Abstract
Redesigning protein-protein interfaces is an important tool for developing therapeutic strategies. Interfaces can be redesigned by in silico screening, which allows for efficient sampling of a large protein space before experimental validation. However, computational costs limit the number of combinations that can be reasonably sampled. Here, we present combinatorial tyrosine (Y)/serine (S) selection (combYSelect), a computational approach combining in silico determination of the change in binding free energy (ΔΔG) of an interface with a highly restricted library composed of just two amino acids, tyrosine and serine. We used combYSelect to design two immunoglobulin G (IgG) heterodimers—combYSelect1 (L368S/D399Y-K409S/T411Y) and combYSelect2 (D399Y/K447S-K409S/T411Y)—that exhibit near-optimal heterodimerization, without affecting IgG stability or function. We solved the crystal structures of these heterodimers and found that dynamic π-stacking interactions and polar contacts drive preferential heterodimeric interactions. Finally, we demonstrated the utility of our combYSelect heterodimers by engineering both a bispecific antibody and a cytokine trap for two unique therapeutic applications.
combYSelect is an in silico screening strategy for redesigning protein-protein interfaces using a restricted amino acid library.
INTRODUCTION
Protein-protein interactions are important for nearly all physiological and pathological processes. One fundamental role of protein-protein interfaces is to facilitate the oligomerization of proteins, which can mediate signal transduction (1), activate or inhibit intracellular enzymes (2), and control gene expression (3). The redesign of protein-protein interfaces has emerged as an important tool for novel therapeutic development, enabling the prevention of antibiotic resistance (4) and the development of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (5) and G protein (guanine nucleotide–binding protein)–coupled receptor (GPCR) nanobodies (6). Advancing approaches to protein-protein interface engineering could expand therapeutic strategies for myriad diseases.
An established experimental tool for redesigning protein-protein interfaces is directed evolution. This approach relies on generating a library of variants, screening the displayed library based on a desired property, and amplifying the variants of interest (7, 8). Directed evolution mimics natural evolution, does not require previous knowledge of protein structure, and is advantageous for uncovering mutations that would otherwise be missed in rational design approaches. However, the large libraries associated with this method can be costly and cumbersome, and inefficiencies, such as low transfection rates, can prohibit the screening of the complete library (9–13). Directed evolution is also unable to encompass all possible combinations of sequences of amino acids in an interface due to the large number of possible combinations (14). Additionally, protein-protein interfaces within obligate homomeric multimers are generally recalcitrant to experimental directed evolution methods since the protein subunits cannot exist or are too destabilized to display and capture in isolation.
Because of these challenges, in silico redesign of protein-protein interfaces represents a promising alternative for certain protein-protein interfaces. In such computational approaches, neural networks generated by machine learning such as AlphaFold (15, 16) and RoseTTAFold (17, 18) can be combined with in silico screening methods to efficiently screen and select a large number of variants based on a specific property before experimental validation. Such a strategy has been used to identify protein-protein interaction hotspots using the interface mode of the Rosetta software package, wherein potentially interacting residues within a protein-protein interface were identified based on distance, mutated in silico to alanine, and the calculated change in binding free energy (ΔΔG) was used as a predictor for the change in binding (19).
Expanding this strategy to sample all 20 amino acids at each residue, while technically possible, is not feasible due to the amount of time and computational power required. For instance, conducting a combinatorial computational search of all 20 amino acids at each of just 10 positions within a protein-protein interface would require some 10 billion ΔΔG calculations, which at 1 s per calculation would require more than 2500 years of computational time using 128 cores. While future advances in computational speed and parallelization will eventually make such in silico screening approaches possible, it is currently necessary to limit computational expense and, therefore, the in silico library of variants tested.
One way in which to reduce the chemical space to be screened, and the resulting computational cost, is to apply previous knowledge of the specific protein-protein interface, or of protein-protein interfaces in general. It has been previously reported that the complementarity determining regions (CDRs) of antibodies are enriched in tyrosines and serines (20–22) and that antibodies with high affinity against the human death receptor (DR5), the human vascular endothelial growth factor (hVEGF), and others could be engineered using a phage display library consisting of only tyrosine and serine residues in their CDR regions (23–28). Other proteins have also been engineered in a similar manner, with the fibronectin type III domain (FN3) being engineered to bind maltose-binding protein (MBP), human small ubiquitin-like modifier 4 (hSUMO4), and yeast small ubiquitin-like modifier (ySUMO) (29, 30).
Inspired by these experimental successes using highly restricted variant libraries, we hypothesized that combining the Rosetta ΔΔG approach described above with an in silico library restricted to tyrosine and serine mutations would allow us to efficiently, and sufficiently, sample the effects of varying multiple positions within a protein-protein interface to identify combinations of mutations that either improve or worsen binding. Specifically, we sought to remodel the binding interface between the fragment crystallizable (Fc) protomers of an immunoglobulin G1 (IgG1) antibody to promote the favorable formation of heterodimers while making any potential resulting homodimers energetically unfavorable. Because engineered IgG heterodimers can exhibit improved therapeutic efficacy and expand the possibilities for constructing Fc-fusion proteins, methods to redesign the IgG Fc homodimer interface to produce Fc heterodimers with unique mutations in either Fc protomer have been previously established, including but not limited to “knobs-in-holes” (KiH), which relies on hydrophobic and steric interactions (31); electrostatic steering, which employs charged side chains (32); and a multistate design strategy using negative state repertoires to screen for mutations that promoted heterodimer formation while disfavoring homodimer formation (33).
Here, we combined restricted libraries with in silico screening to develop a computational approach to redesigning protein-protein interfaces called combinatorial tyrosine (Y)/serine (S) selection (combYSelect). Using our combYSelect approach, we designed two IgG1 heterodimers that exhibit similar heterodimeric chain pairing and stability compared to previously reported IgG Fc heterodimers. We established the molecular basis of heterodimerization for both of these heterodimers and presented two functional applications of these heterodimeric IgG1 antibodies.
RESULTS
Serine and tyrosine mutations at numerous positions promote Fc heterodimer formation
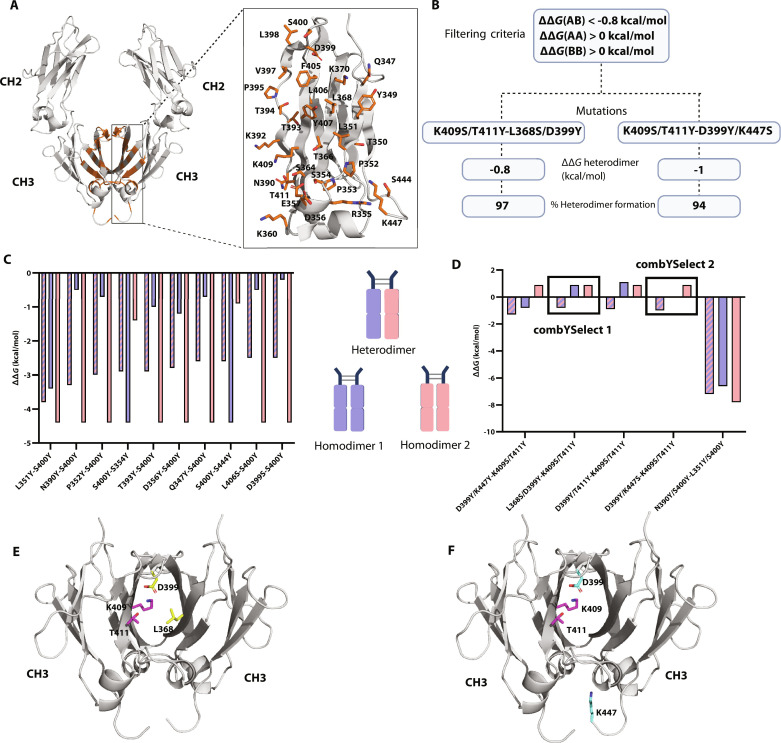
To identify mutations in the CH3 domain of the IgG1 heavy chain (Hc) that would promote heterodimer formation while preventing homodimer formation, we established a computational strategy that we named combYSelect. In this strategy, we identified all residues in the CH3 domain of an IgG1 Fc within 4 Å, as a cutoff for interacting residues, of the opposing CH3 domain. Since the last three to four amino acids of the C terminus are not resolved in any published IgG1 Fc structures, we used an AlphaFold model of an IgG1 Fc to screen all possible residues. We focused exclusively on residues within the CH3 region since these were most likely to be functionally inert compared to residues in the upper regions of the CH2 domains and the hinge, which engage in Fcγ receptor (FcγR) binding as part of the immune cascade (34). We identified a total of 31 residues that met these criteria: Q347, Y349, T350, L351, P352, P353, S354, R355, D356, E357, K360, S364, T366, L368, K370, N390, K392, T393, T394, P395, V397, L398, D399, S400, F405, L406, Y407, K409, T411, S444, and K447 (Fig. 1A). We used a combinatorially restricted library, testing mutations only to serine and tyrosine. We chose these two amino acids based on previous studies that developed high-affinity fragment antigen-binding (Fabs) and scaffold proteins with phage display libraries of only serine and tyrosine residues (23, 26). This allowed us to increase the number of mutations we could assess while maintaining reasonable computational times. We carried out the screen in a combinatorial manner by simultaneously mutating one or two residues in each chain. Mutations of one residue per chain yielded 3844 mutations and took 5 min using 64 central processing unit (CPU) cores. Mutations of two residues per chain yielded 3,459,600 mutations and took 15 hours on 64 CPU cores. We did not test three mutations per chain since, even with the restricted serine/tyrosine library, such a calculation would have taken over 8 months using 64 CPU cores.
