Abstract
During herpes simplex virus type 1 (HSV-1) latent infection in human dorsal root ganglia, limited viral transcription, which has been linked to HSV-1 reactivation ability, takes place. To study the involvement of this transcription in HSV-1 replication in neuronal cells and consequently in viral latency, we constructed stably transfected neuronal cell lines containing (i) the entire HSV-1 latency transcriptionally active DNA fragment, (ii) the same DNA sequence with deletions of the latency-associated transcript (LAT) promoters, or (iii) the DNA coding sequence of the LAT domain. Replication of HSV-1 or a LAT-negative mutant was markedly repressed in the LAT-expressing cells, a phenomenon mediated by the LATs. To study the mechanism responsible for this effect, we examined LAT influence upon expression of HSV-1 immediate-early (IE) genes ICP0, ICP4, and ICP27, by Northern blot analysis. Following infection of a LAT-expressing neuronal cell line with a LAT-negative mutant, the steady-state levels of all three IE mRNAs were reduced compared to those for control cells. Transient transfections into a neuronal cell line indicated that the LAT suppressive effect upon ICP0 mRNA was mediated directly and was not due to the LAT effect upon the ICP0 promoter. We therefore propose that the LATs may repress viral replication in neuronal cells by reducing IE gene mRNA levels and thus facilitate the establishment of HSV-1 latency in nervous tissue.
Herpes simplex virus type 1 (HSV-1) colonizes and establishes latent infection in human dorsal root ganglia (DRG) to produce periodic reactivations (for reviews, see references 59 and 63). No mature virions are detected in latently infected human nervous tissue (13, 62), and restricted gene expression from the repeat segments of the viral genome (Fig. 1A) is the only transcriptional activity present throughout latency. This latency-associated gene expression consists of the more abundant RNAs, the latency-associated transcripts (LATs) (2.0 and 1.5 kb) (49, 56, 64), and the 8.3-kb minor hybridizing RNA (mLAT) (41, 70). Whether the latency-associated gene(s) codes for proteins is yet unclear (summarized in references 17 and 59), but we have recently demonstrated that the LATs are associated with polyribosomes in latently infected trigeminal ganglia (TG) of mice (22).
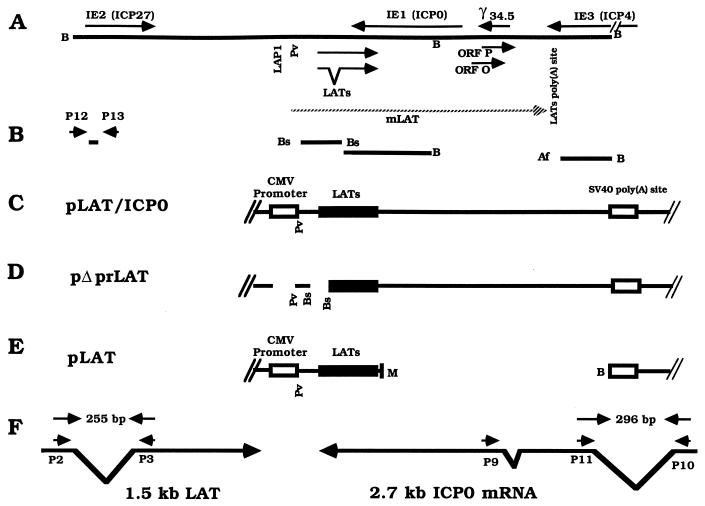
FIG. 1.
Map of the HSV-1 genome region expressing the LATs, structure of plasmids, and location of probes and primers used in this study. (A) The enlarged BamHI fragments B and SP within the repeat region of the HSV-1 genome. Arrows indicate the relevant HSV-1 genes and their transcription orientation. Locations of LAP1 and the LAT polyadenylation site are indicated according to the HSV-1 DNA sequence in GenBank (accession no. X14112). (B) Location of the DNA probes used in this study. (C to E) Structure of plasmids pLAT/ICP0, pΔprLAT, and pLAT (see Materials and Methods for details). (F) Location of the primers used for RT-PCR of the 1.5-kb LAT and of ICP0 mRNA and expected sizes of amplified fragments. Also indicated are locations of the primers used for production of the single-stranded probe for ICP0 mRNA. B, BamHI; LAP1, latency-associated promoter 1; Pv, PvuI; M, MluI; Bs, BstEII; Af, AflIII; SV40, simian virus 40.
