viral envelope components in the process of virus penetration continue to elude our full understanding.
The gB structural gene sequence encodes 904 amino acids (6). Biochemical analysis of gB has demonstrated that it contains a 30-residue N-terminal signal sequence that is cleaved during processing, a 697-residue external domain, a 68-residue transmembrane domain that is predicted to span the membrane three times, and a 109-residue cytoplasmic domain (6, 8, 12, 40). The molecule forms minimally a homodimer during or soon after the process of translation and is subsequently glycosylated through a series of successive stages in the rough endoplasmic reticulum (ER) involving carbohydrate addition to six consensus sites for N-linked glycosylation (10, 11, 32). gB is further processed in the Golgi complex and is transported to the cell surface of infected cells. Inhibitors of Golgi processing (e.g., monensin) do not block functional gB incorporation into cellular infectious virus particles (31). The pathway for gB insertion into the virus envelope is thought to involve diffusion or active transport of immature gB to the inner nuclear membrane where it is initially acquired, along with the other envelope glycoproteins during the process of virion budding (13, 20, 21, 47). At this point, two different pathways are proposed. First, the immature envelope glycoproteins are transported to the Golgi complex, where the precursor glycoproteins are modified in situ as the enveloped particle moves through the Golgi compartment (31, 47). Alternatively, a second model for viral egress involves fusion of the membrane acquired at the inner nuclear membrane with the ER membrane (outer nuclear membrane), releasing nucleocapsids into the cytoplasm which are subsequently reenveloped with Golgi-derived vacuoles containing processed glycoproteins (41, 45).
HSV-1 gB contains 10 cysteine residues which are highly conserved among the gB homologs of different herpesviruses, suggesting a critical role for these cysteines in organizing the three-dimensional structure into stable functional domains (42). Recently, the disulfide bridges among the cysteine residues of HSV-2 gB were analyzed by microsequencing of high-pressure liquid chromatography-purified tryptic peptides in which disulfide bonds were shown to form between cysteines 1 and 8, 2 and 7, 5 and 6, and 9 and 10 (39). A similar pattern of disulfide bond formation is likely to occur in HSV-1-encoded gB, in view of the high degree of sequence conservation between type 1 and type 2 gB (39). The functional form of gB is an oligomer (8), and we recently reported that oligomer formation required a 28-amino-acid movable domain consisting of residues 626 to 653 (34). Because cysteine 10 was located within this domain at residue 633, it was substituted for serine at this position to explore the possibility that the disulfide bridge formed with cysteine 9 (residue 596) might be essential to facilitating the interactions leading to oligomerization. Although oligomerization was reduced within this gB truncated molecule, the domains were functional in the absence of one or both of these cysteine residues (34). Whether the full-length cysteine mutant forms of gB would oligomerize during infection remained to be determined.
To further explore the role of Cys-633 and Cys-596 in gB full-length function and infectious particle production, mutant viruses altered in one or both of these gB residues were studied in cell culture infections. The results of this investigation showed that mutant gB molecules in which serine was substituted for Cys-633 or Cys-633 and Cys-596 were not found in mature virus particles despite their ability to form oligomers. Further analysis of the processing and trafficking of the mutant gB forms demonstrated that they were incompletely processed and remained bound to calnexin, Grp78, and calreticulin, a complex of chaperone molecules for HSV glycoproteins, during processing in the ER. These findings demonstrated that the disulfide bridge formed between cysteines 9 and 10 is essential for proper folding of gB, a prerequisite to gB processing and incorporation into mature virions.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were obtained from the American Type Culture Collection (Rockville, Md.). A Vero cell line stably transfected with the HSV-1 genes encoding gB and ICP 18.5 (A1 [46]) (kindly provided by Fred L. Homa, Upjohn, Kalamazoo, Mich.) was used to propagate a mutant virus, KΔ4BX, deleted for both essential genes (16). KΔ4BX virus was also used as a gB null virus when propagated on a Vero derivative cell line stably transfected with the ICP 18.5 gene (C1 [46]). A Vero cell line stably transfected with the HSV-1 gene encoding gB (D6 [9]) was used to propagate and determine the titers of the gB mutant viruses KgB(C633S) and KgB(C596S/C633S). The cell lines were maintained at 37°C in Dulbecco’s modified essential medium (Gibco-BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum.
Construction of mutant gB plasmids.
Previously, nucleotide sequences encoding a cysteine residue at position 633 of a gB molecule were substituted for nucleotides encoding a serine residue and inserted into pB, a plasmid encoding a truncated gB molecule under control of the SP6 promoter (34). DNA sequence encoding this cysteine mutation was transferred to a plasmid encoding the full-length gB molecule under the control of the SP6 promoter. To transfer this substitution to a full-length gB molecule driven by its natural promoter (encoded by plasmid pKBXX) (8), the fragment containing the serine-for-cysteine substitution was excised as a 3,084-bp BamHI-NotI fragment and cloned into pKBXX digested with the same endonucleases, replacing wild-type sequence with sequences encoding the cysteine substitution and resulting in pKBXX(C633S). The additional substitution of nucleotide sequences encoding a cysteine residue at position 596 for sequences encoding a serine residue was generated by the PCR overlap extension technique (28). pKBXX(C633S) served as template for the creation of pKBXX(C596S/C633S), using primers 1 (5′-GCGGCTGTAGCTAGCCCC-3′), 2 (5′-GCCTCGGTCACCGTGGGC-3′), 3 (5′-CCCGGGGCTAGTTACAGC-3′), and 4 (5′-GCGCATGACCATGTCGG-3′). Amplification with primers 1 and 2 yielded a 200-bp fragment, and amplification with primers 3 and 4 yielded an 800-bp fragment. Amplification of the two PCR products with primers 1 and 4, the external primers, yielded a 1,000-bp fragment which was cloned into pKBXX(C633S) in order to obtain pKBXX(C596S/C633S).
Construction of epitope-tagged mutant gB molecules.
The nonapeptide epitope of influenza virus hemagglutinin (HA) (49) was inserted as previously described (34) at a unique NotI site between codons 41 and 42 of the gB sequence (pKBXX), resulting in pKBXXHA. The HA epitope was transferred to pKBXX(C633S) and pKBXX(C596S/C633S) to create pKBXXHA(C633S) and pKBXXHA(C596S/C633S), respectively.
Construction and isolation of KgB(C633S) and KgB(C596S/C633S) mutant viruses.
Mutant HSV-1 viruses were constructed by standard methods for efficient marker transfer (16) by using LipofectAmine reagent (Gibco-BRL) for cotransfection. KgB(C633S) and KgB(C596S/C633S) mutant viruses were constructed by cotransfection of plasmid pKBXX(C633S) or pKBXX(C596S/C633S) with KΔ4BX viral DNA on the complementing A1 cell line. The mutant viruses were selected for growth on D6 cells and screened by Southern blot hybridization using a gB probe (Fig. 1) and plaque purified three times.
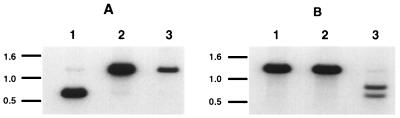
FIG. 1.
Southern blot analysis of the mutant KgB(C633S) and KgB(C596S/C633S) viruses. An NcoI-digested 1,305-bp fragment from gB was 32P labeled and used to Southern blot viral DNA from KOS (lanes 1), KgB(C633S) (lanes 2), and KgB(C596S/C633S) (lanes 3). These viral DNA samples were digested with NcoI and ApaLI (A) or RmaI (B). (A) The [32P]gB probe hybridized with two fragments of 639 and 666 bp in KOS-digested DNA (lane 1, two indistinguishable bands on this gel) and hybridized with one fragment of 1,305 bp in KgB(C633S)- and KgB(C596S/C633S)-digested DNA, indicating the loss of an ApaLI restriction site marking the gB mutation at amino acid 633. (B) The same [32P]gB probe hybridized to a fragment of 1,259 bp in KOS-digested DNA (lane 1) as well as KgB(C633S)-digested DNA (lanes 2) and hybridized to two fragments of 724 and 535 bp in KgB(C596S/C633S)-digested DNA (lane 3), marking the insertion of an RmaI restriction site and the gB mutation at amino acid 596. Positions of migration of the DNA standard are marked in kilobases on each panel.
Immunoprecipitation analysis.
