Abstract
Hepatitis B virus consists of an outer envelope and an inner capsid, or core, that wraps around the small genome plus the viral replication enzyme. The icosahedrally symmetric nucleocapsid is assembled from multiple dimeric subunits of a single 183-residue capsid protein, which must therefore contain interfaces for monomer dimerization and for dimer multimerization. The atomic structure of the protein is not known, but electron microscopy-based image reconstructions suggested a hammerhead shape for the dimer and, very recently, led to a tentative model for the main chain trace. Here we used a combination of interaction screening techniques and functional analyses of core protein variants to define, at the primary sequence level, the regions that mediate capsid assembly. Both the two-hybrid system and the pepscan technique identified a strongly interacting region I between amino acids (aa) 78 and 117 that probably forms part of the dimer interface. Surprisingly, mutations in this region, in the context of a C-terminally truncated but assembly-competent core protein variant, had no detectable effect on assembly. By contrast, mutations in a second region, bordered by aa 113 and 143, markedly influenced capsid stability, strongly suggesting that this region II is the main contributor to dimer multimerization. Based on the electron microscopic data, it must therefore be located at the basal tips of the dimer, experimentally supporting the proposed main chain trace.
Hepatitis B virus (HBV), the causative agent of acute and chronic B-type hepatitis in humans (for a review, see reference 5), is a small enveloped DNA virus that replicates via reverse transcription of an RNA intermediate (for reviews, see references 24 and 25). Its 3.2-kb genome encodes a reverse transcriptase (P), a transactivator (X), three envelope proteins, and a single capsid, or core protein (C) of 183 amino acids (aa); a secreted, processed, and nonparticulate form of the protein is known as HBeAg. Authentic nucleocapsids are generated, in a highly specific reaction, by assembly of the capsid protein around a complex of P protein bound to a structured RNA element on one of the viral transcripts; subsequent conversion of the RNA into DNA yields mature core particles that acquire their outer envelope during export from the cell (for a review, see reference 26).
Heterologously expressed core protein, in the absence of further viral products, still forms particles resembling authentic liver-derived HBV capsids (10, 18). C-terminal truncations up to aa 144 or 140 rendered the protein assembly competent, while no particles were detected with the further truncated and poorly expressed variants ending with aa 138 or 139 (4, 43). These results defined the first 140 aa as the assembly domain of the protein (Fig. 1). Functional studies in the context of the complete viral genome demonstrated that the highly basic, R-rich C terminus, starting at aa 150, acts as a nucleic acid binding domain that is required for RNA encapsidation and proper reverse transcription (17, 22). Further genetic and biochemical analyses showed that the protein forms dimers that can be disulfide linked via the C-61 residues in the two monomers (23, 40) and that dimers are the only detectable assembly intermediates (41). While full-length capsids are extraordinarily stable, probably owing to the additional interactions between the basic C terminus and (nonspecifically) encapsidated RNA (4), capsids from truncated variants can be dissociated into dimers that spontaneously reassemble in vitro (39).
FIG. 1.
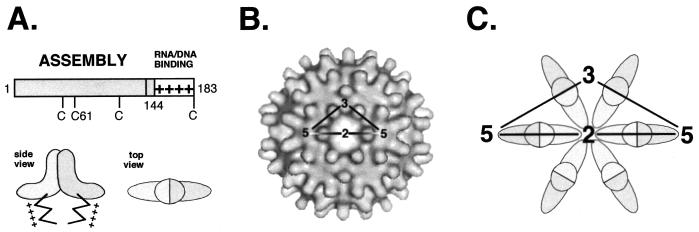
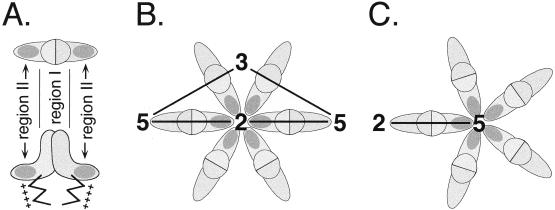
Structural organization of the HBV core protein. (A) Functional domains. The bar represents the primary sequence. The assembly domain is located in the first 144 aa and is followed by a nucleic acid binding region containing four clusters of R residues (indicated by +). The schematic representations of the core protein dimer shown below are based on cryo-electron microscopic reconstructions of capsids. The dimer interface forms the spikes visible on the capsid surface, and the R-rich C termini face its interior. (B) Architecture of the T=4 HBV capsid. A total of 120 dimers are arranged on an icosahedrally symmetric surface lattice; two-, three- and fivefold symmetry axes are indicated (adapted from reference 18). (C) Schematic representation of the arrangement of dimers around a local sixfold (strict twofold) axis of symmetry. The view is the same as in panel B.
After mapping studies of antibody-binding (27) and protease-sensitive (reviewed in reference 26) sites, more detailed structural information came from cryoelectron microscopic analyses of capsids (12, 18, 43). They showed two classes of icosahedrally symmetric particles, containing 90 (triangulation number T=3) and 120 (T=4) hammerhead-shaped, dyad-symmetry related dimers (Fig. 1A). Each dimer forms a prominent spike on the particle surface (Fig. 1B). Very recent data at resolutions below 10 Å revealed that the dimer interface is formed by a four-helix bundle (6, 11), in accord with a theoretical predicition (7). From these data, Böttcher et al. (6) proposed a largely α-helical model for the fold of the core protein, including a tentative main chain trace, that is compatible with the available biochemical data.
In this study, we used a combination of interaction screening techniques and functional analyses of mutant core proteins to identify, at the primary sequence level, the regions in the protein that are involved in mediating the contacts between subunits; the rationale was that each monomer must have at least two interfaces, one mediating the dimerization of monomers and the other mediating the multimerization of dimers. The screening data revealed two prominent interaction regions, one between aa 78 to 117 (region I) and the other between aa 113 to 143 (region II). To functionally test the relevance of these regions, a series of core protein variants, designed according to the interaction data, were expressed in Escherichia coli and assayed for assembly competence. Surprisingly, mutations in region I had no detectable effect, although from the electron microscopy-based model (6), region I would overlap with two of the helices forming the four-helix bundle in the dimer. By contrast, several mutations in region II markedly affected particle stability, strongly suggesting region II to be one of the elements, if not the major element, in multimerization of the dimers. Since the dimer contacts are clearly revealed at the fivefold and quasi-sixfold vertices in the electron microscopy-based reconstructions, primary sequences between aa 113 and 143 are located at each of the two basal tips of the dimer.
MATERIALS AND METHODS
Plasmids.
Plasmids pGAD-HBc1/183 and pGBT-HBc1/183 contain a synthetic full-length HBV C gene from plasmid pPLC4-1 (21) cloned into the polylinker sequences of pGBT9 and pGAD424 (Clontech, Heidelberg, Germany); these, as well as the corresponding HBV X gene control constructs, were kindly provided by L. Runkel.
