Abstract
The risk of developing Alzheimer’s disease (AD) significantly increases in individuals carrying the APOEε4 allele. Elderly cognitively healthy individuals with APOEε4 also exist, suggesting the presence of cellular mechanisms that counteract the pathological effects of APOEε4; however, these mechanisms are unknown. We hypothesized that APOEε4 carriers without dementia might carry genetic variations that could protect them from developing APOEε4-mediated AD pathology. To test this, we leveraged whole-genome sequencing (WGS) data in the National Institute on Aging Alzheimer's Disease Family Based Study (NIA-AD FBS), Washington Heights/Inwood Columbia Aging Project (WHICAP), and Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA) cohorts and identified potentially protective variants segregating exclusively among unaffected APOEε4 carriers. In homozygous unaffected carriers above 70 years old, we identified 510 rare coding variants. Pathway analysis of the genes harboring these variants showed significant enrichment in extracellular matrix (ECM)-related processes, suggesting protective effects of functional modifications in ECM proteins. We prioritized two genes that were highly represented in the ECM-related gene ontology terms, (FN1) and collagen type VI alpha 2 chain (COL6A2) and are known to be expressed at the blood–brain barrier (BBB), for postmortem validation and in vivo functional studies. An independent analysis in a large cohort of 7185 APOEε4 homozygous carriers found that rs140926439 variant in FN1 was protective of AD (OR = 0.29; 95% CI [0.11, 0.78], P = 0.014) and delayed age at onset of disease by 3.37 years (95% CI [0.42, 6.32], P = 0.025). The FN1 and COL6A2 protein levels were increased at the BBB in APOEε4 carriers with AD. Brain expression of cognitively unaffected homozygous APOEε4 carriers had significantly lower FN1 deposition and less reactive gliosis compared to homozygous APOEε4 carriers with AD, suggesting that FN1 might be a downstream driver of APOEε4-mediated AD-related pathology and cognitive decline. To validate our findings, we used zebrafish models with loss-of-function (LOF) mutations in fn1b—the ortholog for human FN1. We found that fibronectin LOF reduced gliosis, enhanced gliovascular remodeling, and potentiated the microglial response, suggesting that pathological accumulation of FN1 could impair toxic protein clearance, which is ameliorated with FN1 LOF. Our study suggests that vascular deposition of FN1 is related to the pathogenicity of APOEε4, and LOF variants in FN1 may reduce APOEε4-related AD risk, providing novel clues to potential therapeutic interventions targeting the ECM to mitigate AD risk.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00401-024-02721-1.
Introduction
Alzheimer’s disease (AD) is typically characterized clinically by progressive memory impairment and decline in other cognitive domains; however, there is a long pre-symptomatic period without clinical manifestations [74]. At death, pathological hallmarks in the brain include extracellular β-amyloid protein in diffuse and neuritic plaques and neurofibrillary tangles made of hyper-phosphorylated tau protein. AD, a progressive neurodegenerative disorder, is currently unpreventable, and, with available drugs only marginally affecting disease severity and progression, remains effectively untreatable. A critical barrier to lessening the impact of late-onset AD (LOAD) is the slow development of drugs that prevent or treat AD due, in part, to an incomplete characterization of the basic pathologic mechanisms. Determining which genes and gene networks contribute to AD could reveal the biological pathways for drug development and inform the development of genetic testing methods for identifying those at greatest risk for AD.
The presence of the APOEε4 allele is among the most prominent genetic risk factors for AD in White, non-Hispanic populations [21], but the associated risks observed in African-Americans and Hispanics are somewhat lower [82]. Relative risk of AD associated with a single copy of APOEε4 is 2.5- to 3.5-fold in Caucasians compared to 1.0–2.4 and 1–1.9 in African-Americans and Hispanics, respectively [9, 82]. However, in every population, homozygosity for the APOEε4 allele is associated with increased risk and nearly complete penetrance [7, 64, 91]. APOE, a critical player in lipid metabolism and transport, has been extensively studied for its role in Alzheimer's disease (AD) and other neurodegenerative disorders [14, 55, 56]. The APOEε4 allele is a well-established risk factor for late-onset AD, with carriers of this allele exhibiting an increased susceptibility to cognitive decline and dementia and earlier age at onset of clinical symptoms. However, within the population of APOEε4 carriers, there is variability in age of onset and severity of AD symptoms. Some "resilient" or "cognitively normal, unaffected" individuals who carry the ε4 allele do not develop AD or experience a delayed onset of symptoms. Several potential factors might contribute to the variability in AD risk and presentation among APOEε4 carriers. Genetic modifier mutations outside of the APOE gene might interact with APOEε4 to influence the risk of AD. APOEε4 carriers might also be influenced by other risk factors for AD, such as vascular health, inflammation, and metabolic conditions. Interactions between APOEε4 and these factors could modify the course of the disease. Certain rare protective variants in other genes could offset the risk posed by APOEε4.
Amid the well-documented association between APOEε4 and AD risk, a growing body of evidence suggests intriguing nuances in the effects of this allele, particularly in certain subsets of individuals who defy the expected trajectory of cognitive decline and remain remarkably resilient to neurodegenerative diseases. Notably, heterozygosity of APOEε4 has incomplete penetrance [31], and the polygenic risk of the rest of the genome could stratify APOEε4 carriers into high- and low-risk strata. In this study, we aimed to identify putative protective mechanisms, influenced by genetic modifiers that might counteract the detrimental effects of the APOEε4 allele. We sought to identify “protective” genetic factors that can modify or reduce the effect of APOEε4 on AD risk and to identify new pathogenic mechanisms, proteins, and pathways that inform development of therapeutic targets and diagnostics.
Results
Whole-genome sequencing identifies putative protective variants in cognitively unaffected elderly APOEε4 carriers
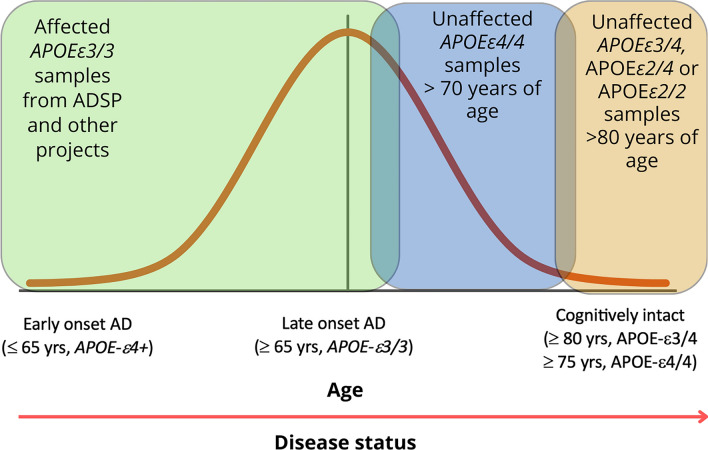
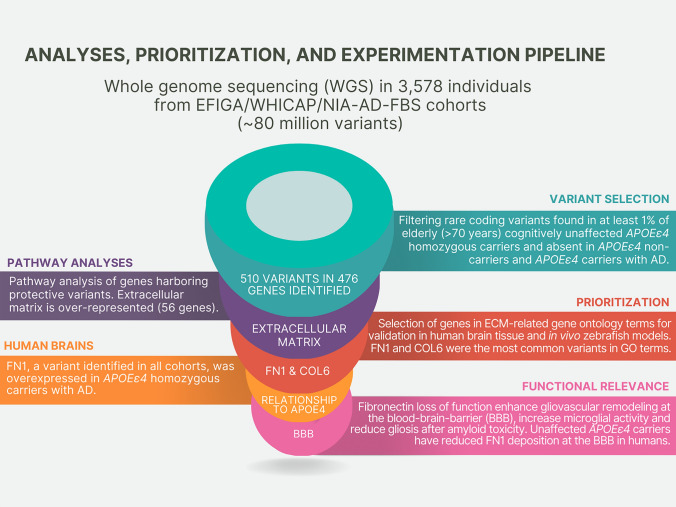
We accessed whole-genome sequencing data in 3,578 individuals from over 700 non-Hispanic White and Caribbean Hispanic families multiply affected by AD (Table 1). After harmonization and QC of the WGS data, we identified rare (MAF < 1% in gnomAD) coding variants in the healthy elderly APOEε4 homozygous (over the age of 70) and heterozygous (over the age of 80) carriers that were absent in non-carriers (Fig. 1). We further prioritized exon coding variants in healthy APOEε4 carriers that bear the potential to be damaging to the resulting protein product. Supplementary Tables 1–3 provide the lists of candidate variants that were identified in cognitively unaffected elderly APOEε4 carriers. Our strategy and analysis pipeline are summarized in Fig. 2. We found 510 variants in 476 genes that were present in at least 1% of APOEε4 unaffected homozygous carriers (388 in EFIGA/WHICAP and 130 in NIA-AD FBS and 8 variants found in both datasets) (Supplementary Table 1 and 2). Two variants (rs116558455 and rs140926439) in the FN1 gene (fibronectin-1) were found in healthy elderly ε4 homozygous carriers in EFIGA/WHICAP and NIA-AD FBS cohorts with MAF = 1.85% and 3.33%, respectively (Table 2). In Hispanics, rs116558455 was absent in all APOEε4 carriers with AD. In non-Hispanic Whites rs140926439 was absent in homozygous APOEε4 AD patients, but found in 1% of heterozygous patients. Pathway analysis of the genes harboring variants segregating in APOEε4 carriers identified several biological pathways and molecular functions such as “actin binding”, “microtubule binding”, and “extracellular matrix structural constituent” (Fig. 3). These results suggested a strong correlation with cellular morphologies and the architectural organization of those cells.
Table 1.
Demographics of samples sequenced
| Hispanics | Non-Hispanic White | |
|---|---|---|
| N | 1840 | 590 |
| AD cases | 693 | 455 |
| AD controls | 1147 | 135 |
| Families with 2 or more individuals | ||
| # APOEε4 heterozygotes | 724 | 438 |
| # APOEε4 homozygotes | 189 | 190 |
| # APOEε4 heterozygote AD cases | 442 | 265 |
| # APOEε4 homozygote AD Cases | 114 | 155 |
| # APOEε4 heterozygote healthy controls | 282 | 161 |
| # APOEε4 homozygote healthy controls | 75 | 30 |
| # APOEε4 homozygote healthy controls > = 70 years of age | 27 | 15 |
| # APOEε4 heterozygote healthy controls > 80 years of age | 75 | 45 |
Fig. 1.
Study design. Comparison of the genomes of elderly APOEε4 carriers with non-carriers
Fig. 2.
Schematic analytical pipeline for this study
Table 2.
