Summary
Acute myeloid leukemia (AML) poses a singular challenge for chimeric antigen receptor (CAR) therapy owing to its phenotypic heterogeneity and similarity to normal hematopoietic stem/progenitor cells (HSPCs). Here we expound a CAR strategy intended to efficiently target AML while minimizing HSPC toxicity. Quantification of target expression in relapsed/refractory patient samples and normal HSPCs reveals a therapeutic window for gated co-targeting of ADGRE2 and CLEC12A: We combine an attenuated ADGRE2-CAR with a CLEC12A-chimeric costimulatory receptor (ADCLEC.syn1) to preferentially engage ADGRE2posCLEC12Apos leukemic stem cells over ADGRE2lowCLEC12Aneg normal HSPCs. ADCLEC.syn1 prevents antigen escape in AML xenograft models, outperforms the ADGRE2-CAR alone and eradicates AML despite proximate myelopoiesis in humanized mice. Off-target HSPC toxicity is similar to that of a CD19-CAR and can be mitigated by reducing CAR T cell-derived interferon-γ. Overall, we demonstrate the ability of target density-adapted cooperative CAR targeting to selectively eliminate AML and potentially obviate the need for hematopoietic rescue.
eTOC Blurb
To address efficacy and safety challenges in AML, Haubner et al. quantify antigen distribution and densities in primary AML and normal cells to guide a cooperative CAR targeting strategy. ADCLEC.syn1 operates via CCR-gated sensitivity and thereby overcomes antigen escape in heterogenous AML and reduces on-target toxicity in humanized mice.
Graphical Abstract

Introduction
Acute myeloid leukemia (AML) remains difficult to treat due to relapsed or refractory (r/r) disease, leading to poor outcomes with approximately 30% 5-year overall survival.1 Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is currently the only therapeutic option with curative potential for r/r AML, but treatment-related mortality and post-transplant relapse rates are high.2 There is an immense clinical need for novel therapies for r/r AML. Chimeric antigen receptor (CAR) therapy has been established as highly potent therapeutic option in the setting of r/r B cell malignancies and multiple myeloma, improving long-term remission rates as well as overall survival.3–12 Current efforts to further enhance CD19- and BCMA-directed CAR therapies for B cell malignancies and multiple myeloma, respectively, aim to maximize tumor targeting and functional CAR T cell persistence. In these settings, the restricted expression profiles of the CAR target antigens limit on-target toxicities to the depletion of normal B cells or plasma cells without incurring the risk of eliminating hematopoietic stem cells. This is not the case in AML where the surface proteome of leukemic cells bears a high degree of similarity to that of normal HSPCs.13 Thus, while CD19 and BCMA stand out as clinically validated CAR targets in FDA-approved CAR T cell products, optimal target choices in AML are still uncertain.
One of the key mechanisms of AML resistance owes to disease heterogeneity. AML is indeed driven by multiple leukemic stem cell (LSC) clones with different trajectories of sequentially acquired mutations leading to an overall AML population that is genomically heterogenous.14,15 The clonal complexity of AML LSCs is associated with differences in surface protein expression of stem/progenitor and myeloid antigens,16,17 and heterogenous expression levels of common CAR targets.13,18,19 A second, perhaps greater, challenge is to achieve selective targeting of AML LSCs without depleting the normal hematopoietic stem/progenitor cell (HSPC) pool. At the transcriptional and surface protein level, AML LSCs have been found to share phenotypic similarities to normal HSPCs, raising the concern for on-target hematotoxicity.13,18,20,21 Not surprisingly, CAR-related myeloablation due to on-target hematotoxicity has been observed both pre-clinically22–26 and clinically27–29 with CD33-, CD123- or CLEC12A-targeted CARs. As emerging clinical CAR reports have demonstrated both poor efficacy and concerning on-target toxicities, CAR therapies are being harnessed before or after obligatory allo-HSCT to rescue normal hematopoiesis. HSCT may either be conventional or a gene-edited transplant lacking expression of the CAR target epitope to protect the HSCT from CAR-related on-target hematotoxicity.30–35
We have not discarded the prospect of a standalone CAR T cell approach in AML and to this end have searched for a combinatorial targeting strategy that can eliminate LSCs with minimal targeting of HSPCs. Our previous work identified a series of candidate surface targets that pass stringent safety criteria based on their expression profiles in normal hematopoietic and non-hematopoietic tissues.13 One of the top candidates with abundant expression in AML and low or absent expression in most normal tissues is ADGRE2, an adhesion G protein-coupled receptor. ADGRE2 expression in AML correlates with poor molecular risk profile as well as shorter overall survival independent of age, molecular risk status and transplant status.36 Given the known heterogeneity in AML and shared target expression in normal tissues, we hypothesized that a quantitative target density profiling of ADGRE2 and other established AML targets such as CD33, CD123 and CLEC12A may inform a strategy for a combinatorial CAR design to target AML safely and efficiently. Based on quantitative measurements of candidate target expression in r/r AML and normal hematopoietic cells we defined a gated, combinatorial strategy targeting ADGRE2 and CLEC12A that achieved efficient elimination of representative AML cell lines and patient-derived xenograft (PDX) models, including in humanized hematopoietic chimeras, with HSPC toxicity that did not exceed that of a control CD19-targeted CAR therapy. These results are the foundation for a phase 1 clinical trial utilizing this design, termed ADCLEC.syn1, in patients with r/r AML (NCT05748197).
Results
Antigen distribution and density inform AML target selection
To rationally select AML targets and an adapted targeting strategy for CAR therapy of AML, we quantitatively profiled surface target protein expression in bone marrow and peripheral blood (PB) samples of AML patients and healthy donors. The AML patient cohort included 39 patients with r/r AML of diverse genetically defined subtypes, including AML with recurrent genetic abnormalities, mutated TP53 and myelodysplasia-related abnormalities (Figure S1A).
We analyzed by flow cytometry both target distribution (percentage positivity) and target density (absolute number of surface molecules/cell) of ADGRE2, CLEC12A, CD33 and CD123. Cell surface expression analysis distinguished between bulk AML and the CD34pos/CD38neg LSC-enriched fraction. Target antigen distribution, represented as median percentage of target-positive AML cells in the patient cohort, was overall similar for ADGRE2 (bulk 82% / LSC 91%), CD33 (bulk 93% / LSC 87%) and CD123 (bulk 88% / LSC 96%) (Figure 1A). CLEC12A distribution was more heterogenous (bulk 54% / LSC 14%) and overlapping with ADGRE2 expression (CLEC12Apos bulk 54% / LSC 14%; ADGRE2pos∩CLEC12Apos bulk 46% / LSC 11%). (Figures 1A-B, S1B).
Figure 1. Antigen distribution and density inform AML target selection.
(A-F) Flow cytometric quantitative profiling of AML surface target antigens ADGRE2, CLEC12A, CD33 and CD123 on AML bulk or LSCs in a relapsed/refractory AML patient cohort (n=39, Figure S1) and on normal hematological cells in a healthy adult donor cohort (n=8). Each dot represents an individual patient/donor sample.
(A-B) Target antigen distribution profiles on AML cells as measured by percentage positivity gating. Horizontal bars represent median percentage target positivity in total patient population. ADGRE2pos∪CLEC12Apos (A∪C) denotes union of ADGRE2 and CLEC12A positivity (positive for either one or both antigens), ADGRE2pos∩CLEC12Apos (A∩C) denotes intersection of ADGRE2 and CLEC12A positivity (positive for both antigens).
(C-E) Target antigen density profiles on AML and normal hematological cells as measured by median absolute numbers of surface molecules/cell. Dashed lines indicate 3×102 molecules/cell and 1×103 molecules/cell.
(F) Heatmap summarizing target protein densities of ADGRE2, CLEC12A, CD33, CD123 and CD19 on normal bone marrow-derived hematological cell populations. Numbers within heatmap indicate median number of surface molecules/cell. Cell frequency of the respective population relative to total live bone marrow cells is represented by horizontal bars and numbers indicating median percentage from n=8 independent healthy donor samples.
(G) Mean target gene expression in different normal cell types of non-hematological origin based on single-cell RNAseq dataset.56
(H) Schematic comparing single-targeting and combinatorial CAR designs (OR gate, AND gate, IF-BETTER gate) and their predicted killing (red filling) and sparing (blank filling) characteristics on tumor (T) and normal (N) cells depending on ADGRE2 and CLEC12A target densities. Axes indicate ADGRE2 and CLEC12A target densities from negative (left and bottom, respectively) to low (middle) and high (right and top, respectively).
Also see Figure S1.
Target antigen density was determined in AML and normal hematopoietic cells (represented as median surface molecule count per cell in Figure 1C-F). In AML LSCs, 95% of patients had detectable ADGRE2 (>3.0×102 molecules/cell), with 79% displaying more than 1.0×103 ADGRE2 molecules per cell. CLEC12A was detectable in AML LSCs of 62% of patients, of whom 96% had detectable ADGRE2 expression. Detectable levels (>3.0×102 molecules/cell) of CD33 and CD123 in LSCs were found in 97% of patients, and CD33/CD123 co-expression of more than 1.0×103 molecules/cell in LSC was found in 87% of patients (Figure 1C).
In healthy donor bone marrow-derived HSPCs and granulocytes, we observed a different pattern of ADGRE2 and CLEC12A expression compared to AML. Early HSPCs (phenotypically hematopoietic stem cells (HSCs), multipotent progenitors (MPPs) and multilymphoid progenitors (MLPs)) were found to express only 9×102, 8×102 and 1.1×103 ADGRE2 molecules per cell, respectively, and no detectable levels of CLEC12A. Committed progenitors (CPs) expressed only low ADGRE2 (7×102 molecules) and CLEC12A (8×102 molecules). Granulocytes expressed 6.5×103 CLEC12A molecules and no detectable ADGRE2. Only monocytes were found to co-express high levels of ADGRE2 and CLEC12A, with a median of 1.6×103 ADGRE2 molecules and 1.2×104 CLEC12A molecules per cell. On the other hand, CD33 and CD123 were co-expressed in all early HSPC subsets (up to 2.5×103 CD33 and 1.9×103 CD123 molecules), granulocytes (3.9×103 CD33 and 5×102 CD123 molecules) and monocytes (2.4×104 CD33 and 1.5×103 CD123 molecules). For comparison, CD19 was only detected in normal B cells, averaging 5.4×103 molecules per cell (Figure 1D-F). ADGRE2 and CD19 thus presented the most restricted pattern, with <4% positivity in total bone marrow cells, while CD33, CD123 and CLEC12A were detected in >60% of total bone marrow cells (Figure 1F).
We further evaluated target expression profiles in non-hematological cells in normal tissues, defining specific cell types based on single-cell transcriptomic profiles. In this analysis, ADGRE2, CLEC12A and CD33 were all found to be undetectable in n=13 different cell types, while CD123 expression was detected in adipocytes and endothelial cells (Figure 1G).
In summary, our target protein quantification in AML vs normal cells revealed differential expression patterns of ADGRE2 and CLEC12A: ADGRE2 and CLEC12A are co-expressed in AML LSCs, but not in normal HSPCs, while ADGRE2 is absent in granulocytes. In contrast, CD33 and CD123 were similarly expressed in AML but also found in most normal stem/progenitor and mature myeloid cells including granulocytes. Furthermore, the target density of ADGRE2 in normal HSPCs is lower than that of CD33 or CD123. Considering the target distribution and density profiles of these 4 cell surface molecules in AML vs various normal tissues, we proceeded to investigate a combinatorial targeting strategy that takes advantage of a potential therapeutic window to distinguish LSCs and HSPCs based on ADGRE2 and CLEC12A expression.
IF-BETTER gated combinatorial targeting of ADGRE2 and CLEC12A
CAR therapy for AML is constrained by the risk of AML escape on one end and on-target toxicities to normal HSPCs on the other. We hypothesized that suitable co-targeting of ADGRE2 and CLEC12A may enhance efficacy against AML including LSCs while limiting on-target toxicity. An OR-gate is the most common strategy to engage two antigens to provide greater tumor coverage in a heterogenous tumor setting. OR-gated CAR designs rely on co-expression of two independent CARs or one CAR with dual specificity, triggering cytolysis if either one of two targeted antigens is sufficiently expressed37–43: This approach however increases the cumulative on-target toxicity potential, thus extending the relatively limited toxicity spectrum of ADGRE2 to the larger spectrum of CLEC12A (Figure 1H, second panel). Targeting ADGRE2 and CLEC12A in OR-gate format may limit clinical tolerability by compounding monocyte depletion with agranulocytosis, increasing the risk of life-threatening bacterial infections. AND-gated CAR designs rely on co-expression of two complementary but individually deficient receptors, triggering cytolysis only if two targeted antigens are co-expressed44,45: This approach may limit on-target toxicity but increase the risk of tumor antigen escape if one of the two targets is absent or scarce (Figure 1H, third panel). We therefore adopted another combinatorial strategy to balance efficacy and safety requirements.
The lower ADGRE2 antigen density found in early HSPCs (<1.0×103 molecules per cell) relative to its higher density in AML LSCs (>1.0×103 molecules per cell) indicated that a killing threshold of approximately 1.0×103 ADGRE2 molecules per cell may be best suited to preferentially target LSCs. As limited sensitivity may allow for antigen escape, we sought to selectively enhance ADGRE2 engagement on LSCs but not HSPCs by co-targeting CLEC12A. We hypothesized that a sensitivity-tuned ADGRE2-CAR co-expressed with a CLEC12A-targeted chimeric costimulatory receptor (CCR)46 may display greater activity against ADGRE2med/highCLEC12Apos LSCs without increasing on-target/off-tumor toxicity against ADGRE2lowCLEC12Aneg HSPCs. This combinatorial receptor design to preferentially direct target cell killing based on the presence or absence of a second target is best described as an IF-BETTER gate (Figure 1H, fourth panel).
A CLEC12A-CCR increases sensitivity of ADGRE2-CAR-1XX
To generate IF-BETTER gated chimeric receptor designs combining an ADGRE2-CAR and CLEC12A-CCR, we first screened different scFv candidates for both targets, aiming for an ADGRE2-CAR with a killing threshold sparing ADGRE2low cells, and a CLEC12A-CCR that efficiently enhances the associated CAR activity (Figure S2A). For the ADGRE2-CAR, a library of n=18 humanized ADGRE2 scFv with different affinities was created by introducing VH/VL framework alterations. These ADGRE2 scFv were then screened in the CAR-28z1XX format, a CAR design with only 1 instead of 3 functional ITAMs in the CD3z domain, associated with calibrated activation and increased in vivo functional T cell persistence.47 We selected lead scFv candidates in vitro and in vivo based on efficient elimination of ADGRE2high target cells (1×104 ADGRE2 molecules) and limited activity against ADGRE2low target cells (1×103 ADGRE2 molecules) achieving the intended targeting threshold under CAR T cell stress dose conditions (injection of 5.0×105 or fewer CAR T cells). For the CLEC12A-CCR, a library of n=16 fully-human CLEC12A scFv with different epitopes was first screened in the CAR-28z1XX format, selecting a lead candidate with high efficacy in vitro and in vivo under CAR T cell stress dose conditions. The top ADGRE2-scFv and CLEC12A-scFv were chosen to construct a combinatorial design co-expressing an ADGRE2-CAR-28z1XX and a CLEC12A-CCR-BB incorporating a 4–1BB costimulatory endodomain, encoded by an SFG-gammaretroviral bicistronic vector, termed ADCLEC.syn1 (Figures 2A-B, S2B-C).
