Abstract
Human cells express distinct but related receptors for the gibbon ape leukemia virus (GALV) and the amphotropic murine leukemia virus (A-MuLV), termed Pit1 and Pit2, respectively. Pit1 is not able to function as a receptor for A-MuLV infection, while Pit2 does not confer susceptibility to GALV. Previous studies of chimeric receptors constructed by interchanging regions of Pit1 and Pit2 failed to clarify the determinants unique to Pit2 which correlate with A-MuLV receptor function. In order to identify which regions of Pit2 are involved in A-MuLV receptor function, we exchanged the putative second and third extracellular domains of Pit1, either individually or together, with the corresponding regions of Pit2. Our functional characterization of these receptors indicates a role for the putative second extracellular domain (domain II) in A-MuLV infection. We further investigated the influence of domain II with respect to A-MuLV receptor function by performing site-specific mutagenesis within this region of Pit2. Many of the mutations had little or no effect on receptor function. However, the substitution of serine for methionine at position 138 (S138M) in a Pit1 chimera containing domain II of Pit2 resulted in a 1,000-fold reduction in A-MuLV receptor function. Additional mutations made within domain II of the nonfunctional S138M mutant restored receptor function to nearly wild-type efficiency. The high degree of tolerance for mutations as well as the compensatory effect of particular substitutions observed within domain II suggests that an element of secondary structure within this region plays a critical role in the interaction of the receptor with A-MuLV.
Most retroviruses initiate infection of host cells through a specific interaction with a cell surface receptor. Among the murine leukemia viruses (MuLVs) there are now six different classes identified based on receptor usage, including ecotropic (E-MuLV), amphotropic (A-MuLV), xenotropic, dualtropic (mink cell focus-forming virus), 10A1-MuLV, and the recently identified (4) Mus dunni endogenous virus. MuLVs belonging to each receptor class use discrete receptors for viral entry into mouse cells except 10A1-MuLV, which can use two different receptors (14, 19), one of which is also used by A-MuLVs. The E-MuLV receptor was the first of the murine type C retrovirus receptors to be identified and has been shown to be a multimembrane-spanning amino acid transporter (2). The human cDNAs encoding the receptors for a primate type C retrovirus, the gibbon ape leukemia virus (GALV) and a second type C retrovirus, A-MuLV, have also been identified (16, 23). These two receptors, originally named Glvr-1 and Glvr-2, respectively, have recently been renamed Pit1 and Pit2, respectively, to reflect their normal function as transporters of inorganic phosphate (10, 17). The Pit receptors not only have comparable cellular functions but also have the same proposed membrane topology and 62% amino acid identity (23). Despite the similarities of these receptors, they exhibit distinct virus recognition properties: Pit1 functions for GALV infection but does not confer susceptibility to A-MuLV, while the reverse is true for Pit2 (14, 20).
The current structural model for the Pit receptors is based on hydropathy analysis (16) and features 10 transmembrane domains, internal N and C termini, a large cytoplasmic region, and five extracellular loops. Although most of the proposed membrane topology has yet to be verified, the location of sections of the cytosolic loop has been confirmed (6). A number of studies using chimeric receptors constructed by interchanging regions of Pit1 and Pit2 (Pit1-Pit2 receptors) (8, 18, 22) have indicated the importance of the putative fourth extracellular domain for GALV receptor function. Indeed, a single mutation in this region of Pit2 rendered it functional for GALV infection (8). However, recent studies have raised the possibility of involvement by other domains in the interaction with GALV (5, 21). A similarly ambiguous picture has emerged with respect to A-MuLV permissivity. Initial results from the assessment of Pit1-Pit2 chimeric receptors with either the first three or the last two extracellular domains of the Pit1 receptor replaced with the corresponding regions of Pit2 suggested that the determinants of A-MuLV receptor function do not reside exclusively in any single region of the Pit2 receptor (8). In an effort to obtain a more complete understanding of A-MuLV-receptor interaction, we have constructed a series of Pit1-Pit2 chimeric and mutant receptors and have assessed their abilities to confer sensitivity to A-MuLV infection on CHO K1 cells. We have observed not only an important role for the putative second extracellular domain in A-MuLV receptor function but also the ability of certain combinations of mutations within the second domain to compensate for other substitutions in this domain that abolish receptor function.
MATERIALS AND METHODS
Cells.
