Abstract
The causative mutation for Huntington disease (HD), an expanded trinucleotide repeat sequence in the first exon of the huntingtin gene (HTT) is naturally polymorphic and inevitably associated with disease symptoms above 39 CAG repeats. Although symptomatic medical therapies for HD can improve the motor and non-motor symptoms for affected patients, these drugs do not stop the ongoing neurodegeneration and progression of the disease, which results in severe motor and cognitive disability and death. To date, there is still an urgent need for the development of effective disease‐modifying therapies to slow or even stop the progression of HD. The increasing ability to intervene directly at the roots of the disease, namely HTT transcription and translation of its mRNA, makes it necessary to understand the pathogenesis of HD as precisely as possible.
In addition to the long-postulated toxicity of the polyglutamine-expanded mutant HTT protein, there is increasing evidence that the CAG repeat-containing RNA might also be directly involved in toxicity. Recent studies have identified cis- (DNA repair genes) and trans- (loss/duplication of CAA interruption) acting variants as major modifiers of age at onset (AO) and disease progression. More and more extensive data indicate that somatic instability functions as a driver for AO as well as disease progression and severity, not only in HD but also in other polyglutamine diseases. Thus, somatic expansions of repetitive DNA sequences may be essential to promote respective repeat lengths to reach a threshold leading to the overt neurodegenerative symptoms of trinucleotide diseases. These findings support somatic expansion as a potential therapeutic target in HD and related repeat expansion disorders.
Keywords: Huntington disease, repeat expansion disorders, somatic repeat expansion, genetic modifiers
Introduction
Huntington disease (HD) is a slowly progressive and ultimately fatal neurodegenerative disorder, which is inherited in an autosomal dominant manner and characterized by movement disorders and changes in behavior and mental state. Typically, the motor defects include chorea and loss of coordination. Neuropsychiatric symptoms cover the entire spectrum of psychiatric illnesses, with depression, psychosis and obsessive-compulsive disorder most frequently occurring in the course of the illness and being particularly stressful for patients, relatives and caregivers [1].
In 1993, the DNA sequence and the precise nature of the HD-associated mutation in the HTT gene on chromosome 4 was determined. The underlying mutation is the expansion of a physiologically polymorphic CAG trinucleotide repeat in exon 1, which is translated into an elongated polyglutamine tract in the huntingtin protein (HTT) [2].
HD is the most common disorder of at least nine CAG/polyglutamine diseases, including several spinocerebellar ataxias (SCAs), in which CAG repeat expansions encode elongated stretches of glutamines in the respective entirely unrelated disease-associated proteins [3]. HD occurs worldwide but its occurrence varies widely, with the highest prevalence rates for HD reported for western populations from Europe with up to 12 per 100,000 [4].
HTT is a soluble largely α-helical 3,144-amino-acid (348-kDa) protein, essential for embryonic development and involved in cellular activities such as vesicular transport and recycling, endocytosis, endosomal trafficking, autophagy and transcription regulation; however, the entire range of its normal function(s) still remains incompletely defined [5].
The average CAG repeat length in the general population comprises 16–20 repeats; 36 or more CAG units are pathogenic, with repeat lengths of 36–39 CAGs considered reduced-penetrance (RP) alleles [6]. Individuals who carry an expanded HTT allele can become symptomatic at any time point in their life and be healthy until then with no apparent signs of the disease. In the vast majority of cases, the clinical course of HD slowly begins in adulthood, typically in the mid-40s; the age of onset (AO) of disease refers to the time where pre-manifest mutation carriers convert into symptomatic HD patients. Formally, this point of time is reached when the first characteristic motor signs like chorea, bradykinesia or dystonia become overt [7].
The dominant genetic transmission and the fact that AO, phenotype and disease progression do not significantly differ between homozygotes and heterozygotes early led to the proposal of a toxic gain-of-function mechanism that acts through augmentation or dysregulation of one or more normal functions of HTT, either at the RNA or the protein level.