Fig. 1. combYSelect screening strategies and workflow.
(A) AlphaFold model of IgG1 Fc that was used for in silico mutation screen, with residues identified to be 4 Å apart from another residue on the opposing chain highlighted in orange stick form. (B) Schematic depicting the combYSelect strategy used to screen for mutations that promote IgG1 heterodimer formation. We used the interface mode of Rosetta coupled with restricting mutations to tyrosine (Y) and serine (S) and filtering based on ΔΔGhomodimer and ΔΔGheterodimer values. (C) Bar graph depicting the computationally determined ΔΔG values for heterodimers (blue-pink stripes) or homodimers (blue, when the mutation listed first is considered, and pink when the mutation listed second is considered) of Fcs, in which only one residue on each chain was mutated. (D) Bar graph depicting the computationally determined ΔΔG values for heterodimers (blue-pink stripes) or homodimers (blue, when the set of mutations listed first is considered, and pink when the set of mutations listed second is considered) of Fcs, in which two residues on each chain were mutated. (E) The two sets of mutations for combYSelect1: K409S/T411Y-L368S/D399Y, one of the heterodimers selected for further testing, are highlighted in purple and yellow, respectively. (F) The two sets of mutations for combYSelect2: K409S/T411Y- D399Y/K447S, one of the heterodimers selected for further testing, are highlighted in purple and cyan, respectively.
To computationally assess the effect of our mutations on heterodimerization, we calculated the ΔΔG for each combination of mutations using the interface mode of Rosetta (19), which outputs a change in ΔG, or ΔΔG value, where positive values indicate that the mutation reduces stability of the protein complex, while negative values signify improved binding relative to the wild-type (WT) model. We chose ΔΔG < −0.8 kcal/mol as the criterion for a potential successful mutation to account for the previously observed variability between experimentally measured and computationally predicted ΔΔG values from that protocol (19, 35). Additionally, while developing and optimizing this strategy, we determined that a predicted favorable value for ΔΔGheterodimer was not sufficient as a selection criterion. As a result, we added a filtering step in which we selected for ΔΔGhomodimer > 0 to ensure that we selected mutants in which homodimer formation was energetically unfavorable (Fig. 1B).
When mutating one chain per residue, we observed that the 10 mutants with the most negative ΔΔGheterodimer values also had negative ΔΔGhomodimer values (Fig. 1C). Thus, these mutants did not fit our criteria and will likely still form an appreciable proportion of homodimers. We therefore assessed the results of the in silico mutations of two residues per chain. We selected a total of five sets of mutations from the screen that resulted in negative ΔΔGheterodimer values (Fig. 1D). The first set of mutations D399Y/K447Y-K409S/T411Y had a ΔΔGheterodimer of −1.3 kcal/mol, and one of the homodimers was energetically unfavorable with a ΔΔGhomodimer 2 of 0.9 kcal/mol; however, ΔΔGhomodimer 1 was −0.8 kcal/mol. Another set of mutations N390Y/S400Y-L351Y/S400Y had the most negative ΔΔGheterodimer of −7.2 kcal/mol. However, the similarly negative ΔΔGhomodimer values made it an unsuitable candidate. Only three of the variants followed all the filtering criteria of negative ΔΔGheterodimer and positive ΔΔGhomodimer values: L368S/D399Y-K409S/T411Y (ΔΔGheterodimer = −0.8 kcal/mol, ΔΔGhomodimer 1 = 0.9 kcal/mol, ΔΔGhomodimer 2 = 0.9 kcal/mol), D399Y/T411Y-K409S/T411Y (ΔΔGheterodimer = −0.9 kcal/mol, ΔΔGhomodimer 1 = 1.1 kcal/mol, ΔΔGhomodimer 2 = 0.9 kcal/mol), and D399Y/K447S-K409S/T411Y (ΔΔGheterodimer = −1 kcal/mol, ΔΔGhomodimer 1 = 0 kcal/mol, ΔΔGhomodimer 2 = 0.9 kcal/mol). We selected for further experimental testing the two candidates that each had four unique mutations: L368S/D399Y-K409S/T411Y—hereafter referred to as “combYSelect1” (Fig. 1E) and D399Y/K447S-K409S/T411Y—hereafter referred to as “combYSelect2” (Fig. 1F).
Mutations identified by combYSelect result in maximal heterodimerization
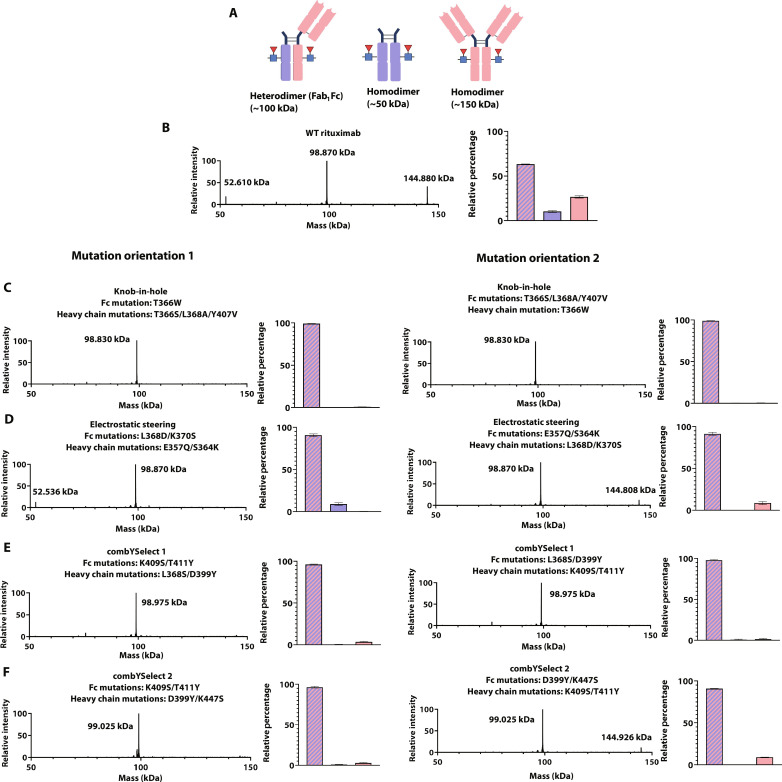
To experimentally assess the relative yields of heterodimer and homodimer formation for our combYSelect designs and compare them to published KiH (T366W-T366S/L368A/Y407V) (36) and electrostatic steering (E357Q/S364K-L368D/K370S) heterodimers (32), we developed a model system involving the coexpression of an IgG1 Fc region and full-length Hc of rituximab, each from distinct protomers in a heterodimer design, as well as the light chain (Lc) of rituximab, our model IgG1 antibody. Rituximab, an anti-CD20 chimeric monoclonal antibody, targets B cells and is often used as a therapeutic for B cell lymphomas (37). After purification by protein A affinity chromatography (38) and treatment with the IgG-specific endoglycosidase EndoS2 (39, 40) to remove heterogeneous glycosylation at residue Asn297, three potential products with unique masses that are easily detectable by liquid chromatography–mass spectrometry (LC-MS) can result from such a coexpression, including an intact Fc (~50 kDa), an intact IgG (~150 kDa), and a monovalent IgG molecule (Fab1Fc; ~100 kDa) (Fig. 2A). The formation of intact Fc regions and IgG antibodies each result from the two possible homodimeric pairs, while the formation of the Fab1Fc molecule arises strictly from heterodimerization. Thus, the ratio of Fab1Fc to Fc and IgG is a direct measure of heterodimer formation.
Fig. 2. Intact LC-MS heterodimer assessment.
(A) Diagram showing the potential constructs formed upon coexpression of the Hc, Lc, and Fc plasmids. MS spectra depicting homodimer (Fc or IgG) and heterodimer formation of Fab1Fc for WT rituximab (B), KiH (C), electrostatic steering (D), combYSelect1 (E), and combYSelect2 (F). The intensity of each of the three peaks is determined relative to each other, with the highest peak normalized to 100. The bar graphs depict the relative percentage formation of the Fab1Fc heterodimer (blue with pink stripes), Fc homodimer (blue), and IgG homodimer (pink).
When we evaluated WT rituximab in this assay, we observed 63% Fab1Fc (Fig. 2B). For the KiH heterodimer (T366W-T366S/L368A/Y407V) (36), we observed 99% heterodimerization when the Hc, containing the T366S/L368A/Y407V mutations, was coexpressed with the Fc containing the T366W mutation, along with the Lc (Fig. 2C, left). Similarly, we observed 99% Fab1Fc formation when the T366W mutation was on the Hc and the T366S/L368A/Y407V mutations were on the Fc (Fig. 2C, right). The electrostatic steering heterodimer we tested resulted in 91% Fab1Fc formation regardless of which chain contained each set of mutations (Fig. 2D). In comparison to the controls above, we observed 97% Fab1Fc formation for both mutation-chain combinations for combYSelect1 (L368S/D399Y-K409S/T411Y) (Fig. 2E). Similarly, combYSelect2 (D399Y/K447S-K409S/T411Y) resulted in 96% Fab1Fc formation when the D399Y/K447S mutations were contained within the Hc (Fig. 2F, left) and 91% when that set of mutations was contained in the Fc (Fig. 2F, right). These results indicate that our combYSelect heterodimers exhibit nearly ideal IgG heterodimerization that is comparable to those of two other previously identified heterodimers that we selected for testing.