Figure 2. A CLEC12A-CCR increases sensitivity of ADGRE2-CAR-1XX.
(A) Schematic depicting ADCLEC.syn1 combinatorial receptor design.
(B) SFG-gammaretroviral bicistronic vector map for ADCLEC.syn1 expression.
(C) EL4 murine lymphoma cell line variants expressing either no target (black), ADGRE2 alone (green), CLEC12A alone (violet), or ADGRE2 and CLEC12A together (orange).
(D) 48h in vitro assay to measure cytotoxic activity of ADCLEC.syn1 vs 19-28z1XX CAR T cells at different effector:target (E:T) ratios in co-culture with EL4 cell line variants providing either no target, ADGRE2 alone, CLEC12A alone or ADGRE2 and CLEC12A together. T cell activation is represented by CD25 median fluorescence intensity (MFI) on T cells. Cytotoxicity was determined via flow cytometric enumeration of target cells. Data are represented as mean ± SEM. p value was determined via unpaired t test.
(E) MOLM13 AML cell line variants with low CLEC12A density (5×102 molecules/cell) and varying ADGRE2 antigen density: high/WT (1×104), low (1×103) and very-low (4×102).
(F) 18h in vitro assay to measure cytotoxic activity of ADCLEC.syn1 vs its single receptor components (ADGRE2–28z1XX CAR or CLEC12A-BB CCR) vs untransduced T cells (UTD) in MOLM13 variants modeling antigen escape. Data are represented as mean ± SEM. p values were determined via unpaired t test.
(G) M13-Alow-Chigh MOLM13 variant with low ADGRE2 (1×103 molecules/cell) and high CLEC12A (2×105).
(H) AML burden (total flux) and Kaplan-Meier survival analysis of M13-Alow-Chigh-bearing mice treated with 5×105 CARpos T cells.
(I) M13-Avery-low-Chigh MOLM13 variant with very-low ADGRE2 (4×102 molecules/cell) and high CLEC12A (1×105 molecules/cell).
(J) AML burden (total flux) and Kaplan-Meier survival analysis of M13-Avery-low-Chigh-bearing mice treated with 5×105 CARpos T cells.
Also see Figures S2, S3, S4.
To investigate how ADCLEC.syn1 operates in the context of individual ADGRE2-CAR and/or CLEC12A-CCR engagement, we set up a prototypic in vitro model of target cells expressing none, either single target, or both targets, co-cultured with ADCLEC.syn1 T cells. T cell activation assessed by CD25 upregulation was only observed in conditions where target cells expressed ADGRE2, enabling CAR engagement (Figure 2C-D). In line with T cell activation patterns, target cell lysis was limited to conditions where target cells expressed ADGRE2. In addition, CLEC12A co-expression enhanced target cell lysis (p=0.0061), but CLEC12A expression alone did not trigger T cell activation or target cell lysis (Figure 2D). To model contexts of cellular heterogeneity, we performed in vitro cytotoxicity assays in the presence of mixtures of cells expressing either only ADGRE2 or only CLEC12A. Bystander-mediated CLEC12A-CCR engagement did not alter ADGRE2-CAR-mediated killing of ADGRE2posCLEC12Aneg AML cells (Figure S3A), and conversely, bystander-mediated ADGRE2-CAR activation did not trigger killing of ADGRE2negCLEC12Apos cells (Figure S3B). Overall, these results support that ADCLEC.syn1 T cells efficiently co-engage CAR and CCR to enhance killing of dual antigen-positive cells, while separate CAR and CCR engagement via single antigen-positive cells in close proximity does not result in unintended cytotoxicity.
Having established proof-of-principle evidence that ADCLEC.syn1 operates as IF-BETTER gate, we next tested its application in an AML model of CAR target antigen escape. For this, we created stable MOLM13 cell line variants with unmodified or lowered ADGRE2 antigen densities: M13-Ahigh (WT, 1×104 ADGRE2 molecules), M13-Alow (1×103 ADGRE2 molecules) and M13-Avery-low (4×102 ADGRE2 molecules), all endogenously expressing low CLEC12A levels (Clow, 5×102 molecules) (Figures 2E, S4A). In the context of high ADGRE2 levels (1×104 molecules), T cells expressing only the ADGRE2-CAR-28z1XX had high MOLM13 killing efficacy with no difference compared to T cells expressing ADCLEC.syn1. However, in the context of low levels of ADGRE2 (1×103 molecules) and CLEC12A (5×102 molecules), T cells expressing only the ADGRE2-CAR-28z1XX failed to kill, whereas ADCLEC.syn1 T cells maintained killing capacity, demonstrating in vitro superiority of ADCLEC.syn1 efficacy over a single ADGRE2-targeted CAR (p=0.0011) (Figures 2F).
To elucidate the functional role of the 4–1BB endodomain of a CCR assisting CAR-1XX-mediated killing in an antigen-low setting, we engineered ADGRE2-CAR-28z1XX-CLEC12A-CCR-del which included a non-signaling truncated CCR but was otherwise identical to ADCLEC.syn1. We compared the efficacy of ADCLEC.syn1 vs ADGRE2-CAR-28z1XX-CLEC12A-CCR-del in a NOD scid gamma (NSG) xenograft model based on a MOLM13 variant expressing low ADGRE2 (1×103 molecules/cell) and high CLEC12A (2×105 molecules/cell) antigen density to ensure potential CCR effects (Figures 2G, S4 B-C); lower CCR target levels were explored in subsequent experiments. At a CAR T cell stress dose of only 5.0×105 CAR T cells, ADCLEC.syn1 outperformed not only a single ADGRE2-CAR-28z1XX (p=0.0017), but also the ADGRE2-CAR-28z1XX-CLEC12A-CCR-del (p=0.0330), demonstrating superior in vivo efficacy of ADCLEC.syn1 over a 28z1XX-CAR alone, owing to 4–1BB costimulation provided by the CCR (Figures 2H). In a more extreme setting of detectable but very-low ADGRE2 (4×102 molecules/cell) and high CLEC12A (1×105 molecules/cell), ADCLEC.syn1 again outperformed both ADGRE2-CAR-28z1XX (p=0.0023) and ADGRE2-CAR-28z1XX-CLEC12A-CCR-del (p=0.0035), demonstrating that a CLEC12A-CCR-BB selectively enhances ADGRE2–28z1XX sensitivity, even at extremely low CAR target levels (<5×102 molecules/cell) (Figure 2I-J).
CCR engagement regulates cytolysis directed to low antigen density
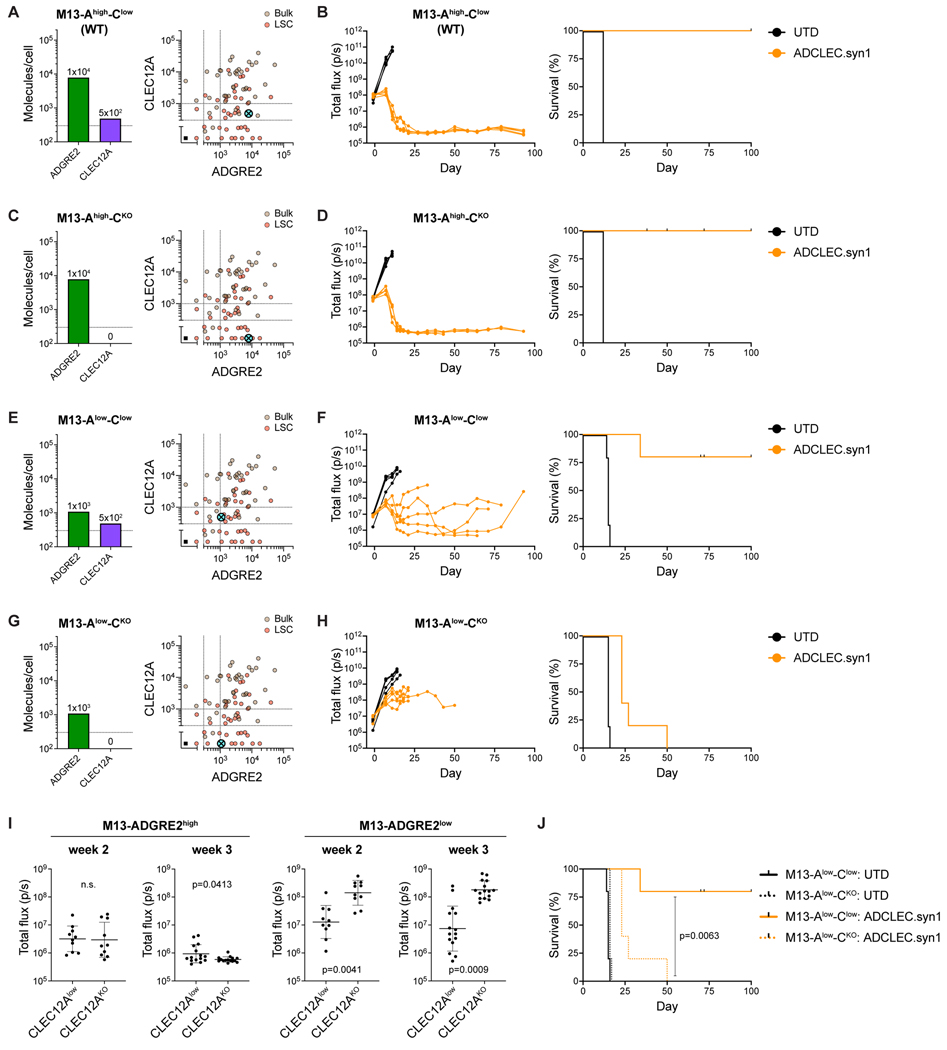
To test the limits of CCR-gated sensitivity of ADCLEC.syn1, we investigated additional settings where either both CAR and CCR targets are expressed at low density (modeling AML antigen escape), or where the CAR target is expressed at low density while the CCR target is negative (modeling normal tissues). For this, we utilized MOLM13 cell line variants with either high (1×104 molecules) or low (1×103 molecules) ADGRE2 antigen density and either endogenously low CLEC12A density (5×102 molecules) or CLEC12A-negativity (KO) (Figures 3A,C,E,G, S4D). In the setting of high ADGRE2 expression (1×104 molecules) representing AML, ADCLEC.syn1 led to long-term remission in all mice regardless of CLEC12A co-expression (Figure 3A-D,I), demonstrating that CCR co-engagement is not required to eliminate AML cells with high CAR target density. In the setting of low ADGRE2 expression (1×103 molecules), CCR co-engagement at low levels of CLEC12A (5×102 molecules) was sufficient for ADCLEC.syn1 to enable durable AML reduction and long-term survival, whereas cells with low ADGRE2 expression (1×103 molecules) and absence of CLEC12A expression, representing normal cells, were not completely eradicated (p=0.0063, Figure 3E-J). Collectively, these results demonstrate the potential of a CCR to regulate cytolytic activity in settings of low CAR and CCR target antigen densities, resulting in a more selective increase of CAR-1XX antitumor efficacy.
Figure 3. CCR engagement regulates cytolysis directed to low antigen density.
(A,C,E,G) MOLM13 variants with modified ADGRE2 (high vs low) and CLEC12A (low vs KO) levels: M13-Ahigh-Clow, M13-Ahigh-CKO, M13-Alow-Clow, M13-Alow-CKO. Bivariate plots illustrate how the respective ADGRE2/CLEC12A antigen densities (absolute number of surface molecules/cell) compare to the analyzed AML patient cohort (n=39) and their AML bulk (brown) as well as LSC (red) populations.
(B,D,F,H) AML burden (total flux) and Kaplan-Meier survival analysis of mice bearing MOLM13 variants treated with 5×105 ADCLEC.syn1 T cells.
(I) AML burden (total flux) in week 2 and week 3 post ADCLEC.syn1 T cell injection in mice bearing MOLM13 variants. Data are represented as individual measurements and geometric mean with geometric SD. p values were determined via unpaired t test.
(J) Kaplan-Meier survival of mice bearing either M13-Alow-Clow or M13-Alow-CKO.
Also see Figure S4.
Low-dose ADCLEC.syn1 efficiently ablates AML with effective recall responses
The ADGRE2 and CLEC12A target antigen quantification in our AML patient cohort allowed us to select AML cell line variants with relevant combinations of high or low ADGRE2 and CLEC12A antigen densities to represent the spectrum of phenotypes observed in the AML patient cohort (Figure 4A,C,E,G). In a xenograft model of ADGRE2high MOLM13 AML, both a single ADGRE2-CAR-28z1XX and ADCLEC.syn1 induced rapid remissions (Figure 4B). In ADGRE2low MOLM13 and U937 AML however, only ADCLEC.syn1 maintained its efficacy, whereas the efficacy of a single-targeted ADGRE2-CAR-28z1XX was limited (Figure 4D,F,H). Significantly, the benefit afforded by the CLEC12A-CCR was observed at both CLEC12Alow (5×102 molecules/cell) and CLEC12Ahigh levels (2×105 molecules/cell), demonstrating functional impact across a range of AML phenotypes. In a stress test model of mice bearing ADGRE2high(1×104)/CLEC12Ahigh(2×104) AML, a dose as low as 5×104 CAR T cells was sufficient to induce remissions and long-term survival, further highlighting the in vivo potency of ADCLEC.syn1 T cells (Figure S5A). To assess the functional persistence of ADCLEC.syn1 T cells, we repeatedly challenged mice with MOLM13 injections two months after a single initial injection of only 1–2.5×105 ADCLEC.syn1 T cells, using either MOLM13-ADGRE2high(1×104)/CLEC12Alow(5×102) or MOLM13-ADGRE2high(1×104)/CLEC12Ahigh(2×104) (Figure 4I-L). All mice re-challenged with ADGRE2high(1×104) MOLM13 formed effective recall responses, successfully averting relapse. Re-challenge with antigen-escape control ADGRE2very-low(4×102)/CLEC12Alow(5×102) MOLM13 (Figure S5B) led to rapid relapse, thereby ruling out a potential allogeneic graft-versus-leukemia (GVL) effect and re-confirming the threshold antigen density required for reactivating persisting ADCLEC.syn1 T cells.
Figure 4. Low-dose ADCLEC.syn1 efficiently ablates AML with effective recall responses.
(A,C,E,G,I,K) AML cell lines with modified ADGRE2 and CLEC12A densities used for xenograft CAR studies. Bivariate plots illustrate how the respective ADGRE2/CLEC12A antigen densities (absolute numbers of surface molecules/cell) compare to the analyzed AML patient cohort (n=39) and their AML bulk (brown) as well as LSC (red) populations.