RNA-dependent mechanisms in the molecular pathogenesis of HD
Although aggregated protein fragments are the histopathological hallmark of not only HD but also several other neurodegenerative diseases, their exact role remains controversial. From early on, a predominant hypothesis of toxicity in HD was protein misfolding and accumulation of insoluble aggregates that trigger neuronal dysfunction and lead to cell death [8], [9], [10], [11]. Therefore, prevention of aggregate formation appeared to be a universal strategy to reduce toxicity in HD and other polyglutamine diseases. Since the observation that N-terminal fragments of HTT accumulate in the nucleus, cytoplasm, dendrites and neurites of neurons in HD affected brains, many studies have supported the idea that the generation of small fragments plays a critical role in HD pathogenesis, whereas the formation of large aggregates might be part of a protective cellular mechanism to sequester soluble toxic species [10], [12]. Initially, the proteolytic cleavage of mutant HTT by caspases, calpains and other endoproteases was identified to be involved in the formation of N-terminal fragments [13], [14]. In the meantime, however, it has also been shown that aberrant splicing events at the RNA level play a role in the formation of these fragments. Beside the full-length HTT transcripts that differ in their 3′ UTR lengths, also CAG repeat length-dependent aberrant splicing of exon 1 resulting in short polyadenylated mRNAs that are translated into highly pathogenic exon 1 HTT proteins, both in human and mouse, has been described [15], [16].
Furthermore, it has previously been shown that CAG and CUG expansion transcripts can undergo a novel type of protein translation in which homopolymeric proteins are expressed in all three reading frames without an ATG initiation codon. These repeat-associated non-AUG (RAN) translated products are abundantly expressed in affected regions of HD autopsy brains [17]. However, whether RAN-translated products contribute to disease pathogenesis remains unknown [18]. But it is becoming apparent that beside the abnormal function of the mutant protein, also direct detrimental effects of RNA or RNA-dependent mechanisms could provide a mechanistic basis for the molecular pathogenesis of HD and should be considered in the development of RNA-targeted therapies to lower HTT levels [19].
Genetic modifiers – DNA repair pathways, especially mismatch repair, play a central role in the pathogenesis of HD
As soon as the number of CAG triplets exceeds 40, the length of the CAG repeat in the expanded HTT disease allele inversely correlates with the AO and explains up to 70 % of its variability. Especially in patients with the typical CAG repeat range (40–55 CAG triplets), which is associated with mid-life adult onset of disease, considerable variation of up to 40 years in AO of neurological symptoms is described, even among individuals with identical repeat lengths. This onset variability in patients bearing the same mutations emphasizes the role of functional genetic differences in the genome of these patients that could modify the rate and onset of pathogenesis.
Genetic research approaches in HD can therefore help to understand the differences in clinical onset observed between patients, as well as to identify novel biomarkers and therapeutic strategies to improve disease management. Various human genetic strategies have been used in the last two decades in order to search for these disease-modifying factors that act before clinical diagnosis [20].
Until some years ago, the effort of discovering genetic modifiers was dominated by targeted approaches in which specific genes and variants were chosen on the basis of known or suspected participation in the disease pathology or to functional interaction with the (elongated) HTT protein. A number of trans modifiers in different genes have since been proposed as genetic HD modifiers, which could, however, not yet be replicated in the newer genome-wide association studies (GWAS) applying the stringent genome-wide significance threshold [20].
Interestingly, the GWAS in large patient cohorts all provide evidence for variation in DNA repair genes that modify the AO [21], [22], [23]. The first GWAS of 4,082 HD patients using the difference between AO predicted by CAG length and actual AO of motor symptoms identified FAN1 (FANCD2 and FANCI associated nuclease 1) on chromosome 15 and RRM2B (ribonucleotide reductase regulatory TP53 inducible subunit M2B) on chromosome 8 [21]. In addition, a suggestive association signal was detected on chromosome 3p22.2 near MLH1 (mutL homolog 1), a tumor suppressor gene involved in DNA mismatch repair.
Further candidate single nucleotide polymorphism (SNP) analyses in a cohort of 3,314 additional HD patients independently confirmed the chromosome 8 and 15 loci and moved the MLH1 association to genome-wide significance [24]. Interestingly, the ortholog of MLH1 has previously been indicated as a potential genetic modifier of strain-specific HTT CAG instability [25]. Furthermore, MSH3 (MutS homolog 3), another member of the DNA mismatch repair proteins that has been extensively implicated in the pathogenesis of HD in both mouse and cell studies, could be identified as a likely modifier of disease progression in HD [22], [26], [27]. The latest HD genetic modifier GWAS with more than 9,000 HD patients found an association with a cis-eQTL for increased MSH3 expression in blood cells and identified additional loci with candidate modifier genes involved in DNA maintenance processes, PMS1 (post-meiotic segregation increased 1 homolog), PMS2 (post-meiotic segregation increased 2 homolog) and LIG1 (DNA Ligase 1) [23].