FN1 minor allele frequencies
| Cohorts | SNP | MAF in elderly* cognitively unaffected APOEε4 homozygotes (%) | MAF in all cognitively unaffected APOEε4 homozygotes (%) | MAF in APOEε4 homozygous AD patients (%) | MAF in cognitively unaffected elderly* APOEε4 heterozygotes (%) | MAF in all healthy APOEε4 heterozygotes (%) | MAF in APOEε4 heterozygous AD patients (%) |
|---|---|---|---|---|---|---|---|
| EFIGA and WHICAP | rs116558455 | 1.85 | 0.67 | 0.00 | 0.67 | 0.18 | 0.00 |
| NIA-AD FBS | rs140926439 | 3.33 | 5.17 | 0.00 | 2.22 | 1.55 | 0.96 |
*Elderly APOEε4 homozygous are over 70 years old and heterozygous are over 80 years old
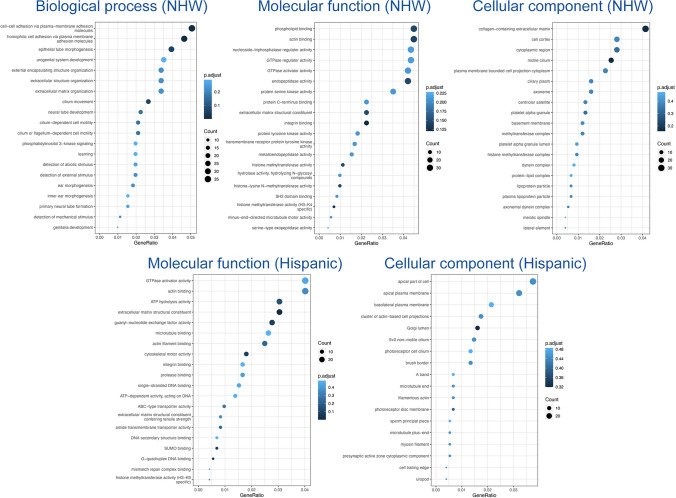
Fig. 3.
Pathway analysis of variants segregating in APOEε4 carriers
Potential protective alleles against APOEε4 enrich extracellular matrix components
To determine the molecular mechanisms enriched in the protective alleles that we identified, gene ontology review was performed with term analyses for biological processes, cellular compartments, and molecular functions (Fig. 3). We found a strong enrichment for extracellular matrix (ECM)-related processes such as cell adhesion, ECM organization, integrin binding, and structural component of the ECM (Fig. 3). This suggested that functional alterations in the ECM composition could act as a protective mechanism in APOEε4 carriers, both heterozygotes and homozygotes without dementia. We hypothesized that APOEε4-related increase in ECM components could be counteracted by loss-of-function (LOF) variants in those genes, leading to protection through rescue of pathological mechanisms that those ECM components partake.
To test our hypothesis, we selected two genes from the variant lists that were common in ECM-related gene ontology classes (Fig. 3), collagen type VI alpha 2 chain (COL6A2) and fibronectin 1 (FN1). These genes are well-known ECM components that harbor putatively protective variants in APOEε4 cognitively unaffected carriers. Additionally, prioritized variants in FN1 were, respectively, present in both Hispanic and non-Hispanic White cohorts (Supplementary Table 1, Supplementary Table 2). COL6A2 variation (rs777822883) generates a substitution of arginine at the 862nd residue to tryptophan, while FN1 variation (rs140926439) converts the glycine at the 357th position to glutamic acid. Since both alterations result in change in charged residues (loss in COL6A2, gain in FN1), we hypothesized that these variations could have detrimental effect on the protein function, as charged interactions are essential for matric proteins and their stability [20, 61, 94]. Therefore, we analyzed the AlphaFold structures of these proteins in Ensembl (http://www.ensembl.org) and found that both variations are potentially detrimental according to SIFT, REVEL, and MetaL R predictions (Supplementary Fig. 1). Arginine in COL6A2 at the 862nd position may coordinate with valine 859 and glutamic acid 858 in the alpha helix structure, while glycine at the 357th position in FN1 may provide structural stability by coordinating with glutamic acid 358 and serine 355 (Supplementary Fig. 1). Therefore, we categorized these variants as likely loss-of-function alleles based on loss of electrostatic interactions.
FN1 variant is protective in an independent cohort of APOEε4 carriers
We queried an independent collection of AD-related genetic cohorts from several sources: the Alzheimer’s Disease Genetic Consortium (ADGC), the Alzheimer’s Disease Sequencing Project (ADSP), and United Kingdom Biobank (UKB) resources, primarily consisting of non-Hispanic White individuals of European ancestry (EU) (Supplementary Table 4 and 5). A total of 465,669 NHW case–control individuals ages 60 and above were available after genetic and phenotypic quality control. Since rs116558455 is very rare in EU (gnomAD non-Finnish EU allele frequency = 0.016%), we focused on rs140926439 which is less rare in EU and thus provides sufficient allele counts to enable replication analyses (gnomAD non-Finnish EU allele frequency = 0.46%). We specifically focused analyses on APOEε4/4 carriers ages 60 and above (N = 7185), to interrogate whether there were any elevated frequencies for the rs140926439 minor allele (T) associated with reduced AD risk and delayed age at onset.
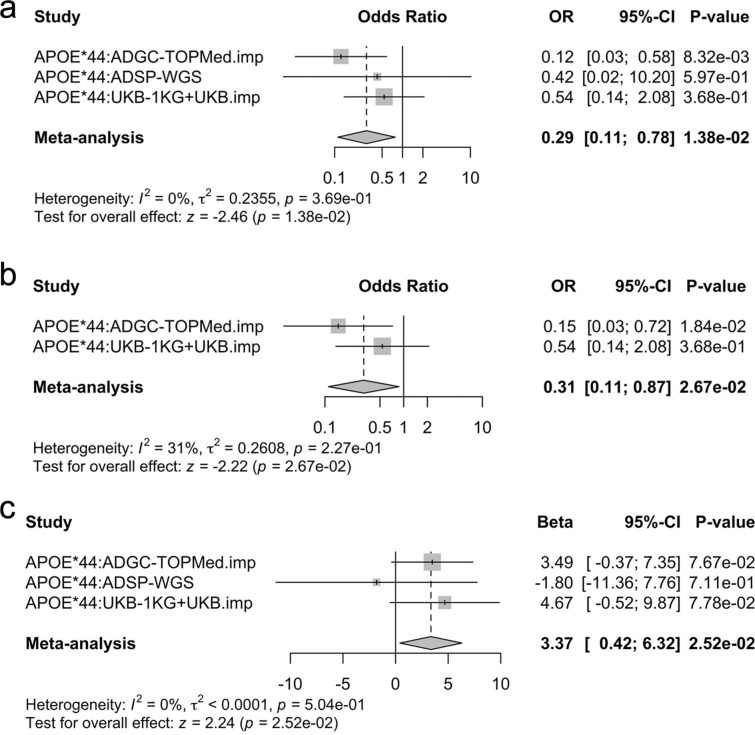
The variant rs140926439 was associated with strongly reduced risk of AD in APOEε4/4 carriers (OR = 0.29; 95% CI [0.11, 0.78], P = 0.014; Table 3, Fig. 4a). Sensitivity analyses were conducted, which ensured that any overlapping samples with our discovery were excluded and corroborated primary findings (OR = 0.31; 95% CI [0.11, 0.87], P = 0.027; Supplementary Table 6, Fig. 4b). Secondary age at onset analyses further showed a significant protective effect in APOEε4/4 carriers, delaying age at onset by 3.4 years for a single copy of the minor allele (beta = 3.37; 95% CI [0.42, 6.32], P = 0.025; Fig. 4c). These analyses represent the largest-to-date genetic association tests in a sample of APOEε4/4 carriers at an age range relevant to AD.
Table 3.
Replication of FN1 variant in the ADSP and UKBB cohorts
| Cohort | Total no. | Total, EAF (%) | CN, carrier no./total no. (%) | AD, carrier no./total no. (%) | CN–AD, EAF (%) | OR [95% CI] | P-value | CN, carrier age, mean (SD) | AD, carrier age, mean (SD) |
|---|---|---|---|---|---|---|---|---|---|
| ADGC | 1510 | 0.43 | 4/129 (3.10%) | 9/1381 (0.65%) | 1.55–0.33% | 0.12 [0.03, 0.58] | 8.3E−03 | 78.8 (11.1) | 72.9 (7.5) |
| ADSP | 356 | 0.42 | 1/81 (1.23%) | 2/275 (0.73%) | 0.62–0.36% | 0.42 [0.02, 10.2] | 0.60 | 74.0 (–) | 65.5 (7.8) |
| UKB | 5319 | 0.54 | 54/4804 (1.12%) | 3/515 (0.58%) | 0.56–0.29% | 0.54 [0.14, 2.08] | 0.37 | 65.2 (4.4) | 78.7 (2.9) |
| Combined | 7185 | 0.51 | 59/5014 (1.18%) | 14/2171 (0.64%) | 0.59–0.32% | 0.29 [0.11, 0.78] | 0.014 | 66.3 (5.5) | 73.1 (6.6) |
Fig. 4.
Replication analyses. a Forest plot showing the association of rs140926439 with Alzheimer’s disease risk in APOEε4/4 carriers. Significance was considered at P < 0.05. Results across the datasets were combined using fixed-effects inverse-variance weighted meta-analysis. Cochran’s Q test indicated no significant heterogeneity. OR, odds ratio; CI, confidence interval. b Case–control regression sensitivity analyses for rs140926439 in APOEε4/4 carriers. To ensure an independent replication of discovery findings, in ADGC, samples from NIA-AD FBS cohort were excluded and ADSP whole-genome sequencing data was fully excluded. Results across the datasets were combined using fixed-effects inverse-variance weighted meta-analysis. Cochran’s Q test indicated no significant heterogeneity. c Age at onset analyses for rs140926439 in APOEε4/4 carriers. The large confidence intervals in ADSP whole-genome sequencing (WGS) individuals reflect that there were only two case carriers and one of those had an age at onset at 60 years (an outlier compared to other case carriers). Results across the datasets were combined using fixed-effects inverse-variance weighted meta-analysis. Cochran’s Q test indicated no significant heterogeneity
FN1 deposition correlates with APOEε4 dosage
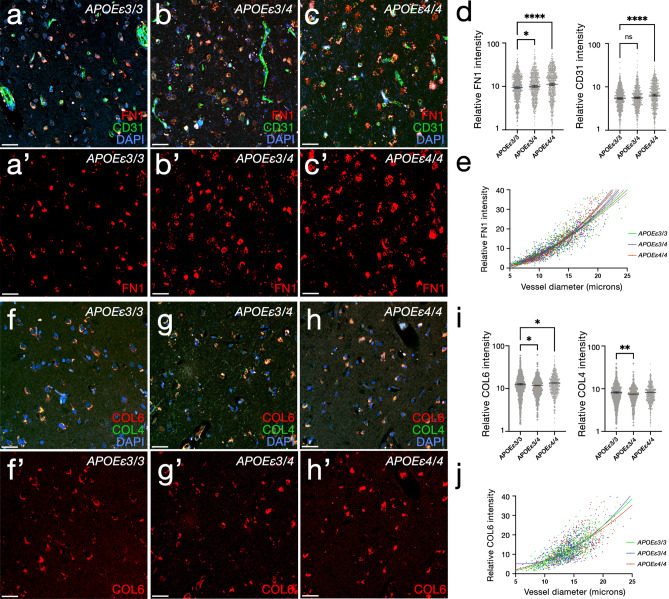
Based on our findings, we hypothesized that APOEε4 dosage might correlate with deposition of COL6A2 and FN1, at the blood–brain barrier (BBB) basement membrane, one of their prominent expression locations, as FN1 is an important signaling molecule that interacts with specific integrins [69] expressed in various vascular niche cell types [52]. We immunostained and analyzed the brains of 27 individuals with known APOE genotypes (8 APOEε4/4 homozygous carriers with AD, 8 APOEε3/4 heterozygote carriers with AD, and 11 APOEε4 non-carriers (APOEε3/3) with AD (Supplementary Table 7) for FN1 and CD31 (endothelial cell marker), and COL6A2 and COL4 (a vascular basement membrane marker) (Fig. 5, Supplementary Dataset 1, Supplementary Dataset 2). We found that FN1 levels (Fig. 5a–c′) significantly increased with APOEε4 dosage (Fig. 5d, Supplementary Fig. 2). Compared to APOEε3/3 individuals, FN1 expression increased significantly in APOEε3/4 (8.1%, P = 3.4e−02) and in APOEε4 homozygous individuals (26.6%, P = 3.1e−09). Least squares linear regression and non-linear fit comparison of FN1 intensities according to the diameter of the vessels showed that compared to APOEε3/3, FN1 expression is more prominent with increasing vessel size in APOEε3/4 and APOEε4/4 individuals (adjusted R2: APOEε3/3: 0.81, APOEε3/4: 0.86, APOEε4/4: 0.89; all P values are less than 1.0xE-15 for non-zero significance of the slopes) (Fig. 5e). Immunostainings for COL6 (Fig. 5f–h′) showed a non-linear relationship between APOEε4 dosage and COL6 expression. APOEε4 heterozygotes showed reduced (7.7%, P = 9.9e−03) homozygotes indicating increased levels of COL6 (6.7%, P = 3.4e−02) (Fig. 5i). COL4 expression is only reduced in APOEε4 heterozygotes (8.6%, P = 1.4e−03), but remain unchanged in homozygotes (Fig. 5i). The changes in COL6 expression with blood vessel size was less pronounced (adjusted R2: APOEε3/3: 0.67, APOEε3/4: 0.50, APOEε4/4: 0.55; all P values are less than 1.0 × E−15 for non-zero significance of the slopes) (Fig. 5j).