Plasmids encoding GAD fusions with C terminally truncated core protein (pGAD/c1–36, pGAD/c1–47, pGAD/c1–81, pGAD/c1–94, pGAD/c1–117, pGAD/c1–136, and pGAD/c1–149), or internal fragments (pGAD/c28–47, pGAD/c36–149, pGAD/c42–117, pGAD/c42–149, pGAD/c78–117, and pGAD/c94–117) were created by excision of the corresponding C gene restriction fragments from pPLC4-1, or existing pGAD derivatives, and insertion, after appropriate modification of the ends, between the SmaI and BamHI sites in plasmid pGAD-3-stop; this pGAD424 derivative carries a synthetic linker introducing stop codons in all three reading frames after the cloning sites between the original BamHI and PstI sites (GGATCCTAGGTGAGTGACCTGCAG; stop codons underlined); hence, the fusion proteins carry only two to four foreign amino acids at their C terminus. The plasmid names indicate the first and last aa of the core protein present in the encoded fusion proteins.
For the construction of the E. coli expression plasmids, an AlwNI site was first created at core nucleotide (nt) 294 by silent exchanges via PCR-mediated mutagenesis (CAA CTC TTG→CAG CTC CTG; mutated residues in bold) of plasmid pPLC/c1–149; this derivative of plasmid pPLC4-1 (21), under the control of the phage lambda pL promoter, produces substantial amounts of particulate protein c1–149 upon heat induction (4). Point mutations at aa T91 and K96 were introduced by replacing the XbaI (nt 241)-AlwNI fragment with synthetic oligonucleotide duplexes that were degenerate at the respective codon positions. Mutations at R127 and P138 were generated by replacing the XbaI (nt 241)-SalI (nt 420) fragment with PCR-derived fragments obtained with an appropriately degenerated primer. Colonies were picked at random, and the individual sequences were verified by sequencing. No attempts were made to isolate all the expected sequences. Plasmid pPLC/c1–124 was obtained, within a series of C-terminal truncations, by Bal 31 nuclease treatment of HindIII-linearized plasmid pPLC/c1–149. All enzymes for molecular cloning experiments were either from Boehringer (Mannheim, Germany) or New England Biolabs (Bad Schwalbach, Germany) and were used as specified by the manufacturer.
Bacterial and yeast strains.
For plasmid preparations, E. coli Top10 (Invitrogen, NV Leek, The Netherlands) was used. Expression experiments were performed either by thermoinduction (42°C) of appropriately transformed E. coli NF1 (29) cells (this strain carries an integrated copy of the temperature-sensitive lambda cI857 repressor) or by tryptophan induction of the corresponding GI724 or GI698 (Invitrogen) cells (in these strains, synthesis of the lambda cI repressor is suppressed by the addition of tryptophan); transformation and protein induction were performed as specified by the manufacturer recommendations; usually, the cells were grown to an optical density of 0.5 (600 nm) and induced for 3 to 4 h at 32°C; the P138G variant could be expressed in soluble form only at 23°C.
For the two-hybrid experiments, yeast strains HF7c and SFY526 (Clontech) grown in YPD medium (2) were used. Yeast transformations were performed by the lithium acetate method essentially as described previously (2), except that the transformed HF7c cells were grown for 2 days in plasmid-selecting liquid medium before an aliquot was plated on His− plates. β-Galactosidase (β-gal) activity was assayed in SFY526 cells grown in His-containing medium or in HF7c cells grown without His, after replica plating the yeast colonies on filters containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside). Double transformants containing pGAD/c1–183 and pGBT/c1–183 were positive in all assays. Negative controls included either plasmid alone, a combination of pGAD/c1–183 and pGBT/HBx, and the reverse combination; all were negative in the growth test and in the β-gal assay. For all constructs scoring positive in the SFY526 cell β-gal assays, specificity was confirmed by demonstrating that no reaction occurred with pGBT/HBx.
Protein purification.
Protein c1–149 capsids were purified as previously described (4). Briefly, bacterial cells from 1 liter of induced culture were lysed by sonication and core protein was precipitated with 40% saturated ammonium sulfate. The precipitate was resuspended in phosphate-buffered saline (PBS) (140 mM NaCl, 2 mM NaH2PO4, 8 mM Na2HPO4 [pH 7.4]) and subjected to sedimentation in 10 to 60% (wt/vol) sucrose gradients in PBS (SW-40 rotor; 30,000 rpm for 15 h at 4°C). Fourteen 0.9-ml fractions were collected, and the presence of core protein was assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (0.1% SDS, 15% polyacrylamide) in the Laemmli system (19) with Coomassie blue staining. The same procedure was used for variants mutated at aa T91, K96, and P138.
For variants R127L and R127Q, no particles were observed on analytical sucrose gradients after the ammonium sulfate precipitation step. The precipitate was dissolved in 9 ml of 25 mM sodium phosphate (pH 8.0) and dialyzed overnight at 4°C against the same buffer containing 2 M urea. Aliquots of 5 ml were subjected to size exclusion chromatography with a fast protein liquid chromatography system (Pharmacia, Freiburg, Germany) on a Hiload 16/60 Superdex 200HR column equilibrated in the same buffer. Both proteins eluted at a volume expected for dimers and were used without further purification. Protein c1–124 was similarly enriched by size exclusion chromatography on Superdex 75HR.
Small-scale sucrose gradients.
Cells from 30-ml cultures were collected by low-speed centrifugation, and the pellets were sonicated in 0.3 ml of PBS. The lysates were cleared by centrifugation, and 0.2 ml of the soluble fraction was loaded onto 10 to 60% (wt/wt) sucrose gradients (six steps of 0.2 ml each) in PBS (TLS55 rotor; 55,000 rpm for 40 min at 20°C), essentially as previously described (42). Fourteen fractions of 0.1 ml were collected from the top, and 10 to 20 μl of each fraction was monitored for core proteins by SDS-PAGE as described above or by Western blotting. Dimeric core protein, together with the bulk of E. coli proteins, was typically present in fractions 1 to 3, and capsids were present in fractions 6 to 8. For protein P138G, the sedimentation analysis was repeated on gradients containing TAE buffer (40 mM Tris-acetic acid [pH 8.1], 0.1 mM EDTA). The same gradient system was also used for analytical purposes.
Agarose gel electrophoresis of core particles.
Agarose gel assays were performed as previously described (4). In brief, fractions containing between 1 and 10 μg of purified or enriched core protein were loaded on 1% (wt/vol) agarose (SeaKem; FMC Bioproducts, Natick, Maine) gels in TAE buffer and electrophoresed for about 3 h at 10 V/cm. In some experiments, encapsidated RNA was visualized by staining with 0.5 μg of ethidium bromide per ml and illumination at 302 nm. Proteins were stained with Coomassie blue. Core protein-specific bands were detected, after blotting the contents of the gel by capillary transfer in 20× SSC (0.3 M sodium citrate [pH 7.0], 3 M NaCl) to a nylon membrane (Porablot NY; Macherey-Nagel, Düren, Germany), with the monoclonal antibodies mc275 and mc312 (31, 32). Differential capsid stabilities were assayed by including 1 M urea in the gel.