Virions released from cells and infected cell lysates were prepared as follows. Confluent 60-mm-diameter dishes of Vero cells at 37°C were methionine-cysteine starved for 4 h. Monolayers were infected with viruses at a multiplicity of infection (MOI) of 10 and incubated for 48 h in the presence of 100 μCi of [35S]methionine-cysteine (NEN-Dupont, Boston, Mass.). Media containing radiolabeled virions were cleared from cell debris by low-speed centrifugation, and the supernatants were brought to a concentration of 1× lysis buffer, using a 10× lysis buffer stock solution (200 mM Tris-HCl [pH 8.0], 1.5 M NaCl, 10% Triton X-100, 10 mM Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK]). Detergent-denatured virions were sonicated and cleared by centrifugation, and the supernatants were subjected to immunoprecipitation. Infected cell monolayers were scraped in Tris-buffered saline (TBS), the cell pellet was lysed in 1× lysis buffer, sonicated, and cleared by centrifugation, and the supernatants were subjected to immunoprecipitation. Immunoprecipitation was carried out at 4°C using gB-specific monoclonal antibodies (MAbs) (gB-1 pool or epitope-specific gB antibodies [36]), a gB MAb which recognizes only dimeric gB (DL16; kindly provided by Gary H. Cohen and Roselyn J. Eisenberg, Philadelphia, Pa.), or the HA epitope-specific antibody (12ca5J; Babco, Berkeley, Calif.). The immunocomplexes were incubated with protein A-Sepharose (Sigma) for 1 h, centrifuged, washed with lysis buffer, and resuspended in Laemmli buffer (33) before separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Following electrophoresis, gels were fixed, treated with En3Hance solution (NEN-Dupont), vacuum dried, and exposed to X-Omat-XAR film (Kodak, Rochester, N.Y.). Films were developed and quantitated by using the 1-D Scan and Report program (Biomed Instruments).
Analysis of released virions.
For detection of virions released from infected cells, supernatants harvested 24 h postinfection (p.i.) were cleared of debris by brief centrifugation at 3,000 rpm for 2 min and centrifuged at 20,000 × g for 45 min to pellet the virus. The virus pellets were resuspended in 0.5 ml of complete medium and applied to 30 to 65% sucrose gradients in 0.5× TBS, which were centrifuged at 23,000 × g in an SWTI-41 rotor (Beckman) for at least 3 h. The virion band, visible as a white cloudy ribbon, was extracted and subjected to immunoprecipitation under native conditions (without detergent) with gC-1 (37a)- or gB-1-specific MAbs in order to capture released mature virions containing that envelope glycoprotein. Protein A complexes were washed with complete medium in order to maintain the integrity of the virus structure, and viral DNA extracted from the antibody-captured virus was digested with BamHI and Southern blotted as described above, or the captured virus was resuspended in Laemmli buffer before separation by SDS-PAGE and Western blotting with a VP5 polyclonal antibody (kindly provided by Gary H. Cohen and Roselyn J. Eisenberg). An anti-rabbit antibody conjugated with horseradish peroxidase (Sigma, St. Louis, Mo.) was used in conjunction with the ECL (enhanced chemiluminescence) system (Amersham, Arlington Heights, Ill.) for detection of bound VP5-specific antibody.
Immunofluorescence.
At 8 h p.i. or 24 h posttransfection, Vero cell monolayers were fixed in ice-cold methanol or left untreated prior to incubation at 34°C with the gB-1 MAb pool (36) or a polyclonal antibody specific for the carboxy-terminal domain of the gB molecule (kindly provided by Thomas C. Holland, Wayne State University, Detroit, Mich.). Monolayers were washed and incubated with a cy3-conjugated anti-mouse or anti-rabbit antibody (Jackson Laboratory, West Grove, Pa.). The monolayers were fixed with ice-cold methanol before photography with a Nikon model 211910 TMS microscope.
Oligomerization of mutant gB molecules with a truncated gB molecule.
Monolayers of Vero cells were transfected with plasmid pKΔ5C and/or plasmid pKBXXHA, pKBXXHA(C633S), or pKBXXHA(C596S/C633S), using LipofectAmine reagent. Twenty-four hours posttransfection, monolayers were infected with KΔ4BX virus at an MOI of 10 in the presence of [35S]methionine-cysteine. Eight hours p.i., cells were harvested in lysis buffer A (150 mM NaCl, 50 mM Tris-HCl [pH 6.8], 1% Nonidet P-40, 1 mM TLCK) and subjected to immunoprecipitation with a gB carboxy-terminal antibody or an influenza HA MAb as described above. Samples were then separated by SDS-PAGE and exposed to X-Ray film.
Endo H treatment of mutant gB molecules.
Confluent monolayers of Vero cells were infected with KOS, KgB(C633S), or KgB(C596S/C633S) virus at an MOI of 10. Six hours p.i., cells were pulsed for 10 min with [35S]methionine-cysteine and harvested in lysis buffer A (150 mM NaCl, 50 mM Tris-HCl [pH 6.8], 1% Nonidet P-40, 1 mM TLCK) or chased for indicated times before harvesting. Lysates were sonicated, centrifuged, and subjected to immunoprecipitation as previously described. Protein A-Sepharose complexes were further washed with phosphate-buffered saline and resuspended in 20 μl of endoglycosidase H (endo H) buffer (0.1 M sodium citrate [pH 5.5], 0.1% SDS) in the presence or absence of 1 mU of endo H (Boehringer Mannheim, Indianapolis, Ind.). Following overnight incubation at 37°C, samples were processed for separation by SDS-PAGE as described above.
Inhibition of glucosidases I and II by CST.
Vero cells were infected at an MOI of 10 with KOS, KgB(C633S), or KgB(C596S/C633S) virus. Five hours p.i., medium was supplemented with 5 mM castanospermine (CST; Sigma) and infected cells were further incubated at 37°C for 45 min. Monolayers were pulsed and chased in the presence of CST, harvested, and immunoprecipitated as described above.
Coimmunoprecipitation of gB molecules with calnexin, calreticulin, and Grp78, using anticalnexin, anticalreticulin, and anti-Grp78 antibodies.
Vero cells were infected with KOS, KgB(C633S), or KgB(C596S/C633S) virus at an MOI of 10. Six hours p.i., cells were pulsed and chased for indicated times, harvested in lysis buffer A, immunoprecipitated with the gB-1 MAb pool or anticalnexin, anticalreticulin, or anti-Grp78 antibodies (StressGen, Victoria, British Columbia, Canada), and separated by SDS-PAGE as described above. Presence of gB molecules within anticalnexin, anti-Grp78, or anticalreticulin coimmunoprecipitates was confirmed by Western blotting using the polyclonal antibody against the carboxy-terminal domain of gB. A second antibody against rabbit immunoglobulin G molecules labeled with alkaline phosphatase was used to detect the presence of the primary antibody against gB and revealed with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate substrates (Promega, Madison, Wis.).
RESULTS
Construction of gB mutant viruses containing altered cysteine residues.
Two mutant viruses were produced by marker rescue for these studies. KgB(C633S) contained a single mutation in which a serine was substituted for cysteine at position 633, and KgB(C596S/C633S) contained a double mutation in which serine residues were substituted for cysteine at both amino acids 596 and 633. The substitution of serine for cysteine at position 633 (Cys-633) resulted in the loss of an ApaLI site, and the substitution at position 596 (Cys-596) resulted in the creation of a novel RmaI site. To confirm these mutant virus genotypes, KgB(C633S) and KgB(C596S/C633S) viral DNA samples were digested with NcoI and ApaLI (Fig. 1A) or with NcoI and RmaI (Fig. 1B) and analyzed by Southern blotting using a 32P-labeled gB probe consisting of an NcoI fragment overlapping both mutation sites. The gB probe hybridized to two fragments, 639 and 666 bp (indistinguishable on this gel [Fig. 1A, lane 1]), from wild-type KOS viral DNA digested with NcoI and ApaLI, while only a 1,305-bp fragment was detected with the gB probe from either similarly digested mutant viral DNA (Fig. 1A, lanes 2 and 3), confirming the serine substitution at Cys-633. The labeled gB probe hybridized to a 1,259-bp fragment of KOS and KgB(C633S) DNA digested with NcoI and RmaI (Fig. 1B, lanes 1 and 2, respectively) but hybridized to two fragments of 535 and 724 bp when KgB(C596S/C633S) DNA was digested with these same endonucleases (Fig. 1B, lane 3), confirming that the double mutant had a serine substitution at the second location.