CHO K1 cells, Chinese hamster ovary cells, were obtained from the American Type Culture Collection (CCL 61). M. dunni tail fibroblast (MDTF) cells, which were derived from the feral mouse M. dunni, were provided by Olivier Danos (and are also available from the American Type Culture Collection [CRL 2017]). MDTF cells expressing Pit1 or Pit2 receptors have been previously described (8). The PA317/G1BgSvN (11, 12, 25) and PG13/G1BgSvN (11, 12) retrovirus packaging cell lines have been described previously. CHO K1 cells were maintained in alpha minimal essential medium supplemented with 5% fetal bovine serum, 4 mM glutamine, 1 mM sodium pyruvate, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. All other cells were maintained in Dulbecco’s modified Eagle’s medium supplemented in a similar manner.
Construction of chimeric and mutant receptor cDNA plasmids.
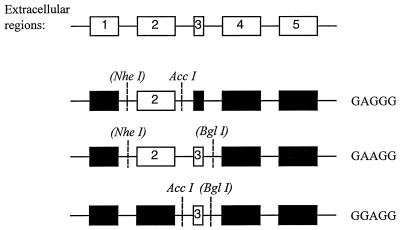
The Pit1-Pit2 chimeras (Fig. 1) are named by using five letters which designate the origin of each of the five extracellular domains, A for those derived from the A-MuLV receptor (Pit2) and G for those derived from the GALV receptor (Pit1). Wherever nucleotide or amino acid positions are indicated, the numbers given correspond to the Pit2 sequence unless otherwise noted (23). The chimeras were constructed in the pSP72 plasmid (Promega, Madison, Wis.) as follows: GAGGG was made by exchange of the cDNA for the putative second extracellular domain of Pit1 with that of Pit2 between the NheI and AccI sites (nucleotides [nt] 151 to 597); GAAGG was made by replacing the cDNA for both the second and third putative extracellular regions of Pit1 between the NheI and BglII sites (nt 151 to 1065) with that for the corresponding regions of Pit2; GGAGG was made by the exchange of the cDNA for the putative third extracellular domain of Pit1 with that of Pit2 between the AccI and BglII sites (nt 597 to 1065). Pit1 cDNA (pSP72-OJ9) and the chimeric and mutant receptor cDNAs constructed in pSP72 were subcloned from pSP72, using the HindIII and EcoRV sites at either end of the receptor sequence, into the pLNSX retroviral vector (13) prepared with HindIII and ClaI (filled in with T4 DNA polymerase [Boehringer Mannheim, Indianapolis, Ind.]) for expression analysis. Pit2 cDNA was subcloned from pSP72 (pSP72-Pit2), using the EcoRI sites at either end of the receptor sequence, into the pLNSX retroviral vector prepared with EcoRI, and the cohesive ends were dephosphorylated with alkaline phosphatase (Boehringer GmbH, Mannheim, Germany). Retroviral expression vectors (pLNSX) encoding the various receptors are generically referred to here as pLNSR.
FIG. 1.
Schematic representation of chimeric receptor cDNAs used to examine virus receptor function. Numbered boxes represent regions encoding the five predicted extracellular domains (left to right, 5′ to 3′). The regions derived from Pit1 are shown in black; those derived from Pit2 are shown in white. Chimeric receptors are named by using single letters to designate the source for each of the five extracellular domains (A for Pit2, G for Pit1). Restriction enzyme sites are given in their appropriate locations in the receptor cDNAs; nucleotide positions of the restriction sites are given in the Materials and Methods section. The NheI and BglII sites are given in parentheses because they were originally introduced as silent mutations in either Pit1 or Pit2 (8).
Mutations were introduced into the second extracellular domain of either Pit1 or GAGGG by using one of two methods of site-directed mutagenesis. In the first method, synthetic oligonucleotides containing nucleotide changes appropriate either to introduce a specific amino acid residue change or to create a restriction site without affecting the encoded amino acid were designed. PCRs were performed in a two-step process described previously (9), and the resulting products were cloned directly into the TA pCRII vector (Invitrogen, San Diego, Calif.). Mutant versions of the Pit1 and GAGGG cDNAs generated in this way were constructed in the pSP72 plasmid by replacing the region between the NheI and AccI sites (nt 151 to 597) of either Pit1 or GAGGG cDNA with the corresponding region containing the mutant sequence from the TA PCRII plasmid. Final subcloning into pLNSX was accomplished as described above for the Pit1 and GAGGG constructs. In the second method, pLNSR plasmid constructs of Pit1 and the chimeric receptor GAGGG were used as templates for mutagenesis according to the QuikChange method (Stratagene, La Jolla, Calif.), with appropriately designed mutant oligonucleotides. Plasmids constructed by either method were sequenced to confirm the presence of desired mutations and to verify the absence of unscheduled mutations.
Expression of chimeric and mutant receptor cDNAs.