Pepscan analysis.
The pepscan filters (Jerini Bio Tools, Berlin, Germany) contained all possible 15-mer peptides spanning the core protein sequence from positions 1 to 161, each shifted by 2 positions. Pretreatment of the filter, incubation with core protein, and detection were performed essentially as recommended by the supplier. In brief, after blocking with 5% nonfat milk powder in Tris-buffered saline (TBS) (100 mM Tris-HCl [pH 8.0], 150 mM NaCl) containing 5% sucrose, the filter was incubated at 4°C overnight with the respective core protein (about 30 μg in 10 ml of the same buffer without milk powder). After two washes with TBS, four sequential electrophoretic transfers of the protein to a polyvinylidene difluoride (PVDF) membrane (Millipore, Eschborn, Germany) were performed. Core protein on the membranes was detected immunologically with the enhanced chemiluminescence system (Amersham, Braunschweig, Germany).
Dissociation and reassociation of capsids.
Dissociation of gradient-purified capsids was performed by overnight dialysis against 25 mM sodium phosphate (pH 8.0) containing 2 M urea. Dimer formation was monitored by using analytical sucrose gradients and size exclusion chromatography in PBS as described above. For reassociation, isolated dimers, stored in phosphate buffer containing 2 M urea, were dialyzed overnight at 37°C against 25 mM sodium phosphate (pH 7.0) containing 100 or 500 mM NaCl.
Immunological techniques.
For Western blotting experiments, the core proteins present on polyacrylamide or agarose gels were transferred to PVDF or nylon membranes and analyzed with monoclonal antibodies mc312, recognizing aa 76 to 84 as a linear epitope (31, 32), and mc275, recognizing only particulate core protein (32). Both antibodies were kindly provided as horseradish peroxidase conjugates by Behringwerke (Marburg, Germany). Detection was performed with the enhanced chemiluminescence substrate.
RESULTS
A two-hybrid-system screen suggests the presence of a central (region I) and an N-proximal (region III) interaction site in the HBV core protein.
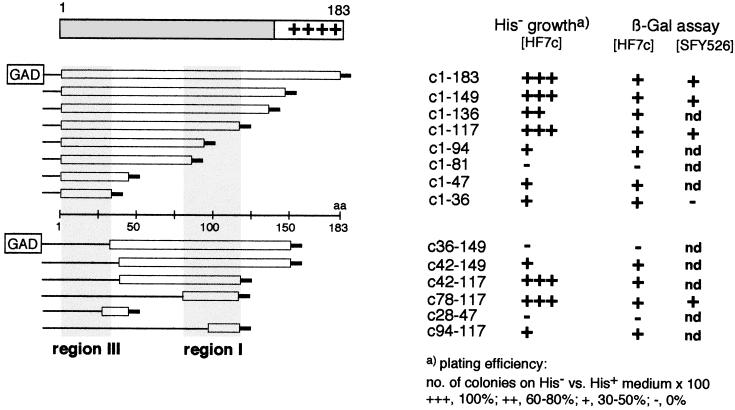
The yeast two-hybrid system (14, 20, 38) is an increasingly popular tool for identifying protein-protein interactions. Here we used a fusion of the complete HBV core gene with the Gal4 DNA binding domain (encoded by plasmid pGBT/c1–183) to screen a series of constructs encoding N-terminal and internal fragments of the core gene fused to the Gal4 activation domain (pGAD derivatives) (Fig. 2). An interaction between the fusion proteins should allow yeast cells containing both plasmids to grow on His− medium and to produce β-gal, giving rise to blue colonies on X-Gal-containing medium. As a positive control, a combination of plasmids pGBT/c1–183 and pGAD/c1–183, both containing the complete core gene, was used and produced the expected phenotypes. Individually or in combination with the respective HBV X gene plasmids, both constructs were negative in both assays, confirming the specificity of the test system. Of the constructs encoding C-terminally truncated core protein fusions, all except c1–81 scored positive in the β-gal and His− growth test in Hf7 cells (Fig. 2), although differences in plating efficiency that probably relate to the strength of the interactions were seen (16). The smallest fragment contains residues 1 to 36, suggesting the presence of an interaction site(s) in the N-terminal region (region III). In SFY526 cells, this construct did not give rise to blue colonies on X-Gal plates. By contrast, plasmid pGAD/c1–117, like pGAD/c1–183 and pGAD/c1–149, was clearly also positive in the SFY526 strain, suggesting the presence of one or more additional stronger interaction sites.
FIG. 2.
Homologous interactions detected with the yeast two-hybrid system. (Left) Overview of the pGAD/core fusion constructs. The bars below the schematic representation of the core protein indicate the core protein fragments present in individual constructs with respect to the primary sequence (central line). The short thick lines at the ends symbolize the presence of two to four vector-derived amino acids. Shaded areas correspond to the detected interaction regions I and III. (Right) Interaction data obtained with double transformants containing pGBT/c1–183 and the corresponding pGAD construct. The numbers indicate the first and last core-specific amino acids present in each construct. All constructs were tested for growth on His− medium and for β-gal activity in HF7c cells; selected constructs were also tested for β-Gal activity in SFY526 cells. nd, not determined.
This was confirmed by using the series of constructs encoding internal core protein fragments. Those containing aa 42 to 149, 42 to 117, 78 to 117, and 94 to 117 conferred growth on His− medium and gave blue colonies in HF7 cells. pGAD/c78–117 was also positive in the β-gal assay in strain SFY526, indicating that this sequence contains a prominent interaction site, which we will refer to as region I; in the electron microscopy-based structure model (6), this region would be part of the sequence forming the dimer interface.
Plasmid pGAD/c28–47 scored negative in all assays, suggesting that this sequence does not contribute to interactions. Surprisingly, construct pGAD/c36–149, but not pGAD/c42–149, was also negative, although it contains region I; a similar effect was seen with pGAD/c1–81, which also contains the sequence 1 to 36, which by itself was positive in Hf7 cells. Possibly, the additional sequences present in the two proteins lead to misfolding and/or instability.
Together, these experiments identified the core sequence 78 to 117, i.e., region I, as a major interaction site. The positive growth test with pGAD/c1–36, in view of the clearly negative results with some other constructs, suggests the presence of an additional, N-proximal interaction site, i.e., region III. To independently confirm these conclusions and to distinguish between regions involved in dimerization and multimerization, which is intrinsically difficult with the two-hybrid system, as well as for a more precise mapping, we turned to the pepscan technique.
The pepscan technique confirms region I as a strong interaction site and suggests the presence of an additional C-proximal interacting region (region II).