Transcriptome-wide association studies (TWAS) integrating gene expression with GWAS data provided additional support for the role of DNA repair in disease onset. Here, the genes FAN1, PMS1, PMS2 and ASNSD1 were associated with later onset and increased expression and MSH3 with decreased expression [23], [28].
CAA-loss and CAACAG-duplication alleles as cis-acting AO modifiers
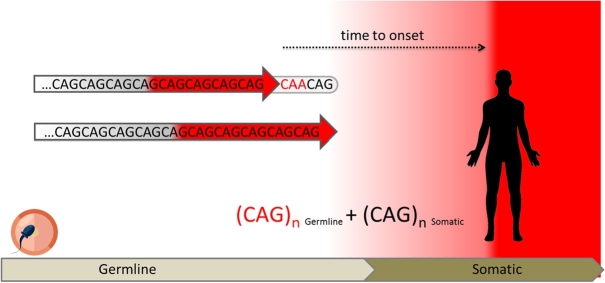
Another consistently significant GWAS signal is on chromosome 4 near HTT. However, since neither HTT promoter SNPs nor significant cis-eQTL SNPs could explain the significant signal, the HTT CAG repeat sequence itself came into focus again. In fact, previously described sequence variations in the HTT repeat sequence appear to be associated with the significant signal on chromosome 4 [29], [30], [31]. Typically, the HTT allele involves a pure CAG repeat that is followed immediately downstream by an additional glutamine-encoding CAA-CAG sequence. Thus, the total number of consecutive glutamines encoded by this region, which is measured by the standard diagnostic HTT PCR fragment-based genotyping assay, is typically equal to the number of pure CAGs, plus two glutamines encoded by the CAA-CAG sequence. Individuals carrying a loss of the penultimate CAA codon (i. e., CAA-CAG to CAG-CAG), therefore have an identical polyglutamine tract length as subjects with the frequent CAA codon, but exhibit a longer uninterrupted CAG sequence. Thus, in carriers of the loss of repeat interruption (LOI) diagnostic testing causes an underestimation of the uninterrupted CAG sequence by two repeats since the polyglutamine repeat length is inferred from fragment sizes based on the assumption of the common interrupting sequence (Figure 1).
Figure 1.
Interplay of inherited and somatic expansions to the progression of HD. The length of the HTT CAG repeat determines the disease phenotype. The disease occurs in individuals who have inherited a repeat tract that has expanded beyond a certain length. Individuals who carry an expanded HTT allele can become symptomatic at any time point in their life and be healthy until then with no apparent signs of the disease. The normal range of CAG repeats is present in unaffected individuals (CAG on a gray background). HTT alleles expanded into the pathological range (CAG on a red background) lead invariably to the onset of HD, and longer alleles are usually associated with an earlier onset of the disease. Although people with reduced-penetrance (RP) alleles have the chance of being asymptomatic throughout lifetime, there are always cases that become symptomatic earlier than expected. Some of these HD patients carry a loss of the penultimate glutamine-encoding CAA codon (in red) in their HTT allele. They have identical polyglutamine tract lengths as subjects with the frequent CAA codon, but exhibit a longer uninterrupted CAG sequence (red arrow), thus indicating that the pure number of uninterrupted CAG repeats (red arrow) makes a more decisive contribution to AO of HD than the number of encoded polyglutamines. Cis- and trans-acting modifiers like the loss of the CAA repeat interruption or variations in DNA repair genes appear to significantly affect the rate at which somatic expansions occur. Under the assumption that the repeat undergoes somatic expansion throughout lifetime, brain cells being most susceptible to disease pathogenesis in particular, HD becomes manifest and the first symptoms appear when the repeat length (the sum of the inherited and somatic expansions, CAGn germline + CAGn somatic) exceeds a disease-specific threshold that may determine the onset of overt toxicity.
