Abstract
The interest in MS-based analysis of modified nucleic acids is increasing due to the application of nucleic acids in therapeutics. However, there are few available integrated platforms for characterizing nucleic acid modifications. Herein, we report a general mass spectrometry-based SWATH platform to identify and quantify both RNA and DNA modifications, which we call SWATH Analysis of Modified Nucleic Acids (SWAMNA). SWAMNA incorporates the search engine, NuMo Finder, enabling the analysis of modifications in native and permethylated form. SWAMNA will aid discoveries that provide new insights into nucleic acid modifications.
Nucleic acids (DNA and RNA) are fundamental biological molecules that encode and convey genetic information within living organisms. Both DNA and RNA can carry modifications, which are involved in critical biological functions, including regulating gene expression and stabilizing nucleic acid structure.1 Nucleic acid modifications are also being investigated as potential therapeutic targets for a variety of diseases, including cancer, neurodegenerative diseases, and infectious diseases.2 Therefore, it is crucial to develop techniques to characterize these modifications. Several next-generation sequencing (NGS) based technologies have been employed for genome-wide studies of nucleic acid modifications.3 However, the high diversity (over 150) of DNA and RNA modifications creates obstacles when using these methods for profiling different modifications in a single analysis. Recently, researchers have demonstrated detecting specific RNA modifications, such as m6A, using the signature signals generated from Nanopore.4–6 The correlation between modification features and their corresponding signatures depends on empirical observations and can be hard to predict, which limits this approach for analyzing diverse modifications.
Liquid chromatography with tandem mass spectrometry (LC-MS/MS) enables simultaneous characterization and quantification of different modifications.7 The ability to identify modifications directly based on their distinct mass and fragmentation patterns distinguishes MS-based techniques from other analytical approaches. LC-MS/MS analysis of nucleic acid modifications has often been performed at the nucleoside level. This can provide novel insights into distribution, abundance, and potential functional roles. Due to the hydrophilicity of the nucleoside molecules, porous graphitic carbon (PGC) or hydrophilic interaction chromatography (HILIC) column is routinely used.8, 9 Chemical derivatization, such as permethylation, allows sensitive characterization using C18 reversed-phase columns.10 Targeted MS analysis has been predominantly used, and the alternation in RNA and DNA modification was observed from different biological conditions.11–13 However, the information regarding the modifications not within the inclusion list is overlooked. Recently, our lab and the Sabidó group applied data-independent acquisition (DIA) to enable the potential detection of unanticipated molecular species.14, 15 These advances captured much attention in using LC-MS/MS to analyze nucleic acid modifications; however, the absence of an comprehensive platform for the characterization of DNA and RNA modifications hampers its potential for broader applications.
Herein, we report a platform, SWAMNA (SWATH analysis of Modified Nucleic Acids), to comprehensively identify and quantify both RNA and DNA modifications (Scheme 1). SWAMNA includes the whole workflow from nucleic acids digestion, chemical derivatization, sequential window acquisition of all theoretical fragment ion spectra (SWATH) analysis, and data analysis. We demonstrated the reproducibility and sensitivity using SWAMNA, and the application of the SWAMNA allows researchers to decipher variations introduced by different biological conditions.
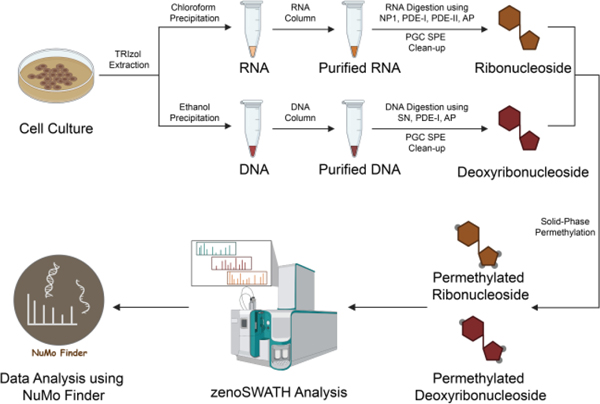
Scheme 1. The SWAMNA platform for analyzing DNA and RNA modifications.
Cellular DNA and RNA were extracted and purified, then enzymatically digested into mononucleosides using a “one-pot” approach. The resulting digested nucleosides underwent purification through PGC solid-phase extraction and were subsequently subjected to derivatization using permethylation. The zenoSWATH was employed for the analysis, and the NuMo Finder search engine was developed to identify and quantify the modifications present.