Fig. 5.
Changes in FN1 and COL6A2 according to APOE genotype. a–c′ Double IFS for CD31 (green) and FN1 (red) with DAPI nuclear counterstain in APOEε3/3 (a, a′), APOEε3/4 (b, b′) and APOEε4/4 (c, c′). d FN1 and CD31 intensity comparisons in 2,044 blood vessels from 28 individuals. e Regression model for FN1 intensity with respect to blood vessel diameter in three APOE genotypes. f–h′ Double IFS for COL4 (green) and COL6A2 (red) with DAPI nuclear counterstain in APOEε3/3 (f, f′), APOEε3/4 (g, g′) and APOEε4/4 (h, h′). i COL4 and COL6A2 intensity comparisons in 1,816 blood vessels from 28 individuals. j Regression model for COL6A2 intensity with respect to blood vessel diameter in three APOE genotypes. Scale bars equal 100 μm
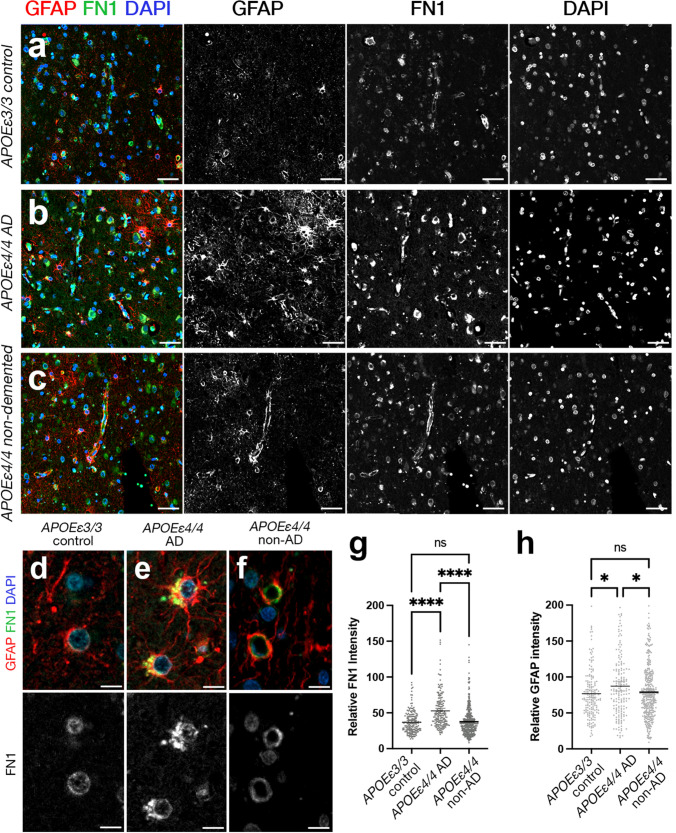
FN1 deposition is different between demented and cognitively unaffected APOEε4/4 carriers
Based on our findings that FN1 deposition is increased in patients with AD and APOE dosage correlates with FN1 levels, we hypothesized that FN1 deposition could be a downstream driver of the pathological effects of APOEε4 in AD. We tested this hypothesis by comparing FN1 and GFAP (marker for reactive gliosis) levels in APOEε3/3 (control, n = 2), APOEε4/4 AD (n = 2), and APOEε4/4 unaffected (n = 6) individuals (Fig. 6, Supplementary Table 8, Supplementary Dataset 3). We found elevated reactive gliosis and FN1 deposition in APOEε4/4 carriers with AD compared to APOEε3/3 controls (ANOVA adjusted P = 1.5E−02 for GFAP intensity, 4.1E-11 for FN1 intensity) (Fig. 6). APOEε4/4 unaffected carriers had FN1 and GFAP levels that were similar to that in controls (ANOVA adjusted P = 0.5245 for GFAP intensity, P = 0.8884 for FN1 intensity) (Fig. 6, Supplementary Fig. 3). This implies that the non-demented APOEε4 carriers are protected from gliosis and FN1 deposition (Fig. 6g, h).
Fig. 6.
FN1 deposition and gliosis reduce to control levels in APOEε4/4 cognitively unaffected individuals, but not in APOEε4/4 AD patients. a–c Double IFS for FN1 (green) and GFAP (red) with DAPI nuclear counterstain in APOEε3/3 (a), APOEε4/4 AD (b), and APOEε4/4 cognitively unaffected individuals (c). Black–white images are individual fluorescent channels for FN1, GFAP, and DAPI. d–f Two blood vessels in every condition are shown in high magnification together with FN1 channel alone. g FN1 intensity comparisons (2 APOEε3/3 individuals without AD, 2 APOEε4/4 individuals with AD, and 6 APOEε4/4 individuals without AD). h GFAP intensity comparisons. Scale bars equal 50 μm (a-c) and 10 μm (d-f)
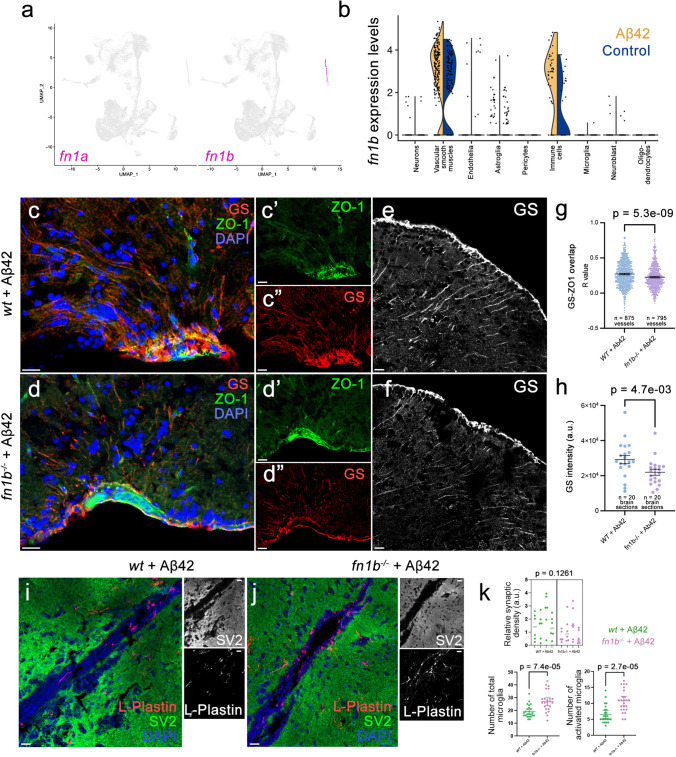
Fibronectin loss-of-function zebrafish model enhances gliovascular endfeet retraction and microglial activity while reducing gliosis after amyloid toxicity
To determine whether fibronectin activity is related to cellular responses after amyloid toxicity, we used our established amyloid toxicity model in the adult zebrafish brain [11, 13, 22, 42, 47, 72]. Zebrafish has two fibronectin 1 genes: fn1a and fn1b [78]. Our single-cell transcriptomics analyses in the zebrafish brain showed that fn1b, but not fn1a is expressed in the zebrafish forebrain (Fig. 7a). fn1b expression is predominantly detected in vascular smooth muscle cells and immune cells, while endothelia and astroglia express fn1b at considerably lower levels (Fig. 7b). Amyloid toxicity results in increased fn1b expression in immune cells and vascular smooth muscle cells (Fig. 7b), similar to what we observed in AD brains (Figs. 5, 6). To determine the effects of fibronectin function in amyloid-induced pathology, we used an fn1b full knockout zebrafish line (fn1b−/−), which was previously functionally validated [33]. After treating wild-type and fn1b−/− animals with Aβ42, we performed immunohistochemical stainings for astroglia (red, GS) and tight junctions that mark vascular structures (green, ZO-1) (Fig. 7c–f, Supplementary Dataset 4). Compared to wild-type animals treated with Aβ42, fn1b−/− animals with Aβ42 showed less colocalization of GS and ZO-1 (− 16.3%, P = 5.3E−09), suggesting that gliovascular interactions were reduced with fibronectin loss of function (LOF) (Fig. 7g). Based on our previous findings that reduced gliovascular contact upon amyloid toxicity is a protective mechanism through enhancing clearance of toxic protein aggregates and immune systems activity [47], our results suggest that fibronectin could negatively regulate amyloid beta clearance and therefore an LOF variant could be protective against disease pathology. By performing intensity measurements for astroglia with GS immunoreactivity, we observed that GS intensity reduces with fn1b LOF (− 24.7%, P = 4.7E−03; Fig. 7h, Supplementary Dataset 5), indicative of reduced gliotic response upon Aβ42. To determine the effect of fibronectin on synaptic density and the number and activation state of microglia, we performed immunostainings (Fig. 7i, j, Supplementary Dataset 6) and found that loss of fibronectin leads to increased numbers of total (41.5%, P = 8.7E−04) and activated microglia (64.3%, P = 2.9E−04). We did not observe change in the synaptic density when Aβ42-treated fn1b−/− was compared to Aβ42-treated wild-type animals (Fig. 7i–k, Supplementary Dataset 7).
Fig. 7.