Heterodimer mutations do not affect IgG stability or FcRn binding
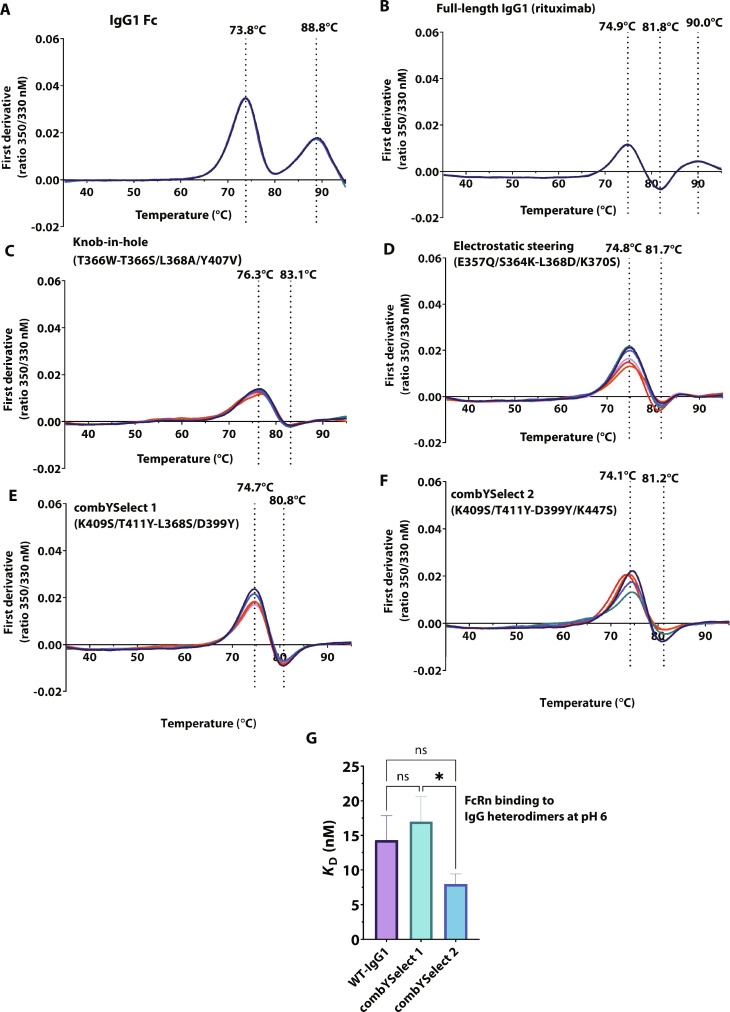
In addition to promoting heterodimer formation, we wanted to ensure that the mutations introduced in the CH3 domains did not adversely affect the functional properties of IgG molecules. We assessed the thermal stability of each construct by determining the melting curves of the resulting heterodimeric antibodies in the Fab1Fc form (i.e., the product resulting from coexpression of the Hc, Lc, and Fc fragment) using nano-differential scanning fluorimetry (nDSF), which, unlike conventional DSF, is a dye-free method that relies on the detection of tryptophan autofluorescence as a function of temperature (41). Our results showed that rituximab had three transition temperatures: 74.9°C, 81.8°C, and 90°C, corresponding to the unfolding of the CH2, Fab, and CH3 domains, respectively (Fig. 3, A and B, and fig. S1A), which is in agreement with previously reported results (42). Additionally, the near-identical transition temperatures between the rituximab IgG and rituximab in the Fab1Fc form confirmed that thermal stability of the latter is representative of that of the full-length IgG. In contrast, the KiH (36) (Fig. 3C) and electrostatic steering constructs (Fig. 3D) previously reported in the literature (32) and combYSelect1 and combYSelect2 (Fig. 3, E and F), all of which are in the Fab1Fc form, only showed two transition temperatures corresponding to the CH2 and Fab domains. The first transition temperature ranged from 74.1°C to 76.3°C, whereas the second was between 80.8°C and 83.1°C. When we assessed the melting temperatures of combYSelect1 and combYSelect2 Fcs, we observed only one melting temperature at approximately 74°C, which usually corresponds to the CH2 domain (fig. S1, B and C). This suggests that the CH3 domain of the heterodimers we tested either unfolds at the same temperature as the CH2 domain or only undergoes reversible unfolding within our tested temperature range.
Fig. 3. Stability and function of IgG heterodimers.
(A to F) First derivative plots of the melting temperatures of the Fab1Fc forms of each of the indicated constructs. The labeled temperatures are the mean of six total replicates, three for each orientation in which the mutations per chain are on the Fc fragment only, or on the Fc and Fab fragment. (G) KD values for WT IgG1, combYSelect1, and combYSelect2 IgG derived from BLI binding assays. Statistical significance was determined by one-way ANOVA with Tukey’s multiple-comparison test (ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
We also assessed the potential effects of these mutations on the overall structure of the CH2-CH3 interface. This region is of particular interest as it is where the neonatal Fc receptor (FcRn) binds in a pH-dependent manner to recycle endogenous IgGs and extend their serum half-life (43). We used biolayer interferometry (BLI) analysis to compare the FcRn binding affinity of rituximab to combYSelect1 and combYSelect2 IgGs with rituximab Fab domains. We obtained nearly identical dissociation constant (KD) values of 14.3, 17.0, and 8.0 nM for rituximab, combYSelect1, and combYSelect2, respectively, at pH 6 (Fig. 3G and fig. S2, A to F). As expected, all of these IgGs showed minimal binding to FcRn at pH 7.4 (fig. S2, G to I). Overall, our data show that the heterodimer forming mutations did not affect the stability or function of the IgG.
Conformationally variant residues exist in the heterodimer interface
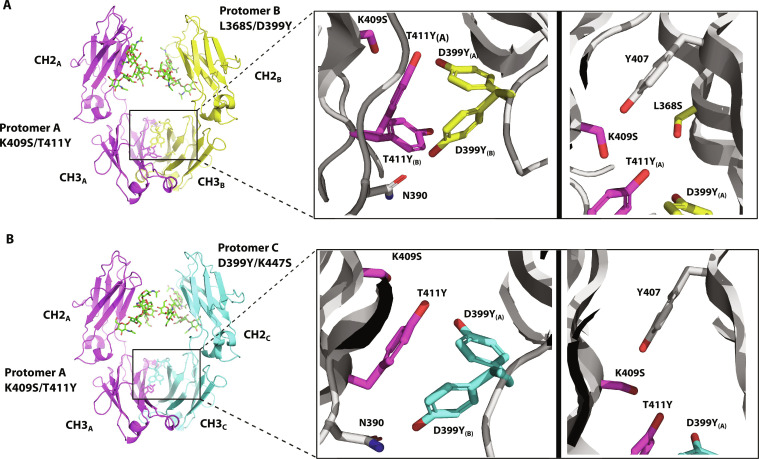
To understand how the introduced mutations promote heterodimer formation, we determined the x-ray crystal structures of combYSelect1 and combYSelect2 Fc regions to resolutions of 2.5 Å and 3 Å, respectively (Fig. 4 and table S1). These structures revealed that the T411Y mutation in the first protomer is involved in π-stacking interactions with the D399Y mutation on the opposing protomer (Fig. 4, A and B, left panels). The electron density map shows continuous unaccounted electron density near the mutated tyrosines, which was best explained by modeling these residues as occupying multiple conformations and positions. We modeled two possible conformations for each mutated tyrosine in combYSelect1 (fig. S3A). In conformation A, the hydroxyl groups of T411Y and D399Y are 2.3 Å apart and likely form a hydrogen bond. Similarly, when D399Y is in conformation B, its hydroxyl group is at a distance from the carboxyl group of N390 on the opposing chain, consistent with the formation of polar contacts with the amide group (Fig. 4A). We observed similar interactions in combYSelect2, in which unmodeled electron density near mutation D399Y also suggested that this residue likely occupies numerous positions. We similarly modeled two possible conformations. However, we observed no such density for the T411Y mutation and therefore modeled only one tyrosine rotamer conformation in the interface. (Fig. 4B and fig. S3B). It is important to note that our data do not enable us to identify specific conformations or pairings of the tyrosine residues. Instead, the modeled conformations represent some of the states that can exist transiently. Still, such alternate conformations of interface residues are consistent with other published structures of IgG heterodimers. The EW-RVT heterodimer was improved by introducing a disulfide bond in the CH3 domain (44), perhaps counteracting the dynamic tendencies of the original heterodimer. The structure of the ZW1 heterodimer, which is formed by four distinct mutations in each chain, was modeled with each of the mutated residues at a 50% occupancy for each of two possible CH3/CH3 heterodimeric orientations (45). This suggests that the Fc chains can consistently fluctuate between multiple possible orientations. Additionally, the hydroxyl moieties of the mutated S409 residue on protomer A and Y407 on protomers B and C are 3.6 Å and 2.8 Å apart, respectively, and form potential hydrogen bonds (Fig. 4, A and B, right panels). Overall, our structures reveal that the introduced residues in our combYSelect IgGs exist in conformations that likely engage in π-π interactions and H-bonds to drive heterodimer formation.