The single- and double-cysteine mutations resulted in the production of noninfectious viruses lacking gB.
The gB mutant viruses were unable to support infectious virus production on a noncomplementing Vero cell line, indicating that the cysteine-to-serine mutations were lethal; therefore, the gB mutant viruses were propagated on gB-complementing Vero (D6) cells. The inability of both mutant viruses to form plaques on normal Vero cells indicated that the formation of disulfide bridges involving these two cysteine residues was essential for functional gB and infectious virus production. This inability could have resulted from enveloped particles either devoid of gB or containing nonfunctional gB. To distinguish between these possibilities, we determined whether extracellular virus contained the mutant forms of gB and whether gB was present in an oligomeric form.
Vero cells infected with either cysteine mutant, a gB null mutant (KΔ4BX grown on C1 cells), or wild-type KOS virus were radiolabeled with [35S]methionine-cysteine for 24 h. Purified extracellular viruses were extracted with nonionic detergent, and gB was immunoprecipitated from these extracts by using a pool of gB-specific MAbs (Fig. 2A, lanes 1, 3, 5, and 7) and a MAb, DL16 (Fig. 2A, lanes 2, 4, 6, and 8), reactive with a gB epitope present only in the oligomeric form of gB. As shown in Fig. 2A, KOS virions contained gB in a mature oligomeric form in the virus envelope (lanes 1 and 2), since gB was immunoprecipitated equally by both MAbs. As expected, no gB polypeptide was immunoprecipitated from gB deletion mutant virus KΔ4BX (lanes 3 and 4). Similarly, the cysteine mutants [KgB(C633S) and KgB(C596S/C633S)] failed to incorporate the mutant gB molecules into virion envelopes (lanes 5 or 6 and 7 or 8, respectively), which accounts for their lack of infectivity on noncomplementing Vero cells.
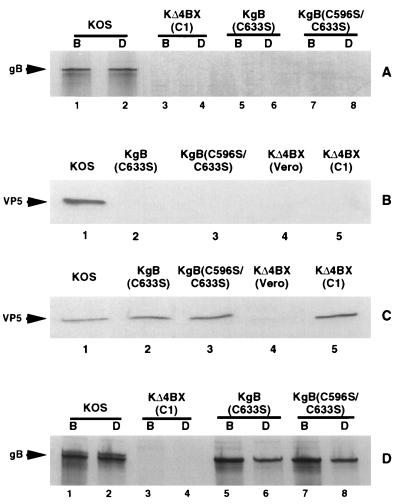
FIG. 2.
Release of virions deficient in gB molecules from cells infected with gB mutant viruses. Vero cells were infected with KOS, KgB(C633S), KgB(C596S/C633S), or KΔ4BX virus, and C1 cells were infected with KΔ4BX virus, in the presence (A and D) or absence (B and C) of [35S]methionine-cysteine. Forty hours p.i., supernatants containing virions (A) and infected cells (D) were harvested, solubilized, and subjected to immunoprecipitation with gB-1 MAb pool (B; lanes 1, 3, 5, and 7) or dimeric antibody (D; lanes 2, 4, 6, and 8), and the protein A-Sepharose immunocomplexes were separated by SDS-PAGE. Twenty-four hours p.i., supernatants were harvested and immunoprecipitated with gB-1 (B) or gC-1 (C) MAb pools before separation by SDS-PAGE and Western blotting with a VP5 polyclonal antibody. The positions of gB and VP5 are indicated by arrows at the left.
To verify that the gB mutations did not preclude virus release, supernatants of infected cells were examined for the presence of virions. Since the gB mutant viruses were noninfectious, immunobiochemical methods were used to detect their presence. We reasoned that immunoprecipitation using glycoprotein-specific antibodies in combination with protein A-Sepharose beads would capture the virus particles if the glycoprotein targeted by the antibody was present in the virus envelope. The presence of mature virus in these immune complexes was confirmed by detection of both viral DNA and the major capsid protein VP5. As shown in Fig. 2B, immune complexes of virus particles from KOS-, KgB(C633S)-, and KgB(C596S/C633S)-infected Vero cell supernatants and from KΔ4BX-infected C1 cell supernatant (C1 cells provide ICP 18.5) demonstrated release of virions from the gB mutants [KgB(C633S) and KgB(C596S/C633S)] or gB-deleted (C1 cells infected with KΔ4BX) infected cells. As expected, no VP5 was detected in immunoprecipitated samples from Vero cells infected with KΔ4BX, due to the absence of the ICP 18.5 gene product, essential for virus egress (46). In agreement with Fig. 2A, Fig. 2C demonstrates that released virions from cells infected with wild-type virus contained gB, while released virions from infections with KgB(C633S) and KgB(C596S/C633S) were deficient in gB, as evidenced by the lack of VP5 in purified released virions immunoprecipitated with the gB-1 MAb pool. As expected, no VP5 was detected in immunoprecipitated samples from KΔ4BX infections on Vero (ICP 18.5, gB deletions) or C1 (gB deletion, ICP 18.5 provided in trans) cells (Fig. 2C) due to the absence of gB. Southern blot analysis for the presence of viral DNA in glycoprotein immunoprecipitated virions gave results similar to those described above for the presence of VP5 (data not shown).
The finding that the cysteine mutant gB molecules were not detected in mature virions could have resulted from the inability either to form oligomers or to undergo proper processing and/or intracellular transport. To distinguish among these possibilities, we first determined whether the mutant molecules were capable of forming oligomers during infection (Fig. 2D). Cells infected with mutant or wild-type virus were radiolabeled as described above, and detergent-soluble infected cell extracts were treated with the pool of gB MAbs and the oligomer-specific antibody DL16 to determine the amount of gB present in an oligomeric form. The results of the SDS-PAGE analysis of the immune complexes shown in Fig. 2D demonstrated that wild-type and cysteine mutant viruses [KgB(C633S) and KgB(C596S/C633S)] produced gB (lanes 1, 5, and 7, respectively) in a dimeric form (lanes 2, 6, and 8, respectively). Immunoprecipitation of mutant gB molecules from cells infected with KgB(C633S) or KgB(C596S/C633S) demonstrated that the quantity of mutant gB immunoprecipitated by MAb DL16 (lanes 6 and 8, respectively) was approximately 50% less abundant than the quantity of mutant gB immunoprecipitated with the gB-1 MAb pool (lanes 5 and 7, respectively), as quantified by densitometric analysis. No gB was detected following immunoprecipitation from C1 cells infected with KΔ4BX (lanes 3 and 4) with either MAb, demonstrating the specificity of the antibodies used. These data showed that the cysteine mutant viruses [KgB(C633S) and KgB(C596S/C633S)] encoded a gB molecule capable of forming homo-oligomers, albeit apparently less efficiently than a wild-type gB molecule. Alternatively, the reduced amount of oligomeric gB detected by antibody DL16 could be due to a change in the structure formed by the mutant oligomer-component epitopes, resulting in reduced quantities of immunoprecipitated product.
To distinguish between these possibilities, a coimmunoprecipitation assay was performed to detect oligomeric complexes of proteins that did not involve antibody recognition of a dimer-specific epitope. In these experiments, Vero cells were cotransfected with plasmids which expressed an oligomerization-competent, truncated form of gB and either the full-length wild-type or cysteine mutant gB molecule. Following infection by a gB null mutant virus in order to transactivate gB expression from the plasmids, the formation of hetero-oligomers between the truncated oligomer partner and normal-length wild-type or cysteine mutant gB molecule could be detected by antibodies reactive with the full-length member of the oligomer pair. If oligomerization occurred, the immune complexes would also contain the truncated second member of the pair. Because the partners were of different lengths, they could be readily distinguished by their different molecular ratios following SDS-PAGE analysis of the immunoprecipitates.