Calcium phosphate-mediated gene transfer of mutant and control receptor cDNA plasmids for both transient and stable expression in CHO K1 cells was carried out by the Profection method (Promega). For transient expression, calcium phosphate precipitate containing 3 μg of pLNSR plasmid DNA was prepared and applied to each of three individual wells of CHO K1 cells in a 12-well dish as described previously (8). For stable receptor cDNA expression, cells transfected with chimeric and mutant receptor pLNSR plasmids were selected for 10 to 14 days in appropriate growth medium supplemented with G418 (450 μg per ml of active substance). Following selection, pooled populations of G418-resistant cells were assayed for receptor function as described below.
Stable expression of receptor cDNAs in MDTF cells was achieved by transfecting each of the pLNSR plasmid DNAs into PA317 packaging cells by the Profection method and selecting transfected cells in medium containing 450 μg of active G418 per ml for 10 to 14 days. The supernatants from each of the pLNSR vector-producing cell lines were then used to infect MDTF cells, as previously described (8). MDTF cells were selected for 7 to 10 days at 600 μg of active G418 per ml. Pooled populations of G418-resistant cells were used in GALV infection assays as described below.
Assays for receptor function.
The recombinant retrovirus genome G1BgSvN, which carries the bacterial lacZ and neomycin resistance genes (11), was used in all infection assays. For the assay of A-MuLV infection of CHO K1 cells, cells were infected with 2 ml of filtered (0.45-μm-pore-size filter) supernatant from PA317/G1BgSvN retrovirus packaging cells containing 3 μg of Polybrene per ml, either 48 h after transfection for transient experiments or 24 h after seeding at 3 × 104 cells/well in 12-well dishes for stable expression. At 48 h postexposure to the vector, cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (24) and the numbers of blue foci per well were quantified. The GALV infection assay and histochemical staining of control and pLNSR-expressing MDTF cells were carried out in a manner similar to that described for A-MuLV with supernatant from PG13/G1BgSvN retrovirus packaging cells.
RESULTS
Functional analysis of Pit1-Pit2 chimeric receptors.
Chimeric receptors were constructed to determine whether a minimum domain in the N-terminal half of Pit2 plays a role in A-MuLV receptor function. Chimeras GAAGG, GAGGG, and GGAGG were constructed by replacement of either or both of the second and third extracellular domains of Pit1 with the corresponding regions of Pit2 (Fig. 1). As shown in Table 1, CHO K1 cells expressing chimera GAAGG, containing the Pit2 sequence in both the second and third extracellular domains, are susceptible to infection by vectors with the A-MuLV envelope, whereas CHO K1 cells expressing GGAGG, featuring only the third domain of Pit2, are not susceptible. The GAGGG construct contains the Pit2 sequence in only the second extracellular domain and functions as a receptor for A-MuLV, demonstrating that the presence of the second extracellular domain (domain II) of Pit2 correlates with A-MuLV receptor function.
TABLE 1.
A-MuLV infection of CHO K1 cells transiently expressing chimeric receptors
| Receptor | % Infectiona |
|---|---|
| Pit2 | 100 |
| Pit1 | <0.1b |
| GGGAA | 55.2 ± 25.4c |
| GAGGG | 18.5 ± 7.4d |
| GAAGG | 75.1 ± 25.3 |
| GGAGG | <0.1 |
| No DNA | <0.1 |
As described in Materials and Methods, data given represent the means ± the standard deviations for three (except where indicated otherwise) independent transfection-infection experiments. Each transfection precipitate was placed in triplicate wells of a 12-well dish. The average number (varying from 200 to 800 per well in different experiments) of β-galactosidase-positive foci in the wells transfected with Pit2 DNA was assigned a value of 100% for each experiment, and all other values were normalized to this.
<0.1, no β-galactosidase-positive foci were observed in three independent experiments, each done in triplicate in 12-well dishes.
Previously reported (8).
Data given represent the mean ± the standard deviation for five independent experiments.
Mutations in extracellular domain II influence A-MuLV receptor function.