In the pepscan technique, originally developed by Geysen et al. (15), the sequence of a protein is represented as a series of relatively short overlapping peptides that are immobilized on a solid support, e.g., cellulose filters (for a review, see reference 28). The filter is then probed with the protein of interest. Detection of the protein on the filter, directly or after transfer to a second membrane, identifies interacting peptides.
Here we used filters containing the core protein sequence 1 to 161 as 15-mer peptides, each shifted by 2 aa, and incubated them with dimers of core protein c1–149 obtained by dissociation of purified capsids. Since the dimer interface should not be accessible in this probe, we expected it to identify mainly peptides involved in dimer multimerization. After incubation, the protein was electrophoretically transferred from the filter to PVDF membranes; usually four sequential transfers were performed to optimize the signal-to-background ratio. Protein c1–149 was then detected with a polyclonal rabbit anti-core antiserum and an anti-rabbit immunoglobulin G-peroxidase conjugate with a chemiluminescent substrate.
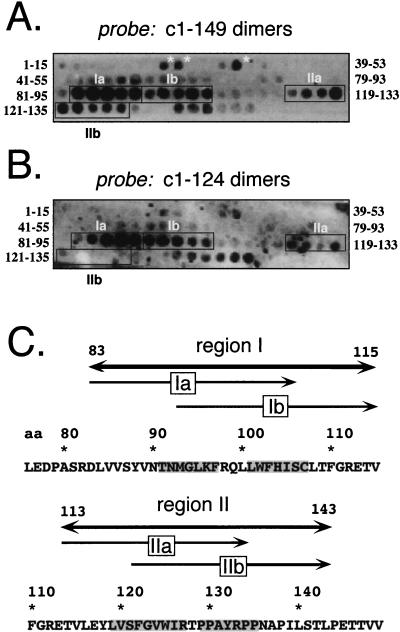
A typical result is shown in Fig. 3A. In addition to a few individual weakly reactive spots, e.g., peptides 15 to 29, 17 to 31, and 25 to 39 and several C-terminal peptides (starting with 137 to 151), two major extended regions comprising adjacent peptides were identified. Region I starts with peptides 83 to 97 and ends with peptides 101 to 115. In several experiments, the first 5 of the 10 peptides reacted more strongly than the following ones; region I was therefore subdivided into regions Ia and Ib (Fig. 3C). The second series of spots started with peptides 113 to 127 and extended to peptides 129 to 143. As outlined below, this region (region II) also appeared to bipartite and hence was divided into subregions IIa and IIb. Together, these data suggested that the sequences from 83 to 115 and 113 to 143 contain important interaction sites. In particular, region I identified by the pepscan technique is almost exactly congruent with the sequence comprising region I in the two-hybrid screen (aa 78 to 117). In view of the electron microscopy-based model, however, the reactivity of region I in the pepscan assay was surprising, since it should be part of the dimer interface that would not be expected to be available for further interactions with the dimeric protein probe.
FIG. 3.
Pepscan analysis of homologous interactions. (A) Dimeric protein c1–149 as a probe. The core protein sequence from aa 1 to 161 was represented on the filter by 15-mer peptides, each shifted by two positions. The sequences of the first and last peptide in each row are given on the left and the right, respectively. Protein bound to individual spots on the filter was electrophoretically transferred to a PVDF membrane and detected immunologically. Three N-proximal peptides that might correspond to region III in the two-hybrid screen are marked by asterisks. The strongly interacting regions I and II, with subregions a and b, are boxed. (B) Core protein c1–124 as a probe. The pepscan filter was treated as in panel A, except that the assembly-deficient variant c1–124 was used as the probe. (C) Correlation of regions I and II with the primary sequence of the core protein. The indicated borders of complete regions I and II and their subregions a and b correspond to the first amino acid of the first and the last amino acid of the last interacting peptide. Residues that are common to all peptides of a subregion are shaded in the primary sequence.
Together, the pepscan data confirmed a strong interaction in region I, as in the two-hybrid assay, and suggested a second important site within the C-proximal region II; whether the signals with three peptides between aa 15 and 39 correspond to the N-proximal region III in the two-hybrid screen is unclear, since they were rather weak and were not contiguous as in regions I and II. Possibly, the putative site encoded by pGAD/c1–36 is discontinuous and may not be properly represented by the 15-mer peptides on the filter. Because of the uncertainties inherent in both interaction screening techniques, we next sought to more directly test the relevance of regions I and II in capsid assembly.
Analysis of the assembly competence of core protein variants mutated in regions I and II.
If the above-defined candidate regions were indeed important for assembly, changes in their primary sequences would be expected to influence capsid formation and/or stability. We therefore expressed, in E. coli, several core protein variants mutated in the respective regions and characterized their quaternary structure by sedimentation in sucrose gradients, by electrophoresis on native agarose gels, and by gel filtration.
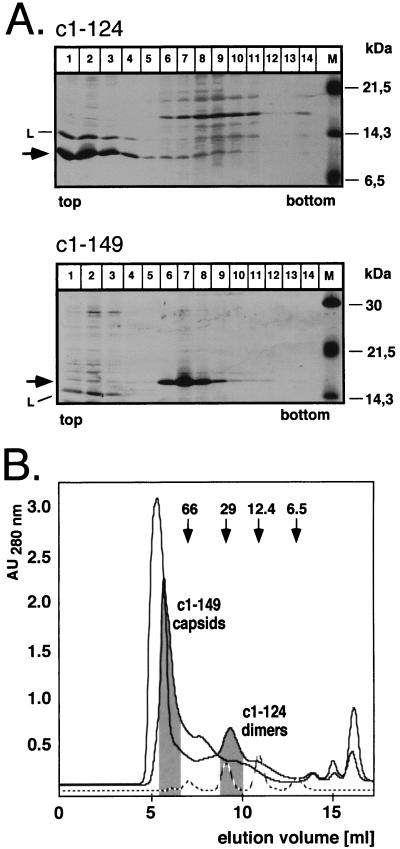
As a first step, we analyzed the effect of deleting most of the C-proximal region II; this was not possible for region I because of its internal localization. Previous experiments had shown that variants truncated after aa 138 and 139 did not form particles (4). However, the corresponding proteins were very poorly expressed. Since capsid assembly is a concentration-dependent process (33, 34), the low concentration of these variants might have substantially contributed to the lack of detectable particle formation. Within a series of further truncation constructs (to be described elsewhere), we found that a variant ending with aa 124 (c1–124) was well expressed in soluble form. When analyzed on sucrose gradients, most of the protein sedimented with the bulk of soluble proteins in fractions 1 to 3, while c1–149, known to form capsids, was present mostly in fractions 6 to 8 (Fig. 4A). Hence, even at concentrations allowing efficient particle formation with c1–149, c1–124 is assembly deficient, suggesting a critical role for the amino acid sequence between residues 125 and 149 in capsid assembly. Next, we used gel filtration on Superdex 75HR to determine whether c1–124 still formed dimers under native conditions. As shown in Fig. 4B, c1–124, with a calculated molecular mass of about 14 kDa, eluted from the column slightly after the 29-kDa marker but clearly ahead of the 12.4-kDa marker; c1–149, as expected, was present in the void volume. With a different set of markers (data not shown), the protein eluted between ovalbumin (44 kDa) and myoglobin (17 kDa), confirming its dimeric nature. This demonstrates that the residues forming the dimer interface are still present in c1–124. When c1–124 was used as a probe in the pepscan analysis (Fig. 3B), the most prominent signals were again detected in region I; in region II, however, only the more N-proximal peptides covering the sequence from 113 to 133, i.e., region IIa, were reactive while the peptides comprising the residues 121 to 143, i.e., region IIb, gave no signals. Hence, the region IIb signals observed with c1–149 but not c1–124 are most probably due to homologous interactions between these C-proximal residues, and these are important for the multimerization of dimers.