Interestingly, LOI carriers show a significantly earlier onset than expected by their CAG length, whereas also existing rare CAA-CAG sequence duplications were associated with a delayed motor onset [23], [32], [33]. Identification of these cis-acting modifiers, therefore, indicates that the pure number of uninterrupted CAG repeats is the most significant contributor to AO of HD and not encoded polyglutamines.
Remarkably, the LOI variant was mainly observed in carriers of alleles with RP (36–39 CAGs), suggesting that the uninterrupted variant partially explains why some individuals who carry RP alleles manifest HD as early as mid-life, while others remain asymptomatic through advanced ages [34].
Thus, the cis-acting AO modifiers have implications for genetic diagnosis and counseling, especially when dealing with an intermediate or RP allele range. The underestimation by two CAG repeats in LOI carriers can have a profound effect on a possible disease manifestation. Therefore, diagnostic tests which currently do not assess the loss of the CAA codon should be adjusted, especially when dealing with individuals that carry alleles in the lower end of the disease-associated CAG range or when including patients in clinical studies with CAG repeat length inclusion criteria.
But what underlies the phenotypic effects of the LOI variant?
Originally, the instability of tandem repeats was explained with the simple DNA slippage model, which postulates that repeats may be lost or gained during local misalignment of DNA during replication [35]. However, this model leaves some questions unanswered, including why repeat interruptions lead to the stabilization of repeat expansions.
Findings that interruptions of the CAG repeat within protein-coding transgenes mitigate toxicity and that an untranslated CAG repeat RNA can cause toxicity on its own further support a role of RNA in polyglutamine diseases. In a Drosophila model of SCA3, altering the CAG repeat sequence to an interrupted CAA-CAG repeat within the polyglutamine-encoding region did not affect ataxin-3 protein accumulation, but dramatically mitigated toxicity and the expression of an untranslated CAG repeat of pathogenic length conferred neuronal degeneration [36]. Naturally occurring interruptions found in several other trinucleotide repeat disorders like SCAs, myotonic dystrophy type 1 (DM1), Friedreich ataxia (FRDA) and Fragile X syndrome (FXS) have all been associated with the stabilization of the respective trinucleotide repeat loci and in some cases even with lower somatic instability and later AO [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48].
As early as 30 years ago, it was postulated that the ability of longer repeat regions to form stable secondary structures could lead to site-specific instability if the structure inhibits the action of replication or repair proteins at the repeat site [49]. It has been shown that trinucleotide repeat instability is mediated by the formation of unusual secondary structures that differ from the canonical B-DNA double helix during DNA replication, repair, recombination and gene transcription [50]. CAG repeats of sufficient length form imperfect stem and loop structures in transcripts [51]. HTT CAGs have been shown to form hairpins in vitro in a tripartite way, where the base is composed of interacting CAG and adjacent CCG repeats, followed by a central motif consisting solely of CAG repeats and a terminal section composed of the fold-back structure from CAG repeats [52]. The folding properties, however, depend on the interplay between repeated and specific flanking sequences, which have a significant influence on the correlation between the repeat length, the stability of the hairpin structure and the ability of this structure to trigger RNA pathogenesis in cells, suggesting a relevant role for the RNA structure towards inducing a toxic effect [51]. Different secondary structures may interfere with the progress of RNA polymerase and involve various DNA repair processes that can impact repeat expansion and contraction. The presence of repeat interruptions in such structures may facilitate their correct repair upon recognizing the mismatches that arise from the misalignment of repeat interruptions [53].