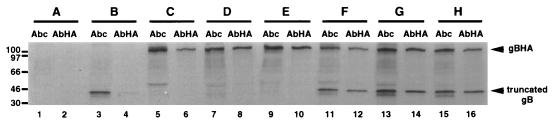
Vero cells were cotransfected with a plasmid that expressed an N-terminal HA-tagged full-length wild-type (gBHA) or cysteine mutant gB molecule [gBHA(C633S) or gBHA(C596S/C633S)] and plasmid pKΔ5C, which expressed a truncated oligomerizing form of gB deleted for residues 43 to 596 (26). The cotransfected cells were subsequently infected with KΔ4BX, and 6 h p.i., the cell monolayers were radiolabeled with [35S]methionine-cysteine for 3 h before solubilization with a nonionic detergent. As shown in Fig. 3, the truncated as well as full-length wild-type and mutant gB molecules were detected by immunoprecipitation using a polyclonal antiserum (Abc) that reacted with their carboxy termini (lanes 3, 5, 7, and 9) while the full-length HA epitope-bearing molecules were detected with an HA-reactive (AbHA) MAb (lanes 6, 8, and 10). No labeled protein was immunoprecipitated from cells that were mock transfected and infected with the null mutant virus (lanes 1 and 2). We first confirmed that wild-type gB formed oligomers with truncated gB by demonstrating that both molecules were present in the immune complexes when the AbHA antibody was used to immunoprecipitate the HA-tagged gB molecule cotransfected with the truncated gB molecule (lane 12). The two proteins were identified by analysis of immunoprecipitates resulting from the use of the Abc antibody (lane 11). We then tested the ability of the HA-tagged single- and double-cysteine mutant forms of gB to oligomerize with the truncated gB molecule. As seen in lanes 14 and 16, the mutant molecules formed oligomers to an extent similar to the wild-type gB molecule, confirming the results using the oligomer specific antibody (Fig. 2D). Quantification by densitometry of the truncated gB molecule coimmunoprecipitated with the full-length wild-type or cysteine-mutated gB molecules demonstrated that the cysteine-mutated gB molecules oligomerized as efficiently as the wild type with the truncated gB peptide and suggested that because less mutant homo-oligomers were immunoprecipitated using antibody DL16 (Fig. 2D), this oligomer-dependent epitope was altered by one or two cysteine mutations. Together, the results of these studies indicate that oligomerization of gB did not require the formation of a disulfide bridge between Cys-596 and Cys-633 and the failure to incorporate gB into extracellular virus was not due to a defect in oligomerization.
FIG. 3.
Ability of mutant gB molecules to form hetero-oligomers. Vero cell monolayers were mock transfected (lanes 1 and 2) or transfected individually with a plasmid encoding a truncated gB molecule (pKΔ5C; lanes 3 and 4), HA-tagged gB molecule (gBHA; lanes 5 and 6), or recombinant HA-tagged gB molecules [gBHA(C633S) or gBHA(C596S/C633S); lanes 7 and 8 or lanes 9 and 10, respectively]. Monolayers were also cotransfected with a plasmid encoding the truncated gB molecule (pKΔ5C) and gBHA (lanes 11 and 12), gBHA(C633S) (lanes 13 and 14), or gBHA(C596S/C633S)S (lanes 15 and 16). Twenty-four hours posttransfection, cells were infected with a gB-deleted virus (KΔ4BX) at an MOI of 10 in the presence of [35S]methionine-cysteine. Seven hours p.i., cell monolayers were harvested and immunoprecipitated with a polyclonal antibody directed against the carboxy-terminal region of gB (Abc; lanes 1, 3, 5, 7, 9, 11, 13, and 15) or anti-HA antibody (AbHA; lanes 2, 4, 6, 8, 10, 12, 14, and 16). The protein A-Sepharose immunocomplexes were separated by SDS-PAGE. The positions of the HA-tagged wild-type and mutant gB molecules (all gBHA) as well as the truncated gB molecule are marked at the right. Molecular size markers are indicated in kilodaltons at the left of the figure.
The cysteine mutant forms of gB are not transported to the surface membrane of infected cells.
Because the cysteine mutant gB molecules were translated and formed oligomers but were absent in mature virus particles, it appeared likely that a defect in processing and/or trafficking of these mutant molecules accounted for their lack of incorporation into virions. In addition, immunoprecipitates from KOS-infected cells showed two forms of gB which are typically the major gB precursor and final products whereas the cysteine mutant viruses appeared to produce only the precursor form, further suggesting that the mutant molecules were incompletely processed (Fig. 2D).
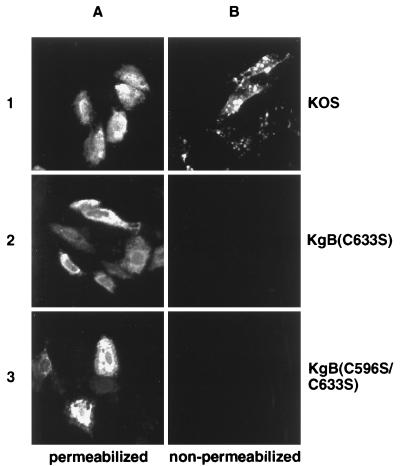
To further examine the processing of the mutant gB molecules, we first determined whether these molecules were transported to the cell surface of infected cells (Fig. 4). Vero cells were infected with KOS, KgB(C633S), or KgB(C596S/C633S) (lane 1, 2, or 3, respectively) for 24 h, and the presence of gB on the infected cell surface membrane was detected by indirect immunofluorescence using a gB-specific MAb pool. The results, shown in Fig. 4B, demonstrated that only wild-type KOS virus-infected cells expressed gB on the cell surface, while all infections [KOS, KgB(C633S) and KgB(C596S/C633S)] contained gB in the cytoplasm, as demonstrated by indirect immunofluorescence using methanol-permeabilized infected cells (Fig. 4A). These data clearly showed that the cysteine mutations interfere with normal trafficking of gB to the cell surface.
FIG. 4.
Failure of the mutant gB molecules to be transported to the cell surface of infected cells. Ten hours p.i., monolayers of Vero cells infected with KOS (row 1), KgB(C633S) (row 2), or KgB(C596S/C633S) (row 3) virus at an MOI of 10 were fixed in ice-cold methanol (A) or left untreated (B). Monolayers were then incubated with the gB-1 MAb pool followed by incubation with cy3-conjugated anti-mouse antibody. Monolayers were visualized with a model 211910 Nikon microscope and photographed.
The cysteine mutant gB molecules are glycosylated in the ER but fail to be transported to the Golgi complex.
Processing of the HSV-1 glycoproteins during infection occurs through successive stages that involve addition of the mannose core sugars, trimming of the glucose residues by glucosidases I and II, and trimming of the mannose residues within the ER. These processed forms become resistant to endo H as the maturing glycoproteins migrate through the Golgi complex. Because the cysteine mutant gB molecules were translated and formed oligomers but were not transported to the cell surface or incorporated into virus envelopes, it appeared that the cysteine mutations prevented complete processing of the mutant gB molecules.
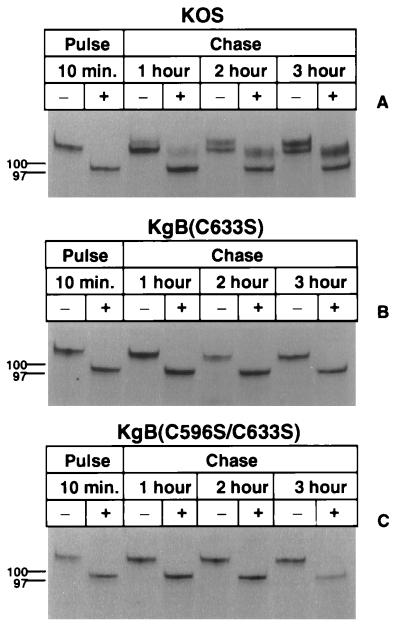
We first examined whether the mutant gB forms were transported to the Golgi complex based on their resistance to endo H treatment. Vero cells infected with KOS or the cysteine mutant viruses were pulse-labeled with [35S]methionine-cysteine for 10 min and chased with isotope-free medium for 1, 2, or 3 h. Infected cell lysates were immunoprecipitated with a gB-specific MAb pool, and a sample of the immune complexes was treated with endo H. Both treated and untreated samples were analyzed by SDS-PAGE and autoradiography to determine the relative sensitivities of the gB mutants to endo H compared with that of wild-type gB. As shown in Fig. 5A, KOS-derived gB was readily digested by endo H (lanes 1 and 2) prior to chase, indicating that the mannose core sugars had not been trimmed and the newly labeled gB still localized to the ER. However, after a 1-h chase, a portion of wild-type gB became resistant to endo H and migrated at a higher molecular ratio than the endo H-sensitive gB product, indicating that the mannose core sugars had been replaced with more complex carbohydrates (lanes 3 and 4) and that wild-type gB had begun its migration to the Golgi complex. The amount of endo H-resistant gB increased at the 2- and 3-h chase periods (lanes 6 and 8). In contrast to these results, experiments involving the cysteine mutants showed that gB remained completely sensitive to endo H digestion even after an extended chase period of 3 h [lanes 7 and 8 in Fig. 5B and C for KgB(C633S) and KgB(C596S/C633S), respectively], demonstrating that the cysteine mutant gB molecules remained associated with the ER. These observations clearly showed that the mutant forms of gB were not transported to the Golgi complex and remained associated with the ER even in experiments where the chase period was extended to 10 h (data not shown). Similar experiments were performed with the dimer-specific antibody (DL16) and similar data were obtained (data not shown), suggesting that wild-type gB and cysteine mutants formed oligomers in the ER soon after synthesis (10-min pulse).