FIG. 4.
Quaternary structure of core protein c1–124. (A) Sedimentation analysis. E. coli-derived protein c1–124 (upper panel) was sedimented on an analytical sucrose gradient. Aliquots from each fraction were analyzed by SDS-PAGE and staining with Coomassie blue. Essentially all of the protein was present in the four top fractions (arrow), cosedimenting with the lysozyme (L) used during preparation of the cell lysate. The molecular masses of the marker proteins (lane M) are indicated on the right. Protein c1–149 was analyzed in parallel (lower panel) and was present mainly in fractions 6 to 8. (B) Size exclusion chromatography. Bacterial lysates containing protein c1–124 or c1–149 were analyzed on a Superdex 75 column equilibrated in PBS. The elution profiles and the positions of the two proteins as determined by SDS-PAGE are indicated. The dashed line shows the elution profile for a set of protein markers with the indicated molecular masses. AU280 nm, absorbance units at 280 nm.
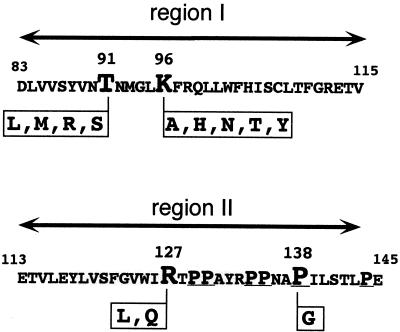
Next we introduced, in the context of protein c1–149, several point mutations into regions I and II (Fig. 5). Based on a mutational pepscan analysis (data not shown) in which the central peptides 86 to 100 and 127 to 139 were resynthesized on the filter with each position being substituted individually with a series of other amino acids (V, I, A, D, F, G, L, P, Q, S, W, or Y), T91 and K96 in region I and R127 and P138 in region II were chosen as targets. At these positions, one or more substitutions reduced the signal intensities when the filter was probed with c1–149. P138, in addition, is one of several clustered P residues in the sequence from 129 to 144 (Fig. 5) and is highly conserved in the core proteins of mammalian HBV. It was also of interest because its replacement by G apparently prevented the production of particulate core protein from a recombinant vaccinia virus (8).
FIG. 5.
Point mutations in regions I and II. In the context of core protein c1–149, T91 and K96 in region I and R127 and P138 in region II were replaced by the indicated amino acids. P138 is located in a P-rich motif; P residues are underlined.
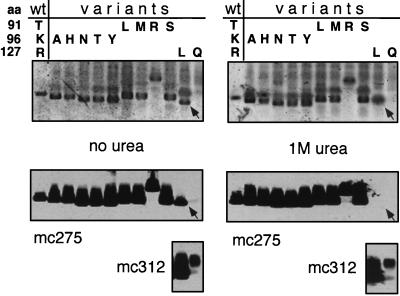
All region I variants tested (T91 replaced by L, M, R, or S; K96 replaced by A, H, N, T, or Y) sedimented essentially like particulate c1–149 (data not shown), indicating that their assembly capability was not significantly affected by the mutations. Particle formation by the variants was confirmed by electrophoresis in native agarose gels (35, 36). In this assay (4), capsids, but not soluble proteins with their higher diffusion coefficients, migrate as distinct sharp-edged bands. The capsid specificity of the assay was further enhanced by blotting the proteins onto a membrane followed by immunological detection with the particle-specific monoclonal antibody mc275, which is essentially nonreactive with nonassembled core protein (32). A representative assay is shown in Fig. 6. All of the T91 and K96 mutants, after staining with Coomassie blue, produced bands with a shape characteristic for capsids; this interpretation was confirmed by an immunoblot with mc275. Interestingly, all K96 variants (substituting the basic lysine side chain with neutral residues) migrated slightly faster than c1–149 capsids, while substitutions of the neutral T91 residue by the positively charged arginine, but not with neutral residues, dramatically retarded migration toward the anode. Since electrophoretic mobility in the gel is a function of size and surface charge (35, 36), these data suggest that the corresponding residues contribute to the surface charge of the particle. Reasoning that the mutations might affect particle stability rather than principal assembly proficiency, we repeated the agarose gel assay in the presence of 1 M urea (Fig. 6, right panels). However, all the variant capsids remained stable. Hence, we were unable to demonstrate, under these conditions, any phenotypic consequences of the mutations in region I. Essentially the same result was obtained with three different double mutants (T91L K96N, T91R K96N, and T91R K96Y [data not shown]).
FIG. 6.
Agarose gel electrophoresis assay of region I and II mutants. Capsid fractions from small-scale sucrose gradients of the indicated core proteins, all in the context of c1–149, were subjected to electrophoresis in agarose gels, in either the absence (left) or the presence (right) of 1 M urea. The amino acids present at positions 91, 96, and 127 in the wild-type protein and the individual variants are indicated at the top. The upper panels show Coomassie blue-stained gels; the lower panels show immunoblots with monoclonal antibodies mc275 and mc312. Only the sharp-edged bands visible in the Coomassie blue stain correspond to particles. Note that variant R127L formed particles only in the absence of urea (arrow), while no particles at all could be detected for variant R127Q.
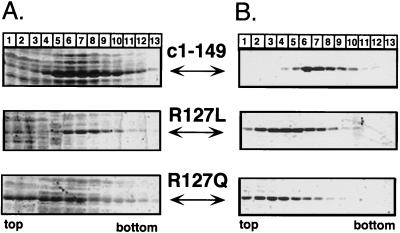
By contrast, the region II mutations R127L and R127Q clearly affected assembly. Sedimentation analysis of the crude E. coli lysates showed that R127L was present in the same fractions (mainly fractions 6 to 9) as particulate c1–149; the bulk of R127Q sedimented slowly, with at most one-third being present in capsid-specific fractions (Fig. 7A). Attempts to purify the variants as capsids by our standard method involving an ammonium sulfate precipitation as an early step were unsuccessful; no particles could be detected by sedimentation after this procedure. Instead, the two variant proteins were enriched in dimeric form by size exclusion chromatography in the presence of 2 M urea. We then tried to reassemble them, in vitro, into particles (see Materials and Methods for details). Under conditions leading to essentially complete reassociation of dimeric c1–149, only a small amount of R127L and almost nothing of R127Q was present in particle-specific gradient fractions (Fig. 7B). The low stability of the mutant capsids was confirmed in the agarose gel assay (Fig. 6, lanes R127 L and Q). Variant R127L, in the absence of urea, showed a typical capsid band that was also detected by mc275. No reaction was seen with the more diffusely distributed slower-migrating material visible in the Coomassie blue stain. With 1 M urea in the gel, however, the capsid-specific band disappeared and no signal was obtained with mc275. The presence of the protein was confirmed by immunological detection with another monoclonal antibody, mc312, which recognizes a linear epitope between aa 76 and 84. Variant R127Q did not produce a particle-specific band even in the absence of urea (lanes 127Q). Hence, mutations of R127, in the center of region II defined by the pepscan technique, significantly reduce particle stability, confirming a critical role for region II residues in multimerization.