Somatic instability
The HTT CAG repeat is unstable in germline and somatic cells, and expansion in both cell types has deleterious consequences. Germ line expansions are responsible for the intergenerational instability of the HTT CAG repeat, a phenomenon called anticipation. Transmission of the mutation to offspring is characterized by the tendency to expansion and the longer the repeat, the more severe the disease and the earlier the onset of symptoms. Already more than 25 years ago a highly significant correlation between CAG instability and the size of the parental CAG repeat as well as somatic mosaicism in sperm was described for paternal transmission in HD [54]. Later investigations in male transgenic mice showed that in germ cells, expansion is limited to the post-meiotic haploid cell, implying DNA repair mechanisms rather than DNA replication in DNA synthesis as the cause of the expansions [55]. Yet, somatic CAG repeat expansion also occurs in several other tissues, including the brain. Studies both in HD patient tissues and in HD mouse models have shown that the expanded HTT CAG repeat has a high tendency to further expand in somatic cells, whereas the degree of somatic instability is highly tissue-specific, repeat length-dependent and age-dependent [27], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70]. In transgenic and knock-in mouse models of HD, increased somatic CAG expansion is described in cells from the striatum, cortex and liver, whereas in cells from the cerebellum, blood and tail the repeat length is relatively stable [57], [63], [66], [71]. Large CAG repeat expansions with a similar distribution pattern like in mice were also demonstrated in post-mortem human tissues [60], [62], [64]. The latest study revealed a similar profile of tissue instability in seven adult and one juvenile HD patient that was also apparent in an individual with SCA1. Somatic CAG expansion was observed in all tissues, but to different degrees, with multiple cortical regions and neostriatum tending to have the greatest instability in the CNS and liver in the periphery [72]. These similar tissue-specific patterns of repeat expansion support the assumption that disease locus-independent trans factors cause the extent of CAG expansion in different cell types. Beside the inherited CAG length also previously identified DNA repair pathway-related modifier variations were shown to mediate somatic expansions in mouse models and humans. This supports the assumption that abnormal structure-dependent interactions of expanded RNA repeats with various cellular proteins and differing expression levels of repair genes might be the main or a contributing factor of the cell-specific vulnerability/CAG expansion [25], [26], [27], [32], [67], [73], [74].
Conclusion
For nearly 30 years, scientists have been trying to uncover the basic principles that underlie the mechanisms of repeat instability. Especially in polyglutamine diseases the progressive, late-onset characteristics were initially attributed to a low accumulating toxicity of the polyglutamine proteins expressed from expanded CAG repeats. However, a synopsis of earlier and more recent studies shows that in addition to the abnormal function of the mutated protein also direct RNA-damaging effects or RNA-dependent mechanisms could represent a mechanistic basis for the molecular pathogenesis of HD.
For HD as well as several types of SCA, variation in DNA repair proteins as well as sequence interruptions have been identified as major modifiers of AO and disease progression, implicating ongoing somatic expansions as a common mechanism of disease. Thus, there is much to suggest that the combination of the number of inherited repeats together with the degree of somatic instability determines the AO for a given disease and patient (Figure 1).
There is currently no cure for HD and pharmacotherapy is limited to symptomatic treatment of movement disorders and psychiatric symptoms. For the moment, the most promising treatment appears to be targeting the pathological processing of HTT mRNA, or upstream. A number of RNA-targeting therapies have recently entered clinical trials, which aim to lower the mutant HTT production by the use of antisense oligonucleotides (ASOs) and RNAi. However, each of the different strategies has specific advantages and disadvantages, as recently seen by the failures of Roche and Wave Therapeutics’s ASO candidates in HD [75].
Considering the influence of somatic instability on disease onset and progression, targeting the repeat instability is a very attractive disease-modifying strategy in HD and related expansion disorders, and possibly suppressing or reversing somatic repeat expansion may halt or delay the progression of the disease.
Funding Statement
Does not apply, no funding.
Footnotes
Author contributions: The authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Competing interests: Authors state no conflict of interest.
Informed consent: Does not apply, review, no subjects involved.
Ethical approval: Does not apply, review, no subjects involved.