Fig. 4. Structure of combYSelect1 and combYSelect2.
(A) X-ray crystal structure (2.5 Å) highlighting the T411Y/K409S and L368S/D399Y mutations. The tyrosines are dynamic with two conformations modeled for T411Y at 67% (T411YA) and 33% (T411YB) occupancy and for D399Y at 42% (D399YA) and 58% (D399YB) occupancy. (B) X-ray crystal structure (3 Å) highlighting the T411Y/K409S and D399Y/K447S mutations. The tyrosines are dynamic with two conformations modeled for D399Y at 38% (D399YA) and 62% (D399YB) occupancy. N390 and Y407 are nearby residues involved in the interactions.
All combYSelect mutations are required for optimal heterodimerization
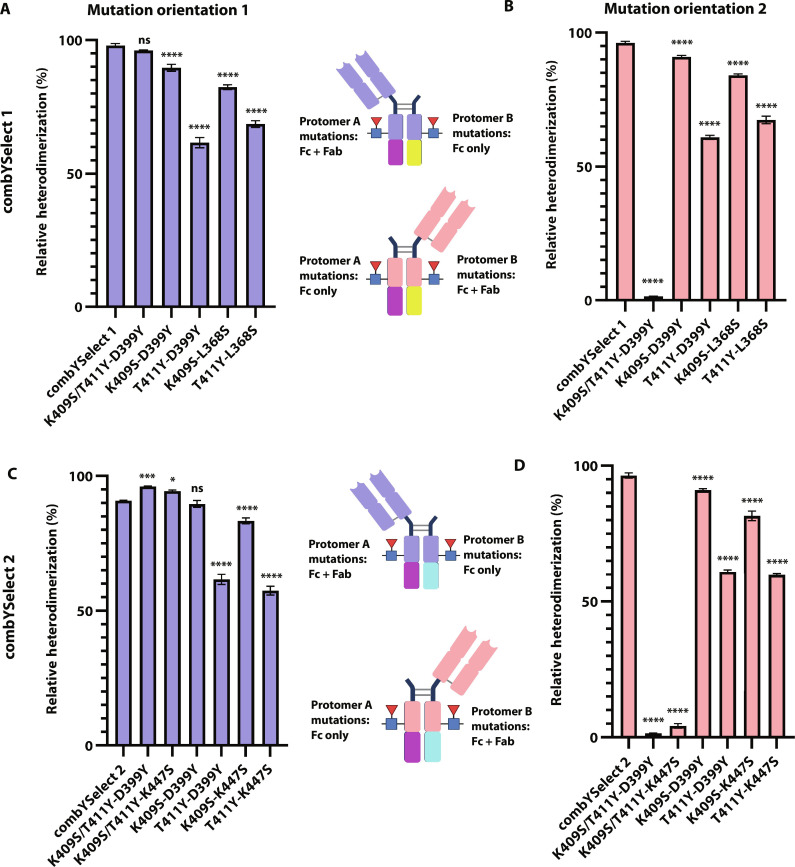
Our structure suggests that the L368S mutation in combYSelect1 is not at a sufficient distance to make any polar contacts with the residues on the opposing chain. Additionally, the electron density in our structure of combYSelect2 was insufficient to model K447S. As a result, we used the same intact LC-MS method for assessing heterodimerization described in Fig. 2 to determine whether those two mutations are essential for optimal heterodimerization. The L368S mutation was dispensable when the D399Y mutation was on an Fc, with protomer A mutations on the Hc. The observed 96% heterodimerization for K409S/T411Y-D399Y was not significantly different from combYSelect1 (Fig. 5A and fig. S4A, right). However, there was no detectable heterodimerization when the D399Y mutation was on the Fc fragment attached to the Fab, suggesting that L368S might play a role in stabilization of the Hc (Fig. 5B and fig. S4A, left). On the other hand, we observed 96% and 94% Fab1Fc formation for the K409S/T411Y-D399Y and K409S/T411Y-K447S Hc-Fc constructs, respectively, which indicated improved heterodimerization compared to combYSelect2 (Fig. 5C and fig. S4, A and B, right). However, when the D399Y and K447S individual mutations were contained in the Hc, they were required for heterodimer formation (Fig. 5D and fig. S4, A and B, left). Finally, we confirmed that constructs with single mutations on each chain in all four possible combinations—K409S-D399Y, T411Y-D399Y, K409S-L368S, and T411Y-L368S for combYSelect1 and K409S-D399Y, T411Y-D399Y, K409S-K447S, and T411Y-K447S for combYSelect2—have significantly reduced heterodimerization regardless of mutation-chain pairing (Fig. 5 and fig. S4, C to J).
Fig. 5. Mutation controls for combYSelect1 and combYSelect2.
(A and B) Histograms of the relative percentage of heterodimerization as determined by intact LC-MS when some or all protomer A mutations are on the fragment containing a Fab and Fc and protomer B mutations are on the fragment containing only an Fc (A) or when the orientation is reversed (B). (C and D) Histograms of the relative percentage of heterodimerization as determined by intact LC-MS when some or all protomer A mutations are on the fragment containing a Fab and Fc and protomer C mutations are on the fragment containing only an Fc (C) or when the orientation is reversed (D). Statistical significance (n = 3 for each construct) was determined by one-way ANOVA with Dunnett’s multiple-comparison test, in which each mutation control was compared to combYSelect1 or combYSelect2 (ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
IgG1 heterodimers predicted by combYSelect have therapeutic potential
IgG heterodimers are often used for the development of bispecific monoclonal antibodies and Fc-fusion proteins that exhibit unique and/or improved therapeutic properties (46). To confirm that combYSelect heterodimers can be used broadly for potential therapeutic applications, we engineered combYSelect-based bispecific antibodies and cytokine traps and tested their abilities to bind simultaneously to two target cell lines expressing distinct antigens and to inhibit cytokine-mediated cellular signaling, respectively.
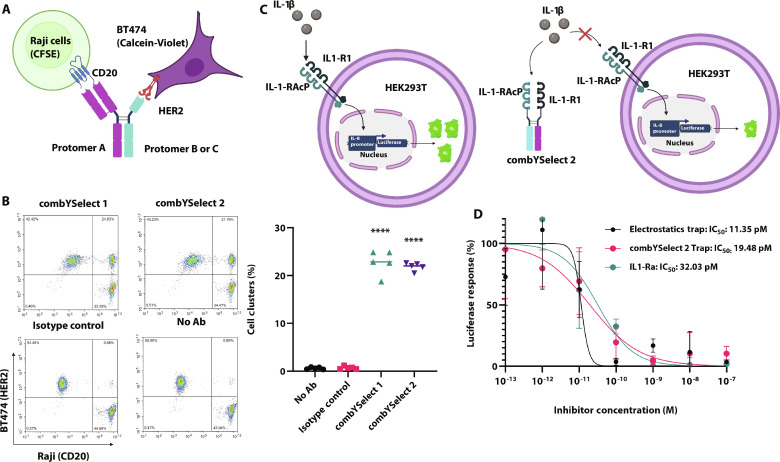
First, we designed a bispecific antibody in which one arm consisted of the rituximab anti-CD20 Fab, while the other was a nanobody specific for human epidermal growth factor receptor 2 (HER2) (Fig. 6A). Both of our bispecific combYSelect heterodimers were able to simultaneously bind the two antigens expressed on Raji (CD20+) and BT474 (HER2+) cells, as shown by the formation of cell clusters that were positive for the distinct dyes used to stain each of the two cell types (Fig. 6B and fig. S5A). This was the case regardless of the antigen specificity of each protomer in versions of these bispecific antibodies in which the anti-CD20 Fab and anti-HER2 nanobody were fused to the opposite Fc protomer (fig. S5, B and E). Conversely, cell-bridging clusters did not form when using a nonspecific IgG1 isotype control (Fig. 6B) or any monospecific heterodimer or homodimer (fig. S5, C to E).
Fig. 6. Functional applications of combYSelect heterodimers.
(A) Representation of the cell-bridging assay, in which the Fc of protomer A (combYSelect1 and combYSelect2) is engineered with an anti-CD20 rituximab Fab and the Fc protomer B (combYSelect1) or protomer C (combYSelect2) is attached to an anti-HER2 nanobody. Binding to CFSE-stained Raji cells and Calcein-Violet BT474 cells will be assessed. (B) Flow cytometry density plots and a scatterplot depicting cell cluster formations of Raji and BT474 cells when combYSelect1, combYSelect2, a nonspecific IgG1 isotype control, or no antibody was added to the cell mixture. Statistical significance (n = 5) was determined by one-way ANOVA with Tukey’s multiple-comparison test (ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). Each combYSelect IgG was compared to both the no antibody and isotype control. (C) Depiction of combYSelect2 cytokine trap and the luciferase inhibition cell-based assay. (D) Percentage of luciferase response, normalized to that when no inhibitor is added, is plotted as a function of varying inhibitor concentration in the presence of 5 pM IL-1β. The IC50 values were determined using a nonlinear least square fit.