To elucidate which, if any, specific residues within the second extracellular domain are responsible for its apparent role in A-MuLV receptor function, site-specific mutagenesis was performed in this region of both native Pit1 and the chimeric GAGGG receptor. These receptors represent nonfunctional (Pit1) and functional (GAGGG) A-MuLV receptors. A comparative analysis of the amino acid sequences of Pit1 and Pit2 in the proposed second extracellular domain (Fig. 2) reveals eight positions occupied by different residues in the two receptors. Seven of the eight differences in the second extracellular domain are clustered in the C-terminal portion of the domain. The goal of mutagenesis in this region was to make changes at each of these seven positions to determine if either a loss of A-MuLV receptor function in the GAGGG backbone or a gain of function in native Pit1 could be achieved. Mutant receptors were tested for their abilities to render CHO K1 cells susceptible to PA317/G1BgSvN in transient transfection-infection experiments. Both Pit1 and GAGGG function as receptors for GALV. Therefore, in order to verify the expression and integrity of receptor proteins, all mutant receptor cell lines were tested for GALV receptor function. All mutant receptors reported here retain GALV receptor function at efficiencies close to that of the wild type, Pit1
FIG. 2.
Comparison of Pit1 and Pit2 sequences in the predicted second extracellular domain (domain II; amino acids 107 to 141 [based on Pit2 numbering]).
The series of Pit1 mutant receptors shown in Table 2 begins with the replacement of the three charged amino acids in domain II of Pit1 with the corresponding Pit2 residues (Pit1-1); progressive substitutions with corresponding Pit2 residues (Pit1-2 and Pit1-3) have been introduced until the sequence in domain II resembles that of Pit2 (Pit1-4). All the Pit1 mutants differ from Pit2 in this region by a single lysine (position 108; arginine in Pit2). Mutant Pit1-4, which is otherwise identical to Pit2 in domain II is functional for A-MuLV infection. The Pit1-3 mutant was found not to function as an A-MuLV receptor when assessed in the transient assay, even though it differs from the functional Pit1-4 mutant by a single residue (the Q132T change). A number of other mutational combinations in this series of mutants have no detectable effect on A-MuLV receptor function (data not shown). Three changes involving charged residues, K130I, E133K, and K136Q, together with mutation S138M, do not appear to affect receptor function (mutant Pit1-1). However, single and double mutations involving only S138M and I141V result in functional receptors, Pit1-5, Pit1-6, and Pit1-7.
TABLE 2.
A-MuLV infection of CHO K1 cells transiently expressing Pit1 mutant receptors
| Receptor | Sequencea (amino acids 120–141) | % Infectionb |
|---|---|---|
| Pit1 | GATIGFSLVAKGQEGVKWSELI | <0.1c |
| Pit1-1 | GATIGFSLVAIGQKGVQWMELI | <0.1 |
| Pit1-2 | GSTIGFSLVAIGTKGVQWSELI | <0.1 |
| Pit1-3 | GSTIGFSLVAIGQKGVQWMELV | <0.1 |
| Pit1-4 | GSTIGFSLVAIGTKGVQWMELV | 6.3 ± 2.2 |
| Pit1-5 | GATIGFSLVAKGQEGVKWMELI | 3.5 ± 0.6 |
| Pit1-6 | GATIGFSLVAKGQEGVKWSELV | 2.8 ± 0.8 |
| Pit1-7 | GATIGFSLVAKGQEGVKWMELV | 1.5 ± 0.5 |
| Pit2 | GSTIGFSLVAIGTKGVQWMELV | 100 |
| No DNA | <0.1 |
The numbering used is that of Pit2. Mutations are shown in boldface. For the Pit2 sequence, residues which differ from those of Pit1 are shown in boldface.
Data given represent the means ± the standard deviations for three independent transfection-infection experiments. In each experiment, the individual transfection precipitate was placed in triplicate wells of a 12-well dish.
<0.1, no β-galactosidase-positive foci were observed in three independent triplicate experiments.
Mutagenesis of the seven residues that differ between Pit1 and Pit2 in the second extracellular domain of functional GAGGG was used to identify receptors that fail to function for A-MuLV infection (Table 3). The two large hydrophobic residues in the domain, F125 and W137, were replaced with alanine in mutants C1-1 and C1-2 without an effect on A-MuLV receptor function. Differences in charge were explored with mutants C1-3, C1-4, and C1-5; the T132Q change was made in C1-6; and a group of three mutations, I130K, T132Q, and K133E, was introduced in mutant C1-7. None of these mutations produced any significant change in the efficiency of A-MuLV receptor function. The single mutation M138S present in receptor C1-8 results in a loss of A-MuLV receptor function, as shown by a transient CHO K1 assay.
TABLE 3.