The function of the LATs was studied mainly with HSV-1 mutants harboring genomic changes that modify LAT transcription. It is clear that HSV-1 latency-associated gene expression is not required for viral lytic replication in vivo (29, 33, 61), nor does a lack of the latency-associated gene(s) prevent viral transport from the periphery to the DRG. The possibility that these RNAs take part in the establishment of latency was suggested, since LAT-negative mutants established latent infection in fewer neuronal cells than did the parental virus (38, 52, 53, 66). It seems also that the LATs are not required for the maintenance of latency (5, 28, 29, 33, 61). The most constant and common phenotypic behavior of HSV-1 mutants that are unable to express the latency-associated gene(s) is a prolonged and asynchronous explant reactivation kinetics (29, 33, 61) and reduced or absent in vivo reactivation from DRG (26, 67).
The latency-associated gene(s) may act through a protein and/or as a specialized RNA. Its molecular action mechanism(s) is yet unknown, but two main possibilities have been proposed. (i) The LATs may act during reactivation. Neurons possess means to inhibit the transactivation of the immediate-early (IE) genes of HSV-1, a mechanism that renders the virus incapable of replication in neuronal cells at low multiplicities of infection (MOIs) (31, 34). This hypothesis assumes that the LATs, the only RNAs that are expressed during latency (56, 62, 64), may enable the viral replication cycle to bypass this inhibition at the initial stages of reactivation. (ii) Alternately, the LATs may act during the establishment of latent infection, by suppressing HSV-1 IE gene expression. This assumption is based on the fact that, since HSV-1 is a lytic virus, one of the early requirements for establishment of latent infection in neuronal cells is prevention of viral replication, before or at the stage of IE gene expression (50, 58).
The aim of the present study was therefore to examine the effect of the latency-associated gene(s) of HSV-1 on viral replication in neuronal cell lines, to map the functional region, and to investigate whether any effect is associated with expression of HSV-1 IE genes ICP0, ICP4, and ICP27. Our findings indicate that the LATs suppress HSV-1 replication in neuronal cell lines and reduce ICP0 and also ICP4 and ICP27 steady-state mRNA levels in these cells.
MATERIALS AND METHODS
Construction of plasmids.
The construction and characteristics of pLAT/ICP0 (Fig. 1C), previously termed pNM3 (22, 37), have been reported before (37). Briefly, the entire HSV-1 DNA fragment that is transcriptionally active during latent infection was inserted into a vector under the control of the cytomegalovirus (CMV) IE promoter. This DNA insert also contains the ICP0 (46), γ34.5 (12), open reading frame (ORF) P (32), and ORF O (48) genes.
pΔprLAT (Fig. 1D) was derived from pLAT/ICP0 by two deletions that removed the CMV IE promoter and the latency-associated promoter 2 (LAP2 [21]) and therefore is incapable of expressing HSV-1 LATs or mLAT but contains the ICP0, γ34.5, ORF P, and ORF O genes. pLAT/ICP0 was cleaved with HindIII and NdeI enzymes to delete the CMV promoter sequence (500 bp). The 14.4-kb vector with a deletion of the promoter was subjected to Klenow fragment (Promega) activity for a filling-in reaction, followed by ligation by T4 ligase (Promega). This vector was then cleaved with BstEII enzyme at nucleotides 119194 and 120091 to delete the LAP2 promoter (897 bp). The 13.5-kb DNA fragment was subjected to ligation reaction to obtain the pΔprLAT vector.
pLAT (Fig. 1E) was derived from pLAT/ICP0 by deletion of the sequences downstream from the 3′ end of the LAT gene; therefore, it is capable of expressing only the LATs. pLAT/ICP0 was cleaved with restriction enzyme MluI at nucleotide 121651 and with BamHI at nucleotides 123459 and 129259. This yielded three DNA fragments, 1.8, 5.8, and 7.3 kb. The 7.3-kb fragment, which contains only the LAT coding sequence, was subjected to Klenow fragment filling-in reaction, followed by ligation by T4 ligase.
Cell lines and viruses.
NA cells (39), a subclone of neuro-2a, a clonal line of C-1300 mouse neuroblastoma cells (1), were kindly provided by A. McMorris, The Wistar Institute, Philadelphia, Pa. Cells were maintained in minimal essential Eagle medium (MEEM) with 10% heat-inactivated fetal calf serum (FCS). CV-1 cells were maintained in Dulbecco’s modified Eagle’s medium with 10% FCS. HSV-1 strain F was obtained from B. Roizman, University of Chicago, Chicago, Ill. FS1001K, a KOS strain-derived LAT-negative mutant (15), was kindly provided by N. Fraser, The Wistar Institute. The viruses were propagated and their titers were determined on CV-1 cells, and results are represented as PFU per milliliter.
Cell transfections.
Transient transfections were performed according to the method of Cullen (14) and as detailed before (37). Cells were harvested for RNA extraction 48 h posttransfection. Stable transfections were performed as described by us before (22). pSV2neo, a plasmid containing the neomycin resistance gene, was used for clonal selection.