FIG. 5.
Endo H sensitivity of mutant gB molecules. Vero cell monolayers were infected with KOS (A), KgB(C633S) (B), or KgB(C596S/C633S) (C) virus at an MOI of 10. Six hours p.i., monolayers were pulse-labeled for 10 min in the presence of [35S]methionine-cysteine and lysed immediately (Pulse; lanes 1 and 2) or further incubated in complete media and harvested after 1, 2, or 3 h of chase (lanes 3 and 4, 5 and 6, or 7 and 8, respectively). Samples were solubilized and subjected to immunoprecipitation with the gB-1 MAb pool, captured with protein A-Sepharose, incubated in absence (−) or presence (+) of endo H, and separated by SDS-PAGE. Molecular size markers are indicated in kilodaltons at the left.
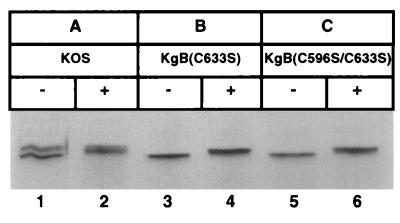
Since the mutant gB products were not transported from the ER to the Golgi complex, we performed experiments to further define the step in processing where the mutant gB forms were retained. Glucosidases I and II are glucose-trimming enzymes located within the ER whose function is inhibited by CST (18). To determine whether the mutant gB molecules were sensitive to glucosidase I and II trimming activity, KOS and cysteine mutant infections were pulse-radiolabeled and chased with cold medium in the presence and absence of CST. Infected cell extracts were immunoprecipitated with anti-gB antibodies, and the immune complexes were analyzed by SDS-PAGE and autoradiography (Fig. 6). KOS gB appeared as a doublet, indicative of a precursor product relationship between gB in the ER and gB which had migrated to the Golgi complex for terminal sugar addition (Fig. 6, lane 1). Immune complexes from wild-type virus infections treated with CST contained a product that migrated at a higher molecular mobility (lane 2) than the untreated gB samples (lower band of doublet seen in lane 1), demonstrating that CST inhibited the trimming of wild-type gB. Similar results were obtained with the cysteine mutant virus infections [lanes 3 to 6, KgB(C633S) and KgB(C596S/C633S)]. These results showed that the mutant forms of gB were modified by glucosidase I and II similarly to wild-type gB.
FIG. 6.
Processing of mutant gB by glucosidases I and II. Vero cell monolayers were infected with KOS (A), KgB(C633S) (B), or KgB(C596S/C633S) (C) virus at an MOI of 10. Five hours p.i., cells were preincubated in the absence (−; lanes 1, 3, and 5) or presence (+; lanes 2, 4, and 6) of CST for 1 h. Infected cells were pulse-labeled for 10 min in the presence of [35S]methionine-cysteine and chased for 2 h in absence or presence of CST. Monolayers were harvested, solubilized, and subjected to immunoprecipitation with a gB-1 MAb pool, and protein A-Sepharose immunocomplexes were separated by SDS-PAGE.
The cysteine mutations prevented release by the molecular chaperones calnexin, Grp78, and calreticulin.
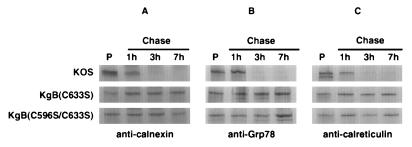
The experiments above demonstrated that the cysteine mutant gB molecules were retained in the ER (Fig. 6) at a step following trimming by glucosidases I and II. Calnexin, Grp78, Grp94, and calreticulin, which are ER-resident chaperone molecules, interact with glycoproteins and mediate protein folding and oligomerization; misfolded proteins are retained by these chaperones, preventing further processing (22a, 22b). Calnexin has been demonstrated to bind to maturing HSV-1 gB (51), while calnexin, Grp78, Grp94, and calreticulin have been demonstrated to associate with maturing human cytomegalovirus gB (52). Together, these proteins may form a quality control complex which ensures proper protein folding and oligomerization (18a, 39a, 45a). To examine the possibility that improperly folded glycoproteins are retained in the ER by calnexin, Grp78, and/or calreticulin, we performed experiments in which antichaperone antibodies were used to determine whether the mutant gB products could be coimmunoprecipitated (Fig. 7). Vero cells were infected with KOS, KgB(C633S), or KgB(C596S/C633S). The infected cells were pulse-radiolabeled for 10 min and chased with complete medium for 1, 3, or 7 h before detergent extraction and treatment with an anticalnexin, anti-Grp78, or anticalreticulin antibody. As shown in Fig. 7, immunoprecipitation of the pulse-labeled samples (lanes P) with each chaperone antibody resulted in the presence of gB in the immune complexes, demonstrating that the chaperones initially associated with gB. The identity of gB was confirmed by Western blot analysis using an anti-gB antibody reactive with the cytoplasmic domain (data not shown). However, pulse-labeled wild-type gB became uncoupled from each chaperone by 3 h of chase, showing that it was released by the chaperone molecule to continue on its pathway to the Golgi complex. Similar to wild-type gB and in agreement with an earlier report (51), gC and gD were also coimmunoprecipitated by the anticalnexin antibody and were uncoupled following a 3-h chase period, indicating that calnexin is involved in chaperoning multiple HSV-1 glycoproteins (data not shown). The results of similar immunoprecipitation experiments using KgB(C633S)- and KgB(C596S/C633S)-infected cells showed that in contrast to wild-type gB, calnexin, Grp78, and calreticulin were continuously associated with the cysteine mutant gB molecules even after a 7-h chase period. These data suggested that the cysteine mutations altered the conformation of gB in a manner to prevent dissociation from ER chaperones.
FIG. 7.
Retention of mutant gB molecules in the ER by calnexin, Grp78, and calreticulin. Vero cell monolayers were infected with KOS, KgB(C633S), or KgB(C596S/C633S) virus at an MOI of 10. Six hours p.i., monolayers were pulse-labeled for 10 min in the presence of [35S]methionine-cysteine and lysed immediately (Pulse; lanes P) or further incubated in complete medium and harvested after 1, 3, or 7 h (Chase). Samples were solubilized and subjected to immunoprecipitation with anticalnexin (A), anti-Grp78 (B), or anticalreticulin (C) polyclonal antibodies. Protein A-Sepharose immunocomplexes were separated by SDS-PAGE in order to visualize gB molecules which coprecipitated with the various chaperones.
The cysteine mutations alter conformationally dependent antigenic sites.
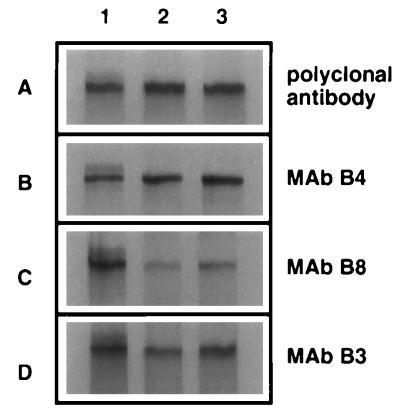
Since gB mutated in Cys-633 alone or in combination with Cys-596 remained bound to molecular chaperones of the ER, it was likely that these mutations affected gB folding at least in the region where these two cysteines form a disulfide bridge, and possibly in other regions of gB as well. These conformational changes might be reflected in the alteration of nonlinear epitopes detected by MAbs which recognize distinct antigenic sites on gB. Previously, we reported that gB contains at least four antigenic sites, three of which were nonlinear conformationally dependent determinants (36). These determinants were designated site I (residues 381 to 441), site II (residues 595 to 737), and site III (residues 283 to 380) recognized by the cognate MAbs B3, B8, and B4, respectively. A polyclonal antibody directed against the carboxy-terminal region of gB (Fig. 8A) could be used to immunoprecipitate wild-type or gB mutant molecules present in each sample as a control for the total amount of gB detectable by immunoprecipitation. The site-specific MAbs (Fig. 8B to D) were exploited to discern possible regional conformational changes within the cysteine mutant forms of gB. If any of these antibodies failed to bind or showed reduced binding compared with the total quantity of gB present in each sample, the retention of the mutant gB molecules in the ER could be associated with a conformational change colocalizing with the affected epitope structure.