(B,D,F,H) AML burden (total flux) and Kaplan-Meier survival analysis of mice bearing MOLM13 variants treated with 5×105 (B,D,F) or 1×106 (H) CAR/CCRpos T cells, comparing ADCLEC.syn1 vs its single receptor components (ADGRE2–28z1XX CAR or CLEC12A-BB CCR) vs untransduced T cells (UTD).
(J,L) AML burden (total flux) of mice bearing M13-Ahigh-Clow (J) or M13-Ahigh-Chigh (L) treated with UTD or ADCLEC.syn1 T cells at the indicated dose (1–2.5×105). Arrows indicate repeated MOLM13 re-challenges (dose 5×105, on d66 and d73 post initial CAR T injection) with either the same MOLM13 variant as on d0 (left graphs) or an antigen escape control variant M13-Avery-low-Clow (right graphs) with minimal target levels (Figure S5B) expected to cause AML relapse.
Also see Figure S5.
Prior research has demonstrated that AML cells may induce T cell dysfunction by impairing T cell activation and leading to their senescence.48,49 We therefore evaluated if an autologous approach to generating ADCLEC.syn1 T cells may be limited by AML-induced T cell dysfunction. To this end, we isolated T cells from PB of an 88-year-old female AML patient with high AML burden (18% blasts in PB manual differential at time of venipuncture for T cell collection). Following T cell isolation and a 7-day T cell transduction and expansion protocol, end-of-production ADCLEC.syn1 T cells were cryopreserved to model the clinical setting. Thawed AML patient-derived ADCLEC.syn1 T cells were then administered to NSG mice engrafted with MOLM13 (WT) AML cells. AML patient-derived ADCLEC.syn1 T cells induced rapid and durable remissions in all mice even at a CAR T cell stress dose of only 2.5×105 CAR T cells per mouse, a dose that is within a 5-fold range of the minimum efficacious dose of fresh, healthy donor-derived ADCLEC.syn1 CAR T cells (Figure S5C). Overall, these results support that T cells derived from AML patients can be a viable source for production of ADCLEC.syn1 CAR T cells with high in vivo potency.
ADCLEC.syn1 eliminates leukemic stem cells in heterogenous AML PDX models
To assess efficacy of ADCLEC.syn1 in the setting of genotypically and phenotypically heterogenous AML, we selected three independent AML PDX models originating from r/r AML patients with prior failure of multiple AML-directed therapies (Figure S6A-D).
PDX#1 originated from an AML patient relapsing after chemotherapy, hypomethylating agent and allo-HSCT. Target antigen density of ADGRE2 and CLEC12A was representative of profiles observed in our studied AML patient cohort and more than 10-fold lower than CD33 (Figure 5A-B). In this PDX model, we evaluated the potential of CAR T cells to eliminate AML LSCs defined by their re-engraftment potential, and chose to compare a reference CD33-targeted CAR-28z1XX (33–28z1XX) against ADCLEC.syn1 (Figure 5C). CD33 was chosen as reference target due to its reported highest percentage of positivity in AML among the most commonly investigated CAR target antigens.50 In a primary mouse cohort, PDX#1-engrafted mice were treated with either ADCLEC.syn1 or 33–28z1XX at a stress dose of 5×105 CAR T cells. 33–28z1XX T cells expanded similarly to ADCLEC.syn1 T cells, but failed to completely eliminate AML cells in PB and bone marrow (Figures 5D and S6E). Persistence of AML LSC post 33–28z1XX T cells was evidenced by serial transplantation of relapsing AML cells leading to AML PDX engraftment in all mice in a secondary mouse cohort (Figure 5E, S6F). While 33–28z1XX T cells failed in both cohorts, ADCLEC.syn1 T cells induced remissions, both in the primary cohort and the secondary cohort of mice with relapsed AML post-33–28z1XX therapy (Figures 5D, 5E, S6G-H). In two additional AML PDX models with adverse risk profile and patient-representative target levels of ADGRE2 and CLEC12A, ADCLEC.syn1 induced durable AML remission and long-term survival at a stress dose of 5×105 CAR T cells (Figure 5F-I).
Figure 5. ADCLEC.syn1 eliminates leukemic stem cells in heterogenous AML PDX models.
Three relapsed/refractory AML PDX models (PDX#1 in A-E, PDX#2 in F-G, PDX#3 in H-I) were utilized to assess CAR T cell efficacy (PDX clinical annotations and target phenotype in Figure S6A-D). T cell expansion and AML PDX burden were serially monitored via flow cytometry of PB and are shown as normalized T cell and AML cell counts per 100ul PB. Survival is shown as Kaplan-Meier analysis.
(A,F,H) ADGRE2/CLEC12A antigen densities (absolute number of surface molecules/cell) of PDX LSCs compared to the analyzed AML patient cohort (n=39) and their AML bulk (brown) as well as LSC (red) populations.
(B) ADGRE2/CLEC12A compared to CD33 antigen densities (absolute number of surface molecules/cell) on PDX#1.
(C) Schematic of PDX#1 experimental setup for results shown in D-E
(D) Primary engraftment of PDX#1 in NSG mice on d-11 was followed by treatment with 5×105 CAR/CCRpos T cells on d0, comparing UTD vs a reference CD33-CAR (33–28z1XX) vs ADCLEC.syn1. Mice receiving the reference CD33-CAR relapsed, and their bone marrow was harvested on d29 for subsequent secondary engraftment in NSG mice on d-16.
(E) PDX#1 post-33–28z1XX failure was secondarily engrafted in NSG mice on d-16 and was followed by treatment with 2.5×105 CAR/CCRpos T cells on d0.
(G) Engraftment of PDX#2 in NSG-SGM3 mice on d-9 was followed by treatment with 5×105 CAR/CCRpos T cells on d0.
(I) Engraftment of PDX#3 in NSG-SGM3 mice on d-18 was followed by treatment with 5×105 CAR/CCRpos T cells on d0.
Also see Figure S6.
Humanized AML mouse model to assess ADCLEC.syn1 efficacy and HSPC toxicity
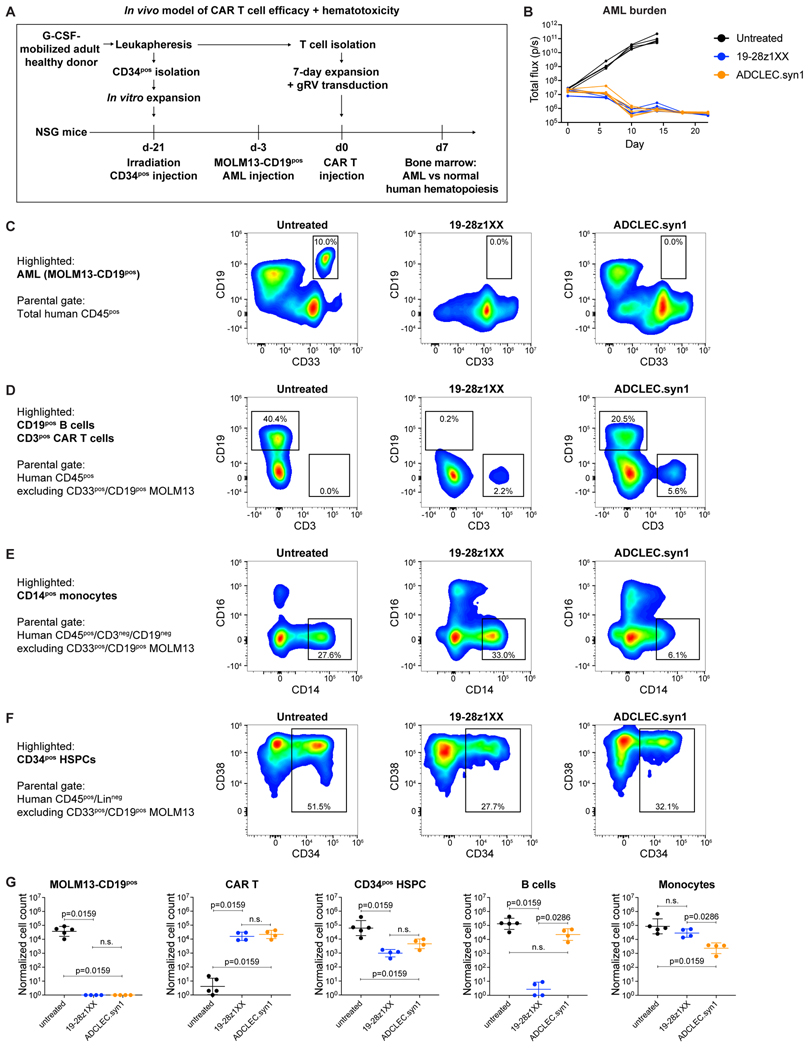
We next evaluated the on-target/off-tumor activity of ADCLEC.syn1 on normal hematologic cells in the context of an in vivo anti-leukemic CAR T cell response. For this, we established a humanized AML xenograft mouse model to assess in vivo CAR T cell efficacy and hematotoxicity simultaneously in the same mouse. A CD19-targeted CAR (19–28z1XX) was selected as positive control to benchmark expected on-target activity resulting in the depletion of CD19pos tumor cells and normal human CD19pos B cells. Furthermore, 19–28z1XX served as an important negative control for on-target activity against normal HSPCs as CD19 is not expressed in HSPCs. G-CSF-mobilized adult healthy donor-derived CD34pos HSPCs were used for humanization of NSG mice, followed by engraftment of MOLM13 AML cells that had been modified to express CD19 at levels similar to NALM6 B-ALL cells (2.7×104 molecules per cell51). Humanized MOLM13-CD19pos AML-engrafted mice received either control 19–28z1XX or ADCLEC.syn1 CAR T cells at a low dose of 2.5×105 CAR T cells (Figure 6A). Both 19–28z1XX and ADCLEC.syn1 led to rapid AML remission in humanized mice; untreated humanized mice did not control AML (Figure 6B). The similar anti-leukemic efficacy of 19–28z1XX and ADCLEC.syn1 allowed to directly assess and compare their respective impact on normal hematologic cells. On day 7 post CAR T cell injection, bone marrow analysis confirmed complete MOLM13-CD19pos AML eradication in mice treated with 19–28z1XX or ADCLEC.syn1 (Figure 6C), consistent with overall bioluminescence results. We found complete B cell depletion only in 19–28z1XX-treated mice, and partial monocyte depletion only in ADCLEC.syn1-treated mice (Figure 6D-E), indicating that both CAR T cell populations were active in these humanized bone marrows. In the same samples, normal human CD34pos cells were detected at similar frequency in mice treated with 19–28z1XX or ADCLEC.syn1, showing the same reduction with both CAR treatments relative to untreated mice (Figure 6F). This experimental setting thus did not reveal on-target/off-tumor activity directed to CD34pos cells but suggested HSPC reduction owing to a target-independent mechanism. These observations were further supported by absolute cell counts (Figure 6G): At time of complete AML eradication and similar CAR T cell expansion, ADCLEC.syn1-treated mice had persistent CD34pos cells at similar absolute numbers compared to 19–28z1XX, demonstrating that high anti-leukemic efficacy of ADCLEC.syn1 occurred without any more HSPC toxicity than a clinically validated CD19-targeted CAR.
Figure 6. Humanized AML mouse model to assess ADCLEC.syn1 efficacy and HSPC toxicity.
Humanized AML xenograft mouse model to assess CAR T cell hematotoxicity in the context of an in vivo anti-leukemic CAR T cell response. Anti-leukemic response and impact on normal human hematopoiesis was assessed upon receiving either no treatment or treatment with a reference CD19-CAR (19–28z1XX) vs ADCLEC.syn1.
(A) Schematic of humanized AML xenograft CAR T cell hematotoxicity model. NSG mice were humanized via sublethal irradiation and injection of G-CSF-mobilized healthy donor-derived CD34pos cells on d-21, followed by MOLM13-CD19pos cell line injection on d-3, either untreated or treated with 2.5×105 CAR/CCRpos T cells on d0.
(B) AML burden (total flux) of humanized mice bearing MOLM13-CD19pos AML.
(C-F) Representative ex vivo bone marrow distribution of AML and normal human hematopoietic cells on d7.
(C) MOLM13-CD19pos AML cells identified via positivity for CD33 and CD19 within total human CD45pos cells
(D) CD19pos normal B cells and CD3pos adoptively transferred CAR T cells within total human CD45pos cells (excluding CD33pos/CD19pos MOLM13)
(E) CD14pos/CD16neg normal classical monocytes within CD3neg/CD19neg human CD45pos cells
(F) CD34pos normal HSPCs within lineage-negative (CD3neg/CD19neg/CD14neg/CD16neg) human CD45pos cells
(G) D7 ex vivo quantification of human bone marrow populations, with n=4–5 mice per group. Data are shown as individual counts and geometric mean with geometric SD. p values were determined via Mann-Whitney test.
Off-target hematotoxicity is mitigated by reducing CAR T cell-derived IFN-γ
To further investigate the potential mechanism of off-target (antigen-independent) HSPC toxicity, we tested in vitro and in vivo whether CAR T cell-derived products may account for HSPC reduction. Since IFN-γ has been reported to mediate differentiation and depletion of myeloid-biased HSCs in murine models of chronic inflammation52, we hypothesized that excessive IFN-γ release in the context of CAR T cell anti-tumor activity could trigger HSC loss due to rapid differentiation. In an in vitro model of T cell-derived IFN-γ off-target hematotoxicity, we first co-cultured ADCLEC.syn1 T cells with MOLM13 target cells, leading to CAR activation (Figure 7A, upper panel). Cell-free supernatant (CAR T + AML-conditioned medium) was then added to healthy donor-derived HSPCs (Figure 7A, lower panel). HSPC phenotype and absolute cell counts were determined after 24h. The potential effect of CAR T cell-derived IFN-γ was evaluated by either IFNG CRISPR/Cas9 T cell editing or addition of anti-IFN-γ blocking antibody to the conditioned medium. We found that addition of conditioned medium from ADCLEC.syn1 IFNG-WT + MOLM13 co-culture led to a striking reduction of CD34pos/CD38neg immature HSPCs while CD34pos/CD38pos progenitors increased, both in absolute and relative numbers (Figure 7B). This was not observed when adding conditioned medium from either MOLM13 alone or ADCLEC.syn1 IFNG-WT alone. Genetic disruption of IFNG in ADCLEC.syn1 T cells or blocking of IFN-γ protein was associated with a significant increase of CD34pos/CD38neg immature HSPCs and decrease of CD34pos/CD38pos progenitors in absolute and relative numbers, compared to HSPCs cultured with conditioned medium from ADCLEC.syn1 IFNG-WT + MOLM13 co-culture.
Figure 7. Off-target hematotoxicity is mitigated by reducing CAR T cell-derived IFN-γ.