Fibronectin loss of function affects gliovascular interactions, gliosis, and microglial activity after amyloid toxicity in zebrafish brain. a Feature plots for fibronectin 1a (fn1a) and fibronectin 1b (fn1b) genes in zebrafish brain. b Violin plots in control and Aβ42-treated brains. fn1b is mainly expressed in vascular smooth muscle cells and immune cells and is upregulated with Aβ42. c, d Double IF for astroglia marker glutamine synthase (GS, red) and tight junction marker (ZO-1, green) in wild-type and fn1b−/− animals. Individual fluorescent channels in c′, c′′, d′, and d′′. e, f Individual GS channels. g Quantification for colocalization of ZO-1 and GS. h Comparison of intensity measurements for GS. i, j Double IF for synaptic marker SV2 (green) and microglial marker l-Plastin (red) in wild-type and fn1b−/− animals treated with Aβ42. Individual fluorescent channels in black–white channel. k Quantifications for synaptic density, total number of microglia, and activated microglia. Scale bars equal 25 µm
Discussion
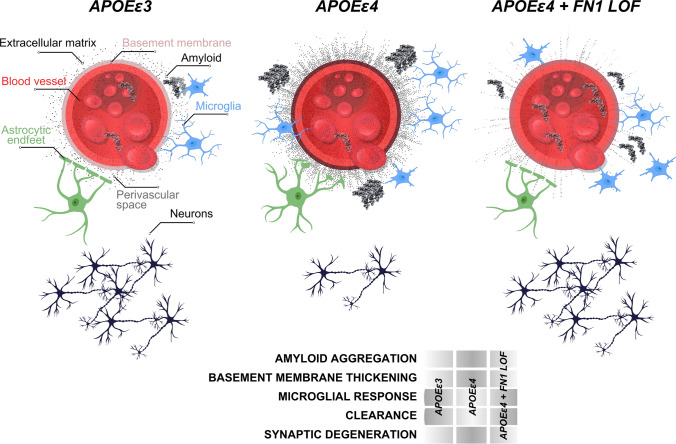
In our study, we found that two missense, potential loss-of-function (LOF) variants in FN1 may protect against APOEε4-mediated AD pathology. We base our conclusions on four main observations: (1) FN1 coding variants were present in cognitively unaffected APOEε4 homozygous carriers, but not in affected carriers with clinically diagnosed AD (Supplementary Table 1), and the protective effect was independently replicated is a large cohort of APOEε4 homozygous carriers. (2) Deposition of FN1 at the BBB basement membrane increases with APOEε4 dosage (Fig. 5). (3) Unaffected/resilient homozygous APOEε4 carriers above the age of 70 without AD have FN1 deposition levels similar to APOEε3 control individuals (Fig. 6). (4) In the zebrafish brain, knockout of fn1b alleviates amyloid toxicity-related pathological changes (Fig. 7). These results suggest that the basement membrane thickening and remodeled ECM composition in the BBB may be a pathological contribution to APOEε4-mediated AD pathology that may be mitigated by variants in FN1 or other ECM genes (Fig. 8). This conclusion is supported by the presence of variants in other BBB-related ECM components, such as LAMA1, LAMA3, and HSPG2, in unaffected elderly APOEε4 carriers but not in carriers with AD (Supplementary Table 1). Therefore, our findings propose a new direction for potential therapeutic interventions reducing the impact of APOEε4-mediated risk of AD by targeting the BBB basement membrane. Thus, we propose that fibronectin loss of function may be a protective mechanism for AD (Fig. 8).
Fig. 8.
Schematic abstract for the protective effect of FN1 variants
APOEε4 has been associated with increased neuroinflammation and neurodegeneration, which can accelerate the progression of AD [63]. Our results in zebrafish fn1b knockout model showed that reduced fibronectin 1 increased the gliovascular (GV) endfeet retraction and reduced gliosis. We previously showed that the relaxed GV contact was a beneficial response to amyloid toxicity [47] as it helps enhance the clearance of toxic aggregates through the bloodstream. Additionally, gliosis is an immediate response in astroglia to insult and prevents functional restoration of neuronal activity in disease [16, 29, 73, 93]. Independent reports showed that astrocytic removal of APOE protects against vascular pathology [89], and gliosis is a mediator of amyloid-dependent tauopathy in late AD [6]. We propose that the relationship of fibronectin with these processes is pathogenic, and reduced fibronectin could be protective by allowing more efficient clearance through the bloodstream and reduced astrogliosis. The enhanced microglial activity supports this hypothesis, as acute activation of microglia is a beneficial response to toxic protein aggregation [36, 62].
Our results are consistent with the previous findings on APOE-dependent vascular pathologies and their relationship to AD [38, 45, 46, 56, 67, 79]. Endothelial fibronectin induces disintegration of endothelial integrity and leads to atherosclerotic vascular pathologies [1, 18, 95], supporting our findings that reduced fibronectin 1 protects the blood–brain barrier integrity disrupted by APOEε4. Our findings are coherent with the previous observations, where AD-related changes in collagen and fibronectin around the blood–brain barrier (BBB) and alterations in the BBB's structure and function were documented [43, 80, 92]. Additionally, the serum levels of fibronectin increase in AD patients in comparison to healthy individuals [15]. Collagen and fibronectin can also be early pathological markers of AD [48], where the increase in the deposition and cross-linking of basement membrane around the cerebral blood vessels lead to a thickening of the basement membrane, potentially compromising its permeability and function [35, 67, 83, 84, 90]. Fibronectin expression levels in brain vasculature increases in AD [22, 41, 49, 79, 88], where remodeling of the BM and replacing ECM with FN1 have been suggested to indicate hypoperfusion and atherosclerosis-prone state [1, 46, 54]. Additionally, APOEε4 might regulate BM remodeling through inhibition of pericyte-mediated matrix proteinase expression [55]. Pericyte degeneration, mural cell dysfunction, and alterations in cerebrospinal flow dynamics are long-term consequences of vascular pathologies in aging and AD and is accelerated with APOEε4 [5, 25, 27, 34, 38, 66]. Therefore, based on our findings, we propose that excess ECM deposition and BM thickening with collagen and fibronectin could promote the blood–brain barrier breakdown. Potential loss-of-function variants in ECM genes are likely to render ECM components non-functional, thus protecting against AD progression. Stronger instructive interactions of collagen and fibronectin with their receptors on various BBB cell types in AD [39, 59, 79, 88] support this hypothesis. Consistently, FN1 provides attachment surface for immune cells, which—when becomes chronic—damages the vascular functions, contribute to BBB breakdown, and loss of synaptic integrity.
We found that despite their APOEε4/4 status, unaffected/resilient individuals who do not develop cognitive decline have lower FN1 deposition and gliosis at the vascular basement membrane that are not different from APOEε3/3 control individuals, but significantly lower than those in APOEε4/4 AD patients (Fig. 6). This demonstrated that FN1 is a critical component of APOEε4-mediated development of AD, and a yet unknown protective mechanism against the effects of APOEε4/4 genotype suppresses FN1 deposition. We propose that FN1 is a critical downstream effector of APOEε4 and reduced FN1 levels, either through rare, protective genetic variations in FN1 or through other resilience mechanisms, promoting protection against AD. An interesting future research could investigate the other rare protective variants of APOE such as APOEε2 [28, 31] and APOEε3 Christchurch [70] and their effects on the BBB basement membrane.
The strength of this study is the cross-species design with pathological and functional validation to show that ECM component fibronectin could be related to key pathological aspects of AD such as toxic protein clearance, blood–brain barrier integrity, and microglial activity. We present the first knockout zebrafish for fibronectin 1 in relation to amyloid toxicity and identified cellular changes that relate to fibronectin activity.
Further studies could address some limitations of our study. First, the mechanism by which APOEε4 enhances FN1 requires further investigations. Although in human and zebrafish brains, fibronectin is upregulated, the longitudinal relationship of amyloid aggregation to FN1 activity needs to be analyzed. Additionally, our genetic studies were conducted in clinically assessed individuals, and given the rarity of the FN1 mutation, we did not have neuropathological assessments of APOEε4/4 individuals with this rare protective mutation. Future studies in large-scale neuropathologic cohorts are necessary to demonstrate the pathological consequences of the rare FN1 variants. Finally, mechanistic studies of FN1 with and without the rare mutation are necessary to demonstrate the nuanced functional consequences.
Materials and methods
Ethics statement
All human samples were de-identified and the researchers could not infer or obtain personal information of the donors. Institutional Review Board approval from Columbia University Irving Medical Center and Mayo Clinic was taken before clinical data generation. Human cohorts and their characteristics are provided below. Animal experiments were carried out in accordance with the animal experimentation permits of the Institutional Animal Care and Use Committee (IACUC) at Columbia University (protocol number AC-AABN3554). Animals were maintained according to the Institutional Animal Care and Use Committee (IACUC) standards of the Institute of Comparative Medicine at the Columbia University Irving Medical Center and the accepted guidelines [2, 30, 44, 77]. The animal care and use program at Columbia University is accredited by the AAALAC International and maintains an Animal Welfare Assurance with the Public Health Service (PHS), Assurance number D16-00003 (A3007-01). Animal experiments were approved by the IACUC at Columbia University (protocol number AC-AABN3554). For zebrafish studies, 8- to 10-month-old wild-type AB strains or fn1b−/− homozygous knockout fish lines of both genders were used. In every experimental set, animals from the same fish clutch were randomly distributed for each experimental condition.
Human cohort information
NIA-AD Family Based Study (NIA-AD FBS): This study recruited multiplex families across the USA. Families were included if at least one member had a diagnosis of definite or probable Alzheimer’s disease [40, 51] with onset after age 60 and a sibling with definite, probable, or possible disease with a similar age at onset. Demographic information, diagnosis, age at onset for patients with Alzheimer’s disease, method of diagnosis, Clinical Dementia Rating Scale [37], and the presence of other relevant health problems were available for each individual. The age at onset for patients was the age at which the family first observed signs of impaired cognition. For unaffected family members, we used their age at the time of their latest examination without impairment. Each recruitment site used standard research criteria for the diagnosis of Alzheimer’s disease [51]. For deceased family members who had undergone autopsy, the results were used to determine the diagnosis. For analyses, clinical Alzheimer’s disease was defined as any individual meeting NINCDS–ADRDA criteria for probable or possible Alzheimer’s disease [51] and definite Alzheimer’s disease when CERAD pathological criteria [53] were met postmortem.
Washington Heights/Inwood Columbia Aging Project (WHICAP): WHICAP is a multiethnic, community-based, prospective cohort study of clinical and genetic risk factors for dementia. Three waves of individuals were recruited in 1992, 1999, and 2009 in WHICAP, all using similar study procedures [32, 60]. Briefly, participants were recruited as representatives of individuals living in the communities of northern Manhattan who were 65 years and older. At the study entry, each person underwent a structured interview of general health and function, followed by a comprehensive assessment including medical, neurological, and psychiatric histories, and standardized physical, neurological, and neuropsychological examinations. Individuals were followed every 18–24 months, repeating examinations that were similar to baseline. All diagnoses were made in a diagnostic consensus conferences attended by a panel consisting of at least one neurologist and one neuropsychologist with expertise in dementia diagnosis, using results from the neuropsychological battery and evidence of impairment in social or occupational function. All-cause dementia which was determined based on Diagnostic and Statistical Manual of Mental Disorders, 4th Edition criteria [4]. Furthermore, we used the criteria from the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer Disease and Related Disorders Association to diagnose probable or possible AD [51].
Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA): We used families from a different ethnic group to identify protective alleles in APOEε4 healthy individuals. This cohort comprises participants from a group of families from the Dominican Republic, Puerto Rico, and New York. Recruitment, study design, adjudication, and clinical assessment of this cohort have been previously described [86] as were details of genome-wide SNP data, quality control, and imputation procedures of the GWAS data [68, 85]. Participants were followed every 2 years and evaluated using a neuropsychological battery [76], a structured medical and neurological examination, and an assessment of depression [65, 75]. The Clinical Dementia Rating Scale (CDR) [57, 58] and functional status were done and the clinical diagnosis of Alzheimer’s disease was based on the NINCDS–ADRDA criteria [10, 50].
Whole-genome sequencing and quality control
The demographics of the individuals selected for sequencing is shown in Table 1. WGS was performed at the New York Genome Center (NYGC) using 1 µg of DNA, an Illumina PCR-free library protocol, and sequencing on the Illumina HiSeq platform. We harmonized the WGS and the EFIGA families (n = 307), and jointly called variants to create a uniform, analysis set. Genomes were sequenced to a mean coverage of 30×. Sequence data analysis was performed using the NYGC automated analysis pipeline which matches the CCDG and TOPMed-recommended best practices [3]. Briefly, sequencing reads were aligned to the human reference, hs38DH, using BWA-MEM v0.7.15. Variant calling was performed using the GATK best practices. Variant filtration was performed using variant quality score recalibration (VQSR at tranche 99.6%) which identified annotation profiles of variants that were likely to be real and assigns a score (VQSLOD) to each variant.
Identification of variants segregating in healthy APOEε4 individuals
First, we filtered high-quality rare (MAF < 0.01 in gnomAD) variants with genotype quality (GQ) ≥ 20 and depth (DP) ≥ 10. We then excluded any variant observed in APOE ε4 non-carriers. Within variants that segregated in APOEε4 carriers, we prioritized those that were observed in at least 1% of APOEε4 homozygous healthy elderly (≥ 70 years) and had additional support in healthy elderly (≥ 80 years) heterozygous carriers. We further prioritized variants that were absent in AD patients carrying an APOEε4 allele. A simplified pipeline is provided in Fig. 2.
Genotyping, amyloid administration, and tissue preparation
A previously generated fn1b knockout line using CRISPR–Cas9 gene editing [33] was used in homozygous form. The full deletion was genotyped as described [33]. Amyloid-β42 was administered to the adult zebrafish brain through cerebroventricular microinjection into the cerebral ventricle [13]. Euthanasia and tissue preparation were performed as per institutional ethics committee approval and international guidelines [13, 44]. 12-µm-thick cryo-sections were prepared from these brain samples using a cryostat and collected onto glass slides which were then stored at − 20 °C.
Replication of FN1 variant
An in-depth overview of the methodology and analyses of replication datasets is provided in the Supplementary Text. The current study followed STREGA reporting guidelines. Participants or their caregivers provided written informed consent in the original studies. The current study protocol was granted an exemption by the Stanford Institutional Review Board because the analyses were carried out on “de-identified, off-the-shelf” data; therefore, additional informed consent was not required. Case–control, family-based, and longitudinal AD genetic cohorts were available through public repositories, with genetic data from high-density single-nucleotide polymorphism microarrays, exome microarrays, whole-exome (WES) and whole-genome sequencing (WGS) (Supplementary Table 4–6). These data pertained to cohorts belonging to the ADGC and the ADSP R3. We additionally used population-based data from the UKB, where we had access to health record information to derive case–control diagnoses [17].
Genetic quality control procedures for the UKB are detailed elsewhere. ADGC and ADSP genetic data underwent extensive quality control, imputation to the TOPMed reference panel (for ADGC array-based samples) [26, 81], and ancestry determination (SNPweights v2.1) [19]. Duplicated individuals were identified and their clinical, diagnostic, and pathological data, as well as age at onset of cognitive symptoms, age at examination for clinical diagnosis, age at last examination, age at death, sex, race, ethnicity, and APOE genotype, were cross-referenced across cohorts. Duplicate entries with irreconcilable phenotypes were excluded. APOE genotypes were adjudicated using state-of-the-art APOE prioritization approaches, filtering out samples where APOE genotypes lacked robustness (prioritizing APOE genotypes from sequencing data and cross-referencing APOE genotypes from high-quality imputation with those provided in study demographics through various protein-based and DNA-based methods) [8].
Finally, in all datasets, samples were filtered to ages 60 years and above, cases or controls, belonging to non-Hispanic White ethnicity and European ancestry, and retaining only a single individual per cryptic relatedness cluster (determined down to third-degree relatedness). Secondary analyses evaluated associations with AD age at onset. Significant discoveries were considered at P < 0.05. All statistical analyses were conducted using R (v.4.2.1).
Immunohistochemistry
Postmortem human brain sections from BA9 prefrontal cortex were obtained from New York Brain Bank at Columbia University and Mayo Clinic Jacksonville as paraffin-embedded blanks and with neuropathology assessments (Supplementary Table 7 and 8). Immunohistochemistry (IHC) was performed as described [47, 87]. As primary antibodies FN1 (Proteintech, catalog number 66042-1-Ig, 1:250), CD31 (Abcam, catalog number ab134168, 1:250), COL6A2 (Thermo Fisher, catalog number PA5-65085, 1:200), COL4 (Thermo Fisher, 14-9871-82, 1:100), and GFAP (Thermo Fisher, catalog number OPA1-06100), and as secondary antibodies goat anti-mouse Alexa Fluor 448 (Thermo Fisher, catalog number A-21131, 1:500) and goat anti-rabbit Alexa Fluor 555 (Thermo Fisher, catalog number A-21137, 1:500) were used. In short, for deparaffinization and hydration, xylene and alcohol were used. Antigen retrieval was performed with citrate buffer (pH: 6.0) or antigen retriever EDTA buffer (pH:8.5) in a pressure cooker or microwave for 18–25 min. Sections were blocked in 10% normal goat serum for 1 h at room temperature and were incubated with primary antibody combinations (FN1-CD31, COL6A2-COL4 or FN1-GFAP) overnight at 4 °C in a humidified chamber. Each secondary antibody to the respective primaries was applied for 2 h at room temperature. Slides were covered by mounting medium with nuclear counterstain DAPI (Thermo Fisher, catalog number D1306, 5 ng/ml). Immunohistochemistry for zebrafish was performed as described [13]. In short, the slides were dried at room temperature for 30 min and washed with PBS with 0.03% Triton X-100 (PBSTx). Primary antibody combinations (ZO-1 + GS and SV2A + l-Plastin) were applied overnight at 4 °C. Next day, after washing three times with PBSTx appropriate secondary antibodies were applied for 2 h at room temperature. The slides were then washed several times before mounting using 70% glycerol in PBS. The following antibodies were used: mouse anti-ZO-1 (1:500, Thermo Fisher Cat. No. 33-9100), rabbit anti-glutamine synthetase (GS) (1:500, Abcam Cat. No. ab176562), mouse anti-SV2A (1:500, DSHB Cat. No. SV2), and rabbit anti-l-plastin (1:3000, gift from Michael Redd), secondary antibodies goat anti-mouse Alexa Fluor 448 (Thermo Fisher, catalog number A-21131, 1:500), and goat anti-rabbit Alexa Fluor 555 (Thermo Fisher, catalog number A-21137, 1:500). For antigen retrieval of ZO-1 and SV2, slides were heated in 10 mM sodium citrate (pH:8.0) at 85 °C for 15 min before primary antibody incubation.
Image acquisition, quantification, and statistical analyses
Five random illumination field images per patient from the immunostained slides were acquired using Zeiss LSM800 confocal microscope equipped with ZEN software (version blue edition, v3.2, Carl Zeiss, Jena, Germany). Based on vascular markers, coronally sectioned blood vessels were delineated with the selection tool of ZEN software. Fluorescence intensity measures, diameter, and area was calculated. Acquisitions were performed in a blinded fashion (sample IDs, neuropathology details, and genotypes were revealed after the acquisition) and no vessels were specifically left out unless their diameters were larger than 50 μm. GraphPad Prism software version 9.2.0. was used for the statistical analyses. For multiple comparisons, one-way Brown–Forsythe and Welch ANOVA test with two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli comparison with individual calculation of variances was used. For non-Gaussian distributions, non-parametric Kruskal–Wallis test with Dunn’s multiple comparison test was performed. For correlation of vessel diameter to fluorescent intensity, simple linear regression model and second-order polynomial robust regression with no weighting was used. Significance is indicated by ∗(P < 0.0332), ∗∗(P < 0.0021), ∗∗∗(P < 0.002), ****(P < 0.0001). No asterisks indicate non-significance. No sample set was excluded from the analyses unless the histological sections were damaged severely during the acquisition of the sections (constitutes less than 3% of all sections analyzed).
For zebrafish studies, the effect sizes for animal groups were calculated using G-Power, and the sample size was estimated with n-Query. Four zebrafish from both sexes were used per group. For quantification of SV2-positive synapses, 3D object counter module of ImageJ software was used with the same standard cutoff threshold for every image. For quantification of activated/resting l-plastin-positive microglial cells, two different microglial states were classified based on their cellular morphology: slender and branched as resting microglia; round and regular as active microglia. Six images each from telencephalon sections were analyzed per animal. For colocalization studies, vascular fields were determined using ZO-1 staining on sections (20 for every group), and colocalization with glial endfeet labeled with GS stainings was performed using ImageJ software (v.2.1.0/1.53c) with its colocalization test. Data acquisition was randomized with Fay (x, y, z translation) to acquire in total 1670 data points from two experimental groups. R(and) correlation values from wild-type and fn1b−/− animals were compared using GraphPad Prism (v.9.2.0). Intensity values for individual fluorescent channels were obtained with modal gray values and integrated density measurements using ImageJ. Comparison of 40 sections from two experimental groups was performed. An unpaired non-parametric Kolmogorov–Smirnov t test was performed to test the statistical significance for all analyses.
In silico structure prediction
Protein structures, interspecies similarities, and the deleterious effects of variants were analyzed by SWISS-MODEL protein structure homology-modeling server through Expasy web server (https://swissmodel.expasy.org). SWISS-MODEL repository entries for respective proteins were retrieved and compared to desired protein orthologs using the superposition function. Deleterious mutation prediction was performed using Ensembl-integrated AlphaFold prediction model with SIFT, MetaLR, and REVEL modules for prediction of deleteriousness.