References
- [1].Rosenblatt A. Neuropsychiatry of Huntington’s disease. Dialogues Clin Neurosci. 2007 doi: 10.31887/dcns.2007.9.2/arosenblatt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].HD collaborative, research group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group [see comments] Cell. 1993;72(6):971–83. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- [3].Paulson H. Handbook of Clinical Neurology. 2018. Repeat expansion diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rawlins MD, Wexler NS, Wexler AR. et al. The prevalence of Huntington’s disease. Neuroepidemiology. 2016 doi: 10.1159/000443738. [DOI] [PubMed] [Google Scholar]
- [5].Saudou F, Humbert S. The biology of huntingtin. Neuron. 2016 doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- [6].McNeil SM, Novelletto A, Srinidhi J. et al. Reduced penetrance of the Huntington’s disease mutation. Hum Mol Genet. 1997 doi: 10.1093/hmg/6.5.775. [DOI] [PubMed] [Google Scholar]
- [7].Kieburtz K. Unified Huntington’s disease rating scale: reliability and consistency. Mov Disord. 1996 doi: 10.1002/mds.870110204. [DOI] [PubMed] [Google Scholar]
- [8].Becher MW, Kotzuk JA, Sharp AH. et al. Intranuclear neuronal inclusions in Huntington’s disease and dentatorubral and pallidoluysian atrophy: correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol Dis. 1998 doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- [9].Davies SW, Turmaine M, Cozens BA. et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997 doi: 10.1016/S0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- [10].DiFiglia M, Sapp E, Chase KO. et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997 doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- [11].Ordway JM, Tallaksen-Greene S, Gutekunst CA. et al. Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell. 1997 doi: 10.1016/S0092-8674(00)80464-X. [DOI] [PubMed] [Google Scholar]
- [12].Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp Neurol. 2012 doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Landles C, Sathasivam K, Weiss A. et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem. 2010 doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goldberg YP, Nicholson DW, Rasper DM. et al. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet. 1996 doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- [15].Neueder A, Dumas AA, Benjamin AC, Bates GP. Regulatory mechanisms of incomplete huntingtin mRNA splicing. Nat Commun. 2018 doi: 10.1038/s41467-018-06281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sathasivam K, Neueder A, Gipson TA. et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bañez-Coronel M, Ayhan F, Tarabochia AD. et al. RAN translation in Huntington disease. Neuron. 2015 doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang S, Yang H, Huang L. et al. Lack of RAN-mediated toxicity in Huntington’s disease knock-in mice. Proc Natl Acad Sci USA. 2020 doi: 10.1073/pnas.1919197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Franich NR, Hickey MA, Zhu C. et al. Phenotype onset in Huntington’s disease knock-in mice is correlated with the incomplete splicing of the mutant huntingtin gene. J Neurosci Res. 2019 doi: 10.1002/jnr.24493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Arning L. The search for modifier genes in Huntington disease – multifactorial aspects of a monogenic disorder. Mol Cell Probes. 2016 doi: 10.1016/j.mcp.2016.06.006. [DOI] [PubMed] [Google Scholar]
- [21].Lee JM, Wheeler VC, Chao MJ. et al. Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell. 2015 doi: 10.1016/j.cell.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Moss DJH, Tabrizi SJ, Mead S. et al. Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol. 2017 doi: 10.1016/S1474-4422(17)30161-8. [DOI] [PubMed] [Google Scholar]
- [23].Lee JM, Correia K, Loupe J. et al. CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell. 2019 doi: 10.1016/j.cell.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee JM, Chao MJ, Harold D. et al. A modifier of Huntington’s disease onset at the MLH1 locus. Hum Mol Genet. 2017 doi: 10.1093/hmg/ddx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pinto RM, Dragileva E, Kirby A. et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: genome-wide and candidate approaches. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tomé S, Manley K, Simard JP. et al. MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington’s disease mice. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dragileva E, Hendricks A, Teed A. et al. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009 doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wright GEB, Caron NS, Ng B. et al. Gene expression profiles complement the analysis of genomic modifiers of the clinical onset of Huntington disease. bioRxiv. 2019 doi: 10.1101/699033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pêcheux C, Mouret JF, Dürr A. et al. Sequence analysis of the CCG polymorphic region adjacent to the CAG triplet repeat of the HD gene in normal and HD chromosomes. J Med Genet. 1995 doi: 10.1136/jmg.32.5.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Goldberg YP, Mcmurray CT, Zeisier J. et al. Increased instability of intermediate alleles in families with sporadic Huntington disease compared to similar sized intermediate alleles in the general population. Hum Mol Genet. 1995 doi: 10.1093/hmg/4.10.1911. [DOI] [PubMed] [Google Scholar]