(A) In vitro assay to evaluate off-target HSPC toxicity due to soluble factors released upon CAR T cells engaging target cells. ADCLEC.syn1 CAR T cells with or without IFNG CRISPR/Cas9 editing were co-cultured with MOLM13-WT AML cell line for 10h at E:T ratio 1:1. Subsequently, cell-free supernatant (conditioned medium from CAR-T + AML-co-culture and individual controls) was collected and added to a separate in vitro culture of normal human CD34pos/CD38neg HSPC, with or without anti-IFN-γ blocking antibody. After 24h, HSPC phenotype (CD34/CD38 expression) and cell counts were measured via flow cytometry.
(B) Absolute cell count and relative distribution of HSPC subsets upon in vitro culture with different CAR T + AML-conditioned media. Horizontal and error bars represent mean value and SD of technical triplicates. p values were determined via unpaired t test. FACS plots show representative CD34/CD38 HSPC phenotypes at time of assay read-out.
(C) Schematic of in vivo model of CAR T cell IFN-γ-mediated off-target hematotoxicity. NSG mice were humanized via sublethal irradiation and injection of G-CSF-mobilized healthy donor-derived CD34pos cells on d-21 (dose 6.0×105), followed by injection of 19–28z1XX or ADCLEC.syn1 CAR T cells derived from the same donor (dose 3.0×105 CAR/CCRpos T cells, ± IFNG editing) on d0.
(D) IFNG editing of 19–28z1XX and ADCLEC.syn1 T cells for in vivo study was demonstrated by intracellular IFN-γ staining 10h after in vitro culture with or without MOLM13-CD19pos target cells at E:T ratio 1:1.
(E) D7 ex vivo quantification of human bone marrow populations (lineage-negative CD34pos/CD38neg HSPCs, CD19pos B cells and CD3pos CAR T cells), with n=5 mice per group. Data are shown as individual counts and geometric mean with geometric SD. p values were determined via Mann-Whitney test.
Also see Figure S7.
Overall, these in vitro observations suggested that soluble factors released upon CAR T cell activation may lead to reduction of immature HSPCs potentially owing to rapid differentiation, and that CAR T cell-derived IFN-γ is a direct mediator of this effect. To investigate whether this effect may explain our observed in vivo reduction of HSPCs in the context of CAR T cell activation, we treated humanized mice with 19–28z1XX CAR or ADCLEC.syn1 T cells with or without IFNG editing, and compared human engrafted cell counts in bone marrow (Figure 7C-D). Consistent with our previous findings, CAR T cell expansion was similar between 19–28z1XX and ADCLEC.syn1, and only 19–28z1XX led to B cell depletion (Figure 7E). Of note, IFNG editing was not associated with any changes in CAR T cell expansion or B cell counts. The partial HSPC reduction associated with 19–28z1XX and ADCLEC.syn1 T cells (IFNG-WT) was not observed when reducing IFN-γ expression (IFNG-KO), even though on-target CAR T cell activity was maintained (Figures 7E, S7).
Discussion
We provide here a quantitative analysis of target expression in r/r AML, based on which we devised a tethered combinatorial targeting strategy for a CAR T cell therapy. Specifically, we address the challenge of achieving selective tumor targeting while limiting on-target activity against tumor-related normal tissues. We demonstrate that a gated, combinatorial chimeric receptor design can achieve anti-tumor efficacy with limited on-target/off-tumor toxicity to tumor-related normal tissue by following below principles: (1) selection of targets with frequent expression in malignant cells (including cancer stem cells where applicable) and limited expression in vital normal tissues; (2) accurate quantification of target antigen densities that distinguish tumor cells from critical normal cells; (3) design of cooperative receptors that afford activation thresholds adapted to target expression signatures that part tumor from normal cells.
There is a growing number of available chimeric receptor designs that require different target antigen densities to elicit T cell activation, ranging from a few hundred molecules per cell for HIT receptors51 to a few thousand molecules per cell for 4–1BB/CD3z CARs.37,53,54 The rational selection of an adapted CAR design thus warrants precise measurement of target antigen densities in tumor and normal cells. This is particularly important in the context of combinatorial receptor formats that aim to skew T cell engagement towards tumor cells and away from normal cells. Here, we report a novel combinatorial CAR approach to target AML, informed by target quantification in AML and normal cells. To select a suitable target combination, we performed quantitative profiling of the target candidates ADGRE2, CD33, CD123 and CLEC12A, based on flow cytometry in a cohort of r/r AML patients (n=39) with diverse AML disease subtypes including AML with recurrent genetic abnormalities, mutated TP53 and myelodysplasia-related abnormalities,55 and healthy donors (n=8). We also evaluated the expression of these targets in normal non-hematopoietic tissues using a cell type-specific RNA expression dataset.56 For each target antigen, we quantified the number of surface molecules in AML bulk, LSC and normal hematopoietic cells. This allowed us to identify a target combination with differential expression in AML LSCs and normal HSPCs.
While all four targets were abundantly expressed in most AML samples, their different expression patterns in normal tissues informed our target selection. High CD33 levels in normal HSPCs (up to 3×103), granulocytes (4×103) and monocytes (2.4×104), and CD123 expression in normal HSPCs (2×103) and endothelium57 complicated the use of differential antigen expression to selectively target CD33 or CD123 in AML. ADGRE2 expression in normal tissues was found to be more restricted, with less than 9×102 molecules per cell on HSC and negativity in non-hematopoietic cell types, allowing us to devise an ADGRE2-targeted CAR strategy with a threshold of activation of approximately 1×103 molecules per cell. CLEC12A expression was found to be absent in early HSPCs and non-hematopoietic cell types, but its high antigen density in normal granulocytes (6.5×103) would make those cells immediate CAR targets. We therefore chose to target ADGRE2 with a CAR and co-engage CLEC12A with a CCR instead of a CAR, to selectively enhance killing of ADGRE2pos CLEC12Apos AML cells without killing ADGRE2neg CLEC12Apos normal cells.
We utilize here a logic-gated targeting approach that we term IF-BETTER gating: CAR target sensitivity is enhanced by CCR target co-engagement, thereby expanding the range of overall targetable cells. This combinatorial CAR approach can be achieved with a variety of different configurations of combined CAR and CCR expression. Its ultimate effect on tumor and normal cells depends on target choice as well as thresholds of activation for CAR and CCR. Our target density profiling in AML and normal cells established that an ideal IF-BETTER gated CAR design targeting ADGRE2 and CLEC12A would trigger killing of any cells with more than 1×103 ADGRE2 molecules while cells with at least 5×102 ADGRE2 molecules would still be efficiently eradicated if CLEC12A is co-expressed (Figure 8A-B). To achieve this, we developed ADCLEC.syn1, which consists in an ADGRE2-targeted CAR-28z1XX co-expressed with a CLEC12A-targeted CCR providing 4–1BB costimulation. To mitigate the risk of ablating ADGRE2low HSPCs, we selected an ADGRE2 scFv with limited target sensitivity in the context of a CD3z ITAM-reduced 1XX format.47 In vitro, deletion of the two distal CD3z ITAMs has been associated with reduced cytotoxicity at low CD19 target antigen density of 1×103 molecules per cell.53 The 1XX format has been previously shown to increase functional CAR T cell persistence while limiting effector functions including IFN-γ production relative to CARs retaining 3 functional ITAMs.47,58,59 For the CCR, a 4–1BB costimulatory domain was selected, building on previous evidence of synergy of combined CD28 and 4–1BB costimulation in the context of endogenous T cell responses60 as well as CAR T cells.61–64
Figure 8. ADCLEC.syn1 enhances distinction between AML and normal cells based on combined target signatures.
(A) Chimeric receptor architecture of single CAR (ADGRE2-CAR) vs IF-BETTER gated CAR+CCR (ADCLEC.syn1).
(B) Schematic outlining CAR T cell activity depending on target densities on AML and normal cells: ADGRE2-CAR kills only ADGRE2high AML cells but fails to kill ADGRE2low AML cells; ADCLEC.syn1 kills both ADGRE2high and ADGRE2low/CLEC12Apos AML cells while sparing ADGRE2low/CLEC12Aneg normal cells
(C) Summary of in vivo activity of ADGRE2–28z1XX-CAR vs ADCLEC.syn1 against AML cell lines or PDX with target antigen densities as shown. Dashed line delineates in vivo target cell killing vs sparing as observed in experiments shown in Figures 2–5.
(D) Projection of line for ADGRE2/CLEC12A in vivo killing threshold onto primary AML target phenotypes from r/r AML patient cohort.
(E) Projection of line for ADGRE2/CLEC12A in vivo killing threshold onto normal hematopoietic cells
We provide experimental evidence that ADCLEC.syn1 operates with killing thresholds that are tuned to allow target killing based on combinatorial antigen density profiles rather than single-antigen expression (Figure 8A-B). In a series of AML cell line xenograft and PDX experiments we applied in vivo stress tests in terms of targeting ADGRE2 at reduced antigen densities, comparing a single CAR vs IF-BETTER gated ADCLEC.syn1. A conventional ADGRE2-CAR-28z1XX failed to eliminate target cells with only 1×103 or 5×102 ADGRE2 molecules (MOLM13-ADGRE2low/CLEC12Alow/high and U937, respectively) while ADCLEC.syn1 enabled efficient in vivo elimination of the same target cells owing to CLEC12A-CCR engagement (Figure 8C). We demonstrated that the ADGRE2 targeting sensitivity of ADCLEC.syn1 is purposefully gated by the coopted CLEC12A-CCR, allowing to spare MOLM13-ADGRE2low/CLEC12Aneg target cells with 1×103 ADGRE2 molecules and CLEC12A negativity, which represent normal cells, addressing an important safety requirement. On the other hand, MOLM13-ADGRE2low/CLEC12Alow target cells with 1×103 ADGRE2 and 5×102 CLEC12A molecules were efficiently eradicated, highlighting that even a low CCR target density of only 5×102 molecules can determine the susceptibility to cytolysis of target cells.
Overall, we demonstrate an in vivo killing pattern of ADCLEC.syn1 that reflects a two-dimensional antigen density threshold for target cell lysis, where CCR engagement determines the CAR sensitivity and therefore killing fate of CAR target-low cells (Figure 8C). The IF-BETTER gated killing thresholds translate into increased potential for complete tumor eradication at limiting CAR target densities (1×103 or fewer molecules) (Figure 8D) while sparing models for vital normal cells (Figure 8C) and normal HSPCs (Figure 8E).
Previous studies have reported on different uses of CCRs to enhance second-generation CARs: Muliaditan et al. used a 4–1BB CCR to enhance functional persistence of 28z CAR T cells in a breast cancer model.63 Katsarou et al. compared different CD38-CCR formats to enhance efficacy of BCMA- or CD19-targeted 28z-CARs in multiple myeloma and B cell precursor acute lymphoblastic leukemia (B-ALL), respectively. In a Nalm6 B-ALL model of CD19 antigen escape, a CD19-CAR-28z+CD38-CCR-28BB design with shared CD28 transmembrane domains induced remissions in mice.64 Hirabayashi et al. reported that CAR and CCR sharing the same transmembrane domain enable CAR/CCR heterodimerization, resulting in CAR activation by the CCR target thus acting as OR-gated killing in settings where on-target toxicity is not a concern.65 Other logic-gated CAR approaches include OR gates based on dual or tandem CARs,37–39,41,43 AND gates based on two individually dysfunctional receptors,44,45 and NOT gates incorporating inhibitory signaling.66,67 Preference for a logic-gate will depend on the malignancy and expression profiles of target antigens. Further investigation of these alternate gates with clinically relevant ranges of target antigen densities is required to determine their relative antigen sensitivities68,69, as we have done here. In previously reported studies, the CAR+CCR design was investigated to maximize anti-tumor activity by increasing avidity and combining costimulation to promote T cell persistence.63,64 The present study investigates a different use of a CCR: to amplify a therapeutic window based on antigen density differences so as to skew CAR activity and cytolysis towards tumor cells and away from closely related normal cells.
We demonstrate high in vivo antileukemic efficacy of ADCLEC.syn1 in several AML cell line xenograft models using stress test CAR T cell doses as low as 5×104, matching clinically feasible dosing if normalized to body weight.70 Furthermore, we demonstrate efficacy of ADCLEC.syn1 in AML of different phenotypes and genomic background. ADCLEC.syn1 induced durable remissions in xenograft models based on MOLM13 variants representing a wide spectrum of ADGRE2 and CLEC12A antigen densities as well as U937, which endogenously represents ADGRE2lowCLEC12Ahigh AML. Of note, ADCLEC.syn1 induced durable remissions in three molecularly distinct AML PDX models. PDX#1 is derived from a patient with KMT2A-rearranged r/r AML and has been reported as highly aggressive AML model that only partially responded to targeted therapy.71–73 In mice engrafted with PDX#1, ADCLEC.syn1 induced durable remissions and outperformed a reference CD33-CAR-28z1XX. We chose CD33 as reference target due to its known abundant expression in AML including PDX#1, and its wide use as immunotarget in AML, e.g. with the FDA-approved antibody-drug-conjugate Gemtuzumab ozogamicin.74
On-target hematotoxicity is one of the key concerns for CAR therapy in AML. Pre-clinical models for CAR hematotoxicity are technically challenging but invaluable to guide safe CAR development, using humanized mouse models to study CAR hematotoxicity in the setting of an in vivo antitumor CAR T cell response. Kim et al. demonstrated the myeloablative potential of CD33-targeted CAR therapy leading to complete depletion of the CD34pos HSPC compartment, which required genetic removal of CD33 from normal human hematopoiesis to avoid on-target hematotoxicity.30 Sugita et al. evaluated hematotoxicity of CD123-targeted CAR therapy and reported ablation of normal myeloid cells in mice treated with CD123-CAR T cells.75 Both pre-clinical studies and the emerging clinical evidence of myeloablation in patients responding to CD33-CAR and CD123-CAR therapies27 underline the potential predictive value of pre-clinical in vivo hematotoxicity models. Other studies reported on alternative CAR approaches targeting GRP7876 or SIGLEC6,77 demonstrating promising results with limited in vitro hematotoxicity. The in vivo hematotoxicity in the context of GRP78- or SIGLEC6targeted CAR T cell responses remains to be investigated.