Isolation of RNA and Northern blot analysis.
Total RNA was extracted from cells with Tri reagent (Molecular Research Center, Inc.) according to the manufacturer’s instructions. For reverse transcription-PCR (RT-PCR), RNAs were treated with 5 U of RQ1 DNase (Promega), extracted with phenol-chloroform and chloroform, and precipitated in ethanol. Northern blot analysis was performed according to the method of Spivack and Fraser (56). RNA markers (281 to 6,583 bases) were purchased from Promega. Computerized image analysis and quantitation were performed by a Bio-Imaging analyzer (Fuji Corp.) (68).
PCR amplification of reverse-transcribed RNA.
RT-PCR was performed as described by us before (37). The following oligonucleotide primers that flank the intron within the 2.0-kb LAT (Fig. 1F) were prepared according to the published sequence of HSV-1 (45): P2, 5′-GACTCTGTTACTTACCCGTCCGAC-3′ (HSV-1 bases 119612 to 119635), and P3, 5′-GAAAGCATCCTGCCACTGGCATGGA-3′ (bases 120426 to 120402). RT was performed with primer P3.
For RT-PCR analysis of the HSV-1 ICP0 gene, the following primers that flank the first 5′ intron within the gene (Fig. 1F) were used: P11, 5′-TCTCGAACAGTTCCGTGTCCGT-3′ (bases 123130 to 123151), and P10, 5′-TCTCCGCATCACCACAGAAG-3′ (bases 124192 to 124173). RT was performed with primer P11.
For PCR analysis of the ICP27 gene, the following primers were used (Fig. 1B): P12, 5′-CCCTTTCTCCAGTGCTACCTGAA-3′ (bases 114919 to 114941), and P13, 5′-GTGCGTGTCTAGGATTTCGATC-3′ (bases 115170 to 115149).
For RT-PCR analysis of the mouse CuZn superoxide dismutase (CuZn SOD) gene (2), the following primers were used: P14, 5′-GAAAGCGGTGTGCGTGCTGAAG-3′ (SOD exon 1), and P15, 5′-GAGTGAGGATTAAAATGAGGTCC-3′ (SOD exon 3). RT was performed with primer P15.
The primers were chosen according to the following criteria: (i) GC content of the primer of not less than 50%, (ii) absence of significant homology with all rodent and human sequences present in the GenBank database or with other HSV-1 sequences, (iii) size of the PCR product of between 0.2 and 1 kb, and (iv) avoidance of sequences which contain polypurine or polypyrimidine base repetition.
Southern blot analysis.
DNA fragments were resolved by 1.5% agarose gel electrophoresis and transferred onto a GeneScreen Plus membrane (New Research Products), by the salt transfer protocol of the manufacturer. Hybridization and washes were also performed according to the manufacturer’s instructions. The filters were autoradiographed with XAR-5 film at −70°C with intensifying screens (Du Pont).
Preparation of radioactively labeled DNA probes.
The DNA fragments that served as probes for the LATs and ICP0 and ICP4 mRNAs were cleaved from the vector pLAT/ICP0 and purified from an agarose gel. We used the following DNA fragments (Fig. 1B): a BstEII-BamHI probe, covering the HSV-1 genomic region from bp 121068 to 123463; a BstEII-BstEII probe consisting of two equimolar fragments, 897 and 977 bp, covering the genomic sequence from bp 119194 to 121068; and an AflIII-BamHI probe covering the genomic sequence from bp 128059 to 129259.
The 251-bp probe for the ICP27 gene was prepared by PCR of BamHI DNA fragment B (47) with primers P12 and P13.
A 198-bp DNA probe for the housekeeping CuZn SOD gene (2) was prepared by RT-PCR of RNA from TG of mice with primers P14 and P15.
All probes were labeled with [α-32P]dCTP by random priming with the Multiprime DNA labeling system of Amersham. Specific activities of the probes were approximately 108 cpm/μg of DNA.
For preparation of the single-stranded DNA probe for ICP0 mRNA, the following primers were used (Fig. 1F): P11 (described above) and P9, 5′-TGGTGTTGGTGTTACTGCTG-3′ (bases 122310 to 122330). Primers were 5′ end labeled with [γ-32P]dATP followed by PCR with the Taq DNA polymerase (sequencing grade; Promega) according to the instructions in the manual. The PCR was performed in the presence of dATP, dGTP, and dTTP (Boehringer) (1.6 mM each); 0.7 mM [α-32P]dCTP; 100 ng of the BstEII-BamHI DNA fragment; and 20 ng of the primer.
For detection of the 18S rRNA, we used the following 5′-end-labeled single-stranded DNA probe: 5′-CTGTCAATCCTGTCCGTGTCCG-3′, according to the 18S rRNA sequence in GenBank (accession no. X00686, coordinates 1273 to 1294).