Amyloid toxicity and single-cell sequencing
Amyloid toxicity was induced as described [13, 47] in the adult telencephalon; the brains were dissected and single-cell suspensions were generated as previously described [23, 24]. Chromium Single-Cell 3’ Gel Bead and Library Kit v3.1 (10X Genomics, 120,237) was used to generate single-cell cDNA libraries. Generated libraries were sequenced via Illumina NovaSeq 6000 as described [12, 13, 23, 24, 71]. The cell clusters were identified using a resolution of 1. In total, 34 clusters were identified. The main cell types were identified by using s100b and gfap for astroglia; sv2a, nrgna, grin1a, grin1b for neurons; pdgfrb and kcne4 for pericytes; cd74a and apoc1 for microglia; mbpa and mpz for oligodendrocytes; myh11a and tagln2 for vascular smooth muscle cells, kdrl for endothelial cells.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1: Structure and deleteriousness prediction for FN1 and COL6A2 (TIF 6441 KB)
Supplementary Figure 2: Individual breakdown of FN1 intensity comparisons in Figure 5d (TIF 2852 KB)
Supplementary Figure 3: Individual breakdown of FN1 intensity comparisons in Figure 6h (TIF 2059 KB)
Supplementary Table 1: The list of coding variants in Hispanics. Supplementary Table 2: The list of coding variants in non-Hispanic Whites. Supplementary Table 3: The list of SNPs. Supplementary Table 4: Overview of genotyping platforms across all available AD-related genetic data from ADSP and NACC. Supplementary Table 5: Overview of ADSP studies with WES or WGS available through NIAGADS DSS (NG00067). Supplementary Table 6: Case-control regression sensitivity analyses for rs140926439 in APOEε4∕4 carriers. To ensure an independent replication of discovery findings, in ADGC, samples from NIA-LOAD were excluded, while ADSP whole genome sequencing data was fully excluded. Combined results reflect a fixed-effects inverse-variance weighted meta-analysis. Supplementary Table 7: Demographics and APOE genotypes of the post-mortem human brains used for the study. Supplementary Table 8: Demographics and APOE genotypes of the post-mortem human brains used for vascular deposition of FN1 (XLSX 526 KB)
Supplementary Dataset 1: Total fluorescent intensity values for COL4, COL6A and DAPI, according to APOE genotype. Supplementary Dataset 2: Total fluorescent intensity values for FN1, CD31 and DAPI, according to APOE genotype. Supplementary Dataset 3: Total fluorescent intensity values for FN1, GFAP and DAPI, according to APOE genotype and Alzheimer’s disease status. Supplementary Dataset 4: Fluorescent intensity for overlapping GS (astrocytes) and ZO-1 (tight junctions) in wild type and fn1b-/- animals treated with Aβ42. Supplementary Dataset 5: Fluorescent intensity for ventricular GS (astrocytes) in wild type and fn1b-/- animals treated with Aβ42. Supplementary Dataset 6: Number of microglia and their activation state in wild type and fn1b-/- animals treated with Aβ42. Supplementary Dataset 7: Automated quantification results of synaptic puncta in wild type and fn1b-/- animals treated with Aβ42 (XLSX 420 KB)
Acknowledgements
We thank the Carol and Gene Ludwig Family Foundation and the Agouron Institute for providing grant support to B.N.V for this work. We would like to thank Taub Institute for Research on Alzheimer’s Disease and the Aging Brain Imaging Platform at Columbia University, Molecular Pathology (MPSR) and Flow Cytometry Core Facility (CCTI, supported in part by the Office of the Director, National Institutes of Health under awards S10OD020056) platforms of the Columbia University Herbert Irving Comprehensive Cancer Center for procedural support, and New York Brain Bank for postmortem human brain sections. We thank the contributors, who collected samples used in this study. We thank the patients and families for their participation, without whom these studies would not have been possible. This work was supported by National Institute on Aging R01 AG067501 (Genetic Epidemiology and Multi-Omics Analyses in Familial and Sporadic Alzheimer's Disease Among Secular Caribbean Hispanics and Religious Order) (R.M, B.N.V, C.K.) and National Institute on Aging RF1 AG066107 Epidemiological Integration of Genetic Variants and Metabolomics Profiles in Washington Heights Columbia Aging Project (R.M, B.N.V, C.K), R01 AG072474 Epidemiological and Genetic Investigations of Blood-Based Biomarkers for Alzheimer's Disease in the Multiethnic, Washington Heights, Inwood, Columbia Aging Project (WHICAP) (R.M, B.N.V, C.K), U01 AG066752 (Gene discovery in multi-ethnic late-onset Alzheimer's disease families (B.N.V), NIGMS grant R35GM148348 (S.A.H), Schaefer Research Scholars Award (C.K), Taub Institute Grants for Emerging Research (TIGER) (C.K) and The Thompson Family Foundation Program for Accelerated Medicine Exploration in Alzheimer’s Disease and Related Disorders of the Nervous System (TAME-AD) (C.K). N.E-T is supported by NIH/NIA grants R01AG061796, U19AG074879, U01 AG046139 and is recipient of the Alzheimer’s Association Zenith Award. M.D.G is supported by NIH/NIA grants AG060747 and AG047366. M.E.B. is supported by NIH/NIA grant R00 AG075238. The National Institute on Aging-AD Family Based Study (NIA-AD FBS; https://www.neurology.columbia.edu/research/research-centers-and-programs/national-institute-aging-alzheimers-disease-family-based-study-nia-ad-fbs) collected the samples used in this study and is supported by National Institute on Aging (NIA) grant U24AG056270. Additional families were contributed to the NIA-AD FBS through NIH grants: R01AG028786, R01AG027944, RO1AG027944, RF1AG054074, U01AG052410. The NIA-AD FBS began in 2003 with the goal of recruiting large, multiply affected families with late-onset Alzheimer's disease (AD) for genetic research. The study created a resource of well-characterized families with late-onset AD. The initial phases of the Alzheimer's Disease Sequencing Project (ADSP) included genotyping of hundreds of participants from NIA-AD FBS. The ADSP Follow-Up Study heavily engages resources provided by the NIA-AD FBS and depends upon the longitudinal follow-up of families, and the collection of additional families, in particular those from diverse populations. Samples include biological materials for genome-wide association studies (GWAS) and whole-genome sequencing (WGS), peripheral blood mononuclear cells (PBMC) for stem cell modeling, plasma for studies of metabolomics, proteomics, and biomarker research, and brain autopsy materials for bulk RNA sequencing. WHICAP Data collection and sharing for this project was supported by the National Institutes on Aging (NIA) of the National Institutes of Health (NIH): R01AG072474, AG066107, AG059013. Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA) is a study of sporadic and familial Alzheimer’s disease among Caribbean Hispanics recruited from clinics in the Dominican Republic and New York (R01 AG067501). The goal of this study is to identify genetic variants that increase late-onset Alzheimer disease risk in this ethnic group. This study was initiated in 1998 and recruited individuals and their families in New York as well as from clinics in the Dominican Republic. Recruitment for the EFIGA began in 1998, to study the genetic architecture of AD in the Caribbean Hispanic population. Patients with familial AD were recruited and if a sibling of the proband had dementia, all other living siblings and available relatives underwent evaluation. Cases were defined as any individual meeting NINCDS–ADRDA criteria for probable or possible AD. EFIGA study is supported by NIA grants R56AG063908, R01AG067501, and RF1AG015473. We acknowledge the services of CEDIMAT for collaborating with sample collection and processing in the EFIGA cohort. UK Biobank data were analyzed under Application Number 45420. Acknowledgements for data used from the ADSP are mentioned in the Supplementary material. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
RM, CK and BNV conceived and designed this study. TIG and BNV generated the variations list. DR-D, RL, MM, DR, PR, and RM provided the AD cohort information. MEB, YLG, and MDG performed the independent replication. GS, CA, EMM, DNH, DB, CLD, and MZ performed sequencing. NE-T, DWD, DF, and AFT provided the human brain samples. DJ and SH provided the fn1b knockout zebrafish. PB and CK performed the human postmortem tissue immunohistochemistry, image acquisition, quantification, and statistical analyses. EY, PB, and CK performed single-cell sequencing, data analyses, and interpretation. PB, HT, BT, and CK performed the amyloid toxicity assessment and relevant quantifications in zebrafish brains and protein structural analyses. RM, BNV, and CK interpreted the results. CK, BNV, and RM wrote the manuscript. All authors read, edited, and approved the final version.
Data availability
Whole Genome Sequencing data (WGS) from EFIGA and NIA-AD FBS cohorts that were analyzed in the study have been deposited to NIAGADS (The National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site) and are shared with the research community as part of the Alzheimer’s Disease Sequencing Project (ADSP). The brain tissue used in this analysis was requested from the New York Brain Bank at Columbia University. Raw data generated from the brain tissue and zebrafish experiments are available upon request. The zebrafish gliovascular single-cell dataset can be accessed at NCBI’s Gene Expression Omnibus (GEO) with the accession number GSE225721.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Prabesh Bhattarai, Tamil Iniyan Gunasekaran, and Michael E. Belloy equally contributed to this work.
Richard Mayeux, Caghan Kizil, and Badri N. Vardarajan equally contributed to this work.
Contributor Information
Caghan Kizil, Email: ck2893@cumc.columbia.edu.