FIG. 8.
Immunoprecipitation of mutant gB molecules with epitope-specific gB antibodies. Vero cell monolayers were infected with KOS (lane 1), KgB(C633S) (lane 2), and KgB(C596S/C633S) (lane 3) viruses at an MOI of 10 in the presence of [35S]methionine-cysteine. Forty hours p.i., infected cells were harvested, solubilized, and subjected to immunoprecipitation with a polyclonal antibody directed against the carboxy terminus of gB (A) or MAb B4 (B), B8 (C), or B3 (D). Protein A-Sepharose complexes were separated by SDS-PAGE.
Vero cells infected in the presence of [35S]methionine-cysteine with either KOS, KgB(C633S), or KgB(C596S/C633S) (lane 1, 2, or 3, respectively) were extracted with detergent and immunoprecipitated with the three representative MAbs. The amount of gB present in these samples was compared to the amount of gB immunoprecipitated with the polyclonal antibody for each virus. As shown in Fig. 8B, MAb B4 immunoprecipitated similar amounts of gB from wild-type-infected (lane 1) and the two gB mutant virus-infected (lanes 2 and 3) cell extracts compared with the quantity of gB molecule immunoprecipitated by the polyclonal antibody. These results indicated that the site III epitope recognized by MAb B4 was undisturbed by the cysteine mutations. In contrast, MAb B8 (which recognizes site II containing the cysteine mutations [Fig. 8C]) immunoprecipitated 30 and 45% of the amount of the mutated gB molecules from KgB(C633S) and KgB(C596S/C633S) viruses, respectively, compared to the amount of gB protein immunoprecipitated by the polyclonal antibody, while wild-type gB was immunoprecipitated to the same extent. These data demonstrated that mutation of Cys-633 or both Cys-596 and Cys-633 altered the conformation of the MAb B8 epitope. Immunoprecipitation with MAb B3 (which recognizes site I, a distal site from the cysteine mutations [Fig. 8D]) also showed a reduction in the amount of gB immunoprecipitated from the gB mutant viruses. The amount of gB immunoprecipitated from KgB(C633S) and KgB(C596S/C633S) with MAb B3, compared to the amount of gB immunoprecipitated with the polyclonal antibody, represented 62 and 89%, respectively, of the product immunoprecipitated by the polyclonal antibody, indicating that a distal epitope from the cysteine mutations was also altered in the mutant gB molecules. Together, these data demonstrated that epitopes I and II were altered by interrupting the Cys-633 to Cys-596 disulfide bridge, confirming that the cysteine mutations caused gB to be misfolded in several distinct molecular regions. These disruptions in normal folding of gB most likely accounted for chaperone-mediated retention of these molecules in the ER.
Hetero-oligomers between mutant and wild-type gB molecules are not properly transported to the cell surface of infected cells.
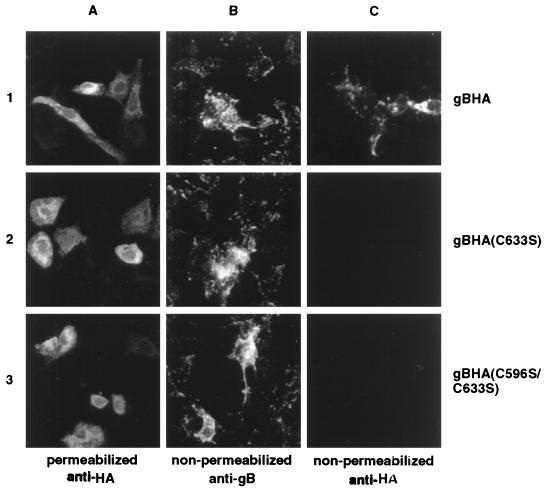
We demonstrated above that the cysteine mutant forms of gB were capable of forming homo-oligomers (Fig. 2) and hetero-oligomers with truncated gB molecule (Fig. 3). However, in Fig. 4 we showed that the mutated gB oligomers were not transported to the cell surface of infected cells due to a conformational misfolding of the mutated proteins (Fig. 8), blocking their release from the ER (Fig. 5) by chaperone molecules (Fig. 7). To determine whether hetero-oligomerization of these mutant gB molecules with wild-type gB might restore their conformational structure, allowing escape from chaperone retention, we tested for the presence of mutated gB molecules at the cell surface of infected cells following hetero-oligomerization with wild-type gB. Cells were transfected with expression plasmids for HA-tagged KOS or the cysteine mutant gB molecules, followed by infection with wild-type virus to provide a source of conformationally intact gB. The presence of mutant and wild-type gB forms at the cell surface was examined on nonpermeabilized infected cells by indirect immunofluorescence using a polyclonal anti-HA antibody. As shown in Fig. 9A, wild-type and cysteine mutant HA-tagged gB molecules were readily detected in the cytoplasm of methanol-fixed cells by the HA epitope-reactive antibody, demonstrating that the transfections of plasmid encoding HA-tagged wild-type (row 1) and mutated (rows 2 and 3) gB resulted in gB production. Moreover, gB was detected at the cell surface in all infections since a pool of gB-specific MAbs detected gB (from wild-type virus production and potentially plasmid expression) on nonpermeabilized cells (Fig. 9B). However, in contrast to gBHA (Fig. 9C, row 1), both mutant gB molecules failed to reach the cell surface (Fig. 9C, rows 2 and 3), as demonstrated by the absence of HA epitope detection at the cell surface of nonpermeabilized cells. Therefore, despite the ability of the cysteine mutants to oligomerize with wild-type gB, the hetero-oligomers were not transported to the cell surface. Although not tested, these results suggested that the hetero-oligomers were not incorporated into virions and indeed the presence of these nonfunctional oligomeric forms can inhibit efficient complementation by the wild-type molecule (9, 16, 26).
FIG. 9.
Failure of the mutant gB molecules to be transported to the cell surface in the presence of wild-type gB as detected by immunofluorescence analysis. Vero cell monolayers were individually transfected with HA-tagged gB (gBHA; row 1), gB(C633S) [gBHA(C633S); row 2], or gB(C596S/C633S) [gBHA(C596S/C633S); row 3]. Twenty-four hours posttransfection, monolayers were infected with KOS at an MOI of 5. Eight hours p.i., cells were fixed in ice-cold methanol (A) or left untreated (B and C) and incubated with anti-HA (A and C) or gB-1 MAb pool (B) antibodies followed by incubation with a cy3-conjugated anti-mouse antibody. Cells were visualized with a model 211910 Nikon microscope and photographed.
DISCUSSION
gB is a multifunctional essential virus envelope component which has been extensively studied using genetic and biochemical approaches. Experiments described in this report were designed to evaluate the role of two highly conserved cysteine residues of gB in oligomer formation, processing in the ER, transport to the cell surface, and incorporation into mature infectious virus particles. We created a single mutant in which a cysteine residue (Cys-633) located within the gB oligomerization domain was replaced by serine and studied it during infection to determine whether oligomers would form during the virus lytic cycle and support active virus production. The double mutant was created to prevent the possible formation of aberrant disulfide bonds by the unpaired cysteine partner at position 596 (Cys-596). A single mutant with a substitution at Cys-596 was not generated since this mutation was located outside the oligomerization domain. The results of this study support the conclusion that cysteines 633 and 596 are not required for oligomerization but are critical to the structural integrity of gB and essential for the development of a three-dimensional structure that allows this molecule to be normally processed and incorporated into mature virions.
gB was first determined to oligomerize shortly after synthesis as detergent-stable, heat-dissociable dimeric forms on PAGE (11). Homo-dimers have also been detected by sedimentation analysis of gB from purified virus, suggesting that the active form of gB is an oligomer (8). The tight association of wild-type gB monomer subunits can form unique epitopes which are recognized by MAbs that bind only to the oligomer. These antibodies have proven valuable in detecting the timing of oligomer formation following gB translation. In this study, oligomers were detected very early (within 10 min) following gB synthesis and likely occur concurrent with translation. This conclusion is consistent with the demonstration that the glycosylation inhibitor tunicamycin does not block oligomerization (26). Oligomerization of gB is also supported by evidence that hetero-oligomers composed of defective and wild-type gB monomer subunits were not functionally active and moreover interfere with the active form by reducing the formation of functional oligomers (wild type-wild type), whereas a mutant form which could not form oligomers did not interfere with the production of active oligomers (8, 16). This form of complementation interference was also observed with the cysteine gB mutant molecules constructed in this study since we demonstrated that hetero-oligomers composed of wild-type and mutant gB molecules were not processed and transported to the cell surface. These observations indicated that the gB mutant monomer subunits were not functionally corrected by association with a wild-type monomer subunit.