FIG. 7.
Sedimentation analysis of R127 variants. (A) Analytical sucrose gradients of crude E. coli lysates. Aliquots of crude lysates from bacteria transformed with the appropriate expression plasmids encoding proteins c1–149 and its R127 variants were subjected to sedimentation in 10 to 60% sucrose gradients without prior ammonium sulfate precipitation. Aliquots of each fraction were analyzed by SDS-PAGE and Coomassie blue staining. R127L sedimented essentially to the same central fractions as did wild-type protein c1–149, while most of protein R127Q was present in the top fractions. (B) Reassociation of isolated dimers. Variants R127L and R127Q, purified as dimers by size exclusion chromatography, were subjected to reassociation conditions, in parallel with isolated c1–149 dimers (see Materials and Methods for details), and the reaction products were analyzed on analytical sucrose gradients. These conditions led to efficient reassociation of the wild-type protein (top), while only small amounts of the variant proteins were detected in particle-specific fractions.
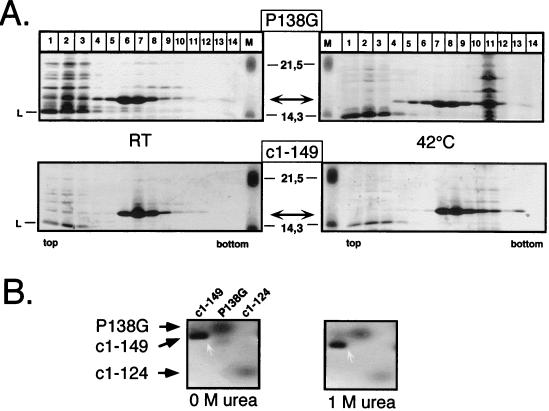
Finally, we subjected variant P138G to similar tests. When expressed at 42°C from a thermoinducible vector system (21), essentially all of the protein was present in the insoluble fraction, suggesting nonspecific aggregation; the R127 variants, by contrast, were mostly soluble under these conditions. Expression at 23°C, however, with a system in which induction is chemically induced by the addition of tryptophan, yielded mainly soluble P138G in particulate form, as shown by sedimentation analysis (Fig. 8A). Suspecting that the variant capsids might be thermolabile, we incubated an aliquot of the preparation side by side with c1–149 capsids at 42°C overnight and reanalyzed it on a sucrose gradient run in PBS. Most of the contaminating E. coli proteins aggregated under these conditions to fast-sedimenting material (fraction 11); however, the variant protein was still present in the same capsid-specific fractions as c1–149. Interestingly, however, P138G capsids were not detectable in the agarose gel mobility assay, even in the absence of urea (Fig. 8B). This apparent discrepancy is probably related to the higher pH (8.1) and lower ionic strength (40 mM Tris-acetic acid) of the TAE buffer used in our standard agarose gel assay than those used in the sedimentation analysis (pH 7.4, PBS); both parameters discourage the assembly of c1–149 particles (39). Indeed, when the sucrose gradients were run in TAE buffer, P138G was completely present in the nonparticulate fraction.
FIG. 8.
Particle formation by variant P138G. (A) Sedimentation analysis. E. coli-derived variant P138G particles were sedimented, in parallel with protein c1–149, on 10 to 60% sucrose gradients after overnight incubation at room temperature (left) or at 42°C (right). Proteins in individual fractions were detected by SDS-PAGE and Coomassie blue staining. The molecular masses of the marker proteins (lanes M) are indicated in the middle; L, lysozyme. Note that both core proteins are present mainly in the central fractions while most of the contaminating E. coli proteins formed fast-sedimenting material (fraction 11) at 42°C. (B) Agarose gel assay. Purified particles from protein c1–149 and its variant P138G, as well as the assembly-deficient variant c1–124, were subjected to electrophoresis in agarose gels containing no urea or 1 M urea (see the legend to Fig. 6). The proteins were stained with Coomassie blue. While c1–149 particles remained stable at 1 M urea (white arrows), no particle-specific bands could be detected for the variants.
Together, the characterization of the above-described core protein derivatives clearly revealed a major contribution to the assembly of residues located in region II, while mutations in region I had surprisingly little effect.
DISCUSSION
Until recently, direct structural information about the HBV core protein remained scarce and theoretical predictions yielded controversial results (1, 7). Here we used a combination of screening procedures to experimentally define primary sequences within the protein that are involved in mediating the contacts between the 240 or 180 subunits forming the complete shell of the HBV capsid. Below we discuss the results in the context of the structural model proposed for the core protein by very recent high-resolution electron microscopy-based reconstructions (6, 11). Our data provide experimental evidence that the electron density visible at the fivefold and local sixfold symmetry axes between the core protein dimers in the capsid is provided mainly by residues from the sequence 113 to 143, i.e., region II, in accord with proposed model. The data for residues 83 to 115, i.e., region I from the two-hybrid and the pepscan screening, however, are not immediately understood in terms of the model.
Comparison of the two-hybrid system and the pepscan results.
Since the HBV capsid is assembled from multiple core protein dimers, one subunit must have at least one interface mediating the dimerization of monomers and one mediating the multimerization of dimers. Both the two-hybrid system (14, 20, 38) and the pepscan technique (28) appeared able to define specific sequences involved in these interactions. In the two-hybrid screen, the interaction between full-length core protein fused to the GAD and GBT domains was clearly detectable in both the His− growth assay and the β-Gal assay. Within a series of terminal deletion constructs (Fig. 2), c78–117 was the smallest core protein fragment that gave the same phenotype, suggesting the presence of a strong interaction site in this region (region I). These results are compatible with the electron microscopy-based (6) as well as the theoretical (7) structural models, since region I overlaps largely with residues predicted to be part of the central helices at the dimer interface. Evidence for an additional interaction site (region III) within the first 36 aa was provided by fragment c1–36, which scored positive in both tests in HF7c cells but displayed a decreased plating efficiency on His− medium and no detectable β-Gal activity in the SFY526 strain, suggesting a weaker interaction (13, 14). That N-terminal residues are important for capsid formation is supported by the known detrimental effect of even short N-terminal deletions or insertions on assembly (3, 37).