IgG heterodimers are also commonly used in Fc-fusion–based therapeutics. Accordingly, in a second potential therapeutic scenario, we used the combYSelect2 Fc to construct a cytokine trap composed of the interleukin-1 (IL-1) cytokine cognate receptor IL-1 receptor I (IL-1RI) and secondary receptor IL-1 receptor accessory protein (IL-1RAcP) to sequester IL-1β (Fig. 6C). We measured the ability of our combYSelect2 cytokine trap to sequester IL-1β, and in turn inhibit luciferase expression driven by the IL-8 promoter in human embryonic kidney (HEK) 293T cells, which express IL-1RI and IL-1RAcP endogenously (47). Inhibition by combYSelect2 trap was comparable to that of a trap fused to a published electrostatic heterodimer E357Q/S364K-L368D/K370S (32). Both traps had slightly improved inhibition compared to IL-1 receptor antagonist (IL-1Ra; Fig. 6D), the natural antagonist of IL-1 cytokine signaling, (48) the recombinant form of which is Anakinra that is used to treat rheumatoid arthritis and other chronic inflammatory conditions (49, 50).
DISCUSSION
Protein-protein interfaces are large and complex, involving many individual amino acid residues that collectively exhibit heterogeneous energetic landscapes to dictate binding affinities, kinetics, and thermodynamics. The ability to accurately model and predict energetic interactions between protein interfaces in silico has led to great success in the protein engineering field. However, the sample space of most proteins currently remains too large to be screened comprehensively. Although increases in computing power are expected, it is unlikely that they will be able to sample the complete diversity of protein sequences in a timely manner. The recent increase in the number of artificial intelligence–based models for protein design (51) and predicting the consequences of mutations in proteins (52) may enable us to bridge the gap between current computational capabilities and efficiently sampling the complete range of protein diversity, potentially leading to routine de novo protein design and engineering in the future. However, the design and redesign of protein-protein interfaces currently remains a challenge.
One strategy to overcome this challenge is to reduce the number of calculations to be made by reducing the combinations of amino acid positions within a given interface and the number of possible amino acid mutations to be made at each position. While sampling too many combinations is computationally infeasible, sampling too few combinations is meaningless as it will insufficiently interrogate the interface. What then is a logical and reasonable way to reduce the number of variables in such calculations and still arrive at solutions that could achieve novel and improved redesigned protein-protein interfaces? Here, we introduce combYSelect, a computational approach to redesigning protein-protein interfaces that is one way, of many possible ways, to strike a balance between oversampling (i.e., prohibitive computational expense) and undersampling (i.e., insufficient interrogation of the features that contribute to protein-protein interactions).
combYSelect is an in silico screening method that relies on restricting the number of possible amino acid mutations at any position to only tyrosine and serine. The choice of these two amino acid options was inspired by previous protein engineering studies showing that phage display libraries of antibody CDR loops with only serine and tyrosine residues gave rise to high-affinity Fabs to a diversity of antigens (26). Our results using combYSelect support the extensive body of literature that describes tyrosine as being beneficial for mediating formation of contacts to promote both affinity and specificity (24). These properties are likely due to the chemical diversity of tyrosine and its ability to form both polar contacts and hydrophobic π interactions. The combination with serine then affords the interface flexibility to form contacts with a variety of residues, as well as additional hydrogen bonding potential. Our results highlight that the properties of tyrosine and serine can also be applied to the redesign of planar interfaces and/or obligate homodimers, i.e., interfaces that have evolved to form highly specific and stable interactions. The flexibility of a Tyr/Ser-populated interface was also evident in the x-ray crystal structures of the combYSelect heterodimers in which electron density of our x-ray crystal structures indicated that the introduced tyrosines occupy multiple conformations. It is possible that the disordered tyrosines contribute to the stability of the interface due to the occurrence of local entropy-enthalpy reinforcement, in which the binding of the two Fc monomers not only is stabilized by favorable enthalpy but also exhibits favorable entropy at the tyrosine interface due to the existence of multiple possible states. This has been previously described on a larger scale as a thermodynamic driver of complex formation between intrinsically disordered proteins (53). We also restricted the number of simultaneous mutations within a single chain to two. Together, these restrictions to the number of variables resulted in 3,459,600 possible variants with which we performed a single Rosetta ΔΔG calculation. This was completed in approximately 15 hours, a manageable amount of time to comprehensively interrogate a protein-protein interface formed by two Ig-fold subunits.
We applied combYSelect to redesign the interface between the CH3 domains on either protomer in the IgG1 Fc homodimer, with an eye toward developing heterodimeric Fc regions that could serve as platforms for bispecific antibodies (bsAbs). The importance of bsAbs is rapidly growing due to their improved clinical efficacy as compared to monospecific antibodies or combination therapy, as well as for their potential as diagnostic tools (46, 54), particularly for antitumor monoclonal antibodies (mAbs) (55–57). Commonly, bsAbs are engineered to simultaneously target T cell–specific antigens and tumor-associated antigens (TAAs), bridging the two cell types together and enhancing the immune response to tumor cells (58). Another common use of bsAbs is for the targeting of two different TAAs simultaneously, which improves specificity and decreases the likelihood of tumors escaping by down-regulating specific epitopes or reverting to redundant pathways (59).
We describe a method that is highly effective for the design of IgG-like bsAbs, which should retain long serum half-lives due to physiological binding to FcRn, compared to non–IgG-like bsAbs that lack the Fc region (46, 60). To functionally evaluate our combYSelect bsAbs, we replaced one of the Fabs with a nanobody to avoid mismatches between Hcs and Lcs. We did so because Lc association problems have long hindered bsAb development and production. One or both Lcs incorrectly pairing with the Hcs can lead to the formation of unwanted IgGs with aberrant antigen binding capabilities, thus reducing the yield of the desired bsAb. As a result, similar to the strategies we have previously described to promote proper Hc association, several strategies have been developed to circumvent the Lc pairing problem. Such methods include using a common Lc for both Fab Hcs. However, such chains are difficult to identify and may restrict the diversity of antigens that can be targeted. Other strategies are more similar to Hc heterodimerization methods such as introducing KiH or electrostatic mutations in either the VH-VL interface or the CH1-CL interface. Finally, others have resorted to swapping certain Hc and Lc Fab domains in a strategy known as CrossMab (61). Considering the similarities in folds and interactions between the CH1 domains of both Hc and Lc and the CH3 domains of IgG1, it would be reasonable to suspect that applying combYSelect to promote specific associations between an Lc and its correct Hc pair could lead to the development of a bsAb that most closely resembles the functionality, stability, and long half-life of a WT antibody while retaining the advantages of dual-antigen specificity.
Additionally, combYSelect could potentially be used as a platform for designing bsAbs of other antibody isotypes, considering that conformations and homodimeric interfaces of the Fcs of other antibody isotypes are similar to IgG Fcs. Although all current approved mAb therapeutics are IgGs, progress in the protein engineering field has expanded the possibility of using alternative isotypes such as IgA and IgE. Protein and glyco-engineering efforts have improved the producibility, stability, and half-life of IgA, as monomeric IgA has shown efficacy in preclinical models (62). IgA has unique properties that include its ability to engage neutrophils, which highly express Fc α receptor I (FcαRI), making it potentially effective as an antitumor therapeutic. Additionally, IgA displays increased stability in mucosal surfaces compared to IgGs, improving potential for mAbs directed at lung- or gastrointestinal-associated inflammation (63, 64). Similarly, monoclonal IgE binds with extremely high affinity to Fcε receptor I (FcεRI), which is expressed on tumor-associated macrophages, and can improve molecular allergy diagnostics (65, 66). Thus, engineering bispecific IgA and IgE could further contribute to improving the clinical efficacy of such mAbs. A bispecific IgE antibody has been engineered using the KiH strategy, confirming that heterodimeric strategies originally developed for IgGs can be expanded to other antibody isotypes (65).
We developed combYSelect using a library restricted to tyrosine and serine residues motivated by experimental evidence that such a restricted amino acid library could be effective in protein design, albeit for antibody CDR loops binding to antigens. Although we applied it successfully to redesigning the CH3-CH3 interface in IgG1 antibodies, there is no reason that libraries restricted to other combinations of amino acids would not also be useful in protein interface redesign. In the context of structured protein folds, protein-protein interfaces are often hydrophobic, containing a large nonpolar surface area. The formation of these interfaces is largely driven by hydrophobic interactions, such as van der Waals forces (67) and electrostatic forces (68, 69). This merits the consideration of whether different amino acids might be more suitable when redesigning Fcs. A systematic analysis of protein-protein interaction hotspots showed that tryptophan, tyrosine, and arginine are most commonly found in hotspots likely due to the aromatic nature of tryptophan and tyrosine, allowing them to form π-stacking interactions and the ability of all three to form hydrogen bonds. Leucine, serine, threonine, and valine are largely absent from protein hotspots. Additionally, analysis of various protein-protein complexes has found that amino acids aspartate and asparagine are favored over glutamate and glutamine, and isoleucine is favored over leucine (70). These findings suggest that, within the context of redesign of protein-protein interfaces, amino acids such as arginine, aspartate, asparagine, and isoleucine should also be considered along with tyrosine and serine when deciding the chemical space to be explored.