A-MuLV infection of CHO K1 cells transiently expressing GAGGG mutant receptors
| Receptor | Sequencea (amino acids 120–141) | % Infectionb |
|---|---|---|
| GAGGG | GSTIGFSLVAIGTKGVQWMELV | 18.5 ± 7.4 |
| Pit2 | GSTIGFSLVAIGTKGVQWMELV | 100 |
| C1-1 | GSTIGASLVAIGTKGVQWMELV | 7.9 ± 0.4 |
| C1-2 | GSTIGASLVAIGTKGVQAMELV | 20.2 ± 4.9 |
| C1-3 | GSTIGFSLVAKGTKGVQWMELV | 90.7 ± 7.7 |
| C1-4 | GSTIGFSLVAIGTKGVKWMELV | 72.3 ± 7.1 |
| C1-5 | GSTIGFSLVAIGTEGVQWMELV | 36.4 ± 6.9 |
| C1-6 | GSTIGFSLVAIGQKGVQWMELV | 74.8 ± 9.3 |
| C1-7 | GSTIGFSLVAKGQEGVQWMELV | 80.3 ± 8.4 |
| C1-8 | GSTIGFSLVAIGTKGVQWSELV | <0.01c |
| Pit1 | GATIGFSLVAKGQEGVKWSELI | <0.01 |
| No DNA | <0.1 |
The numbering used is that of Pit2. Mutations are shown in boldface. For the Pit2 sequence, residues which differ from those of Pit1 are shown in boldface.
Data given represent the means ± the standard deviations for two independent transfection-infection experiments. In each experiment, the individual transfection precipitate was placed in triplicate wells of a 12-well dish.
<0.1, no β-galactosidase-positive foci were observed in three independent triplicate experiments.
An analysis of GAGGG mutants indicated that receptor function is not compromised in spite of a number of nonconservative amino acid changes, suggesting that receptor function can tolerate a high degree of flexibility in this region. To complement these findings, the nonfunctional C1-8 mutant was used as a receptor template on which to make other mutations in the second extracellular domain which could restore A-MuLV receptor function. The trio of changes, I130K, T132Q, K133E, did not affect the function of the GAGGG receptor (C1-7) yet were able to overcome the effect of the M138S mutation, yielding functional receptor C1-9. To determine which, if any, of the three residues is primarily responsible for the observed compensation, each mutation was made individually in the C1-8 receptor (Table 4). Mutants C1-10 and C1-11, featuring the I130K and K133E mutations, respectively, encode functional A-MuLV receptors, rendering CHO K1 cells permissive to A-MuLV, while the T132Q mutation in the C-12 mutant has no observable effect on A-MuLV receptor function.
TABLE 4.
A-MuLV infection of CHO K1 cells transiently expressing C1-8 mutant receptors
| Receptor | Sequencea (amino acids 120–141) | % Infectionb |
|---|---|---|
| C1-8 | GSTIGFSLVAIGTKGVQWSELV | <0.1c |
| C1-9 | GSTIGFSLVAKGQEGVQWSELV | 79.6 ± 3.4 |
| C1-10 | GSTIGFSLVAKGTKGVQWSELV | 42.1 ± 12.1 |
| C1-11 | GSTIGFSLVAIGTEGVQWSELV | 2.7 ± 1.4 |
| C1-12 | GSTIGFSLVAIGQKGVQWSELV | <0.1 |
| Pit2 | GSTIGFSLVAIGTKGVQWMELV | 100 |
| No DNA | 0 |
The numbering used is that of Pit2. Mutations are shown in boldface. For the Pit2 sequence, residues which differ from those of Pit1 are shown in boldface.
Data given represent the means ± the standard deviations for two independent transfection-infection experiments. In each experiment, the individual transfection precipitate was placed in triplicate wells of a 12-well dish.
<0.1, no β-galactosidase-positive foci were observed in three independent triplicate experiments.
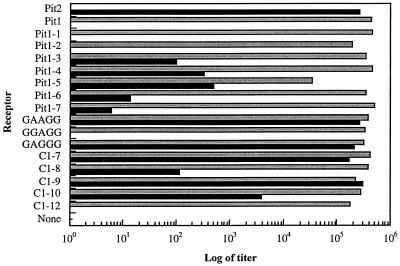
To improve the sensitivity of the A-MuLV infection assay and thereby fully characterize the receptors which give negative results in the transient transfection-infection experiments, stable expression of selected mutant receptors was established in CHO K1 cells. For transient experiments, the upper limit of detection in the Pit2 positive control wells ranges from 2 × 102 to 8 × 102 per well, compared with 8 × 105 to 1 × 106 per well for assays of the stable Pit2 receptor cell line. In the functional analysis of stable receptor-bearing cell lines, several of the mutant receptors which appeared nonfunctional for A-MuLV infection in transient experiments revealed limited receptor function in established cell lines. Specifically, established CHO K1 cell lines expressing Pit1-3, C1-8, and C1-10 (Fig. 3 and Table 2) demonstrated susceptibility to A-MuLV but the titer for each was 1,000-fold less than that for the positive-control Pit2 cell line. These results are consistent with the diminished A-MuLV receptor function observed with these receptors in the transient assay. Receptors which scored lower than 50% of the Pit2 control in the transient infection assay typically produced titers approximately 100-fold lower than those produced by Pit2 in the assay of stable cell lines. Three receptors, GGAGG, Pit1-1, and Pit1-2, do not confer susceptibility to PA317/G1BgSvN when tested in either transient- or stable-expression experiments. All of these mutant receptors efficiently mediate GALV entry when expressed in MDTF cells, demonstrating that the mutations have not caused improper folding or cell membrane targeting (Fig. 3 and Table 2).