- [31].Gellera C, Meoni C, Castellotti B, et al. Errors in Huntington disease diagnostic test caused by trinucleotide deletion in the IT15 gene [1]. Am J Hum Genet. 1996. [PMC free article] [PubMed]
- [32].Ciosi M, Maxwell A, Cumming SA. et al. A genetic association study of glutamine-encoding DNA sequence structures, somatic CAG expansion, and DNA repair gene variants, with Huntington disease clinical outcomes. EBioMedicine. 2019 doi: 10.1016/j.ebiom.2019.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wright GEB, Collins JA, Kay C. et al. Length of uninterrupted CAG, independent of polyglutamine size, results in increased somatic instability, hastening onset of Huntington disease. Am J Hum Genet. 2019 doi: 10.1016/j.ajhg.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Findlay Black H, Wright GEB, Collins JA. et al. Frequency of the loss of CAA interruption in the HTT CAG tract and implications for Huntington disease in the reduced penetrance range. Genet Med. 2020 doi: 10.1038/s41436-020-0917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG)·(CAG) repeat hairpins in human cells. Nat Chem Biol. 2010 doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008 doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chung MY, Ranum LPW, Duvick LA, Servadio A, Zoghbi HY, Orr HT. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993 doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- [38].Ohshima K, Sakamoto N, Labuda M. et al. A nonpathogenic GAAGGA repeat in the Friedreich gene: implications for pathogenesis. Neurology. 1999 doi: 10.1212/wnl.53.8.1854. [DOI] [PubMed] [Google Scholar]
- [39].Eichler EE, Holden JJA, Popovich BW. et al. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994 doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- [40].Tomé S, Dandelot E, Dogan C. et al. Unusual association of a unique CAG interruption in 5′ of DM1 CTG repeats with intergenerational contractions and low somatic mosaicism. Human Mutat. 2018 doi: 10.1002/humu.23531. [DOI] [PubMed] [Google Scholar]
- [41].Gao R, Matsuura T, Coolbaugh M. et al. Instability of expanded CAG/CAA repeats in spinocerebellar ataxia type 17. Eur J Hum Genet. 2008 doi: 10.1038/sj.ejhg.5201954. [DOI] [PubMed] [Google Scholar]
- [42].Pešović J, Perić S, Brkušanin M, Brajušković G, Rakočević-Stojanović V, Savić-Pavićević D. Repeat interruptions modify age at onset in myotonic dystrophy type 1 by stabilizing DMPK expansions in somatic cells. Front Genet. 2018 doi: 10.3389/fgene.2018.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Moseley ML, Schut LJ, Bird TD, Koob MD, Day JW, Ranum LPW. SCA8 CTG repeat: en masse contractions in sperm and intergenerational sequence changes may play a role in reduced penetrance. Hum Mol Genet. 2000 doi: 10.1093/hmg/9.14.2125. [DOI] [PubMed] [Google Scholar]
- [44].Matsuura T, Fang P, Pearson CE. et al. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: repeat purity as a disease modifier? Am J Hum Genet. 2006 doi: 10.1086/498654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hu Y, Hashimoto Y, Ishii T. et al. Sequence configuration of spinocerebellar ataxia type 8 repeat expansions in a Japanese cohort of 797 ataxia subjects. J Neurol Sci. 2017 doi: 10.1016/j.jns.2017.08.3256. [DOI] [PubMed] [Google Scholar]
- [46].Landrian I, McFarland KN, Liu J, Mulligan CJ, Rasmussen A, Ashizawa T. Inheritance patterns of ATCCT repeat interruptions in spinocerebellar ataxia type 10 (SCA10) expansions. PLoS ONE. 2017 doi: 10.1371/journal.pone.0175958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Musova Z, Mazanec R, Krepelova A. et al. Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. Am J Med Genet, Part A. 2009 doi: 10.1002/ajmg.a.32987. [DOI] [PubMed] [Google Scholar]
- [48].Botta A, Rossi G, Marcaurelio M. et al. Identification and characterization of 5′ CCG interruptions in complex DMPK expanded alleles. Eur J Hum Genet. 2017 doi: 10.1038/ejhg.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Moore H, Greenwell PW, Liu CP, Arnheim N, Petes TD. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc Natl Acad Sci USA. 1999 doi: 10.1073/pnas.96.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010 doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sobczak K, Michlewski G, De Mezer M. et al. Structural diversity of triplet repeat RNAs. J Biol Chem. 2010 doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].De Mezer M, Wojciechowska M, Napierala M, Sobczak K, Mutant KWJ. CAG repeats of huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rolfsmeier ML, Dixon MJ, Pessoa-Brandão L, Pelletier R, Miret JJ, Lahue RS. Cis-elements governing trinucleotide repeat instability in Saccharomyces cerevisiae. Genetics. 2001. [DOI] [PMC free article] [PubMed]