Using a coimmunoprecipitation assay, two oligomer-forming regions were identified within the external domain of gB (26): an upstream site between residues 93 and 282 and a downstream site located between residues 595 and 711. The upstream site was shown to provide only a weak interaction between gB monomers, while the downstream site caused a stronger association between monomers (16, 26) and was the only site identified by a functional inhibitory antibody approach where antibodies directed against this site blocked oligomerization (42). We recently reported studies to further characterize the downstream site by using an in vitro coimmunoprecipitation assay (34). This site was shown to be limited to 28 residues (amino acids 626 through 653) and could form oligomers when relocated to the carboxy terminus of gB, demonstrating that the oligomer-forming domain is a movable element and can self-associate in a manner independent of its local protein environment. An interesting feature of this domain was the presence of a cysteine residue at position 633 that was recently reported to form a disulfide bridge with cysteine 596 located outside the oligomer domain in HSV-2 gB (39). Because the type 1 and type 2 molecules are highly homologous and their cysteines are located in precisely the same positions, it is reasonable to propose that the type 1 molecule forms similar if not identical disulfide bridges (39). Conversion of cysteine 633 to serine did not block dimer formation; however, oligomerization was approximately half as efficient (34). We also found that oligomerization did not depend on the predicted disulfide bridge between Cys-596 and Cys-633 but that oligomerization was not sufficient to allow for correct gB processing and incorporation into virus particles. These data suggested that Cys-633 and Cys-596 were essential for proper gB folding, processing, intracellular trafficking, and virion incorporation.
Calnexin and calreticulin are two homologous, lectin-like chaperone molecules located in the ER that bind to partially trimmed, monoglucosylated forms of the N-linked core glycans present on maturing proteins (3, 22, 37a). They promote proper folding, prevent premature oligomerization, inhibit degradation, and function as quality control mediators for a variety of glycoproteins (22a, 22b, 23, 30, 48). Grp78 and Grp94 are soluble proteins also located in the ER which assist protein folding and prevent the exit of molecules that fail to attain the proper conformation (22a, 22b). In our study, we demonstrated that wild-type gB transiently associates with calnexin, Grp78, and calreticulin, in contrast to the cysteine mutant gB forms, which remained associated with calnexin, Grp78, and calreticulin for at least 7 h, accounting for their arrested maturation in the ER. These data are in agreement with previous reports demonstrating that HSV-1 glycoproteins gB, gC, and gD associate with calnexin following partial trimming of N-linked oligosaccharides (51) and that mutant forms of gB from HSV-1 and human cytomegalovirus formed complexes with Grp78 and Grp94, causing their retention in the ER (38, 52). Taken together, these data suggested that the ER-resident chaperones recognized misfolded proteins not based on a sequence-specific mutation but rather based on the entire protein conformation. The fact that the cysteine gB mutants were retained simultaneously instead of sequentially in the ER by calnexin, Grp78, and calreticulin suggested that in agreement with findings of others (18a, 39b, 45a), the chaperone proteins may form a complex as part of an extended network of molecules in the ER that direct the processing of proteins.
Direct evidence for altered conformation of the cysteine mutant gB forms was provided by their reduced recognition by MAbs that recognize epitopes located within the affected domains. The predicted disulfide bridge between cysteines 9 and 10 (Cys-596 and Cys-633, respectively) was required to maintain the structure of these conformational epitopes, and the impact of these mutations had a far-reaching impact on the molecule since two antigenic sites were altered. The alterations were nevertheless subtle since the antibodies could still recognize the molecules but with substantially less avidity. These cysteine mutations did not affect the temperature-dependent stability of these molecules since they were not functional in processing at temperatures ranging from 25 to 39°C (unpublished observation).
Despite the fact that the cysteine mutant forms of gB remained associated with chaperone molecules in the ER, mature virus was released from infected cells completely lacking gB but had the normal complement of the other glycoproteins (gC and gD) as measured by Western blot analysis of purified extracellular virus envelope components. Since gB is essential to virus entry, the absence of gB in the virus envelope accounted for the failure of these mutant viruses to produce infectious particles. Browne et al. (5) recently reported that an ER-restricted gH mutant of HSV-1 lacked gH in virus particles and virus produced was noninfectious. Consistent with this report, biochemical analysis of the cysteine mutant viruses revealed the presence of the mutant gB proteins within the infected cells but their absence in the envelope of mature extracellular virions. Both the gH experimentation and the present study suggest that envelopment may occur in a post-ER compartment such as the Golgi complex, Golgi-derived vacuoles, or the plasma membrane. Alternatively, mutant gB forms are never incorporated into budded particles at the nuclear membrane and the double-enveloped viruses released from the ER undergo membrane exchange at the Golgi complex, where maturing viruses acquire their final envelope. Studies to define the trafficking patterns of these cysteine gB mutant molecules and envelope contents of intracellular mutant virus particles are under way and may help to better define the pathway(s) leading to mature virus production.
ACKNOWLEDGMENTS
Sylvie Laquerre and Dina B. Anderson contributed equally to this research.
We thank Darren P. Wolfe for discussion of the data and Thomas C. Holland for critical reading of the manuscript.
This work was supported by Public Health Service grant R01 CA66141-07 from the National Institutes of Health, by l’Association Française contre les Myopathies, and by GenVec Inc.
REFERENCES
- 1.Albrecht J-C, Fleckenstein B. Structural organization of the conserved block or Herpesvirus saimiri coding for DNA polymerase, glycoprotein B, and major DNA binding protein. Virology. 1990;174:533–542. doi: 10.1016/0042-6822(90)90107-3. [DOI] [PubMed] [Google Scholar]
- 2.Anderson K S, Cresswell P. A role for calnexin (IP90) in the assembly of class II MHC molecules. EMBO J. 1994;13:675–682. doi: 10.1002/j.1460-2075.1994.tb06306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron J J M, Brenner M B, Thomas D Y, Williams D B. Calnexin: a membrane-bound chaperone in the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 4.Bond V C, Person S, Warner S C. The isolation and characterization of mutants of herpes simplex virus type 1 that induce cell fusion. J Gen Virol. 1982;61:245–254. doi: 10.1099/0022-1317-61-2-245. [DOI] [PubMed] [Google Scholar]
- 5.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bzik D J, Fox B A, DeLuca N A, Person S. Nucleotide sequence specifying the glycoprotein gene, gB, of herpes simplex virus type 1. Virology. 1984;133:301–314. doi: 10.1016/0042-6822(84)90397-0. [DOI] [PubMed] [Google Scholar]
- 7.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai W, Person S, DebRoy C, Gu B. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1: an analysis of linker insertion mutants. J Mol Biol. 1988;201:575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- 9.Cai W, Person S, Warner S C, Zhou J, DeLuca N A. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J Virol. 1987;61:714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapsal J M, Pereira L. Characterization of epitopes on native and denatured forms of herpes simplex virus glycoprotein B. Virology. 1988;164:427–434. doi: 10.1016/0042-6822(88)90556-9. [DOI] [PubMed] [Google Scholar]
- 11.Claesson-Welsh L, Spear P G. Oligomerization of herpes simplex virus glycoprotein B. J Virol. 1986;60:803–806. doi: 10.1128/jvi.60.2.803-806.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claesson-Welsh L, Spear P G. Amino-terminal sequence, synthesis, and membrane insertion of glycoprotein B of herpes simplex virus type 1. J Virol. 1987;61:1–7. doi: 10.1128/jvi.61.1.1-7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darlington R W, Moss L H. Herpesvirus envelopment. J Virol. 1968;2:48–55. doi: 10.1128/jvi.2.1.48-55.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David V, Hochstenbach F, Rajagopalan S, Brenner M. Interaction with newly synthesized and retained proteins in the endoplasmic reticulum suggests a chaperone function for human integral membrane protein IP90 (calnexin) J Biol Chem. 1993;268:9585–9592. [PubMed] [Google Scholar]