FIG. 3.
Analysis of A-MuLV and GALV receptor function after stable expression in either CHO K1 or MDTF cells, respectively. CHO K1 cells expressing receptor cDNAs were exposed to PA317/G1BgSvN retroviral vectors at dilutions of supernatant ranging from 1:2 to 1:1,000 (2 ml per well). MDTF cells expressing receptor cDNAs were exposed to PG13/G1BgSvN retroviral vectors at two dilutions of supernatant, 1:100 and 1:1,000 (2 ml per well). Infection assay procedures for both vectors are described in the Materials and Methods section. Mean titers are represented graphically for PA317/G1BgSvN infection of receptor-expressing CHO K1 cells (black bars) and PG13/G1BgSvN infection of receptor-expressing MDTF cells (gray bars).
DISCUSSION
Chimeric receptors have been invaluable in mapping the important regions of virus-receptor interaction for several retroviral receptors, including those for E-MuLV (1) and avian leukosis virus (26), as well as the human immunodeficiency virus (HIV) receptor (3, 20). However, the results obtained using chimeric receptors have been less revealing for both A-MuLV and the HIV coreceptors (3, 20). In the case of A-MuLV, two reciprocal chimeras with either the first three or last two domains of Pit2, AAAGG and GGGAA, are functional for A-MuLV infection (8, 18), indicating that neither the N-terminal nor the C-terminal domains of Pit2 can exclusively account for A-MuLV receptor function. Supporting these findings is the observation that similar reciprocal chimeras constructed by interchanging regions either of Pit1 and HaPit2, the Pit2 homolog expressed in E36 hamster cells (25), or of Pit1 and RaPit2 (rat homolog of Pit2; formerly termed Ram-1) (15), are also functional for A-MuLV infection (8). The finding that both GAGGG and GAAGG chimeras function as receptors for A-MuLV and feature domain II of Pit2, while GGAGG lacks the second extracellular domain of Pit2 and is nonfunctional for A-MuLV infection, is the first unambiguous indication of influence by any particular domain of Pit2 on the A-MuLV receptor interaction. Although it remains evident that no single domain is both necessary and sufficient, the putative second extracellular domain clearly plays a critical role in the interaction with A-MuLV.
We have used a transient assay system for an initial assessment of A-MuLV receptor function, followed by stable expression of the receptors in CHO K1 cells. Evaluation of infection in stable cell lines provides an expanded range with which to more accurately distinguish impaired receptor function from a complete loss of receptor function, and is therefore necessary for accurate quantitation of relative A-MuLV receptor efficiency. Several receptors which routinely failed to function for A-MuLV infection in the transient assay are functional when assayed in established cell lines, although consistently 1,000-fold less efficient than the positive control, Pit2. None of the mutant receptors tested exhibit any significant change in GALV receptor function relative to Pit1, indicating that all mutant and chimeric receptors are capable of being appropriately expressed in cells. The results with stable receptor-bearing cell lines confirm that receptors which were less than 5% of the Pit2 control in the transient assay have impaired A-MuLV receptor function. The infection experiments reported here rely on viral entry and expression. Our system does not distinguish whether a 1,000-fold decrease in A-MuLV infection efficiency correlates with defective entry or with a decrease in receptor binding affinity.
In examining the potential role of the second extracellular domain in A-MuLV infection, several possibilities can be considered unlikely based on the current results. First, the dramatic effects involving charged residues that have been reported for the interaction between GALV and the fourth extracellular domain of Pit1 (5, 8) are not evident as part of the interaction between the second extracellular domain of Pit2 and A-MuLV, as mutations of charged residues alone in this domain neither restored nor abrogated function. In addition, the impact of large hydrophobic residues upon virus-receptor interaction, observed by Zingler and Young (27) with the avian leukosis virus and its receptor, does not appear to be a factor in the loss of A-MuLV receptor function (mutants C1-1 and C1-2). Finally, the presence of a linear virus recognition sequence in the second extracellular domain is unlikely, considering both the large number of different mutations and the combinations of mutations tolerated in this domain without a loss of A-MuLV receptor function.