In SWAMNA, total RNA and DNA were sequentially extracted using the conventional TRIzol protocol, and the high-quality RNA and DNA were further purified using respective RNA and DNA columns. The purity of RNA and DNA was confirmed using nanodrop and gel electrophoresis. To digest the nucleic acid into mononucleosides, different chemical and enzymatic methods have been developed.16 Previously, we found that RNA can be efficiently digested using the “one-pot” reaction by mixing the nuclease P1 (NP1), phosphodiesterase-I (PDE-I), phosphodiesterase II (PDE-II), and alkaline phosphatase (AP).14 At the same time, Wang and co-workers demonstrated the workflow for DNA digestion.17 Therefore, we incorporated the enzymes into our “one-pot” reaction strategy and tested its feasibility. As shown in Figure S1, the DNA was completely digested after overnight incubation, and the deoxyribonucleosides can be purified using PGC solid-phase extraction (SPE).
The variable window data-independent acquisition SWATH provides consistent fragments with pre-defined window selections, and the novel Zeno trap can deliver sensitivity gains for the analysis. In order to establish the zenoSWATH method that would provide unbiased mononucleoside data, different isolation window schemes were designed depending on the mass distribution of different RNA and DNA modifications that are derived from Modomics and DNAmod databases.18, 19 Notably, because 2-O’-methylated ribonucleoside fragmentations are identical to the respective canonical nucleoside product ions, which leads to misidentifications, we thus kept the methylated ribonucleoside and canonical ribonucleoside in the different isolation windows. Meanwhile, considering the potential of large modification on RNAs (such as glycosylation),20 we extended the mass window with fixed window size to cover the potential large modification on nucleosides. Consequently, the total cycle time was about 1 second, which is optimal with enough data points across the peak to obtain decent quantitative results. In addition, regarding the alternative studies in the characterization of nucleosides in their forms, we also included the isolation window scheme for analyzing native RNA and DNA modifications. Details about the isolation schemes are listed in Table S1.
We have demonstrated the permethylation reaction replaced the hydrogens on hydrophilic groups (such as hydroxyl groups, amine groups, and carboxyl groups) with d3-methyl groups and with nearly 100% efficiency to yield hydrophobic permethylated nucleosides that are ideal for reverse-phase retention and separation.10 In SWAMNA, great chromatography in terms of retention and separation was also observed in using the microflow LC system (Figure S2). To investigate the reproducibility and stability of SWAMNA, the biological and instrumental replicate samples were characterized. These results showed that the analysis from SWAMNA is reproducible and stable (Figure S3 and S4). To evaluate the performance in quantification, we mixed standard nucleosides and analyzed them by SWAMNA using three replicates with different dilution factors (from pmol/μL to fmol/μL level). The product ions of nucleosides from each standard were selected, and the quantification at the MS2 level was used to generate the linear calibration curve (Figure S5). Notably, the Zeno trap enhances sensitivity to enable as low as 10 fmol/μl as the limit of quantitation (LOQ) with capillary flow rate. Additionally, quantitative results demonstrated good linearity for all the compounds with correlation coefficients (R2) higher than 0.991, which indicates good reproducibility with the overall coefficients of variation (CVs). Overall, our results demonstrated the sensitivity and accuracy of quantitative analysis using SWAMNA.
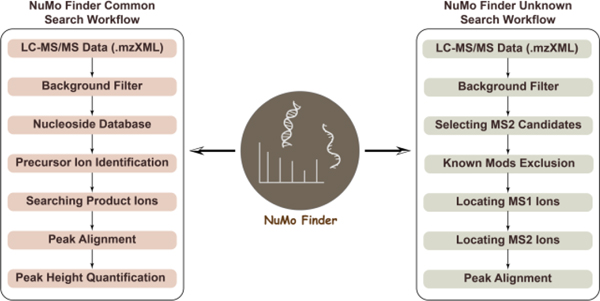
There is an urgent need for bioinformatic tools that allow quicker and easier data interpretation of MS data. Recently, Gosset-Erard et al. reported the first untargeted RNA Post-transcriptional modifications search engine, Nucleos’ID.21 However, the limitation of data type does not allow the input of SWATH results. To make our platform more accessible, we developed a new search engine, NuMo Finder (available at: https://github.com/ChenfengZhao/NuMoFinder). By entering different parameters, such as MS1 and MS2 mass tolerances, intensity threshold, m/z range, and retention time range, the software extracts the signature peaks from MS1 and MS2 data to identify modifications. As shown in Figure 1, two functions, common search and unknown search, were incorporated. The common mods search includes most of the already known nucleic acid modifications referenced in the established Modomics and DNAmod databases. At the same time, because collision-induced dissociation (CID) generates the major ribose/deoxyribose lost product ions of nucleosides, we thus took advantage of this feature for the identification of potential unknown modifications. Specifically, the fragment mass above a certain threshold is considered generated from ribose/deoxyribose loss, followed by adding back the ribose or deoxyribose mass, checking the calculated precursor mass from MS1, and evaluating the alignment between the precursor and product ions. The m/z of potential nucleic acid modifications can be further exported, and its chemical formula can be predicted using elemental composition analysis. As shown in Figure S6, we found some low-level oxidized guanosine using this function, which were generated due to the induced current from PGC column.22 Overall, the NuMo Finder took advantage of the feature of SWATH that fragment all the precursors in certain mass ranges during the acquisition, demonstrating the results from SWAMNA can be revisited in future studies and offer valuable insights about currently unknown modifications.