Badri N. Vardarajan, Email: bnv2103@cumc.columbia.edu
References
- 1.Al-Yafeai Z, Yurdagul A, Jr, Peretik JM, Alfaidi M, Murphy PA, Orr AW. Endothelial FN (fibronectin) deposition by alpha5beta1 integrins drives atherogenic inflammation. Arterioscler Thromb Vasc Biol. 2018;38:2601–2614. doi: 10.1161/ATVBAHA.118.311705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alestrom P, D'Angelo L, Midtlyng PJ, Schorderet DF, Schulte-Merker S, Sohm F, et al. Zebrafish: housing and husbandry recommendations. Lab Anim. 2019 doi: 10.1177/0023677219869037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora K, Shah M, Johnson M, Sanghvi R, Shelton J, Nagulapalli K, et al. Deep whole-genome sequencing of 3 cancer cell lines on 2 sequencing platforms. Sci Rep. 2019;9:19123. doi: 10.1038/s41598-019-55636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell CC. DSM-IV: diagnostic and statistical manual of mental disorders. JAMA. 1994;272:828–829. doi: 10.1001/jama.1994.03520100096046. [DOI] [Google Scholar]
- 5.Bell RD, Winkler EA, Sagare AP, Singh I, LaRue B, Deane R, et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron. 2010;68:409–427. doi: 10.1016/j.neuron.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellaver B, Povala G, Ferreira PCL, Ferrari-Souza JP, Leffa DT, Lussier FZ, et al. Astrocyte reactivity influences amyloid-beta effects on tau pathology in preclinical Alzheimer's disease. Nat Med. 2023;29:1775–1781. doi: 10.1038/s41591-023-02380-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellenguez C, Kucukali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, et al. New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat Genet. 2022 doi: 10.1038/s41588-022-01024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belloy ME, Eger SJ, Le Guen Y, Damotte V, Ahmad S, Ikram MA, et al. Challenges at the APOE locus: a robust quality control approach for accurate APOE genotyping. Alzheimers Res Ther. 2022;14:22. doi: 10.1186/s13195-022-00962-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belloy ME, Andrews SJ, Le Guen Y, Cuccaro M, Farrer LA, Napolioni V, et al. APOE genotype and Alzheimer disease risk across age, sex, and population ancestry. JAMA Neurol. 2023;80:1284–1294. doi: 10.1001/jamaneurol.2023.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett DA, Schneider JA, Aggarwal NT, Arvanitakis Z, Shah RC, Kelly JF, et al. Decision rules guiding the clinical diagnosis of Alzheimer's disease in two community-based cohort studies compared to standard practice in a clinic-based cohort study. Neuroepidemiology. 2006;27:169–176. doi: 10.1159/000096129. [DOI] [PubMed] [Google Scholar]
- 11.Bhattarai P, Cosacak MI, Mashkaryan V, Demir S, Popova S, Govindarajan N, et al. Neuron-glia interaction through Serotonin-BDNF-NGFR axis enables regenerative neurogenesis in Alzheimer’s model of adult zebrafish brain. PLoS Biol. 2020;18:e3000585. doi: 10.1371/journal.pbio.3000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattarai P, Thomas AK, Cosacak MI, Papadimitriou C, Mashkaryan V, Zhang Y, et al. Modeling amyloid-β42 toxicity and neurodegeneration in adult zebrafish brain. J Vis Exp. 2017 doi: 10.3791/56014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattarai P, Thomas AK, Papadimitriou C, Cosacak MI, Mashkaryan V, Froc C, et al. IL4/STAT6 signaling activates neural stem cell proliferation and neurogenesis upon Amyloid-β42 aggregation in adult zebrafish brain. Cell Rep. 2016;17:941–948. doi: 10.1016/j.celrep.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 14.Blanchard JW, Akay LA, Davila-Velderrain J, von Maydell D, Mathys H, Davidson SM, et al. APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes. Nature. 2022;611:769–779. doi: 10.1038/s41586-022-05439-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdan S, Puscion-Jakubik A, Klimiuk K, Socha K, Kochanowicz J, Gorodkiewicz E (2022) The concentration of fibronectin and MMP-1 in patients with Alzheimer's disease in relation to the selected antioxidant elements and eating habits. J Clin Med. 10.3390/jcm11216360 [DOI] [PMC free article] [PubMed]
- 16.Burda JE, Sofroniew MV. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron. 2014;81:229–248. doi: 10.1016/j.neuron.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao L, Nicosia J, Larouche J, Zhang Y, Bachman H, Brown AC, et al. Detection of an integrin-binding mechanoswitch within fibronectin during tissue formation and fibrosis. ACS Nano. 2017;11:7110–7117. doi: 10.1021/acsnano.7b02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CY, Pollack S, Hunter DJ, Hirschhorn JN, Kraft P, Price AL. Improved ancestry inference using weights from external reference panels. Bioinformatics. 2013;29(11):1399–1406. doi: 10.1093/bioinformatics/btt144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chhum P, Yu H, An B, Doyon BR, Lin YS, Brodsky B. Consequences of glycine mutations in the fibronectin-binding sequence of collagen. J Biol Chem. 2016;291:27073–27086. doi: 10.1074/jbc.M116.753566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 22.Cosacak MI, Bhattarai P, De Jager PL, Menon V, Tosto G, Kizil C (2022) Single cell/nucleus transcriptomics comparison in zebrafish and humans reveals common and distinct molecular responses to Alzheimer’s disease. Cells 11:1807. 10.3390/cells11111807 [DOI] [PMC free article] [PubMed]
- 23.Cosacak MI, Bhattarai P, Kizil C. Protocol for dissection and dissociation of zebrafish telencephalon for single-cell sequencing. STAR Protoc. 2020;1:100042. doi: 10.1016/j.xpro.2020.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cosacak MI, Bhattarai P, Reinhardt S, Petzold A, Dahl A, Zhang Y, et al. Single-cell transcriptomics analyses of neural stem cell heterogeneity and contextual plasticity in a zebrafish brain model of amyloid toxicity. Cell Rep. 2019;27(1307–1318):e1303. doi: 10.1016/j.celrep.2019.03.090. [DOI] [PubMed] [Google Scholar]
- 25.Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010;468:562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drieu A, Du S, Storck SE, Rustenhoven J, Papadopoulos Z, Dykstra T, Zhong F, Kim K, Blackburn S, Mamuladze T, et al. Parenchymal border macrophages regulate the flow dynamics of the cerebrospinal fluid. Nature. 2022;611:585–593. doi: 10.1038/s41586-022-05397-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. doi: 10.1001/jama.1997.03550160069041. [DOI] [PubMed] [Google Scholar]
- 29.Fitch MT, Silver J. CNS injury, glial scars, and inflammation: inhibitory extracellular matrices and regeneration failure. Exp Neurol. 2008;209:294–301. doi: 10.1016/j.expneurol.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisler R, Borel N, Ferg M, Maier JV, Strahle U. Maintenance of zebrafish lines at the European Zebrafish Resource Center. Zebrafish. 2016;13(Suppl 1):S19–23. doi: 10.1089/zeb.2015.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry. 2011;16:903–907. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu Y, Beato JM, Amarante E, Chesebro AG, Manly JJ, Schupf N, et al. Assessment of leisure time physical activity and brain health in a multiethnic cohort of older adults. JAMA Netw Open. 2020;3:e2026506. doi: 10.1001/jamanetworkopen.2020.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillon E, Das D, Julich D, Hassan AR, Geller H, Holley S. Fibronectin is a smart adhesive that both influences and responds to the mechanics of early spinal column development. Elife. 2020 doi: 10.7554/eLife.48964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halliday MR, Rege SV, Ma Q, Zhao Z, Miller CA, Winkler EA, et al. Accelerated pericyte degeneration and blood-brain barrier breakdown in apolipoprotein E4 carriers with Alzheimer's disease. J Cereb Blood Flow Metab. 2016;36:216–227. doi: 10.1038/jcbfm.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawkes CA, Gatherer M, Sharp MM, Dorr A, Yuen HM, Kalaria R, et al. Regional differences in the morphological and functional effects of aging on cerebral basement membranes and perivascular drainage of amyloid-beta from the mouse brain. Aging Cell. 2013;12:224–236. doi: 10.1111/acel.12045. [DOI] [PubMed] [Google Scholar]
- 36.Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, et al. Neuroinflammation in Alzheimer's disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- 38.Iadecola C. The neurovascular unit coming of age: a journey through neurovascular coupling in health and disease. Neuron. 2017;96:17–42. doi: 10.1016/j.neuron.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.İs O, Wang X, Patel T, Quicksall Z, Heckman M, White L, Lewis-Tuffin L, et al. Single nuclei transcriptome reveals perturbed brain vascular molecules in Alzheimer’s disease. Biorxiv. 2022 doi: 10.1101/2021.12.28.474255. [DOI] [Google Scholar]
- 40.Jack CR, Jr, Albert MS, Knopman DS, McKhann GM, Sperling RA, Carrillo MC, et al. Introduction to the recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:257–262. doi: 10.1016/j.jalz.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang J, Wang C, Qi R, Fu H, Ma Q. scREAD: a single-cell RNA-Seq database for Alzheimer's disease. iScience. 2020;23:101769. doi: 10.1016/j.isci.2020.101769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kizil C, Sariya S, Kim YA, Rajabli F, Martin E, Reyes-Dumeyer D, Vardarajan B, et al. Admixture Mapping of Alzheimer's disease in Caribbean Hispanics identifies a new locus on 22q13.1. Mol Psychiatry. 2022 doi: 10.1038/s41380-022-01526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knox EG, Aburto MR, Clarke G, Cryan JF, O'Driscoll CM. The blood-brain barrier in aging and neurodegeneration. Mol Psychiatry. 2022;27:2659–2673. doi: 10.1038/s41380-022-01511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohler A, Collymore C, Finger-Baier K, Geisler R, Kaufmann L, Pounder KC, et al. Report of workshop on euthanasia for zebrafish—a matter of welfare and science. Zebrafish. 2017;14:547–551. doi: 10.1089/zeb.2017.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koizumi K, Hattori Y, Ahn SJ, Buendia I, Ciacciarelli A, Uekawa K, et al. Apoepsilon4 disrupts neurovascular regulation and undermines white matter integrity and cognitive function. Nat Commun. 2018;9:3816. doi: 10.1038/s41467-018-06301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lathe R, Sapronova A, Kotelevtsev Y. Atherosclerosis and Alzheimer–diseases with a common cause? Inflammation, oxysterols, vasculature. BMC Geriatr. 2014;14:36. doi: 10.1186/1471-2318-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee AJ, Raghavan NS, Bhattarai P, Siddiqui T, Sariya S, Reyes-Dumeyer D, Flowers XE, Cardoso SAL, De Jager PL, Bennett DA et al (2022) FMNL2 regulates gliovascular interactions and is associated with vascular risk factors and cerebrovascular pathology in Alzheimer’s disease. Acta Neuropathol. 10.1007/s00401-022-02431-6 [DOI] [PMC free article] [PubMed]
- 48.Lepelletier FX, Mann DM, Robinson AC, Pinteaux E, Boutin H. Early changes in extracellular matrix in Alzheimer's disease. Neuropathol Appl Neurobiol. 2017;43:167–182. doi: 10.1111/nan.12295. [DOI] [PubMed] [Google Scholar]
- 49.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer's disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS–ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 51.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michalski D, Spielvogel E, Puchta J, Reimann W, Barthel H, Nitzsche B, et al. Increased immunosignals of collagen IV and fibronectin indicate ischemic consequences for the neurovascular matrix adhesion zone in various animal models and human stroke tissue. Front Physiol. 2020;11:575598. doi: 10.3389/fphys.2020.575598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 54.Mo FE. Shear-regulated extracellular microenvironments and endothelial cell surface integrin receptors intertwine in atherosclerosis. Front Cell Dev Biol. 2021;9:640781. doi: 10.3389/fcell.2021.640781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montagne A, Nation DA, Sagare AP, Barisano G, Sweeney MD, Chakhoyan A, et al. APOE4 leads to blood-brain barrier dysfunction predicting cognitive decline. Nature. 2020;581:71–76. doi: 10.1038/s41586-020-2247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montagne A, Nikolakopoulou AM, Huuskonen MT, Sagare AP, Lawson EJ, Lazic D, et al. APOE4 accelerates advanced-stage vascular and neurodegenerative disorder in old Alzheimer's mice via cyclophilin A independently of amyloid-beta. Nat Aging. 2021;1:506–520. doi: 10.1038/s43587-021-00073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 58.Morris JC, Ernesto C, Schafer K, Coats M, Leon S, Sano M, et al. Clinical dementia rating training and reliability in multicenter studies: the Alzheimer's Disease Cooperative Study experience. Neurology. 1997;48:1508–1510. doi: 10.1212/WNL.48.6.1508. [DOI] [PubMed] [Google Scholar]