The predominant pepscan signals obtained with dimeric c1–149 as the probe spanned aa 83 to 115 (region I) and aa 113 to 143 (region II). The striking agreement with region I defined by the two-hybrid screen suggested that both data sets reflect the same strong interaction. In view of the structural model, however, these pepscan data created a paradox because the dimer interface should not be accessible on the dimeric c1–149 protein probe. One speculative explanation is that the filter-bound peptides, owing to their high local concentration (about 1 nmol per spot of about 5 mm2), are able to penetrate the dimer interface by substituting for authentic interactions within the four-helix bundle. Further experiments are required to substantiate the implied partial or complete dissociation, or at least structural alteration, of the dimer interface and to clarify the exact nature of these interactions; however, the data provided a guideline for the introduction of site-directed mutations into the core protein that allowed us to directly analyze their effects on particle formation.
Functional analysis of the relevance of regions I and II in HBV capsid assembly.
As surprising as the reactivity of region I peptides with dimeric c1–149 protein was the lack of a detectable assembly defect of region I variants at T91 and/or K96 when analyzed in the context of c1–149. The proteins formed particles in E. coli, and these capsids were stable in the agarose gel assay even in the presence of 1 M urea. Unless one doubts the electron microscopy-based assignment of the corresponding residues to the dimer interface (6) this suggests that the mutations were not radical enough to significantly perturb the monomer-monomer interaction. In keeping with such a very robust nature of the dimer interface, which is also predicted by the four-helix-bundle model, is our observation that mutants with C61 replaced by W or R form stable particles (data not shown) although C61 is clearly located in the dimer interface, since it readily forms a homologous disulfide bridge with the same residue in a second subunit (23, 40). While these questions will probably be resolved only by direct higher-resolution structural analyses, a coherent picture for the role of region II emerged from our data.
First, variant c1–124, lacking most of region II, still formed dimers but was assembly deficient even at concentrations of about 0.1 mM, i.e., 10- to 100-fold higher than the critical concentration for assembly estimated from expression of full-length and C-terminally truncated core protein in Xenopus laevis oocytes (around 1 and 10 μM [34]). Hence, the lack of assembly competence correlated directly with the absence of aa 125 to 149. The data also confirmed that the dimer interface is located within the first 124 aa of the protein (Fig. 9A), pointing in turn to an important role for region II in dimer multimerization. In view of the earlier deletion analyses and the selective of loss of pepscan signals in region IIb (peptides 121 to 135 to peptides 129 to 143) with c1–124 as a probe, it is highly likely that majority of the region II residues is involved in this process.
FIG. 9.
Location of regions I and II in the core protein dimer. (A) Top and side view of the dimer. Region II is represented as a dark-shaded area at each of the two basal tips of the dimer. Region I most probably corresponds to residues forming the dimer interface. (B) Arrangement of dimers around a local sixfold axis. According to the mapping of region II (A), the contacts between individual dimers rely mainly on residues located between positions 113 and 143, forming a network around the local sixfold axes. Only one of the two contact sites in each dimer is shown. (C) Arrangement of dimers around a fivefold axis. Based on its icosahedral symmetry, the T=4 capsid must contain 12 pentameric arrangements of dimers. The contacts mediated by region II are similar to those at the local sixfold axes but not identical, implying a certain structural flexibility.
The results obtained with the variants at R127 and P138 further substantiate an important role for region II in dimer multimerization. On sucrose gradients, only a fraction of variant R127Q was present in particulate form in crude E. coli lysates; purified R127Q dimers could not be reassembled under conditions allowing the efficient reassociation of c1–149 dimers, and no particles could be detected by the agarose gel assay. A similar though less pronounced phenotype was observed for variant R127L, which formed particles that were detectable in the agarose gel assay but disintegrated in the presence of 1 M urea. P138G particles were stable on sucrose gradients run in PBS, even after prolonged incubation at 42°C, but could not be detected in the agarose gel assay. Reanalysis by sedimentation in TAE-buffered sucrose revealed that, in contrast to c1–149, all the variant protein was present in the nonparticulate fraction, indicating that P138G capsids are unstable under these conditions.
Since both deletion of region II (c1–124) and mutation of residues in its central (R127) or C-terminal (P138) part profoundly affected multimerization but did not abolish dimerization, we conclude that region II, encompassing the sequence from 113 to 143, contains at least one element if not the major element, driving the assembly of HBV core protein dimers (Fig. 9A) by forming a framework of homologous interactions between region II residues from multiple dimers. Hence, our data provide experimental evidence that the electron density seen in the electron microscopy-based image reconstructions around the fivefold and twofold axes of symmetry is contributed mainly by region II residues (Fig. 9B), corroborating the assignment in the model by Böttcher et al. (6). They are also in accord with the recent localization of residue 150, by electron microscopic visualization of a covalently attached undecagold cluster (44), underneath the outer tips of the dimer; this residue is only a few positions away from the C-terminal border of our region II.
Based on this assignment, the assembly deficiency of c1–124 is self-evident. The more subtle effects of the point mutations, e.g., at R127 and P138, suggest that these changes introduce perturbations in the network of interacting C-proximal residues that, depending on the specific substitution, have gradually different effects on capsid formation and stability. That they are tolerated at all probably reflects a certain flexibility in the structure of region II, which is naturally required to accommodate the quasi-equivalent but nonidentical contacts where five (at the fivefold axes) and six (at the strict twofold axes) subunits meet (Fig. 9B and C). Since these contacts are nonetheless similar, they will respond similarly to dissociating conditions, explaining why dimers rather than higher-order multimers such as pentamers (as seen, for instance, with poliovirus VP1-VP3-VP0 protomers [30]), are the only stable intermediates in HBV capsid assembly (41). The distinct role of region II in assembly also sheds light on the formation of mixed capsids (9) from core protein dimers of HBV and of woodchuck hepatitis B virus (WHV): overall, the WHV core protein sequence from 1 to 143 is 66% identical to that of HBV, while in region II (aa 117 to 143) the proteins exhibit 85% identity.
In summary, the approach of using interaction screens with protein segments and peptides, combined with functional analyses of variant proteins to map individual contact sites on the HBV core protein, has yielded valuable information, in particular on the importance of the C-proximal region II in multimerization. However, the shortcomings of the individual methods revealed in this study also strongly suggest a cautious interpretation of data that are derived by a single-interaction technique.
ACKNOWLEDGMENTS
We thank Andrea Frank for excellent technical assistance and Heinz Schaller for valuable discussions.
This work was supported by grants from the Bundesministerium für Bildung und Forschung and from the “protein-protein interactions” program of the Land of Baden-Württemberg.