Figure 1. Workflows of identifying RNA and DNA modifications using NuMo Finder.
Two search functions, common search and unknown search, were developed. The modifications in Modomics and DNAmod databases were referenced. Unknown modifications were searched based on the feature of ribose and deoxyribose loss in nucleoside fragmentation, and more details about the NuMo Finder can be found in Supplementary Information 2.
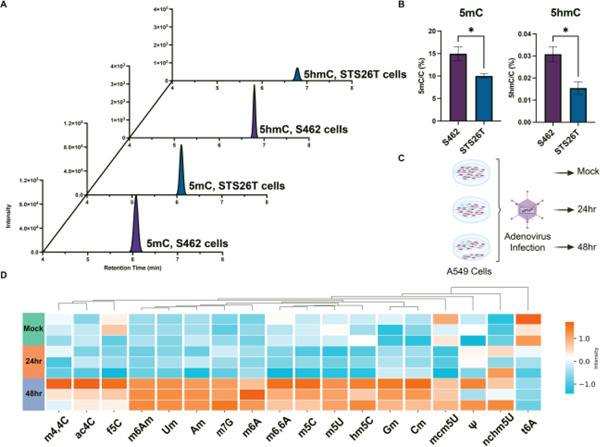
As our SWAMNA platform showed good performance in identification and quantification, we further applied it to examine the DNA modification changes in malignant peripheral nerve sheath tumor (MPNST) cells. Previously, it was reported that global loss of polycomb-repressive complex 2 (PRC2)-mediated repression renders MPNST differentially dependent on DNA methylation to maintain transcriptional integrity and makes them susceptible to therapeutics that promote aberrant transcription initiation with PRC2 loss.23 However, the role of other epigenetic marks has not been fully investigated. Therefore, we employed SWAMNA to monitor the level of different DNA modifications in two MPNST cell lines, STS26T (PRC2 intact) and S462 (PRC2 loss). In line with previous findings, S462 showed a higher degree of 5mC (Figure 2A). Interestingly, 5hmC, an oxidation product of 5mC, was also upregulated in S462 cells (Figure 2B). The data suggest that increased levels of 5mC undergo oxidation in these cells.
Figure 2. SWAMNA was applied to characterize DNA and RNA modifications in different biological conditions.
(A) and (B) SWAMNA monitored the increasing of 5mC and 5hmC levels in MPNST S462 cells due to the PRC-2 loss. (C) RNA extracted from Adenovirus infected A549 cells were analyzed at 24 and 48 hours post-infection. (D) The modification with significant change was shown and the amount of methylated nucleosides generally increased during adenovirus infection. Asterisks indicate the statistical significance between groups compared using the t-test (*p< 0.05%).
SWAMNA is also a powerful tool to assess RNA modifications in cells. As an example, we have applied the technology to study the process of viral infection. Adenovirus is a nuclear replicating DNA virus reliant on host RNA processing machinery. Previous studies discovered that adenoviral RNA undergoes modifications in host cells, which are proposed to contribute to the productive infection of adenoviruses. One of the modifications, m6A, has been proven to play a critical role in the effective splicing of viral late transcripts.5 Nevertheless, little is known about global changes in different RNA modifications during infection, as it is challenging for conventional antibody-based methods. Here, we infected A549 cells with adenovirus and extracted total RNAs at different time intervals (0hr, 24hr, and 48hr) for analysis of the different RNA modifications using SWAMNA (Figure 2C). Over 20 types of modifications were quantified, and the infection significantly altered the levels of several modifications (Figure 2D). Noticeably, we observed an overall increase of methylated nucleosides, including m6A, at 48hr after infection. Interestingly we also observed a decrease in the t6A mark during infection. These results from SWAMNA decipher the landscape of RNA modification changes during the process of adenoviral infection (Figure 2C). Results suggest an increase in RNA modifications at late times of infection (Figure 2D), providing valuable information for further study to dissect the function of each altered modification.