MATERIALS AND METHODS
Residue selection for screening
Residues were selected based on a distance cutoff. An in-house Perl script was used to identify all residues within the homodimeric interface of an AlphaFold model of IgG1 within 4 Å of the opposing chain. Only residues that were located within the CH3 domain of the IgG were used for screening. A total of 31 residues were selected using this method.
Rosetta interface mode
In silico screening of potential mutations to promote heterodimer formation was carried out using the “interface” mode of Rosetta2.3 (19). Command line options were specified to include extra chi1, chi2, and chi3 rotamers (-extrachi_cutoff 1 -ex1 -ex2 -ex3). Only the mutant side chains were repacked (the default behavior of this mode), while the protein backbone from the WT structure was retained.
Cloning
The sequence for Hc and Lc of rituximab was ordered from Thermo Fisher Scientific in the pcDNA3.4-TOPO vector. The Fc plasmid was subcloned out of the Hc plasmid via polymerase chain reaction (PCR). Primers to introduce single or double mutations into the rituximab Hc or Fc plasmids were designed using the NebBaseChanger website (https://nebasechanger.neb.com/), and PCR was carried out using the NEB High-Fidelity 2X Master Mix and the manufacturer’s instructions. All sequences were confirmed by Sanger sequencing at Genewiz (https://www.genewiz.com/).
Recombinant IgG heterodimer expression and purification
All combinations of single-armed IgG proteins were expressed in Expi293 cells. The rituximab Hc, Lc, and Fc plasmids were transfected in a 1:1:1 ratio by weight. The plasmids were transfected as per the manufacturer’s protocol (MAN0007814, Thermo Fisher Scientific) with the addition of penicillin/streptomycin mix 24 hours after transfection. The cells were cultured for 96 hours before harvesting. The proteins were purified using protein A resin (Thermo Fisher Scientific), with phosphate-buffered saline (PBS) (pH 7.4) being used as the binding buffer and 100 mM sodium citrate buffer (pH 3.0) as the elution buffer. The fractions were neutralized with 1 M tris (pH 9.3). The proteins were buffer-exchanged into PBS and stored at 4°C until ready for use. All combinations of potential mutations were tested with the same mutation being tested in both the Hc and Fc plasmids. All proteins were expressed in triplicate in separate transfections.
Analysis of heterodimer formation
Samples for each of the heterodimer constructs were analyzed via LC-MS to determine the percentage of heterodimer formation. The samples were set up in 96-well plates with a final volume of 20 μl and a concentration of 5 μM. Each expressed protein was analyzed in the absence and presence of EndoS2 (final concentration, 50 nM). EndoS2 is an IgG-specific endoglycosidase from Streptococcus pyogenes, which removes the glycans from the conserved Asn297 site and simplifies data analysis. The samples were analyzed using an Agilent 1290 Infinity II LC System equipped with a 50-mm polymeric reverse-phase column from Agilent with 1000-Å pore size. The LC system is attached to an Agilent 6560 Ion Mobility (IM) Quadrupole Time-Of-Flight mass spectrometer (Agilent, Santa Clara, CA). Relative amounts of each of the deglycosylated peaks were quantified after deconvolution of the raw data and identification of the corresponding peaks using BioConfirm (Agilent, Santa Clara, CA).
Thermal stability testing
nDSF was carried out using a TychoNT.6 instrument (NanoTemper Technologies, Munich, Germany) to determine the melting temperatures of the monovalent IgG constructs. Samples (20 μl) at 5 μM were loaded into glass capillaries and placed into the sample folder. For unfolding parameters, samples were heated to 95°C at a ramp rate of 30°C/min. Melting temperature Tm was determined using the instrument software (v1.1.5.668) using first derivative analysis of 350-nm/330-nm fluorescent ratios plotted against temperature. The inflection points that are converted to peaks and troughs are automatically assigned as Tm1/Tm2/Tm3 by the software. All samples were analyzed in triplicate.
Binding of heterodimer constructs to FcRn
For the two sets of mutations that showed successful heterodimer formation, full-length IgG constructs using only Hc and Lc plasmids were recombinantly expressed and purified in the same manner as above for FcRn binding assays.
BLI was conducted on an OCTETRed384 system at 30°C. A human FcRn-b2m protein construct containing an AviTag was recombinantly expressed in Expi293F cells and biotinylated using BirA. The biotinylated FcRn was loaded onto streptavidin-coated biosensors at a concentration of 10 μg/ml. Immobilization levels between 0.6 and 1.0 nm were reached. The assay buffer was 100 mM phosphate and 150 mM NaCl (pH 6).
For association phase monitoring, IgG samples were diluted with assay buffer (pH 6) with a starting concentration of 1000 nM. Samples (40 μl) composed of seven threefold dilutions were made and transferred into OCTET 384 tilted well plates. The eighth sample was left at 0 nM to provide a baseline measurement. The IgG samples were allowed to bind to the FcRn-loaded biosensors for 300 s. The dissociation phase was recorded in a solid black 96-well plate for 150 s. The biosensors were regenerated between runs for 30 s with 10 mM Hepes (pH 8). The regeneration steps were carried out twice, with the sensors washed with pH 6 buffer for 30 s. The experiments were carried out the same way at neutral pH, except the pH of the assay buffer was adjusted to pH 7.4. All experiments were carried out in triplicate.
All data were referenced with the FcRn-loaded streptavidin-coated biosensors loaded only in assay buffer. The sensograms were collected using the ForteBio Data Acquisition software v11.1 and plotted and analyzed using steady-state fits with the ForteBio Data Analysis Software v 11.1.
Crystallization of combYSelect1 and combYSelect2 Fcs
combYSelect1 and combYSelect2 Fcs were expressed as described above and concentrated to 5 mg/ml. For crystal generation, the reservoir of 48-3 Intelliplates (Art Robbins 102-0003-00) was filled with 150 μl of solution, and 0.5 μl of protein was combined with 0.5 μl of mother liquor. combYSelect1 Fcs were crystallized in 0.1 M bis-tris (pH 6.5) and 21% PEG MME 5000 (Hampton Research). The crystals were harvested in mother liquor supplemented with 20% (v/v) glycerol for cryoprotection and flash-frozen in liquid nitrogen. combYSelect2 Fcs were crystallized in 0.1 M bis-tris (pH 6.5) and 29% PEG MME 2000 (Hampton Research). The crystals were harvested in mother liquor supplemented with 30% (v/v) glycerol for cryoprotection and flash-frozen in liquid nitrogen.
Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory on Eiger16 detector and processed on HKL2000. Both structures were solved with molecular replacement with an IgG1 Fc [Protein Data Bank (PDB) ID: 7LBL] as a search model using PHENIX Phaser-MR (71). The structures were built and refined using Coot (72) and phenix.refine (73). The models were also further refined with the PDB-REDO server (74).
Cell-bridging assay
For the cell staining assay, highlighting the potential utility of bsAbs, the previously characterized 5F7 nanobody, which is specific for HER2, was used. The nanobody string was purchased from Twist Bioscience and subcloned into the Hc vector replacing the CH1 domain of the Lc. Like the heterodimers, these constructs were transfected in a 1:1:1 ratio and purified as above.
BT474 and Raji cells were resuspended to a density of 1 × 107 cells/ml in 1× PBS. The BT474 cells were stained with Calcein-Violet-AM (Thermo Fisher Scientific, catalog no. C34858) and the Raji cells were stained with CellTrace CFSE (carboxyfluorescein diacetate succinimidyl ester) (Thermo Fisher Scientific, catalog no. C34554) according to the manufacturer’s protocols, centrifuged, and resuspended in staining buffer (BD Biosciences, catalog no. 554656). The cells were combined in equal volume/density, and human BD Fc block (catalog no. 564220) was added according to the manufacturer’s protocol. The combined cells (200 μl) were added to wells of a 96-well flat bottom plate (Corning, catalog no. 3917). The cells were then stained with primary antibody (50 μg/ml) for 45 min on ice, washed three times with staining buffer, and resuspended in the same buffer before collecting data on an Agilent NovoCyte Penteon flow cytometer. This protocol was adapted from a previously published version of this assay (65).
IL-1 cytokine trap inhibition assay
HEK293T-derived cells grown in serum-free F17 medium [American Type Culture Collection (ATCC), ACS-4500] were transiently transfected with a luciferase gene under the control of the IL-8 promoter (nano-luc, plasmid pNL2.2, Promega) using FugeneHD (Promega). After 18 hours, cells were harvested and seeded into 96-well plates at a concentration of 40,000 cells per well. Tenfold serial dilutions of the antagonist (electrostatics trap, combYSelect2 trap, or IL-1Ra) were prepared with the concentrations ranging from 0.1 μM to 0.1 pM. The cells were pretreated with the antagonist for 15 min and stimulated with 5 pM IL-1β. Cells that were stimulated with cytokine in the absence of antagonists and cells that did not receive any stimulation were used as controls. After a 5-hour incubation at 37°C, the cells were lysed and the luciferase activity was determined using a BioTek luminescence reader. The activity of cells stimulated with cytokine in the absence of the antagonist was used to normalize luminescence data. This protocol was adapted from a previously published version of this assay (48, 75).