REFERENCES
- 1.Argos P, Fuller S D. A model for the hepatitis B virus core protein: prediction of antigenic sites and relationship to RNA virus capsid proteins. EMBO J. 1988;7:819–824. doi: 10.1002/j.1460-2075.1988.tb02880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman I G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 3.Beames B, Lanford R E. Insertions within the hepatitis B virus capsid protein influence capsid formation and RNA encapsidation. J Virol. 1995;69:6833–6838. doi: 10.1128/jvi.69.11.6833-6838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnbaum F, Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumberg B S. Hepatitis B virus, the vaccine, and the control of primary cancer of the liver. Proc Natl Acad Sci USA. 1997;94:7121–7125. doi: 10.1073/pnas.94.14.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böttcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 7.Bringas R. Folding and assembly of hepatitis B virus core protein: a new model proposal. J Struct Biol. 1997;118:189–196. doi: 10.1006/jsbi.1997.3846. [DOI] [PubMed] [Google Scholar]
- 8.Bringas, R. Personal communication.
- 9.Chang C, Zhou S, Ganem D, Standring D N. Phenotypic mixing between different hepadnavirus nucleocapsid proteins reveals C protein dimerization to be cis preferential. J Virol. 1994;68:5225–5231. doi: 10.1128/jvi.68.8.5225-5231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen B J, Richmond J E. Electron microscopy of hepatitis B core antigen synthesized in E. coli. Nature. 1982;296:677–678. doi: 10.1038/296677a0. [DOI] [PubMed] [Google Scholar]
- 11.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 12.Crowther R A, Kiselev N A, Böttcher B, Berriman J A, Borisova G P, Ose V, Pumpens P. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 13.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 15.Geysen H M, Rodda S J, Mason T J, Tribbick G, Schoofs P G. Strategies for epitope analysis using peptide synthesis. J Immunol Methods. 1987;102:259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- 16.Golemis E A, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman I G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 13.14.1–13.14.17. [Google Scholar]
- 17.Hatton T, Zhou S, Standring D N. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J Virol. 1992;66:5232–5241. doi: 10.1128/jvi.66.9.5232-5241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenney J M, von Bonsdorff C-H, Nassal M, Fuller S D. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure. 1995;3:1009–1019. doi: 10.1016/s0969-2126(01)00237-4. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.McNabb D S, Guarente L. Genetic and biochemical probes for protein-protein interactions. Curr Opin Biotechnol. 1996;7:554–559. doi: 10.1016/s0958-1669(96)80061-9. [DOI] [PubMed] [Google Scholar]
- 21.Nassal M. Total chemical synthesis of a gene for hepatitis B virus core protein and its functional characterization. Gene. 1988;66:279–294. doi: 10.1016/0378-1119(88)90364-2. [DOI] [PubMed] [Google Scholar]
- 22.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassal M, Rieger A, Steinau O. Topological analysis of the hepatitis B virus core particle by cysteine-cysteine cross-linking. J Mol Biol. 1992;225:1013–1025. doi: 10.1016/0022-2836(92)90101-o. [DOI] [PubMed] [Google Scholar]
- 24.Nassal M, Schaller H. Hepatitis B virus replication. Trends Microbiol. 1993;1:221–226. doi: 10.1016/0966-842x(93)90136-f. [DOI] [PubMed] [Google Scholar]
- 25.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepatitis. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 26.Nassal M. Hepatitis B virus morphogenesis. Curr Top Microbiol Immunol. 1996;214:297–337. doi: 10.1007/978-3-642-80145-7_10. [DOI] [PubMed] [Google Scholar]
- 27.Pushko P, Sällberg M, Borisova G, Rudén U, Bichko V, Wahren B, Pumpens P, Magnius L. Identification of hepatitis B virus core protein regions exposed or internalized at the surface of HBcAg particles by scanning with monoclonal antibodies. Virology. 1994;202:912–920. doi: 10.1006/viro.1994.1413. [DOI] [PubMed] [Google Scholar]
- 28.Reineke U, Sabat R, Kramer A, Stigler R D, Seifert M, Michel T, Volk H, Schneider-Mergener J. Mapping protein-protein contact sites using cellulose-bound peptide scans. Mol Diversity. 1996;1:141–148. doi: 10.1007/BF01544952. [DOI] [PubMed] [Google Scholar]
- 29.Remaut E, Tsao H, Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981;15:81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- 30.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 609–654. [Google Scholar]
- 31.Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J Virol. 1989;63:798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sällberg M, Ruden U, Magnius L O, Harthus H P, Noah M, Wahren B. Characterisation of a linear binding site for a monoclonal antibody to hepatitis B core antigen. J Med Virol. 1991;33:248–252. doi: 10.1002/jmv.1890330407. [DOI] [PubMed] [Google Scholar]
- 33.Seifer M, Zhou S, Standring D N. A micromolar pool of antigenically distinct precursors is required to initiate cooperative assembly of hepatitis B virus capsids in Xenopus oocytes. J Virol. 1993;67:249–257. doi: 10.1128/jvi.67.1.249-257.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seifer M, Standring D N. Assembly and antigenicity of hepatitis B virus core particles. Intervirology. 1995;38:47–62. doi: 10.1159/000150414. [DOI] [PubMed] [Google Scholar]
- 35.Serwer P. Use of gel electrophoresis to characterize multimolecular aggregates. Methods Enzymol. 1986;130:116–132. doi: 10.1016/0076-6879(86)30010-7. [DOI] [PubMed] [Google Scholar]
- 36.Serwer P, Khan S A, Griess G A. Non-denaturing gel electrophoresis of biological nanoparticles: viruses. J Chromatogr A. 1995;698:251–261. doi: 10.1016/0021-9673(94)01259-h. [DOI] [PubMed] [Google Scholar]
- 37.Ulrich R, Nassal M, Meisel H, Krüger D H. Core particles of hepatitis B virus as carrier for foreign epitopes. Adv Virus Res. 1998;50:141–182. doi: 10.1016/s0065-3527(08)60808-8. [DOI] [PubMed] [Google Scholar]
- 38.Warbrick E. Two’s company, three’s a crowd: the yeast two hybrid system for mapping molecular interactions. Structure. 1997;5:13–17. doi: 10.1016/s0969-2126(97)00162-7. [DOI] [PubMed] [Google Scholar]
- 39.Wingfield P T, Stahl S J, Williams R W, Steven A C. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 40.Zheng J, Schödel F, Peterson D L. The structure of hepadnaviral core antigens. Identification of free thiols and determination of the disulfide bonding pattern. J Biol Chem. 1992;267:9422–9429. [PubMed] [Google Scholar]
- 41.Zhou S, Standring D N. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou S, Yang S Q, Standring D N. Characterization of hepatitis B virus capsid particle assembly in Xenopus oocytes. J Virol. 1992;66:3086–3092. doi: 10.1128/jvi.66.5.3086-3092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 44.Zlotnick A, Cheng N, Stahl S J, Conway J F, Steven A C, Wingfield P T. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc Natl Acad Sci USA. 1997;94:9556–9561. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]