In our work presented here, we used normal CD34pos cells to engraft human hematopoietic cells in NSG mice, which were then co-engrafted with AML cells to assess CAR T cell efficacy and toxicity in the context of normal human bystander cells and an anti-leukemic CAR T cell response. In this model, we compared ADCLEC.syn1 to CD19-CAR-28z1XX as control for on-target toxicity. CD19 is clinically validated as CAR target with a B cell-restricted and overall acceptable on-target hematologic toxicity profile in the context of 28z, BBz or 28z1XX CAR formats.5,78–81 Therefore, comparing 19–28z1XX vs ADCLEC.syn1 allows to benchmark the on-target HSPC toxicity profile in the context of a potent in vivo anti-leukemic CAR T cell response. We found that ADCLEC.syn1 led to efficient eradication of AML cells without reducing normal HSPCs any more than 19–28z1XX. Notably, efficacy of ADCLEC.syn1 was maintained in a bone marrow environment with proximate normal myelopoiesis which may limit efficacy of CARs targeting antigens of higher density on more frequent normal cells, i.e. due to potential competition for CAR engagement. Consistent with our observed target profiles and independent reports of ADGRE2 and CLEC12A antigen expression,13,18,82 on-target hematotoxicity of ADCLEC.syn1 was limited to a reduction of normal monocytes. Of note, the clinical benchmark, 19–28z1XX, was associated with a reduction of HSPC compared to untreated mice, suggesting antigen-independent (off-target) HSPC toxicity. Clinically, off-target hematotoxicity is increasingly recognized in patients treated with CD19- or BCMA-targeted FDA-approved CAR T cell therapies. A correlation with CRS severity and pro-inflammatory cytokine levels has been established while the pathophysiology remains to be elucidated.83–87
We describe here a humanized mouse model in which both on-target and off-target toxicities could be studied, and demonstrate in vivo for both 19–28z1XX and ADCLEC.syn1 that CAR T cell-derived IFN-γ is an important mediator of off-target hematotoxicity. By diminishing IFN-γ expression in CAR T cells, we were able to minimize in vivo off-target hematotoxicity of T cells expressing either a CD19-CAR or ADCLEC.syn1. We thereby provide functional evidence for mitigating off-target CAR hematotoxicity through T cell engineering for limited release of IFN-γ. Off-target hematotoxicity or Immune Effector Cell-Associated Hematotoxicity (ICAHT) has been recognized as one of the most common adverse immune effector cell-associated toxicities, and is distinct from other common toxicities such as cytokine release syndrome (CRS) and immune effector cell-associated neurotoxicity syndrome (ICANS).88 We identify here IFN-γ as a directly actionable mediator of ICAHT, warranting further investigation of reducing IFN-γ to alleviate ICAHT and long-term cytopenias in addition to CRS.89,90
Our study highlights that cooperative CAR design can be adapted to target expression profiles, requiring quantification of surface target molecules in tumor and normal tissues to identify therapeutic windows. We rationally co-target ADGRE2 and CLEC12A with a CAR design termed ADCLEC.syn1, and demonstrate its potential for selective anti-leukemic activity in stringent and controlled pre-clinical models of CAR T cell efficacy and hematotoxicity. We further demonstrate the potential of controlling IFN-γ production to mitigate off-target immune effector cell-associated hematotoxicity (ICAHT) in CAR therapies including CD19-CARs. ADCLEC.syn1 may thus preempt the need for post-CAR T cell allo-HSCT rescue and allow for potential use as a standalone therapy including in consolidation or for treatment of post-transplant AML relapse. A phase 1 clinical trial evaluating ADCLEC.syn1 T cells in patients with r/r AML (NCT05748197) is slated to open in 2023.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michel Sadelain (m-sadelain@ski.mskcc.org).
Materials Availability
Plasmid requests can be directed to the lead contact.
Data and Code Availability
This paper does not report standardized datatypes or original code.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
AML patient and normal donor samples
AML patient samples were collected as peripheral blood or bone marrow aspirates from r/r AML patients at MSKCC, after receiving informed consent under IRB-approved protocols (#06–107 and #14–091). Unlike in previous studies13,18, the present analysis focused exclusively on r/r AML to represent a potential phase 1 CAR T cell trial patient population. Normal donor whole bone marrow aspirates were purchased from AllCells. AML and normal donor samples were received and stored in fresh and EDTA-anticoagulated condition on a tube rocker at room temperature and were processed for flow cytometric analysis within 24h. Samples were filtered through a 70μm cell strainer and red blood cell lysis was performed with a subsequent washing step and Fc receptor blocking before antibody staining per standard flow cytometry protocol. Clinically available patient information on AML burden, cytogenetic and molecular aberrations was used to identify relapsed/refractory cases and their AML subtype according to the ELN 2022 classification.55
Cell lines
Unmodified MOLM13 AML cell line was received as gift from Dr. Marion Subklewe (LMU Munich). Unmodified U937 AML cell line was obtained from ATCC. Both AML cell lines were then transduced with FFLuc-GFP-expressing retroviral vector and subsequently sorted for GFPpos fraction. MOLM13 and U937 cells were cultured in RPMI-1640 medium supplemented with 10% FBS (HyClone FetalClone I), 2mM L-glutamine (Invitrogen), 10U/ml penicillin and 10μg/ml streptomycin (Gibco). MOLM13 cells were CRISPR/Cas9-edited to disrupt ADGRE2 or CLEC12A, and transduced with a retroviral vector expressing human CLEC12A to generate variants with high CLEC12A levels. Limiting dilution was used to identify clones with low and very-low ADGRE2 levels (~1,000 and ~500 molecules per cell, respectively), absent CLEC12A expression (CLEC12A-KO) or high CLEC12A expression (5×104 to 2×105 molecules per cell). ADGRE2 and CLEC12A surface protein densities were determined via flow cytometry as described below. For use in the humanized AML xenograft model, MOLM13 with endogenous ADGRE2 and CLEC12A levels was transduced with a retroviral vector expressing human CD19 and a clone was isolated with CD19 levels matching those found in NALM6 cells. Unmodified EL4 murine lymphoma cell line was obtained from ATCC. EL4 cells were transduced with retroviral vectors expressing human ADGRE2 or CLEC12A. EL4 cells were cultured in DMEM medium supplemented with 10% FBS (HyClone FetalClone I), 2mM L-glutamine (Invitrogen), 10U/ml penicillin and 10μg/ml streptomycin (Gibco). All cell lines were split every 2–3 days, plated at 0.5×106 to 2×106 cells/ml and cultured at 37°C with 5% carbon dioxide (CO2).
AML cell line xenograft models
We used male 8 to 12-week-old NOD/SCID/IL-2Rγ-null (NSG) mice (Jackson Laboratory), under a protocol approved by the MSKCC Institutional Animal Care and Use Committee. In the MOLM13 AML model, a total of 1×106 FFLuc-GFP MOLM13 (WT or target-variant) cells per mouse were administered via tail vein injection (d-5). Five days later (d0), untransduced (UTD) or CAR-transduced T cells were administered via tail vein injection, at a given dose based on transgene-expressing T cells. In the U937 AML model, a total of 0.5×106 FFLuc-GFP U937 cells per mouse were administered via tail vein injection (d-4). Four days later (d0), untransduced (UTD) or CAR-transduced T cells were administered via tail vein injection, at a given dose based on transgene-expressing T cells. AML rechallenge experiments were performed by tail vein injection of 1×106 MOLM13 (WT or CLEC12A-high) cells per mouse at the indicated time points. AML burden was measured by bioluminescence imaging using the Xenogen IVIS Imaging System (Xenogen). Living Image software (Xenogen) was used to analyze acquired bioluminescence data. Survival was monitored for a minimum of 100 days post T cell injection. Mice were euthanized when showing clear clinical signs of distress or when reaching maximum AML burden as defined by hindlimb paralysis. There were no instances at which this maximum was exceeded.
AML patient-derived xenograft models
Peripheral blood samples were collected from AML patients after receiving informed consent under an IRB-approved protocol (#14–091). Samples were processed via isolation of mononuclear cells by Ficoll density centrifugation followed by red blood cell lysis, and engrafted via tail vein injection into immunodeficient mice, as previously described and under a protocol approved by the MSKCC Institutional Animal Care and Use Committee.91 For PDX#1, female 6 to 8-week-old NSG (Jackson Laboratory) mice were used. For PDX#2,3, female 6 to 8-week-old NOD-scid IL2Rgnull-3/GM/SF (NSG-SGM3, Jackson Laboratory) mice were used. AML PDX models were serially transplanted three times before being deemed established. Clinically annotated cytogenetic and molecular aberrations were confirmed on PDX samples. AML PDX-engrafted mice were treated with TRAC-KO untransduced or CAR T cells to avoid any potential allogeneic graft-versus-leukemia effects. Serial flow cytometry of peripheral blood and bone marrow was performed to monitor AML burden (live/hCD45dim/CD4neg/CD8neg) and TRAC-KO CAR T cell expansion (live/hCD45pos/CD3neg/CD4pos or CD8pos). For serial AML transplantation, bone marrow was harvested, followed by red blood cell lysis and re-engraftment into new mice of the same strain, age and sex. Survival was monitored for a minimum of 100 days post T cell injection. Mice were euthanized when showing clear clinical signs of distress or when reaching maximum AML burden as defined by hindlimb paralysis. There were no instances at which this maximum was exceeded.
Normal HSPC collection and in vitro culture
Leukapheresis material from G-CSF-mobilized adult healthy donors (Miltenyi Biotec) was used to isolate CD34pos cells via CliniMACS CD34 GMP MicroBeads (Miltenyi Biotec) and a CliniMACS device (Miltenyi Biotec). T cells from the CD34neg fraction were isolated using a Pan T Cell Isolation Kit (Miltenyi Biotec). Matched donor CD34pos HSPCs and CD34neg pan T cells were separately cryopreserved in CryoStor CS10 (Stemcell Technologies) freezing medium. HSPCs were thawed using thawing buffer (X-vivo 15, 1% human serum albumin, 10U/ml heparin) and subsequently cultured in HSPC cytokine-rich medium (StemSpan SFEM II (Stemcell Technologies), SCF 100ng/ml (R&D Systems), TPO 100ng/ml (Stemcell Technologies), FLT3L 100ng/ml (R&D Systems), IL6 100ng/ml (Peprotech), UM729 0.5μM (Stemcell Technologies), SR1 0.75μM (Cellagen Technology), streptomycin 20mg/ml, penicillin 20U/ml) under hypoxic conditions (2% O2), at 37°C and 5% CO2, maintaining a cell concentration of 0.5×106/ml.92,93 For off-target hematotoxicity in vitro assays, conditioned media were added at 1:1 volume ratio to the above HSPC culture conditions, and anti-IFN-γ blocking antibody (clone B133.5, Bioxcell) was used at a final concentration of 20 μg/ml.
Humanized AML xenograft model
Humanized AML-engrafted mice were used under a protocol approved by the MSKCC Institutional Animal Care and Use Committee, to evaluate the impact of in vivo activated CAR T cells on normal human hematopoietic cells (model outline described in Figure 6A). CAR T cells were generated from cryopreserved T cells originating from the same donor as HSPCs, using the γ-retroviral T cell transduction protocol as described above. 5 days after HSPC thawing (d-21), in vitro expanded HSPCs were injected via tail vein into sublethally (2Gy) irradiated female 4-week-old NSG mice (7×105 per mouse). 18 days after HSPC injection (d-3), FFLuc-GFP MOLM13-CD19pos AML cells (5×105 per mouse) were injected via tail vein. 21 days after HSPC injection (d0), CAR T cells (19–28z1XX or ADCLEC.syn1, 2.5×105 per mouse) were injected via tail vein. Antileukemic efficacy was monitored via bioluminescence imaging. On d7, bone marrow aspirates from untreated, 19–28z1XX-treated and ADCLEC.syn1-treated mice were collected and analyzed via flow cytometry. MOLM13 AML cells and normal human hematopoietic cell populations were defined as follows: MOLM13-CD19pos (hCD45pos CD33pos CD19pos), HSPCs (hCD45pos CD3neg CD19neg CD14neg CD16neg CD34pos), B cells (hCD45pos CD33neg CD19pos), monocytes (hCD45pos CD14pos CD16neg), CAR T cells (hCD45pos CD3pos).
METHOD DETAILS
Flow cytometry
All flow cytometry experiments were performed using standard sample processing and staining protocols, and data was acquired on a spectral flow cytometer (Cytek Aurora, 5-laser configuration). Transgene-positive T cells were identified by detection of ADGRE2-CAR, CLEC12A-CCR or EGFRt reporter, wherever applicable, using the following reagents: ADGRE2-CAR anti-idiotype (custom clone 23D5.G5.D4.C10, Rockland Immunochemicals, Inc.), CLEC12A-CCR anti-idiotype (custom clone 18F11.E11.C6, Rockland Immunochemicals, Inc.), EGFRt (clone AY13, Biolegend). Live cells were identified as cells staining negative for DAPI (BD Biosciences). Human hematopoietic cells were characterized using the following reagents: CD3 (UCHT1, BD Biosciences), CD4 (L200, BD Biosciences), CD8 (SK1, BD Biosciences), CD14 (63D3, Biolegend), CD16 (CB16, ThermoFisher), CD19 (SJ25C1, BD Biosciences), CD25 (2A3, BD Biosciences), CD34 (581, Biolegend), CD38 (HB-7, Biolegend), hCD45 (HI30, Invitrogen), mCD45 (30-F11, Biolegend), CD45RA (HI100, BD Biosciences), CD71 (M-A712, BD Biosciences), CD90 (5E10, Biolegend), IFN-γ (B27, Biolegend). AML target expression was detected using the following reagents: CD33 (P67.6, Biolegend), CD123 (6H6, Biolegend), ADGRE2 (2A1, Bio-Rad), CLEC12A (50C1, BD Biosciences).
Flow cytometric gating and quantification of surface target antigen densities
For AML patient and normal donor samples, the following gating strategies were performed to identify relevant cell populations: AML bulk CD45dim SSClow CD3neg CD19neg. AML LSC CD45dim SSClow CD3neg CD19neg CD14neg CD16neg CD71neg CD34pos CD38neg. HSC (=HSC-enriched MPP fraction) CD45dim SSClow CD3neg CD19neg CD14neg CD16neg CD71neg CD34pos CD38neg CD45RAneg CD90pos. MPP CD45dim SSClow CD3neg CD19neg CD14neg CD16neg CD71neg CD34pos CD38neg CD45RAneg CD90neg. MLP CD45dim SSClow CD3neg CD19neg CD14neg CD16neg CD71neg CD34pos CD38neg CD45RApos CD90neg. CP CD45dim SSClow CD3neg CD19neg CD14neg CD16neg CD71neg CD34pos CD38pos. Granulocytes CD45med SSChigh. Monocytes CD45high SSCmed CD14pos CD16neg. B cells CD45high SSClow CD19pos CD3neg. T cells CD45high SSClow CD19neg CD3pos. Antigen distribution analysis was defined as negative/positive percentages (wherein positive indicates ≥1,000 molecules per cell), as limiting resolution at antigen densities <1,000 molecules per cell did not allow for reliable gating of negative vs low subpopulations. Absolute surface antigen densities of CAR target candidates were determined by reading the median fluorescence intensity of relevant samples against a calibration curve based on external standard microsphere beads saturated with the relevant staining antibody at known antibody binding capacity. Quantum™ Simply Cellular® anti-Mouse IgG (Bangs Laboratories, Inc.) beads were stained and acquired in parallel with the sample at identical flow cytometry instrument settings. Quantitation bead staining, acquisition and subsequent calculations were performed according to the manufacturer’s protocol and the QuickCal® (Bangs Laboratories, Inc.) analysis template.