Statistical analysis
All statistical analysis was done in GraphPad Prism. Ordinary one-way analyses of variance (ANOVAs) were used for comparison of multiple groups with one independent variable, followed by Tukey’s multiple-comparison tests when comparing each mean with all other means or Dunnett’s multiple-comparison tests when comparing each mean with one control mean. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. For the IL-1 cytokine trap inhibition assay, the percentage of luciferase response was normalized to that when no inhibitor is added. The IC50 (median inhibitory concentration) values were determined using a nonlinear least square fit. All intact LC-MS analysis, FcRn binding, thermal stability testing, and IL-1 cytokine trap inhibition assay were performed in independent triplicates.
Acknowledgments
X-ray diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory.
Funding: This work was supported by NIH grants R01AI149297 (E.J.S.) and R35GM144083 (B.G.P.). SER-CAT is supported by its member institutions and NIH equipment grants S10_RR25528, S10_RR028976, and S10_OD027000.
Author contributions: Conceptualization: T.A., J.J.D., E.J.S., and J.K.F. Methodology: T.A., J.J.D., J.K.F., A.M., B.G.P., R.K., E.J.S., N.V., M.W.F., and J.C.H. Investigation: T.A., J.J.D., M.W.F., A.V.A., J.C.H., N.V., R.K., M.B., K.E.M., and D.E.S. Validation: R.K., E.J.S., T.A., A.V.A., B.G.P., M.W.F., J.J.D., and J.C.H. Formal analysis: R.K., T.A., A.V.A., A.M., N.V., M.W.F., and J.J.D. Software: R.K., A.M., B.G.P., and J.J.D. Visualization: T.A., J.J.D., E.J.S., and A.V.A. Supervision: E.J.S. and J.J.D. Project administration: E.J.S. and J.J.D. Resources: E.J.S., A.V.A., M.W.F., and J.J.D. Funding acquisition: E.J.S. and B.G.P. Data curation: E.J.S. and J.J.D. Writing—original draft: T.A., J.J.D., and E.J.S. Writing—review and editing: T.A., J.J.D., E.J.S., and B.G.P.
Competing interests: T.A., J.J.D., and E.J.S. are inventors on a provisional patent application filed with the United States Patent and Trademark Office by Emory University relevant to the work in this manuscript. All other authors declare that they have no competing interests.
Data and materials availability: The structures of combYSelect1 and combYSelect2 are available in the PDB under the accession codes 8TTM and 8TUD, respectively. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Table S1
REFERENCES AND NOTES
- 1.Boriack-Sjodin P. A., Margarit S. M., Bar-Sagi D., Kuriyan J., The structural basis of the activation of Ras by Sos. Nature 394, 337–343 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Guillet V., Lapthorn A., Hartley R., Mauguen Y., Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure 1, 165–176 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Andel F. III, Ladurner A. G., Inouye C., Tjian R., Nogales E., Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286, 2153–2156 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Badran A. H., Guzov V. M., Huai Q., Kemp M. M., Vishwanath P., Kain W., Nance A. M., Evdokimov A., Moshiri F., Turner K. H., Wang P., Malvar T., Liu D. R., Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 533, 58–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wellner A., McMahon C., Gilman M. S. A., Clements J. R., Clark S., Nguyen K. M., Ho M. H., Hu V. J., Shin J. E., Feldman J., Hauser B. M., Caradonna T. M., Wingler L. M., Schmidt A. G., Marks D. S., Abraham J., Kruse A. C., Liu C. C., Rapid generation of potent antibodies by autonomous hypermutation in yeast. Nat. Chem. Biol. 17, 1057–1064 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.English J. G., Olsen R. H., Lansu K., Patel M., White K., Cockrell A. S., Singh D., Strachan R. T., Wacker D., Roth B. L., VEGAS as a platform for facile directed evolution in mammalian cells. Cell 178, 748–761.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonsor D. A., Sundberg E. J., Dissecting protein-protein interactions using directed evolution. Biochemistry 50, 2394–2402 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Labrou N. E., Random mutagenesis methods for in vitro directed enzyme evolution. Curr. Protein Pept. Sci. 11, 91–100 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Akbulut N., Tuzlakoglu Ozturk M., Pijning T., Issever Ozturk S., Gumusel F., Improved activity and thermostability of Bacillus pumilus lipase by directed evolution. J. Biotechnol. 164, 123–129 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Brustad E. M., Arnold F. H., Optimizing non-natural protein function with directed evolution. Curr. Opin. Chem. Biol. 15, 201–210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougherty M. J., Arnold F. H., Directed evolution: New parts and optimized function. Curr. Opin. Biotechnol. 20, 486–491 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo F., Xu H., Xu H., Yu H., Compensation of the enantioselectivity-activity trade-off in the directed evolution of an esterase from Rhodobacter sphaeroides by site-directed saturation mutagenesis. Appl. Microbiol. Biotechnol. 97, 3355–3362 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Urvoas A., Valerio-Lepiniec M., Minard P., Artificial proteins from combinatorial approaches. Trends Biotechnol. 30, 512–520 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Yuen C. M., Liu D. R., Dissecting protein structure and function using directed evolution. Nat. Methods 4, 995–997 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu W., Shenoy A., Kundrotas P., Elofsson A., Evaluation of AlphaFold-Multimer prediction on multi-chain protein complexes. Bioinformatics 39, btad424 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphreys I. R., Pei J., Baek M., Krishnakumar A., Anishchenko I., Ovchinnikov S., Zhang J., Ness T. J., Banjade S., Bagde S. R., Stancheva V. G., Li X. H., Liu K., Zheng Z., Barrero D. J., Roy U., Kuper J., Fernandez I. S., Szakal B., Branzei D., Rizo J., Kisker C., Greene E. C., Biggins S., Keeney S., Miller E. A., Fromme J. C., Hendrickson T. L., Cong Q., Baker D., Computed structures of core eukaryotic protein complexes. Science 374, eabm4805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G. R., Wang J., Cong Q., Kinch L. N., Schaeffer R. D., Millan C., Park H., Adams C., Glassman C. R., DeGiovanni A., Pereira J. H., Rodrigues A. V., van Dijk A. A., Ebrecht A. C., Opperman D. J., Sagmeister T., Buhlheller C., Pavkov-Keller T., Rathinaswamy M. K., Dalwadi U., Yip C. K., Burke J. E., Garcia K. C., Grishin N. V., Adams P. D., Read R. J., Baker D., Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kortemme T., Baker D., A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies D. R., Cohen G. H., Interactions of protein antigens with antibodies. Proc. Natl. Acad. Sci. U.S.A. 93, 7–12 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mian I. S., Bradwell A. R., Olson A. J., Structure, function and properties of antibody binding sites. J. Mol. Biol. 217, 133–151 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Padlan E. A., Anatomy of the antibody molecule. Mol. Immunol. 31, 169–217 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Koide A., Gilbreth R. N., Esaki K., Tereshko V., Koide S., High-affinity single-domain binding proteins with a binary-code interface. Proc. Natl. Acad. Sci. U.S.A. 104, 6632–6637 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birtalan S., Zhang Y., Fellouse F. A., Shao L., Schaefer G., Sidhu S. S., The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J. Mol. Biol. 377, 1518–1528 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Fellouse F. A., Esaki K., Birtalan S., Raptis D., Cancasci V. J., Koide A., Jhurani P., Vasser M., Wiesmann C., Kossiakoff A. A., Koide S., Sidhu S. S., High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 373, 924–940 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Fellouse F. A., Li B., Compaan D. M., Peden A. A., Hymowitz S. G., Sidhu S. S., Molecular recognition by a binary code. J. Mol. Biol. 348, 1153–1162 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Nelson B., Sidhu S. S., Synthetic antibody libraries. Methods Mol. Biol. 899, 27–41 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Rajan S., Sidhu S. S., Simplified synthetic antibody libraries. Methods Enzymol. 502, 3–23 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Koide S., Sidhu S. S., The importance of being tyrosine: Lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem. Biol. 4, 325–334 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbreth R. N., Esaki K., Koide A., Sidhu S. S., Koide S., A dominant conformational role for amino acid diversity in minimalist protein-protein interfaces. J. Mol. Biol. 381, 407–418 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridgway J. B., Presta L. G., Carter P., ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 9, 617–621 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Moore G. L., Bernett M. J., Rashid R., Pong E. W., Nguyen D.-T., Jacinto J., Eivazi A., Nisthal A., Diaz J. E., Chu S. Y., Muchhal U. S., Desjarlais J. R., A robust heterodimeric Fc platform engineered for efficient development of bispecific antibodies of multiple formats. Methods 154, 38–50 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Leaver-Fay A., Froning K. J., Atwell S., Aldaz H., Pustilnik A., Lu F., Huang F., Yuan R., Hassanali S., Chamberlain A. K., Fitchett J. R., Demarest S. J., Kuhlman B., Computationally designed bispecific antibodies using negative state repertoires. Structure 24, 641–651 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nimmerjahn F., Ravetch J. V., Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34–47 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Pierce B. G., Hellman L. M., Hossain M., Singh N. K., Vander Kooi C. W., Weng Z., Baker B. M., Computational design of the affinity and specificity of a therapeutic T cell receptor. PLOS Comput. Biol. 10, e1003478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merchant A. M., Zhu Z., Yuan J. Q., Goddard A., Adams C. W., Presta L. G., Carter P., An efficient route to human bispecific IgG. Nat. Biotechnol. 