- 59.Murdock MH, Tsai LH. Insights into Alzheimer's disease from single-cell genomic approaches. Nat Neurosci. 2023;26:181–195. doi: 10.1038/s41593-022-01222-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogino E, Manly JJ, Schupf N, Mayeux R, Gu Y. Current and past leisure time physical activity in relation to risk of Alzheimer's disease in older adults. Alzheimers Dement. 2019;15:1603–1611. doi: 10.1016/j.jalz.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okuyama K, Miyama K, Morimoto T, Masakiyo K, Mizuno K, Bachinger HP. Stabilization of triple-helical structures of collagen peptides containing a Hyp-Thr-Gly, Hyp-Val-Gly, or Hyp-Ser-Gly sequence. Biopolymers. 2011;95:628–640. doi: 10.1002/bip.21625. [DOI] [PubMed] [Google Scholar]
- 62.Paresce DM, Ghosh RN, Maxfield FR. Microglial cells internalize aggregates of the Alzheimer's disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17:553–565. doi: 10.1016/S0896-6273(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 63.Patel T, Carnwath TP, Wang X, Allen M, Lincoln SJ, Lewis-Tuffin LJ, et al. Transcriptional landscape of human microglia implicates age, sex, and APOE-related immunometabolic pathway perturbations. Aging Cell. 2022;21:e13606. doi: 10.1111/acel.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reitz C, Pericak-Vance MA, Foroud T, Mayeux R. A global view of the genetic basis of Alzheimer disease. Nat Rev Neurol. 2023 doi: 10.1038/s41582-023-00789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruiz-Grosso P, Loret de Mola C, Vega-Dienstmaier JM, Arevalo JM, Chavez K, Vilela A, et al. Validation of the Spanish Center for Epidemiological Studies Depression and Zung Self-Rating Depression Scales: a comparative validation study. PLoS ONE. 2012;7:e45413. doi: 10.1371/journal.pone.0045413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sagare AP, Bell RD, Zhao Z, Ma Q, Winkler EA, Ramanathan A, et al. Pericyte loss influences Alzheimer-like neurodegeneration in mice. Nat Commun. 2013;4:2932. doi: 10.1038/ncomms3932. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Salloway S, Gur T, Berzin T, Tavares R, Zipser B, Correia S, et al. Effect of APOE genotype on microvascular basement membrane in Alzheimer's disease. J Neurol Sci. 2002;203–204:183–187. doi: 10.1016/s0022-510x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 68.Sariya S, Felsky D, Reyes-Dumeyer D, Lali R, Lantigua RA, Vardarajan B, et al. Polygenic risk score for Alzheimer's disease in Caribbean Hispanics. Ann Neurol. 2021;90:366–376. doi: 10.1002/ana.26131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schumacher S, Dedden D, Nunez RV, Matoba K, Takagi J, Biertumpfel C, et al. Structural insights into integrin alpha(5)beta(1) opening by fibronectin ligand. Sci Adv. 2021 doi: 10.1126/sciadv.abe9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sepulveda-Falla D, Sanchez JS, Almeida MC, Boassa D, Acosta-Uribe J, Vila-Castelar C, et al. Distinct tau neuropathology and cellular profiles of an APOE3 Christchurch homozygote protected against autosomal dominant Alzheimer's dementia. Acta Neuropathol. 2022;144:589–601. doi: 10.1007/s00401-022-02467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siddiqui T, Cosacak MI, Popova S, Bhattarai P, Santa-Maria I, Tosto G, et al. Neurogenic induction by Ngfr reduces reactive astrocytic Lcn2/Slc22a17 signalling, amyloid deposition and Tau phosphorylation. bioRxiv. 2022 doi: 10.1101/2022.08.20.504608. [DOI] [Google Scholar]
- 72.Siddiqui T, Cosacak MI, Popova S, Bhattarai P, Yilmaz E, Lee AJ, et al. Nerve growth factor receptor (Ngfr) induces neurogenic plasticity by suppressing reactive astroglial Lcn2/Slc22a17 signaling in Alzheimer's disease. NPJ Regener Med. 2023;8:33. doi: 10.1038/s41536-023-00311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 74.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dementia. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stallones L, Marx MB, Garrity TF. Prevalence and correlates of depressive symptoms among older U.S. adults. Am J Prev Med. 1990;6:295–303. doi: 10.1016/S0749-3797(18)30998-X. [DOI] [PubMed] [Google Scholar]
- 76.Stern Y, Andrews H, Pittman J, Sano M, Tatemichi T, Lantigua R, et al. Diagnosis of dementia in a heterogeneous population. Development of a neuropsychological paradigm-based diagnosis of dementia and quantified correction for the effects of education. Arch Neurol. 1992;49:453–460. doi: 10.1001/archneur.1992.00530290035009. [DOI] [PubMed] [Google Scholar]
- 77.Strahle U, Scholz S, Geisler R, Greiner P, Hollert H, Rastegar S, et al. Zebrafish embryos as an alternative to animal experiments–a commentary on the definition of the onset of protected life stages in animal welfare regulations. Reprod Toxicol. 2012;33:128–132. doi: 10.1016/j.reprotox.2011.06.121. [DOI] [PubMed] [Google Scholar]
- 78.Sun L, Zou Z, Collodi P, Xu F, Xu X, Zhao Q. Identification and characterization of a second fibronectin gene in zebrafish. Matrix Biol. 2005;24:69–77. doi: 10.1016/j.matbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 79.Sun N, Akay LA, Murdock MH, Park Y, Galiana-Melendez F, Bubnys A, et al. Single-nucleus multiregion transcriptomic analysis of brain vasculature in Alzheimer's disease. Nat Neurosci. 2023;26:970–982. doi: 10.1038/s41593-023-01334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun Y, Xu S, Jiang M, Liu X, Yang L, Bai Z, et al. Role of the extracellular matrix in Alzheimer's disease. Front Aging Neurosci. 2021;13:707466. doi: 10.3389/fnagi.2021.707466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590(7845):290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279:751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 83.Thomsen MS, Birkelund S, Burkhart A, Stensballe A, Moos T. Synthesis and deposition of basement membrane proteins by primary brain capillary endothelial cells in a murine model of the blood-brain barrier. J Neurochem. 2017;140:741–754. doi: 10.1111/jnc.13747. [DOI] [PubMed] [Google Scholar]
- 84.Thomsen MS, Routhe LJ, Moos T. The vascular basement membrane in the healthy and pathological brain. J Cereb Blood Flow Metab. 2017;37:3300–3317. doi: 10.1177/0271678X17722436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tosto G, Fu H, Vardarajan BN, Lee JH, Cheng R, Reyes-Dumeyer D, et al. F-box/LRR-repeat protein 7 is genetically associated with Alzheimer's disease. Ann Clin Transl Neurol. 2015;2:810–820. doi: 10.1002/acn3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vardarajan BN, Faber KM, Bird TD, Bennett DA, Rosenberg R, Boeve BF, et al. Age-specific incidence rates for dementia and Alzheimer disease in NIA-LOAD/NCRAD and EFIGA families: National Institute on Aging Genetics Initiative for Late-Onset Alzheimer Disease/National Cell Repository for Alzheimer Disease (NIA-LOAD/NCRAD) and Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA) JAMA Neurol. 2014;71:315–323. doi: 10.1001/jamaneurol.2013.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Waldvogel HJ, Curtis MA, Baer K, Rees MI, Faull RL. Immunohistochemical staining of post-mortem adult human brain sections. Nat Protoc. 2006;1:2719–2732. doi: 10.1038/nprot.2006.354. [DOI] [PubMed] [Google Scholar]
- 88.Wang X, Allen M, Li S, Quicksall ZS, Patel TA, Carnwath TP, et al. Deciphering cellular transcriptional alterations in Alzheimer's disease brains. Mol Neurodegener. 2020;15:38. doi: 10.1186/s13024-020-00392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiong M, Wang C, Gratuze M, Saadi F, Bao X, Bosch ME, et al. Astrocytic APOE4 removal confers cerebrovascular protection despite increased cerebral amyloid angiopathy. Mol Neurodegener. 2023;18:17. doi: 10.1186/s13024-023-00610-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu L, Nirwane A, Yao Y. Basement membrane and blood-brain barrier. Stroke Vasc Neurol. 2019;4:78–82. doi: 10.1136/svn-2018-000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamazaki Y, Zhao N, Caulfield TR, Liu CC, Bu G. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat Rev Neurol. 2019;15:501–518. doi: 10.1038/s41582-019-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang AC, Vest RT, Kern F, Lee DP, Agam M, Maat CA, et al. A human brain vascular atlas reveals diverse mediators of Alzheimer's risk. Nature. 2022;603:885–892. doi: 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng H, Lu C, Lan J, Fan S, Nanda V, Xu F. How electrostatic networks modulate specificity and stability of collagen. Proc Natl Acad Sci USA. 2018;115:6207–6212. doi: 10.1073/pnas.1802171115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zou L, Cao S, Kang N, Huebert RC, Shah VH. Fibronectin induces endothelial cell migration through beta1 integrin and Src-dependent phosphorylation of fibroblast growth factor receptor-1 at tyrosines 653/654 and 766. J Biol Chem. 2012;287:7190–7202. doi: 10.1074/jbc.M111.304972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Structure and deleteriousness prediction for FN1 and COL6A2 (TIF 6441 KB)
Supplementary Figure 2: Individual breakdown of FN1 intensity comparisons in Figure 5d (TIF 2852 KB)
Supplementary Figure 3: Individual breakdown of FN1 intensity comparisons in Figure 6h (TIF 2059 KB)
Supplementary Table 1: The list of coding variants in Hispanics. Supplementary Table 2: The list of coding variants in non-Hispanic Whites. Supplementary Table 3: The list of SNPs. Supplementary Table 4: Overview of genotyping platforms across all available AD-related genetic data from ADSP and NACC. Supplementary Table 5: Overview of ADSP studies with WES or WGS available through NIAGADS DSS (NG00067). Supplementary Table 6: Case-control regression sensitivity analyses for rs140926439 in APOEε4∕4 carriers. To ensure an independent replication of discovery findings, in ADGC, samples from NIA-LOAD were excluded, while ADSP whole genome sequencing data was fully excluded. Combined results reflect a fixed-effects inverse-variance weighted meta-analysis. Supplementary Table 7: Demographics and APOE genotypes of the post-mortem human brains used for the study. Supplementary Table 8: Demographics and APOE genotypes of the post-mortem human brains used for vascular deposition of FN1 (XLSX 526 KB)
Supplementary Dataset 1: Total fluorescent intensity values for COL4, COL6A and DAPI, according to APOE genotype. Supplementary Dataset 2: Total fluorescent intensity values for FN1, CD31 and DAPI, according to APOE genotype. Supplementary Dataset 3: Total fluorescent intensity values for FN1, GFAP and DAPI, according to APOE genotype and Alzheimer’s disease status. Supplementary Dataset 4: Fluorescent intensity for overlapping GS (astrocytes) and ZO-1 (tight junctions) in wild type and fn1b-/- animals treated with Aβ42. Supplementary Dataset 5: Fluorescent intensity for ventricular GS (astrocytes) in wild type and fn1b-/- animals treated with Aβ42. Supplementary Dataset 6: Number of microglia and their activation state in wild type and fn1b-/- animals treated with Aβ42. Supplementary Dataset 7: Automated quantification results of synaptic puncta in wild type and fn1b-/- animals treated with Aβ42 (XLSX 420 KB)
Data Availability Statement
Whole Genome Sequencing data (WGS) from EFIGA and NIA-AD FBS cohorts that were analyzed in the study have been deposited to NIAGADS (The National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site) and are shared with the research community as part of the Alzheimer’s Disease Sequencing Project (ADSP). The brain tissue used in this analysis was requested from the New York Brain Bank at Columbia University. Raw data generated from the brain tissue and zebrafish experiments are available upon request. The zebrafish gliovascular single-cell dataset can be accessed at NCBI’s Gene Expression Omnibus (GEO) with the accession number GSE225721.