Single-cell target RNA expression profile in defined cell types of normal tissues
The data for single-cell target RNA expression in normal tissues and their associated cell types were obtained from the Human Protein Atlas (version 21.1).56 Single-cell data and their annotated cell types are available for download through the Human Protein Atlas (https://www.proteinatlas.org/download/rna_single_cell_type_tissue.tsv.zip).
T cell isolation, activation and culture
Buffy coats from healthy volunteer donors were obtained from the New York Blood Center. Peripheral blood mononuclear cells were isolated by density gradient centrifugation. T cells were purified using a Pan T Cell Isolation Kit (Miltenyi Biotec) and activated with Human T-Activator CD3/CD28 Dynabeads (Thermo Fisher) at 1:1 bead:cell ratio in X-vivo 15 medium (Lonza) supplemented with 5% human serum (Gemini Bioproducts) with 5ng/ml human recombinant interleukin-7 (Miltenyi Biotec) and 5ng/ml human recombinant interleukin-15 (Miltenyi Biotec) at a density of 1×106 cells/ml. The medium was changed every 2 days, and cells were replated at 1–1.5×106 cells/ml. T cells were incubated at 37°C, at 5% carbon dioxide (CO2). For AML patient-derived CAR T cells, clinical information and peripheral blood of an MSKCC AML patient were obtained under IRB-approved protocols (protocols #14–091 and #06–107).
gRNA, Cas9 protein and RNP formation
TRAC gRNA target sequence: 5′-CAGGGTTCTGGATATCTGT as described in Eyquem, Mansilla-Soto et al.94 IFNG gRNA target sequence: 5’-CCAGAGCATCCAAAAGAGTG as described in Bailey et al.89 TRAC and IFNG gRNAs were ordered from Synthego with 2′-O-methyl 3′-phosphorothioate modifications in the first and last three nucleotides.95 Guide RNA was resuspended with TE buffer at 40μM. Cas9 protein (40μM) was obtained from QB3-Berkeley Macrolab core facility. TRAC ribonucleoprotein (RNP) was prepared by mixing Cas9 protein and TRAC gRNA at 1:1 molar ratio, incubating at 37°C for 15 min and immediately using it for T cell editing experiments.
Gene editing
CRISPR/Cas9 editing was used to disrupt the TCRα subunit constant gene (TRAC) in T cells for PDX experiments and IFNG in T cells for off-target hematotoxicity experiments. 48h after initiating T cell activation, the CD3/CD28 beads were magnetically removed, and T cells were transfected by electrotransfer of TRAC or IFNG RNP using a 4D-Nucleofector device (Lonza). Then, 2×106 or 10×106 cells were resuspended in P3 buffer (Lonza) and mixed with 60 or 300pmol TRAC or IFNG RNP in a total volume of 20 or 100μl, respectively. Following electroporation and assuming 66.7% recovery, cells were diluted and incubated in culture medium at 1×106 cells/ml. 12 to 24h post electroporation, T cells were transduced via SFG-γ-retroviral vector following the transduction protocol as described for non-edited T cells. Subsequently, TRAC or IFNG-edited and SFG γ-retrovirally-transduced T cells were cultured using T cell growth medium, replenished at least every two days and as needed to maintain a density of 1–1.5×106 cells/ml.
Gammaretroviral vector construction, production and transduction
Plasmids encoding the SFG γ-retroviral vector96 were used to clone bicistronic constructs for screening of candidate CAR and CAR+CCR designs, control CARs and ADCLEC.syn1, as outlined in Figure S2A-B. The CAR hinge, transmembrane and endodomain have previously been described and incorporate a modified CD3ζ domain with a single proximal immunoreceptor tyrosine-based activation motif (ITAM).47 CARs were either co-expressed with a truncated EGFR (EGFRt) reporter, a chimeric costimulatory receptor (CCR) or a CCR-del control, as previously described.44,46 SFG γ-retrovirus encoding CAR or CCR constructs was produced via transfection of amphotropic Phoenix-AMPHO cell line (ATCC) with SFG vector plasmids, pCMV-gag-pol plasmid (Cell Biolabs) and pCMV-VSV-G envelope plasmid (Cell Biolabs), using FuGene HD transfection reagent (Promega), followed by virus collection 24h post transfection. T cells were transduced with retroviral supernatants by centrifugation on Retronectin-coated plates (Takara), as described previously.97
CAR scFv origin and selection
ADGRE2 scFv candidates were generated based on humanization of VH and VL sequences of a murine ADGRE2 antibody (clone 2A1), using Xoma and Molecular Operating Environment (MOE) humanization methods, leading to amino acid changes in the VH and VL framework and associated changes in affinities (parental clone 2A1 scFv: KD 2.66×10−11M, lead humanized scFv: KD 2.10×10−10M). The parental murine antibody clone 2A1 has previously been reported.98 CLEC12A scFv candidates were generated based on VH and VL sequences from human anti-CLEC12A antibodies generated in Trianni mice immunized with recombinant human CLEC12A protein. Candidate scFv targeting ADGRE2 (n=18 scFv) and CLEC12A (n=16 scFv) were screened for in vitro and in vivo efficacy in the 28z1XX CAR format. The lead ADGRE2 scFv was chosen based on preferential killing of ADGRE2high and relative sparing of ADGRE2low AML cell lines. The lead CLEC12A scFv was chosen based on highest killing of CLEC12Ahigh and CLEC12Alow AML cell lines. ADGRE2 and CLEC12A scFv sequences are listed in patent WO2022232016A2. The CD33 scFv was generated using heavy (VH)- and light (VL)-chain variable regions from a published anti-CD33 antibody sequence (clone 280–31-01, as described in patent WO2012045752A1). The CD19 scFv is based on clone SJ25C1, as previously reported.99
Cytotoxicity assays
In vitro anti-leukemic cytotoxicity of CAR T cells was determined via standard FFLuc-based assay. FFLuc-expressing MOLM13 cells served as target cells. The effector (E) and target (T) cells were co-cultured in triplicates at the indicated E:T ratio using black-walled flat-bottom 96-well plates with 5×104 target cells in a total volume of 100μl per well in T cell medium. 18h later, 50μl D-luciferin (Goldbio) at 1.5μg/μl was directly added to each well. Immediately after, emitted light was detected in a luminescence plate reader, and % cytotoxicity was calculated using the formula 100x(1−(RLUsample)/(RLUtarget alone)). For studies using EL4 cells, in vitro cytotoxicity was determined via flow cytometric quantification of target cell counts. EL4 cells (T) were co-cultured with CAR T cells (E) at the indicated E:T ratio using flat-bottom 96-well plates with 5×104 target cells in a total volume of 100μl per well in T cell medium. 48h later, flow cytometry was used to determine the residual total target cell counts (live/CD3neg cells) as well as T cell activation (CD25 MFI). % cytotoxicity was calculated using the formula 100x(1−(RLUsample)/(RLUtarget alone)).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics
All experimental data are presented as described in the figure legends. In vitro experiments were performed in technical triplicates and replicated with at least two independent donors for CAR T cell production. For in vivo experiments, n=5 mice per group were used unless indicated otherwise. Across all in vivo experiments, CAR T cells from a total of n=13 different adult healthy T cell donors were used. For all Kaplan-Meier survival analyses, log-rank (Mantel-Cox) testing was performed. Statistical tests are described in the figure legends. Statistical analysis was performed on GraphPad Prism v10.0.1 software. Significance was set at p<0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| ADGRE2-CAR (anti-idiotype, 23D5.G5.D4.C10, Dylight405) | This paper | N/A |
| CLEC12A-CCR (anti-idiotype, 18F11.E11.C6, PE) | This paper | N/A |
| human EGFRt (AY13, BV711) | Biolegend | Cat# 352920; RRID: AB_2687123 |
| human CD3 (UCHT1, BUV615) | BD Biosciences | Cat# 612992; RRID: AB_2870263 |
| human CD4 (L200, BUV395) | BD Biosciences | Cat# 564107; RRID: AB_2738596 |
| human CD8 (SK1, BUV805) | BD Biosciences | Cat# 612889; RRID: AB_2833078 |
| human CD14 (63D3, SparkBlue550) | Biolegend | Cat# 367148; RRID: AB_2832724 |
| human CD16 (CB16, SuperBright600) | ThermoFisher | Cat# 63-0168-42; RRID: AB_2662479 |
| human CD19 (SJ25C1, BUV563) | BD Biosciences | Cat# 612916; RRID: AB_2870201 |
| human CD25 (BC96, PE) | Biolegend | Cat# 302606; RRID: AB_314276 |
| human CD34 (581, APC/Fire750) | Biolegend | Cat# 343536; RRID: AB_2650736 |
| human CD38 (HB-7, BV650) | Biolegend | Cat# 356620; RRID: AB_2566233 |
| human CD45 (HI30, PacificOrange) | ThermoFisher | Cat# MHCD4530; RRID: AB_10376143 |
| murine CD45 (30-F11, AF488) | Biolegend | Cat# 103122; RRID: AB_493531 |
| human CD45RA (HI100, BV480) | BD Biosciences | Cat# 566114; RRID: AB_2739516 |
| human CD71 (M-A712, BV711) | BD Biosciences | Cat# 563767; RRID: AB_2738413 |
| human CD90 (5E10, PE/Dazzle594) | Biolegend | Cat# 328134; RRID: AB_2566343 |
| human CD33 (P67.6, PE/Cy7) | Biolegend | Cat# 366618; RRID: AB_2566420 |
| human CD123 (6H6, BV785) | Biolegend | Cat# 306032; RRID: AB_2566448 |
| human ADGRE2 (2A1, AF647) | Bio-Rad | Cat# MCA2330A647T; RRID: AB_2231142 |
| human CLEC12A (50C1, BB515) | BD Biosciences | Cat# 565325; RRID: AB_2739182 |
| human IFNg (B27, PE) | Biolegend | Cat# 506507; RRID: AB_315440 |
| human IFNg (blocking) | Bio X Cell | Cat# BE0235; RRID: AB_2687717 |
|
| ||
| Biological samples | ||
|
| ||
| Mobilized Leukopaks®, G-CSF x 5 Days - Apheresis Day 6, Fresh | Miltenyi Biotec | Cat# 150-000-461 |
| Normal donor whole bone marrow, fresh | Allcells | N/A |
| Buffy coats | New York Blood Center | N/A |
| AML patient samples | MSKCC | IRB# 06-107 and IRB# 14-091 |
| AML patient-derived xenograft models | MSKCC, Antitumor Assessment Core Facility | IRB# 14-091 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Cas9-NLS purified protein | QB3-Berkeley Macrolab | N/A |
| P3 Primary Cell 4D-Nucleofector® X Kit | Lonza | Cat# V4XP-3024 |
| Human IL-7, premium grade | Miltenyi Biotec | Cat# 130-095-363 |
| Human IL-15, premium grade | Miltenyi Biotec | Cat# 130-095-765 |
| Dynabeads™ CD3 | ThermoFisher | Cat# 11151D |
|
| ||
| Critical commercial assays | ||
|
| ||
| Quantum™ Simply Cellular® antiMouse IgG | Bangs Laboratories | Cat# 815 |
| Pan T Cell Isolation Kit, human | Miltenyi Biotec | Cat# 130-096-535 |
|
| ||
| Deposited data | ||
|
| ||
| Normal tissue scRNAseq data | Human Protein Atlas (v21.1) | RRID: SCR_006710; Download:https://www.proteinatlas.org/download/rna_single_cell_type_tissue.tsv.zip |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Phoenix-AMPHO | ATCC | Cat# CRL-3213; RRID: CVCL_H716 |
| EL4 | ATCC | Cat# TIB-39; RRID: CVCL_0255 |
| MOLM-13 | DSMZ | Cat# ACC 554 |
| U-937 | ATCC | Cat# CRL-1593.2; RRID: CVCL_0007 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| NSG mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | Jackson Laboratory | RRID: IMSR_JAX:005557 |
| NSG-SGM3 mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1 Eav/MloySzJ | Jackson Laboratory | RRID: IMSR_JAX:013062 |
|
| ||
| Oligonucleotides | ||
|
| ||
| TRAC sgRNA target sequence: 5′-CAGGGTTCTGGATATCTGT | Synthego | N/A |
| IFNG sgRNA target sequence: 5′-CCAGAGCATCCAAAAGAGTG | Synthego | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV-gag-pol | Cell Biolabs | Cat# RV-111 |
| pCMV-VSV-G | Cell Biolabs | Cat# RV-110 |
| pSFG-ADCLEC.syn1 | This paper | N/A |
| pSFG-ADGRE2-28z1XX-P2A-EGFRt | This paper | N/A |
| pSFG-CLEC12A-BB-P2A-EGFRt | This paper | N/A |
| pSFG-ADGRE2-28z1XX-P2A-CLEC12A-del | This paper | N/A |
| pSFG-CD33-28z 1XX-P2A-EGFRt | This paper | N/A |
| pSFG-CD19-28z1XX-P2A-EGFRt | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism v10.0.1 | GraphPad Software | N/A |
| FlowJo v10.8.1 | BD Biosciences | N/A |
| Living Image | Perkin Elmer | N/A |
| Adobe Illustrator v27.8.1 | Adobe | N/A |
Highlights.
Antigen co-expression signatures guide cooperative CAR T cell design
CAR sensitivity is controlled by combining a calibrated CAR and a CCR (IF-BETTER gate)
ADCLEC.syn1 overcomes AML antigen escape and HSPC on-target toxicity
Reducing CAR T cell interferon-γ release mitigates off-target hematotoxicity
Acknowledgments
We thank Gertrude Gunset from the Sadelain Lab and Oriana Borquez-Ojeda, Jolanta Stefanski, Jagruti Chaudhari and Vladimir Bermudez from the MSKCC Cell Therapy and Cell Engineering Facility (CTCEF) for logistical and technical assistance. We thank Nayan Jain, Ivan Kotchetkov, Karlo Perica and Michelle Saetersmoen for helpful scientific discussions. We also thank the MSKCC core facilities for Antitumor Assessment, Flow Cytometry, Molecular Cytogenetics and Pathology for their expert assistance. S.H. was supported in part by NIH/NCI P50 CA254838-01. M.Su. acknowledges support from DFG (SFB-TRR 388/1 2021 – 452881907 and DFG research grant 451580403), the Else-Kröner-Fresenius Stiftung and the Bavarian Center for Cancer Research (BZKF). This work was supported in part by Takeda Pharmaceuticals, the Fondation ARC Léopold Griffuel Award and the NCI Cancer Center Support Grant no. P30 CA008748.