The amino acid at position 138, methionine in Pit2 and serine in Pit1, appears to have a pivotal influence on A-MuLV receptor function. The single mutation of M138S, or its reverse, affects receptor function significantly in both Pit1 and GAGGG: the Pit1-5 mutant is functional, although at reduced efficiency relative to Pit2, and the C1-8 mutant is 1,000-fold less efficient than Pit2. The apparent importance of this single difference between the receptors is perhaps less surprising when viewed in terms of its possible structural implications. The occurrence of serine in protein sequences is most often associated with turn structures, while methionine is found more often in β-sheets (7). Mutation of the amino acid at position 141 from isoleucine to valine resulted in marginal receptor function in both the transient assay and the assay of the stable Pit1-6 cell line. While the I141V change seems a relatively conservative mutation, the difference in the size of the side chains, owing to the additional methyl group in isoleucine relative to valine, and the statistically significant association of valine with β-sheet structure (7) are possible explanations for the apparent effect on A-MuLV receptor function. The observation that the S138M and I141V changes, either together or alone, confer A-MuLV receptor function on Pit1 may indicate that a specific element of secondary structure, such as a β-sheet, is required in this portion of domain II for proper interaction with A-MuLV or with some other part of the receptor. Whether it is because the residues at positions 138 and 141 of domain II participate in direct interaction with the viral SU or with elements elsewhere in the receptor or because these residues strongly influence secondary structure in this region of the receptor, which in turn affects the interaction with A-MuLV, these residues seem to be an integral part of the role played by the second extracellular domain in A-MuLV receptor function.
The flexibility of the putative second extracellular domain with respect to A-MuLV receptor function is evidenced not only by toleration of a significant number of mutations in the region but also by compensation involving specific combinations of residues in this domain. The type and arrangement of mutational combinations which do not alter A-MuLV receptor function in either native Pit1 or GAGGG suggest that the topology of this domain may have more impact on A-MuLV receptor function than specific interactions involving amino acid side chains and argue against the possibility that a linear epitope of viral interaction exists in this region of the receptor. A compensatory effect on C1-8 mutant receptor function has been observed with mutants C1-9, C1-10, and C1-11, in which the negative effect of the M138S (C1-8) mutation is overcome by other mutations in the domain, and with Pit1-1 and Pit1-5, in which the positive effect of the S138M change is not evident when it is introduced in combination with other changes. Compensation within this region of the second extracellular domain, in the absence of any other obvious correlations between charge or hydrophobicity and A-MuLV receptor function, is further support for the possible importance of the domain II secondary structure in the interaction with A-MuLV rather than a requirement for specific residues. This finding is not unexpected considering the results from chimeric receptor studies, in which C-terminal domains derived from Pit2 are able to compensate in terms of A-MuLV receptor function when N-terminal domains have been derived from Pit1 and vice versa (Table 1), indicating compensation on the domain level.
Although the involvement by domain II in A-MuLV infection has been effectively demonstrated, the nature of its involvement remains unclear. It is reasonable to speculate that neither a concise viral recognition sequence nor a lock-and-key mechanism which involves the second extracellular domain is in operation. The interaction between domain II and A-MuLV might instead be more of a “loose fit,” with a minimum number of interchangeable contact points required. Alternatively, domain II may not be involved in a direct interaction with A-MuLV but rather may participate either as one of several domains required to create the necessary topological features for A-MuLV binding or as a stabilizing element in an interaction between A-MuLV and other regions of the Pit2 receptor. Studies with other retroviral systems have provided similar conclusions. The discovery that HaPit2, the hamster homolog of Pit2, functions as a receptor for GALV even though it differs from Pit1 in seven of the nine residues in the proposed GALV binding site (25) suggests that overall conformational determinants rather than a particular sequence of amino acid residues are acting to influence GALV entry. Our chimera and mutational results with A-MuLV are in accord with these findings, indicating both a high degree of tolerance for sequence variation and the capacity for compensation within the second extracellular domain. A recent mutational analysis of the GALV binding site, in which mutant forms were substituted for the corresponding sequence in Pit1 and several chimeric receptors, has demonstrated that the ability of mutations in the binding site to alter GALV receptor function is dependent upon the receptor into which they are introduced (5). In support of these conclusions, chimeric receptor studies have indicated that more than one domain of the Pit receptors is involved in both GALV (21) and A-MuLV infection (8, 21). Involvement of regions other than domain II of the Pit2 protein in A-MuLV entry is consistent with the observed effects of domain II on viral infection efficiency presented here.