- [54].Telenius H, Almqvist E, Kremer B. et al. Somatic mosaicism in sperm is associated with intergenerational (CAG)n changes in Huntington disease. Hum Mol Genet. 1995 doi: 10.1093/hmg/4.2.189. [DOI] [PubMed] [Google Scholar]
- [55].Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001 doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- [56].De Rooij KE, De Koning Gans PAM, Roos RAC, Van Ommen GJB, Den Dunnen JT. Somatic expansion of the (CAG)n repeat in Huntington disease brains. Hum Genet. 1995 doi: 10.1007/BF00225192. [DOI] [PubMed] [Google Scholar]
- [57].Mangiarini L, Sathasivam K, Mahal A, Mott R, Seller M, Bates GP. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat Genet. 1997 doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- [58].Wheeler VC, Auerbach W, White JK. et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum Mol Genet. 1999 doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- [59].Kennedy L, Shelbourne PF. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Hum Mol Genet. 2000 doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- [60].Kennedy L, Evans E, Chen CM. et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet. 2003 doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- [61].Veitch NJ, Ennis M, McAbney JP, Shelbourne PF, Monckton DG. Inherited CAG·CTG allele length is a major modifier of somatic mutation length variability in Huntington disease. DNA Repair (Amst) 2007 doi: 10.1016/j.dnarep.2007.01.002. [DOI] [PubMed] [Google Scholar]
- [62].Shelbourne PF, Keller-McGandy C, Bi WL. et al. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- [63].Gonitel R, Moffitt H, Sathasivam K. et al. DNA instability in postmitotic neurons. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Swami M, Hendricks AE, Gillis T. et al. Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lee JM, Pinto RM, Gillis T, Claire JC St, Wheeler VC. Quantification of age-dependent somatic CAG repeat instability in Hdh CAG knock-in mice reveals different expansion dynamics in striatum and liver. PLoS ONE. 2011 doi: 10.1371/journal.pone.0023647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lee JM, Zhang J, Su AI. et al. HA novel approach to investigate tissue-specific trinucleotide repeat instability. BMC Syst Biol. 2010 doi: 10.1186/1752-0509-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kovalenko M, Dragileva E, St Claire J. et al. Msh2 acts in medium-spiny striatal neurons as an enhancer of CAG instability and mutant huntingtin phenotypes in Huntington’s disease knock-in mice. PLoS ONE. 2012 doi: 10.1371/journal.pone.0044273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Larson E, Fyfe I, Morton AJ, Monckton DG. Age-, tissue- and length-dependent bidirectional somatic CAG•CTG repeat instability in an allelic series of R6/2 Huntington disease mice. Neurobiol Dis. 2015 doi: 10.1016/j.nbd.2015.01.004. [DOI] [PubMed] [Google Scholar]
- [69].Geraerts FCA, Snell RG, Faull RLM, Williams L, Jacobsen JC, Reid SJ. Comparison of Huntington’s disease CAG repeat length stability in human motor cortex and cingulate gyrus. J Huntingtons Dis. 2016 doi: 10.3233/JHD-160203. [DOI] [PubMed] [Google Scholar]
- [70].Ament SA, Pearl JR, Grindeland A. et al. High resolution time-course mapping of early transcriptomic, molecular and cellular phenotypes in Huntington’s disease CAG knock-in mice across multiple genetic backgrounds. Hum Mol Genet. 2017 doi: 10.1093/hmg/ddx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Møllersen L, Rowe AD, Larsen E, Rognes T, Klungland A. Continuous and periodic expansion of CAG repeats in Huntington’s disease R6/1 mice. PLoS Genet. 2010 doi: 10.1371/journal.pgen.1001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Pinto RM, Arning L, Giordano JV. et al. Patterns of CAG repeat instability in the central nervous system and periphery in Huntington’s disease and in spinocerebellar ataxia type 1. Hum Mol Genet. 2020 doi: 10.1093/hmg/ddaa139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wheeler VC. Mismatch repair gene Msh2 modifies the timing of early disease in HdhQ111 striatum. Hum Mol Genet. 2003 doi: 10.1093/hmg/12.3.273. [DOI] [PubMed] [Google Scholar]
- [74].Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999 doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- [75].Kingwell K. Double setback for ASO trials in Huntington disease. Nat Rev Drug Discov. 2021 doi: 10.1038/d41573-021-00088-6. [DOI] [PubMed] [Google Scholar]