Memorial Sloan Kettering has submitted a patent application (WO2022232016A2) based in part on results presented in this manuscript (M.Sa., S.H. and J.M.-S. are listed among the inventors). M.Sa. and S.H. report research support and research funding from Takeda Pharmaceuticals related to the present research. M.Sa. reports research funding from Atara Biotherapeutics, Fate Therapeutics and Mnemo Therapeutics unrelated to the present research. M.Sa. and I.R. are scientific cofounders of Mnemo Therapeutics. I.R. reports research funding from Atara Biotherapeutics, Takeda Pharmaceuticals; ownership/equity interests at Fate Therapeutics and Mnemo Therapeutics; intellectual property rights at Juno Therapeutics. K.F., M.R.N., and I.R. report employment at Takeda Pharmaceuticals. M.Su. declares the following competing interests: Novartis: Consultancy, Research Funding; Janssen: Consultancy; Seattle Genetics: Research Funding; Amgen: Consultancy, Honoraria, Research Funding; Celgene: Consultancy, Honoraria; Kite/Gilead: Consultancy, Honoraria, Research Funding; Roche AG: Consultancy, Research Funding. J.H.P. declares the following competing interest: research funding support from Takeda Pharmaceuticals, Fate Therapeutics, Genentech, InCyte and Servier; Consultancy from Amgen, Autolus, BMS, Curocel, Kite, Legend Biotech, Minerva, Pfizer, Servier, Sobi, and Takeda Pharmaceuticals; and serves on Scientific Advisory Board of Allogene, Artiva Biotherapeutics, and GC Cell Corporation.
Footnotes
Declaration of Interests
All other authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.National Cancer Institute: Acute Myeloid Leukemia (AML) 5-Year Relative Survival Rates, 2012–2018. Accessed 4 February 2023. Available at: https://seer.cancer.gov/statistics-network/.
- 2.Mohty R, El Hamed R, Brissot E, Bazarbachi A, and Mohty M. (2023). New drugs before, during, and after hematopoietic stem cell transplantation for patients with acute myeloid leukemia. Haematologica 108, 321–341. 10.3324/haematol.2022.280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locke FL, Miklos DB, Jacobson CA, Perales MA, Kersten MJ, Oluwole OO, Ghobadi A, Rapoport AP, McGuirk J, Pagel JM, et al. (2022). Axicabtagene Ciloleucel as Second-Line Therapy for Large B-Cell Lymphoma. N Engl J Med 386, 640–654. 10.1056/NEJMoa2116133. [DOI] [PubMed] [Google Scholar]
- 4.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, et al. (2014). Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 371, 1507–1517. 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park JH, Riviere I, Gonen M, Wang X, Senechal B, Curran KJ, Sauter C, Wang Y, Santomasso B, Mead E, et al. (2018). Long-Term Follow-up of CD19 CAR Therapy in Acute Lymphoblastic Leukemia. N Engl J Med 378, 449–459. 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Otero P, Ailawadhi S, Arnulf B, Patel K, Cavo M, Nooka AK, Manier S, Callander N, Costa LJ, Vij R, et al. (2023). Ide-cel or Standard Regimens in Relapsed and Refractory Multiple Myeloma. N Engl J Med. 10.1056/NEJMoa2213614. [DOI] [PubMed]
- 7.Schuster SJ, Svoboda J, Chong EA, Nasta SD, Mato AR, Anak O, Brogdon JL, Pruteanu-Malinici I, Bhoj V, Landsburg D, et al. (2017). Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. N Engl J Med 377, 2545–2554. 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen AD, Garfall AL, Stadtmauer EA, Melenhorst JJ, Lacey SF, Lancaster E, Vogl DT, Weiss BM, Dengel K, Nelson A, et al. (2019). B cell maturation antigen-specific CAR T cells are clinically active in multiple myeloma. J Clin Invest 129, 2210–2221. 10.1172/JCI126397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, Sommermeyer D, Melville K, Pender B, Budiarto TM, et al. (2016). CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 126, 2123–2138. 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brudno JN, Maric I, Hartman SD, Rose JJ, Wang M, Lam N, Stetler-Stevenson M, Salem D, Yuan C, Pavletic S, et al. (2018). T Cells Genetically Modified to Express an Anti-B-Cell Maturation Antigen Chimeric Antigen Receptor Cause Remissions of Poor-Prognosis Relapsed Multiple Myeloma. J Clin Oncol 36, 2267–2280. 10.1200/JCO.2018.77.8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, et al. (2015). T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528. 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, Liedtke M, Rosenblatt J, Maus MV, Turka A, et al. (2019). Anti-BCMA CAR T-Cell Therapy bb2121 in Relapsed or Refractory Multiple Myeloma. N Engl J Med 380, 1726–1737. 10.1056/NEJMoa1817226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perna F, Berman SH, Soni RK, Mansilla-Soto J, Eyquem J, Hamieh M, Hendrickson RC, Brennan CW, and Sadelain M. (2017). Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell 32, 506–519 e505. 10.1016/j.ccell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ediriwickrema A, Gentles AJ, and Majeti R. (2023). Single-cell genomics in AML: extending the frontiers of AML research. Blood 141, 345–355. 10.1182/blood.2021014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnet D, and Dick JE (1997). Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3, 730–737. 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 16.Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, Bowman M, Famulare C, Patel MA, Mendez P, et al. (2020). Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 587, 477–482. 10.1038/s41586-020-2864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, Kuipers J, Loghavi S, Wang SA, Yan Y, et al. (2020). Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun 11, 5327. 10.1038/s41467-020-19119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haubner S, Perna F, Kohnke T, Schmidt C, Berman S, Augsberger C, Schnorfeil FM, Krupka C, Lichtenegger FS, Liu X, et al. (2019). Coexpression profile of leukemic stem cell markers for combinatorial targeted therapy in AML. Leukemia 33, 64–74. 10.1038/s41375-018-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willier S, Rothamel P, Hastreiter M, Wilhelm J, Stenger D, Blaeschke F, Rohlfs M, Kaeuferle T, Schmid I, Albert MH, et al. (2021). CLEC12A and CD33 coexpression as a preferential target for pediatric AML combinatorial immunotherapy. Blood 137, 1037–1049. 10.1182/blood.2020006921. [DOI] [PubMed] [Google Scholar]
- 20.Taussig DC, Pearce DJ, Simpson C, Rohatiner AZ, Lister TA, Kelly G, Luongo JL, Danet-Desnoyers GA, and Bonnet D. (2005). Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood 106, 4086–4092. 10.1182/blood-2005-03-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, Verga JA, Stephansky J, Pastika TJ, Lombardi Story J, et al. (2019). Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell 176, 1265–1281 e1224. 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baroni ML, Sanchez Martinez D, Gutierrez Aguera F, Roca Ho H, Castella M, Zanetti SR, Velasco Hernandez T, Diaz de la Guardia R, Castano J, Anguita E, et al. (2020). 41BB-based and CD28-based CD123-redirected T-cells ablate human normal hematopoiesis in vivo. J Immunother Cancer 8. 10.1136/jitc-2020-000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenderian SS, Ruella M, Shestova O, Klichinsky M, Aikawa V, Morrissette JJ, Scholler J, Song D, Porter DL, Carroll M, et al. (2015). CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia 29, 1637–1647. 10.1038/leu.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill S, Tasian SK, Ruella M, Shestova O, Li Y, Porter DL, Carroll M, Danet-Desnoyers G, Scholler J, Grupp SA, et al. (2014). Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood 123, 2343–2354. 10.1182/blood-2013-09-529537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tashiro H, Sauer T, Shum T, Parikh K, Mamonkin M, Omer B, Rouce RH, Lulla P, Rooney CM, Gottschalk S, and Brenner MK (2017). Treatment of Acute Myeloid Leukemia with T Cells Expressing Chimeric Antigen Receptors Directed to C-type Lectin-like Molecule 1. Mol Ther 25, 2202–2213. 10.1016/j.ymthe.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizzitola I, Anjos-Afonso F, Rouault-Pierre K, Lassailly F, Tettamanti S, Spinelli O, Biondi A, Biagi E, and Bonnet D. (2014). Chimeric antigen receptors against CD33/CD123 antigens efficiently target primary acute myeloid leukemia cells in vivo. Leukemia 28, 1596–1605. 10.1038/leu.2014.62. [DOI] [PubMed] [Google Scholar]
- 27.Koedam J, Wermke M, Ehninger A, Cartellieri M, and Ehninger G. (2022). Chimeric antigen receptor T-cell therapy in acute myeloid leukemia. Curr Opin Hematol 29, 74–83. 10.1097/MOH.0000000000000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin X, Zhang M, Sun R, Lyu H, Xiao X, Zhang X, Li F, Xie D, Xiong X, Wang J, et al. (2022). First-in-human phase I study of CLL-1 CAR-T cells in adults with relapsed/refractory acute myeloid leukemia. J Hematol Oncol 15, 88. 10.1186/s13045-022-01308-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wermke M, Kraus S, Ehninger A, Bargou RC, Goebeler ME, Middeke JM, Kreissig C, von Bonin M, Koedam J, Pehl M, et al. (2021). Proof of concept for a rapidly switchable universal CAR-T platform with UniCAR-T-CD123 in relapsed/refractory AML. Blood 137, 3145–3148. 10.1182/blood.2020009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim MY, Yu KR, Kenderian SS, Ruella M, Chen S, Shin TH, Aljanahi AA, Schreeder D, Klichinsky M, Shestova O, et al. (2018). Genetic Inactivation of CD33 in Hematopoietic Stem Cells to Enable CAR T Cell Immunotherapy for Acute Myeloid Leukemia. Cell 173, 1439–1453 e1419. 10.1016/j.cell.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borot F, Wang H, Ma Y, Jafarov T, Raza A, Ali AM, and Mukherjee S. (2019). Gene-edited stem cells enable CD33-directed immune therapy for myeloid malignancies. Proc Natl Acad Sci U S A 116, 11978–11987. 10.1073/pnas.1819992116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humbert O, Laszlo GS, Sichel S, Ironside C, Haworth KG, Bates OM, Beddoe ME, Carrillo RR, Kiem HP, and Walter RB (2019). Engineering resistance to CD33-targeted immunotherapy in normal hematopoiesis by CRISPR/Cas9-deletion of CD33 exon 2. Leukemia 33, 762–808. 10.1038/s41375-018-0277-8. [DOI] [PubMed] [Google Scholar]
- 33.Casirati G, Cosentino A, Mucci A, Salah Mahmoud M, Ugarte Zabala I, Zeng J, Ficarro SB, Klatt D, Brendel C, Rambaldi A, et al. (2023). Epitope editing enables targeted immunotherapy of acute myeloid leukaemia. Nature. 10.1038/s41586-023-06496-5. [DOI] [PMC free article] [PubMed]
- 34.Wellhausen N, O’Connell RP, Lesch S, Engel NW, Rennels AK, Gonzales D, Herbst F, Young RM, Garcia KC, Weiner D, et al. Epitope base editing CD45 in hematopoietic cells enables universal blood cancer immune therapy. Science Translational Medicine 0, eadi1145. doi: 10.1126/scitranslmed.adi1145. [DOI] [PMC free article] [PubMed]
- 35.Marone R, Landmann E, Devaux A, Lepore R, Seyres D, Zuin J, Burgold T, Engdahl C, Capoferri G, Dell’Aglio A, et al. (2023). Epitope-engineered human hematopoietic stem cells are shielded from CD123-targeted immunotherapy. J. Exp. Med 220, e2023123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Wu S, and Alachkar H. (2019). Characterization of upregulated adhesion GPCRs in acute myeloid leukemia. Transl Res 212, 26–35. 10.1016/j.trsl.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamieh M, Dobrin A, Cabriolu A, van der Stegen SJC, Giavridis T, Mansilla-Soto J, Eyquem J, Zhao Z, Whitlock BM, Miele MM, et al. (2019). CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature 568, 112–116. 10.1038/s41586-019-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hegde M, Corder A, Chow KK, Mukherjee M, Ashoori A, Kew Y, Zhang YJ, Baskin DS, Merchant FA, Brawley VS, et al. (2013). Combinational targeting offsets antigen escape and enhances effector functions of adoptively transferred T cells in glioblastoma. Mol Ther 21, 2087–2101. 10.1038/mt.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegde M, Mukherjee M, Grada Z, Pignata A, Landi D, Navai SA, Wakefield A, Fousek K, Bielamowicz K, Chow KK, et al. (2016). Tandem CAR T cells targeting HER2 and IL13Ralpha2 mitigate tumor antigen escape. J Clin Invest 126, 3036–3052. 10.1172/JCI83416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin H, Ramakrishna S, Nguyen S, Fountaine TJ, Ponduri A, Stetler-Stevenson M, Yuan CM, Haso W, Shern JF, Shah NN, and Fry TJ (2018). Preclinical Development of Bivalent Chimeric Antigen Receptors Targeting Both CD19 and CD22. Mol Ther Oncolytics 11, 127–137. 10.1016/j.omto.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruella M, Barrett DM, Kenderian SS, Shestova O, Hofmann TJ, Perazzelli J, Klichinsky M, Aikawa V, Nazimuddin F, Kozlowski M, et al. (2016). Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. J Clin Invest 126, 3814–3826. 10.1172/JCI87366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shalabi H, Qin H, Su A, Yates B, Wolters PL, Steinberg SM, Ligon JA, Silbert S, DeDe K, Benzaoui M, et al. (2022). CD19/22 CAR T cells in children and young adults with B-ALL: phase 1 results and development of a novel bicistronic CAR. Blood 140, 451–463. 10.1182/blood.2022015795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zah E, Lin MY, Silva-Benedict A, Jensen MC, and Chen YY (2016). T Cells Expressing CD19/CD20 Bispecific Chimeric Antigen Receptors Prevent Antigen Escape by Malignant B Cells. Cancer Immunol Res 4, 498–508. 10.1158/2326-6066.CIR-15-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kloss CC, Condomines M, Cartellieri M, Bachmann M, and Sadelain M. (2013). Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat Biotechnol 31, 71–75. 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tousley AM, Rotiroti MC, Labanieh L, Rysavy LW, Kim WJ, Lareau C, Sotillo E, Weber EW, Rietberg SP, Dalton GN, et al. (2023). Co-opting signalling molecules enables logic-gated control of CAR T cells. Nature 615, 507–516. 10.1038/s41586-023-05778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krause A, Guo HF, Latouche JB, Tan C, Cheung NK, and Sadelain M. (1998). Antigen-dependent CD28 signaling selectively enhances survival and proliferation in genetically modified activated human primary T lymphocytes. J Exp Med 188, 619–626. 10.1084/jem.188.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feucht J, Sun J, Eyquem J, Ho YJ, Zhao Z, Leibold J, Dobrin A, Cabriolu A, Hamieh M, and Sadelain M. (2019). Calibration of CAR activation potential directs alternative T cell fates and therapeutic potency. Nat Med 25, 82–88. 