In summary, a comprehensive MS-based platform, SWAMNA, for characterizing nucleic acid modifications was developed. Permethylation and zenoSWATH analysis offer reproducible quantitative results, while NuMo Finder meets the need for computational tools that streamline data interpretation and extend the application of nucleic acid analysis. Additionally, the application of SWAMNA demonstrates that this approach can monitor the low-abundant but crucial epigenetic and epitranscriptomic modifications in biological samples. SWAMNA can assist in gaining insights about the diverse and dynamic landscape of chemical alterations within DNA and RNA molecules, potentially opening doors to novel therapeutics.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by grants from the NIH AI118891 (M.D.W. and B.A.G.), AI145266 (M.D.W), AI167545 (R.T.S.), CA196539 (B.A.G.), and AG031862 (B.A.G.). J. K. L. was supported by the Sigrid Jusélius Foundation and Emil Aaltonen Foundation.
The authors thank the instrumental support from SCIEX, especially Patrick Pribil, Scott Leech, and Sahana Mollah, on this research topic.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/x0xx00000x.
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Chen K, Zhao Boxuan S. and He C, Cell Chemical Biology, 2016, 23, 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbieri I. and Kouzarides T, Nature Reviews Cancer, 2020, 20, 303–322. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz S. and Motorin Y, RNA Biology, 2017, 14, 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abebe JS, Price AM, Hayer KE, Mohr I, Weitzman MD, Wilson AC and Depledge DP, Bioinformatics, 2022, 38, 3113–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price AM, Hayer KE, McIntyre ABR, Gokhale NS, Abebe JS, Della Fera AN, Mason CE, Horner SM, Wilson AC, Depledge DP and Weitzman MD, Nature Communications, 2020, 11, 6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA and Novoa EM, Nature Communications, 2019, 10, 4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng L, Kumar J, Rose R, McIntyre W. and Fabris D, Mass Spectrometry Reviews, 2022, n/a, e21798. [DOI] [PubMed] [Google Scholar]
- 8.Sakaguchi Y, Miyauchi K, Kang B.-i. and Suzuki T, in Methods in Enzymology, ed. He C, Academic Press, 2015, vol. 560, pp. 19–28. [DOI] [PubMed] [Google Scholar]
- 9.Sarin LP, Kienast SD, Leufken J, Ross RL, Dziergowska A, Debiec K, Sochacka E, Limbach PA, Fufezan C, Drexler HCA and Leidel SA, RNA, 2018, 24, 1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Y, Janssen KA, Scacchetti A, Porter EG, Lin Z, Bonasio R. and Garcia BA, Analytical Chemistry, 2022, 94, 7246–7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M-Y, Gui Z, Chen K-K, Ding J-H, He J-G, Xiong J, Li J-L, Wang J, Yuan B-F and Feng Y-Q, Chinese Chemical Letters, 2022, 33, 2086–2090. [Google Scholar]
- 12.Chen M-Y, Qi C-B, Tang X-M, Ding J-H, Yuan B-F and Feng Y-Q, Chinese Chemical Letters, 2022, 33, 3772–3776. [Google Scholar]
- 13.Lan M-D, Xiong J, You X-J, Weng X-C, Zhou X, Yuan B-F and Feng Y-Q, Chemistry – A European Journal, 2018, 24, 9949–9956. [DOI] [PubMed] [Google Scholar]
- 14.Janssen KA, Xie Y, Kramer MC, Gregory BD and Garcia BA, Journal of the American Society for Mass Spectrometry, 2022, 33, 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espadas G, Morales-Sanfrutos J, Medina R, Lucas MC, Novoa EM and Sabidó E, Journal of Chromatography A, 2022, 1665, 462803. [DOI] [PubMed] [Google Scholar]
- 16.Jones JD, Grassmyer KT, Kennedy RT, Koutmou KS and Maloney TD, Analytical Chemistry, 2023, 95, 4404–4411. [DOI] [PubMed] [Google Scholar]
- 17.Lai W, Lyu C. and Wang H, Analytical Chemistry, 2018, 90, 6859–6866. [DOI] [PubMed] [Google Scholar]
- 18.Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, Kurkowska M, Shirvanizadeh N, Destefanis E, Groza P, Avşar G, Romitelli A, Pir P, Dassi E, Conticello SG, Aguilo F. and Bujnicki JM, Nucleic Acids Research, 2022, 50, D231–D235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sood AJ, Viner C. and Hoffman MM, Journal of Cheminformatics, 2019, 11, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, George BM, Majzoub K, Villalta PW, Carette JE and Bertozzi CR, Cell, 2021, 184, 3109–3124.e3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosset-Erard C, Didierjean M, Pansanel J, Lechner A, Wolff P, Kuhn L, Aubriet F, Leize-Wagner E, Chaimbault P. and François Y-N, Analytical Chemistry, 2023, 95, 1608–1617. [DOI] [PubMed] [Google Scholar]
- 22.Bapiro TE, Richards FM and Jodrell DI, Analytical Chemistry, 2016, 88, 6190–6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojcik JB, Marchione DM, Sidoli S, Djedid A, Lisby A, Majewski J. and Garcia BA, Cancer Research, 2019, 79, 3205–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.