TABLE 5.
Numerical presentation of titer values shown graphically in Fig. 3
| Receptor | Mean titera ± SD for infection with:
|
|
|---|---|---|
| PA317/G1BgSvN | PG13/G1BgSvN | |
| Pit2 | 259,667 ± 53,049 | <0.001 |
| Pit1 | <0.001 | 420,250 ± 29,788 |
| Pit1-1 | <0.001 | 450,100 ± 80,752 |
| Pit1-2 | <0.001 | 191,550 ± 29,808 |
| Pit1-3 | 104.4 ± 11.5 | 333,700 ± 8,910 |
| Pit1-4 | 324.8 ± 127 | 442,000 ± 63,922 |
| Pit1-5 | 496.8 ± 10.2 | 33,100 ± 8,061 |
| Pit1-6 | 14.6 ± 2.0 | 342,600 ± 51,760 |
| Pit1-7 | 6.9 ± 3.0 | 494,350 ± 133,926 |
| GAAGG | 255,950 ± 27,365 | 375,000 ± 69,296 |
| GGAGG | <0.001 | 331,900 ± 80,469 |
| GAGGG | 215,100 ± 39,834 | 314,700 ± 62,650 |
| C1-7 | 170,750 ± 18,738 | 408,600 ± 105,500 |
| C1-8 | 116.9 ± 4.0 | 376,400 ± 10,748 |
| C1-9 | 304,133 ± 74,105 | 223,300 ± 5,657 |
| C1-10 | 3,907 ± 740 | 279,100 ± 164,190 |
| C1-12 | <0.001 | 174,100 ± 21,920 |
| None | <0.001 | <0.001 |
PA317/G1BgSvN and PG13/G1BgSvN titers represent the average numbers of blue foci per triplicate well and per duplicate well, respectively, in each of two dilutions.
ACKNOWLEDGMENTS
We thank Keith Peden for critical review of the manuscript and the Protein and Nucleic Acid Lab Core facility in the Center for Biologics Evaluation and Research for synthesis of oligonucleotides.
REFERENCES
- 1.Albritton L M, Kim J W, Tseng L, Cunninghan J M. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 3.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 4.Bonham L, Wolgamot G, Miller A D. Molecular cloning of Mus dunni endogenous virus: an unusual retrovirus in a new murine viral interference group with a wide host range. J Virol. 1997;71:4663–4670. doi: 10.1128/jvi.71.6.4663-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudry G J, Eiden M V. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J Virol. 1997;71:8078–8081. doi: 10.1128/jvi.71.10.8078-8081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien M-L, Foster J L, Douglas J L, Garcia J V. The amphotropic murine leukemia virus receptor gene encodes a 71-kilodalton protein that is induced by phosphate depletion. J Virol. 1997;71:4564–4570. doi: 10.1128/jvi.71.6.4564-4570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou P Y, Fasman G D. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 8.Eiden M V, Farrell K B, Wilson C A. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J Virol. 1996;70:1080–1085. doi: 10.1128/jvi.70.2.1080-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: a study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLachlin J R, Mittereder N, Daucher M B, Kadan M, Eglitis M A. Factors affecting retroviral vector function and structural integrity. Virology. 1993;195:1–5. doi: 10.1006/viro.1993.1340. [DOI] [PubMed] [Google Scholar]
- 12.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 14.Miller A D, Wolgamot G. Murine retroviruses use at least six different receptors for entry into Mus dunni cells. J Virol. 1997;71:4531–4535. doi: 10.1128/jvi.71.6.4531-4535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunne K J, Sass P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 17.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 18.Pederson L, Johann S V, van Zeijl M, Pedersen F S, O’Hara B. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J Virol. 1995;69:2401–2405. doi: 10.1128/jvi.69.4.2401-2405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rein A. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982;120:251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- 20.Rucker J, Samson M, Doranz B J, et al. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 21.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tailor C S, Takeuchi Y, O’Hara B, Johann S V, Weiss R A, Collins M K L. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O’Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson C, Eiden M. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J Virol. 1991;65:5975–5982. doi: 10.1128/jvi.65.11.5975-5982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young J A T, Varmus H E, Bates P F. A protein related to the LDL receptor is a cellular receptor specific for subgroup A avian leukosis and sarcoma viruses. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 61–73. [Google Scholar]
- 27.Zingler K, Young J A. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J Virol. 1996;70:7510–7516. doi: 10.1128/jvi.70.11.7510-7516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]