- 15.DeLuca N, Bzik D J, Bond V C, Person S, Snipes W. Nucleotide sequences of herpes simplex type 1 (HSV-1) affecting virus entry, cell fusion, and production of glycoprotein gB (VP7) Virology. 1982;122:411–423. doi: 10.1016/0042-6822(82)90240-9. [DOI] [PubMed] [Google Scholar]
- 16.Desai P, Homa F L, Person S, Glorioso J C. A genetic selection method for the transfer of HSV-1 glycoprotein B mutations from plasmid to the viral genome: preliminary characterization of transdominance and entry kinetics of mutant viruses. Virology. 1994;204:312–322. doi: 10.1006/viro.1994.1536. [DOI] [PubMed] [Google Scholar]
- 17.Desai P J, Schaffer P A, Minson A C. Excretion of non-infectious virus particles lacking glycoprotein H by a temperature-sensitive mutant of herpes simplex virus type 1: evidence that gH is essential for virion infectivity. J Gen Virol. 1988;69:1147–1156. doi: 10.1099/0022-1317-69-6-1147. [DOI] [PubMed] [Google Scholar]
- 18.Elbein A D. Inhibitors of biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem. 1987;56:497–534. doi: 10.1146/annurev.bi.56.070187.002433. [DOI] [PubMed] [Google Scholar]
- 18a.Ferreira L, Norris K, Smith T, Hebert C, Sauk J. Hsp47 and other ER-resident molecular chaperones form heterocomplexes with each other and with collagen type IV chains. Connect Tissue Res. 1996;33:265–273. doi: 10.3109/03008209609028884. [DOI] [PubMed] [Google Scholar]
- 19.Gage P J, Levine M, Glorioso J C. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J Virol. 1993;67:2191–2201. doi: 10.1128/jvi.67.4.2191-2201.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert R, Ghosh H P. Immunoelectron microscopic localization of herpes simplex virus glycoprotein gB in the nuclear envelope of infected cells. Virus Res. 1993;28:217–231. doi: 10.1016/0168-1702(93)90023-g. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert R, Ghosh K, Rasile L, Ghosh H P. Membrane anchoring domain of herpes simplex virus glycoprotein gB is sufficient for nuclear envelope localization. J Virol. 1994;68:2272–2285. doi: 10.1128/jvi.68.4.2272-2285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond C, Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- 22a.Hammond C, Helenius A. A chaperone with a sweet tooth. Curr Biol. 1993;3:884–885. doi: 10.1016/0960-9822(93)90226-e. [DOI] [PubMed] [Google Scholar]
- 22b.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 23.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herold B C, Visalli R J, Sumarski N, Brandt C, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 25.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Highlander S L, Goins W F, Person S, Holland T C, Levine M, Glorioso J C. Oligomer formation of the gB glycoprotein of herpes simplex virus type 1. J Virol. 1991;65:4275–4283. doi: 10.1128/jvi.65.8.4275-4283.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Highlander S L, Sutherland S L, Gage P J, Johnson D C, Levine M, Glorioso J C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho S N, Hunt H D, Horton R M, Pellen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson A C, Johnson D C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson M R, Cohen-Doyle M F, Peterson P A, Williams D B. Regulation of MHC class I transport by the molecular chaperone, calnexin (p88, IP90) Science. 1994;263:384–387. doi: 10.1126/science.8278813. [DOI] [PubMed] [Google Scholar]
- 31.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Kim P S, Arvan P. Calnexin and BiP act as sequential molecular chaperones during thyroglobulin folding in the endoplasmic reticulum. J Cell Biol. 1995;128:29–38. doi: 10.1083/jcb.128.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kousoulas K G, Bzik D J, DeLuca N, Person S. The effect of ammonium chloride and tunicamycin on the glycoprotein content and infectivity of herpes simplex virus type 1. Virology. 1983;125:468–474. doi: 10.1016/0042-6822(83)90217-9. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Laquerre S, Person S, Glorioso J C. Glycoprotein B of herpes simplex virus type 1 oligomerizes through the intermolecular interaction of a 28-amino-acid domain. J Virol. 1996;70:1640–1650. doi: 10.1128/jvi.70.3.1640-1650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marlin S D, Highlander S L, Holland T C, Levine M, Glorioso J C. Antigenic variation (mar mutations) in herpes simplex virus glycoprotein B can induce temperature-dependent alterations in gB processing and virus production. J Virol. 1986;59:142–153. doi: 10.1128/jvi.59.1.142-153.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marlin S D, Holland T C, Levine M, Glorioso J C. Epitopes of herpes simplex virus type 1 glycoprotein gC are clustered in two distinct antigenic sites. J Virol. 1985;53:128–136. doi: 10.1128/jvi.53.1.128-136.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navarro D, Paz P, Pereira L. Domains of herpes simplex virus 1 glycoprotein B that function in virus penetration, cell-to-cell spread, and cell fusion. Virology. 1992;186:99–112. doi: 10.1016/0042-6822(92)90064-v. [DOI] [PubMed] [Google Scholar]
- 39.Norais N, Tang D, Kaur S, Chamberlain S H, Masiarz F R, Burke R L, Marcus F. Disulfide bonds of herpes simplex virus type 2 glycoprotein B. J Virol. 1996;70:7379–7387. doi: 10.1128/jvi.70.11.7379-7387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Otteken A, Moss B. Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperone function similar to that of calnexin. J Biol Chem. 1996;271:97–103. doi: 10.1074/jbc.271.1.97. [DOI] [PubMed] [Google Scholar]
- 40.Pellet P E, Kousoulas K G, Pereira L, Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J Virol. 1985;53:243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penfold M E T, Armati P, Cunningham A L. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc Natl Acad Sci USA. 1994;91:6529–6533. doi: 10.1073/pnas.91.14.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qadri I, Gimeno C, Navarro D, Pereira L. Mutations in conformation-dependent domains of herpes simplex virus 1 glycoprotein B affect the antigenic properties, dimerization, and transport of the molecule. Virology. 1991;180:135–152. doi: 10.1016/0042-6822(91)90017-6. [DOI] [PubMed] [Google Scholar]
- 43.Rasile L, Ghosh K, Raviprakash K, Ghosh H P. Effects of deletions in the carboxy-terminal hydrophobic region of herpes simplex virus glycoprotein gB on intracellular transport and membrane anchoring. J Virol. 1993;67:4856–4866. doi: 10.1128/jvi.67.8.4856-4866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 201–232. [Google Scholar]
- 45.Stackpole C W. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J Virol. 1969;4:75–93. doi: 10.1128/jvi.4.1.75-93.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Tatu, U., and A. Helenius. Interactions between newly synthesized glycoproteins, calnexin and a network of resident chaperones in the endoplasmic reticulum. J. Cell Biol. 136:555–565. [DOI] [PMC free article] [PubMed]
- 46.Tengelson L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torrisi M R, Di Lazzaro C, Pavan A, Pereira L, Campadelli-Fiume G. Herpes simplex virus envelopment and maturation studied by fracture label. J Virol. 1992;66:554–561. doi: 10.1128/jvi.66.1.554-561.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wada I, Rindress D, Cameron P H, Ou W-J, Doherty II J J, Louvard D, Bell A W, Dignard D, Thomas D Y, Bergeron J J M. Ssrα and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;226:19599–19610. [PubMed] [Google Scholar]
- 49.Wilson I A, Niman H L, Houghten R A, Connolly A R, M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita Y, Shimokata K, Mizuno S, Daikoku T, Tsurumi T, Nishiyama Y. Calnexin acts as a molecular chaperon during the folding of glycoprotein B of human cytomegalovirus. J Virol. 1996;70:2237–2246. doi: 10.1128/jvi.70.4.2237-2246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamashita Y, Yamada M, Daikoku T, Yamada H, Tadauchi A, Tsurumi T, Nishiyama Y. Calnexin associates with the precursors of glycoproteins B, C, and D of herpes simplex virus type 1. Virology. 1996;225:216–222. doi: 10.1006/viro.1996.0590. [DOI] [PubMed] [Google Scholar]
- 52.Zheng Z, Maidji E, Tugizov S, Pereira L. Mutations in the carboxyl-terminal hydrophobic sequence of human cytomegalovirus glycoprotein B alter transport and protein chaperone binding. J Virol. 1996;70:8029–8040. doi: 10.1128/jvi.70.11.8029-8040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]