16, 677–681 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Smith M. R., Rituximab (monoclonal anti-CD20 antibody): Mechanisms of action and resistance. Oncogene 22, 7359–7368 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Hober S., Nord K., Linhult M., Protein A chromatography for antibody purification. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 848, 40–47 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Trastoy B., Du J. J., Cifuente J. O., Rudolph L., Garcia-Alija M., Klontz E. H., Deredge D., Sultana N., Huynh C. G., Flowers M. W., Li C., Sastre D. E., Wang L. X., Corzana F., Mallagaray A., Sundberg E. J., Guerin M. E., Mechanism of antibody-specific deglycosylation and immune evasion by Streptococcal IgG-specific endoglycosidases. Nat. Commun. 14, 1705 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klontz E. H., Trastoy B., Deredge D., Fields J. K., Li C., Orwenyo J., Marina A., Beadenkopf R., Gunther S., Flores J., Wintrode P. L., Wang L. X., Guerin M. E., Sundberg E. J., Molecular basis of broad spectrum N-glycan specificity and processing of therapeutic IgG monoclonal antibodies by endoglycosidase S2. ACS Cent. Sci. 5, 524–538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim S. H., Yoo H. J., Park E. J., Na D. H., Nano differential scanning fluorimetry-based thermal stability screening and optimal buffer selection for immunoglobulin G. Pharmaceuticals 15, 29 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joshi S., Maharana C., Rathore A. S., An application of nano differential scanning fluorimetry for higher order structure assessment between mAb originator and biosimilars: Trastuzumab and rituximab as case studies. J. Pharm. Biomed. Anal. 186, 113270 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Roopenian D. C., Akilesh S., FcRn: The neonatal Fc receptor comes of age. Nat. Rev. Immunol. 7, 715–725 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Choi H. J., Seok S. H., Kim Y. J., Seo M. D., Kim Y. S., Crystal structures of immunoglobulin Fc heterodimers reveal the molecular basis for heterodimer formation. Mol. Immunol. 65, 377–383 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Von Kreudenstein T. S., Escobar-Carbrera E., Lario P. I., D’Angelo I., Brault K., Kelly J. F., Durocher Y., Baardsnes J., Woods R. J., Xie M. H., Girod P.-A., Suits M. D. L., Boulanger M. J., Poon D. K. Y., Ng G. Y., Dixit S. B., Improving biophysical properties of a bispecific antibody scaffold to aid developability. MAbs 5, 646–654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma J., Mo Y., Tang M., Shen J., Qi Y., Zhao W., Huang Y., Xu Y., Qian C., Bispecific antibodies: From research to clinical application. Front. Immunol. 12, 626616 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J., Gao X., Li S., Cao Z., Recruitment of IRAK to the interleukin 1 receptor complex requires interleukin 1 receptor accessory protein. Proc. Natl. Acad. Sci. U.S.A. 94, 12829–12832 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fields J. K., Kihn K., Birkedal G. S., Klontz E. H., Sjostrom K., Gunther S., Beadenkopf R., Forsberg G., Liberg D., Snyder G. A., Deredge D., Sundberg E. J., Molecular basis of selective cytokine signaling inhibition by antibodies targeting a shared receptor. Front. Immunol. 12, 779100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeria-Navarro S., Gomez-Gomez A., Park H. S., Calvo-Aranda E., Corominas H., Pou M. A., Diaz-Torne C., Effectiveness and safety of anakinra in gouty arthritis: A case series and review of the literature. Front. Med. 9, 1089993 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mertens M., Singh J. A., Anakinra for rheumatoid arthritis: A systematic review. J. Rheumatol. 36, 1118–1125 (2009). [DOI] [PubMed] [Google Scholar]
- 51.A. Shanehsazzadeh, S. Bachas, M. McPartlon, G. Kasun, J. M. Sutton, A. K. Steiger, R. Shuai, C. Kohnert, G. Rakocevic, J. M. Gutierrez, C. Chung, B. K. Luton, N. Diaz, S. Levine, J. Alverio, B. Knight, M. Radach, A. Morehead, K. Bateman, D. A. Spencer, Z. McDargh, J. Cejovic, G. Kopec-Belliveau, R. Haile, E. Yassine, C. McCloskey, M. Natividad, D. Chapman, J. Bennett, J. Hossain, A. B. Ventura, G. M. Canales, M. Gowda, K. A. Jackson, J. T. Stanton, M. Ura, L. Stojanovic, E. Yapici, K. Moran, R. Caguiat, A. Brown, S. Abdulhaqq, Z. Guo, L. R. Klug, M. Gander, J. Meier, Unlocking de novo antibody design with generative artificial intelligence. bioRxiv 2023.01.08.523187 [Preprint] (2023); 10.1101/2023.01.08.523187. [DOI]
- 52.Diaz D. J., Kulikova A. V., Ellington A. D., Wilke C. O., Using machine learning to predict the effects and consequences of mutations in proteins. Curr. Opin. Struct. Biol. 78, 102518 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hazra M. K., Levy Y., Affinity of disordered protein complexes is modulated by entropy-energy reinforcement. Proc. Natl. Acad. Sci. U.S.A. 119, e2120456119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarkar S., Tang X. L., Das D., Spencer J. S., Lowary T. L., Suresh M. R., A bispecific antibody based assay shows potential for detecting tuberculosis in resource constrained laboratory settings. PLOS ONE 7, e32340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardillo T. M., Karacay H., Goldenberg D. M., Yeldell D., Chang C. H., Modrak D. E., Sharkey R. M., Gold D. V., Improved targeting of pancreatic cancer. Clin. Cancer Res. 10, 3552–3561 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Chames P., Baty D., Bispecific antibodies for cancer therapy: The light at the end of the tunnel? MAbs 1, 539–547 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gold D. V., Goldenberg D. M., Karacay H., Rossi E. A., Chang C. H., Cardillo T. M., McBride W. J., Sharkey R. M., A novel bispecific, trivalent antibody construct for targeting pancreatic carcinoma. Cancer Res. 68, 4819–4826 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Wolf E., Hofmeister R., Kufer P., Schlereth B., Baeuerle P. A., BiTEs: Bispecific antibody constructs with unique anti-tumor activity. Drug Discov. Today 10, 1237–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Huang S., van Duijnhoven S. M. J., Sijts A., van Elsas A., Bispecific antibodies targeting dual tumor-associated antigens in cancer therapy. J. Cancer Res. Clin. Oncol. 146, 3111–3122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kontermann R. E., Brinkmann U., Bispecific antibodies. Drug Discov. Today 20, 838–847 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Klein C., Schaefer W., Regula J. T., Dumontet C., Brinkmann U., Bacac M., Umana P., Engineering therapeutic bispecific antibodies using CrossMab technology. Methods 154, 21–31 (2019). [DOI] [PubMed] [Google Scholar]
- 62.van Tetering G., Evers M., Chan C., Stip M., Leusen J., Fc engineering strategies to advance IgA antibodies as therapeutic agents. Antibodies 9, 70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wallace A. L., Schneider M. I., Toomey J. R., Schneider R. M., Klempner M. S., Wang Y., Cavacini L. A., IgA as a potential candidate for enteric monoclonal antibody therapeutics with improved gastrointestinal stability. Vaccine 38, 7490–7497 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bohlander F., A new hope? Possibilities of therapeutic IgA antibodies in the treatment of inflammatory lung diseases. Front. Immunol. 14, 1127339 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vukovic N., Halabi S., Russo-Cabrera J. S., Blokhuis B., Berraondo P., Redegeld F. A. M., Zaiss D. M. W., A human IgE bispecific antibody shows potent cytotoxic capacity mediated by monocytes. J. Biol. Chem. 298, 102153 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith S. A., Chruszcz M., Chapman M. D., Pomes A., Human monoclonal IgE antibodies-a major milestone in allergy. Curr. Allergy Asthma Rep. 23, 53–65 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young L., Jernigan R. L., Covell D. G., A role for surface hydrophobicity in protein-protein recognition. Protein Sci. 3, 717–729 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stevens J. M., Armstrong R. N., Dirr H. W., Electrostatic interactions affecting the active site of class sigma glutathione S-transferase. Biochem. J. 347 (Pt. 1), 193–197 (2000). [PMC free article] [PubMed] [Google Scholar]
- 69.Ivanov Y. D., Kanaeva I. P., Karuzina I. I., Archakov A. I., Hoa G. H., Sligar S. G., Molecular recognition in the p450cam monooxygenase system: Direct monitoring of protein-protein interactions by using optical biosensor. Arch. Biochem. Biophys. 391, 255–264 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Bogan A. A., Thorn K. S., Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998). [DOI] [PubMed] [Google Scholar]
- 71.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68 (Pt. 4), 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joosten R. P., Salzemann J., Bloch V., Stockinger H., Berglund A. C., Blanchet C., Bongcam-Rudloff E., Combet C., Da Costa A. L., Deleage G., Diarena M., Fabbretti R., Fettahi G., Flegel V., Gisel A., Kasam V., Kervinen T., Korpelainen E., Mattila K., Pagni M., Reichstadt M., Breton V., Tickle I. J., Vriend G., PDB_REDO: Automated re-refinement of X-ray structure models in the PDB. J. Appl. Cryst. 42, 376–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gunther S., Sundberg E. J., Molecular determinants of agonist and antagonist signaling through the IL-36 receptor. J. Immunol. 193, 921–930 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Table S1