10.1038/s41591-018-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knaus HA, Berglund S, Hackl H, Blackford AL, Zeidner JF, Montiel-Esparza R, Mukhopadhyay R, Vanura K, Blazar BR, Karp JE, et al. (2018). Signatures of CD8+ T cell dysfunction in AML patients and their reversibility with response to chemotherapy. JCI Insight 3. 10.1172/jci.insight.120974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rutella S, Vadakekolathu J, Mazziotta F, Reeder S, Yau TO, Mukhopadhyay R, Dickins B, Altmann H, Kramer M, Knaus HA, et al. (2022). Immune dysfunction signatures predict outcomes and define checkpoint blockade-unresponsive microenvironments in acute myeloid leukemia. J Clin Invest 132. 10.1172/JCI159579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daver N, Alotaibi AS, Bucklein V, and Subklewe M. (2021). T-cell-based immunotherapy of acute myeloid leukemia: current concepts and future developments. Leukemia 35, 1843–1863. 10.1038/s41375-021-01253-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansilla-Soto J, Eyquem J, Haubner S, Hamieh M, Feucht J, Paillon N, Zucchetti AE, Li Z, Sjostrand M, Lindenbergh PL, et al. (2022). HLA-independent T cell receptors for targeting tumors with low antigen density. Nat Med 28, 345–352. 10.1038/s41591-021-01621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matatall KA, Shen CC, Challen GA, and King KY (2014). Type II interferon promotes differentiation of myeloid-biased hematopoietic stem cells. Stem Cells 32, 3023–3030. 10.1002/stem.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majzner RG, Rietberg SP, Sotillo E, Dong R, Vachharajani VT, Labanieh L, Myklebust JH, Kadapakkam M, Weber EW, Tousley AM, et al. (2020). Tuning the Antigen Density Requirement for CAR T-cell Activity. Cancer Discov 10, 702–723. 10.1158/2159-8290.CD-19-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heitzeneder S, Bosse KR, Zhu Z, Zhelev D, Majzner RG, Radosevich MT, Dhingra S, Sotillo E, Buongervino S, Pascual-Pasto G, et al. (2022). GPC2-CAR T cells tuned for low antigen density mediate potent activity against neuroblastoma without toxicity. Cancer Cell 40, 53–69 e59. 10.1016/j.ccell.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dohner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, Ebert BL, Fenaux P, Godley LA, Hasserjian RP, et al. (2022). Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 140, 1345–1377. 10.1182/blood.2022016867. [DOI] [PubMed] [Google Scholar]
- 56.Karlsson M, Zhang C, Mear L, Zhong W, Digre A, Katona B, Sjostedt E, Butler L, Odeberg J, Dusart P, et al. (2021). A single-cell type transcriptomics map of human tissues. Sci Adv 7. 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caruso S, De Angelis B, Del Bufalo F, Ciccone R, Donsante S, Volpe G, Manni S, Guercio M, Pezzella M, Iaffaldano L, et al. (2022). Safe and effective off-the-shelf immunotherapy based on CAR.CD123-NK cells for the treatment of acute myeloid leukaemia. J Hematol Oncol 15, 163. 10.1186/s13045-022-01376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duan Y, Chen J, Meng X, Liu L, Shang K, Wu X, Wang Y, Huang Z, Liu H, Huang Y, et al. (2023). Balancing activation and co-stimulation of CAR tunes signaling dynamics and enhances therapeutic potency. Mol Ther 31, 35–47. 10.1016/j.ymthe.2022.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoutrop E, Poiret T, El-Serafi I, Zhao Y, He R, Moter A, Henriksson J, Hassan M, Magalhaes I, and Mattsson J. (2023). Tuned activation of MSLN-CAR T cells induces superior antitumor responses in ovarian cancer models. J Immunother Cancer 11. 10.1136/jitc-2022-005691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melero I, Bach N, Hellstrom KE, Aruffo A, Mittler RS, and Chen L. (1998). Amplification of tumor immunity by gene transfer of the co-stimulatory 4-1BB ligand: synergy with the CD28 co-stimulatory pathway. Eur J Immunol 28, 1116–1121. . [DOI] [PubMed] [Google Scholar]
- 61.Zhao Z, Condomines M, van der Stegen SJC, Perna F, Kloss CC, Gunset G, Plotkin J, and Sadelain M. (2015). Structural Design of Engineered Costimulation Determines Tumor Rejection Kinetics and Persistence of CAR T Cells. Cancer Cell 28, 415–428. 10.1016/j.ccell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drent E, Poels R, Ruiter R, van de Donk N, Zweegman S, Yuan H, de Bruijn J, Sadelain M, Lokhorst HM, Groen RWJ, et al. (2019). Combined CD28 and 4–1BB Costimulation Potentiates Affinity-tuned Chimeric Antigen Receptor-engineered T Cells. Clin Cancer Res 25, 4014–4025. 10.1158/1078-0432.CCR-18-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muliaditan T, Halim L, Whilding LM, Draper B, Achkova DY, Kausar F, Glover M, Bechman N, Arulappu A, Sanchez J, et al. (2021). Synergistic T cell signaling by 41BB and CD28 is optimally achieved by membrane proximal positioning within parallel chimeric antigen receptors. Cell Rep Med 2, 100457. 10.1016/j.xcrm.2021.100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katsarou A, Sjostrand M, Naik J, Mansilla-Soto J, Kefala D, Kladis G, Nianias A, Ruiter R, Poels R, Sarkar I, et al. (2021). Combining a CAR and a chimeric costimulatory receptor enhances T cell sensitivity to low antigen density and promotes persistence. Sci Transl Med 13, eabh1962. 10.1126/scitranslmed.abh1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirabayashi K, Du H, Xu Y, Shou P, Zhou X, Fuca G, Landoni E, Sun C, Chen Y, Savoldo B, and Dotti G. (2021). Dual Targeting CAR-T Cells with Optimal Costimulation and Metabolic Fitness enhance Antitumor Activity and Prevent Escape in Solid Tumors. Nat Cancer 2, 904–918. 10.1038/s43018-021-00244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fedorov VD, Themeli M, and Sadelain M. (2013). PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci Transl Med 5, 215ra172. 10.1126/scitranslmed.3006597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richards RM, Zhao F, Freitas KA, Parker KR, Xu P, Fan A, Sotillo E, Daugaard M, Oo HZ, Liu J, et al. (2021). NOT-Gated CD93 CAR T Cells Effectively Target AML with Minimized Endothelial Cross-Reactivity. Blood Cancer Discov 2, 648–665. 10.1158/2643-3230.BCD-20-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamieh M, Mansilla-Soto J, Riviere I, and Sadelain M. (2023). Programming CAR T Cell Tumor Recognition: Tuned Antigen Sensing and Logic Gating. Cancer Discov 13, 829–843. 10.1158/2159-8290.CD-23-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Z, and Sadelain M. (2023). CAR T cell design: approaching the elusive AND-gate. Cell Res. 10.1038/s41422-023-00828-w. [DOI] [PMC free article] [PubMed]
- 70.Duncan BB, Dunbar CE, and Ishii K. (2022). Applying a clinical lens to animal models of CAR-T cell therapies. Mol Ther Methods Clin Dev 27, 17–31. 10.1016/j.omtm.2022.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K, Sanchez-Martin M, Wang X, Knapp KM, Koche R, Vu L, Nahas MK, He J, Hadler M, Stein EM, et al. (2017). Patient-derived xenotransplants can recapitulate the genetic driver landscape of acute leukemias. Leukemia 31, 151–158. 10.1038/leu.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramaswamy K, Forbes L, Minuesa G, Gindin T, Brown F, Kharas MG, Krivtsov AV, Armstrong SA, Still E, de Stanchina E, et al. (2018). Peptidomimetic blockade of MYB in acute myeloid leukemia. Nat Commun 9, 110. 10.1038/s41467-017-02618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bill M, Mrozek K, Kohlschmidt J, Eisfeld AK, Walker CJ, Nicolet D, Papaioannou D, Blachly JS, Orwick S, Carroll AJ, et al. (2020). Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/KMT2A. Proc Natl Acad Sci U S A 117, 26340–26346. 10.1073/pnas.2014732117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walter RB (2018). Investigational CD33-targeted therapeutics for acute myeloid leukemia. Expert Opin Investig Drugs 27, 339–348. 10.1080/13543784.2018.1452911. [DOI] [PubMed] [Google Scholar]
- 75.Sugita M, Galetto R, Zong H, Ewing-Crystal N, Trujillo-Alonso V, Mencia-Trinchant N, Yip W, Filipe S, Lebuhotel C, Gouble A, et al. (2022). Allogeneic TCRalphabeta deficient CAR T-cells targeting CD123 in acute myeloid leukemia. Nat Commun 13, 2227. 10.1038/s41467-022-29668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hebbar N, Epperly R, Vaidya A, Thanekar U, Moore SE, Umeda M, Ma J, Patil SL, Langfitt D, Huang S, et al. (2022). CAR T cells redirected to cell surface GRP78 display robust anti-acute myeloid leukemia activity and do not target hematopoietic progenitor cells. Nat Commun 13, 587. 10.1038/s41467-022-28243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jetani H, Navarro-Bailon A, Maucher M, Frenz S, Verbruggen C, Yeguas A, Vidriales MB, Gonzalez M, Rial Saborido J, Kraus S, et al. (2021). Siglec-6 is a novel target for CAR T-cell therapy in acute myeloid leukemia. Blood 138, 1830–1842. 10.1182/blood.2020009192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mansson R, Zandi S, Anderson K, Martensson IL, Jacobsen SE, Bryder D, and Sigvardsson M. (2008). B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood 112, 1048–1055. 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- 79.Sigvardsson M. (2018). Molecular Regulation of Differentiation in Early B-Lymphocyte Development. Int J Mol Sci 19. 10.3390/ijms19071928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, Bader P, Verneris MR, Stefanski HE, Myers GD, et al. (2018). Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N Engl J Med 378, 439–448. 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park JH, Palomba ML, Rivière I, Sikder DS, Senechal B, Wang X, Bermudez VP, Cathcart ER, Liotta K, Joseph J, et al. (2022). A Phase I Study of CD19-Targeted 19(T2)28z1xx CAR T Cells in Adult Patients with Relapsed or Refractory Diffuse Large B-Cell Lymphoma. Blood 140, 403–404. 10.1182/blood-2022-168206. [DOI] [Google Scholar]
- 82.I KY, Tseng WY, Wang WC, Gordon S, Ng KF, and Lin HH (2020). Stimulation of Vibratory Urticaria-Associated Adhesion-GPCR, EMR2/ADGRE2, Triggers the NLRP3 Inflammasome Activation Signal in Human Monocytes. Front Immunol 11, 602016. 10.3389/fimmu.2020.602016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Juluri KR, Wu QV, Voutsinas J, Hou J, Hirayama AV, Mullane E, Miles N, Maloney DG, Turtle CJ, Bar M, and Gauthier J. (2022). Severe cytokine release syndrome is associated with hematologic toxicity following CD19 CAR T-cell therapy. Blood Adv 6, 2055–2068. 10.1182/bloodadvances.2020004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rejeski K, Perez A, Sesques P, Hoster E, Berger C, Jentzsch L, Mougiakakos D, Frolich L, Ackermann J, Bucklein V, et al. (2021). CAR-HEMATOTOX: a model for CAR T-cell-related hematologic toxicity in relapsed/refractory large B-cell lymphoma. Blood 138, 2499–2513. 10.1182/blood.2020010543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Read JA, Rouce RH, Mo F, Mamonkin M, and King KY (2022). Apoptosis of HSCs contributes to bone marrow suppression following CAR-T therapy. Transplant Cell Ther. 10.1016/j.jtct.2022.12.020. [DOI] [PMC free article] [PubMed]
- 86.Wang L, Hong R, Zhou L, Ni F, Zhang M, Zhao H, Wu W, Wang Y, Ding S, Chang AH, et al. (2021). New-Onset Severe Cytopenia After CAR-T Cell Therapy: Analysis of 76 Patients With Relapsed or Refractory Acute Lymphoblastic Leukemia. Front Oncol 11, 702644. 10.3389/fonc.2021.702644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fried S, Avigdor A, Bielorai B, Meir A, Besser MJ, Schachter J, Shimoni A, Nagler A, Toren A, and Jacoby E. (2019). Early and late hematologic toxicity following CD19 CAR-T cells. Bone Marrow Transplant 54, 1643–1650. 10.1038/s41409-019-0487-3. [DOI] [PubMed] [Google Scholar]
- 88.Rejeski K, Subklewe M, Aljurf M, Bachy E, Balduzzi AC, Barba P, Bruno B, Benjamin R, Carrabba MG, Chabannon C, et al. (2023). Immune Effector Cell-Associated Hematotoxicity (ICAHT): EHA/EBMT Consensus Grading and Best Practice Recommendations. Blood. 10.1182/blood.2023020578. [DOI] [PubMed]
- 89.Bailey SR, Vatsa S, Larson RC, Bouffard AA, Scarfo I, Kann MC, Berger TR, Leick MB, Wehrli M, Schmidts A, et al. (2022). Blockade or Deletion of IFNgamma Reduces Macrophage Activation without Compromising CAR T-cell Function in Hematologic Malignancies. Blood Cancer Discov 3, 136–153. 10.1158/2643-3230.BCD-21-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manni S, Del Bufalo F, Merli P, Silvestris DA, Guercio M, Caruso S, Reddel S, Iaffaldano L, Pezzella M, Di Cecca S, et al. (2023). Neutralizing IFNgamma improves safety without compromising efficacy of CAR-T cell therapy in B-cell malignancies. Nat Commun 14, 3423. 10.1038/s41467-023-38723-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mattar M, McCarthy CR, Kulick AR, Qeriqi B, Guzman S, and de Stanchina E. (2018). Establishing and Maintaining an Extensive Library of Patient-Derived Xenograft Models. Front Oncol 8, 19. 10.3389/fonc.2018.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bak RO, Dever DP, and Porteus MH (2018). CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nat Protoc 13, 358–376. 10.1038/nprot.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Ravin SS, Li L, Wu X, Choi U, Allen C, Koontz S, Lee J, Theobald-Whiting N, Chu J, Garofalo M, et al. (2017). CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci Transl Med 9. 10.1126/scitranslmed.aah3480. [DOI] [PubMed] [Google Scholar]
- 94.Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, and Sadelain M. (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, et al. (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33, 985–989. 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Riviere I, Brose K, and Mulligan RC (1995). Effects of retroviral vector design on expression of human adenosine deaminase in murine bone marrow transplant recipients engrafted with genetically modified cells. Proc Natl Acad Sci U S A 92, 6733–6737. 10.1073/pnas.92.15.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.June CH, and Sadelain M. (2018). Chimeric Antigen Receptor Therapy. N Engl J Med 379, 64–73. 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kwakkenbos MJ, Chang GW, Lin HH, Pouwels W, de Jong EC, van Lier RA, Gordon S, and Hamann J. (2002). The human EGF-TM7 family member EMR2 is a heterodimeric receptor expressed on myeloid cells. J Leukoc Biol 71, 854–862. [PubMed] [Google Scholar]
- 99.Brentjens RJ, Latouche JB, Santos E, Marti F, Gong MC, Lyddane C, King PD, Larson S, Weiss M, Riviere I, and Sadelain M. (2003). Eradication of systemic B-cell tumors by genetically targeted human T lymphocytes co-stimulated by CD80 and interleukin-15. Nat Med 9, 279–286. 10.1038/nm827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report standardized datatypes or original code.