Cell infection and virus titration.
NA cells or clones derived from them (2 × 104 cells per well) were seeded in triplicate in 24-well plates for 24 h. Cells were infected at MOIs of 0.1, 1, and 10 in 250 μl of MEEM without serum. After 2 h, the medium was removed and 1 ml of fresh medium with serum was added to every well. Twenty-four hours later, the plates were stored at −70°C.
For HSV-1 titration, the cells and medium of each well were removed and sonicated. CV-1 cells (1.3 × 105 per well) were seeded in 24-well dishes in 0.5 ml of Dulbecco’s modified Eagle’s medium including 10% FCS and 48 h later were used for titration as described elsewhere (60), with 0.5 mg of human gamma globulin per ml.
For RNA extraction, cell lines NA-neo and NA-LAT#1, 2 × 106 cells each, were infected at an MOI of 1 with LAT-negative mutant in 4 ml of MEEM without serum. After 1 h, the medium was removed and 10 ml of fresh medium with serum was added to every plate. Six hours later, the cells were extracted for RNA isolation as described above.
Cell viability assay (MTT).
This method enabled us to estimate the number of viable cells seeded for infection and to calibrate the required numbers of viral PFU, to reach similar MOIs during infection. The assay was based upon the ability of cellular mitochondria to convert MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide [Sigma]) into blue formazan product and was performed according to the procedure of Miller and McDevitt (40). In parallel with seeding of the different NA clones for infection, an extra plate was seeded for the MTT assay with the same cells. The optical density (492 nm) was measured in an enzyme-linked immunosorbent assay plate reader (Organon Teknika) with a reference wavelength of 630 nm.
Chloramphenicol acetyltransferase (CAT) assay.
Cell lysates were analyzed for CAT activity at 48 h after transfection by the technique of Gorman et al. (24). The pIE-CAT construct, containing the CAT gene sequence under the control of the HSV-1 ICP0 promoter (from −585 to +150 [19]), and the pRSV-CAT construct, which contains the CAT gene under the control of the long terminal repeat promoter of Rous sarcoma virus (23), were kindly provided by D. Latchman, London, United Kingdom. Cells (0.5 × 106) were transfected with 1.25 and 0.25 μg of DNA, respectively, as described before (37). Following transfections, cells were harvested and the protein content was determined by the method of Bradford (7). Samples equalized for protein content were assayed for CAT activity. The vector pRSV-CAT was used as a control plasmid to correct for differences in transfection efficiencies. This was done in duplicate for each experiment.
DNA dot blot.
Plasmid DNA was extracted from equal amounts of transfected cells by the Hirt procedure (27), and identical volumes were denatured in 0.4 N NaOH–10 mM EDTA for 10 min at 96°C. The samples were then added to the slot blot apparatus, transferred to a GeneScreen Plus membrane (New Research Products), hybridized, and washed according to the manufacturer’s instructions. Computerized image analysis and quantitation were performed by a Bio-Imaging analyzer as described for RNA.
RESULTS
Establishing neuronal cell lines that stably express the HSV-1 latency-associated gene(s).
In order to examine the effect of the gene(s) expressed during HSV-1 latent infection upon viral replication in neuronal cells and to delineate the DNA sequences which are responsible for any observed effect, we have constructed three vectors. (i) pLAT/ICP0 (Fig. 1C [37]) is a plasmid containing an HSV-1 10.4-kb DNA fragment which includes the entire sequences that are transcriptionally active during latency, under the control of the constitutive IE CMV promoter. This fragment also contains the coding sequences of the ICP0, γ34.5, ORF P, and ORF O genes. (ii) pΔprLAT (Fig. 1D) is a plasmid derived from the pLAT/ICP0 vector by deletion of the CMV and LAP2 (21) promoters. This vector is unable to express the LATs or the mLAT but still contains the coding sequences of ICP0, γ34.5, ORF P, and ORF O. (iii) pLAT (Fig. 1E) is a plasmid derived from pLAT/ICP0 and having deletions of all the sequences downstream from the DNA fragment that transcribes the LATs.
These vectors and pSV2neo were stably transfected into the NA neuronal cell line to produce three G418-resistant populations, NA-LAT/ICP0, NA-ΔprLAT, and NA-LAT. Sixteen cell clones were randomly isolated from each population and were examined by RT-PCR and Northern blot analysis for expression of the relevant transcripts. Clones NA-LAT/ICP0#1 (previously termed NA4 [22]), NA-ΔprLAT#1, and NA-LAT#1, which express these transcripts, were randomly chosen. NA-neo, a negative clone harboring only pSV2neo, was selected as a negative control.
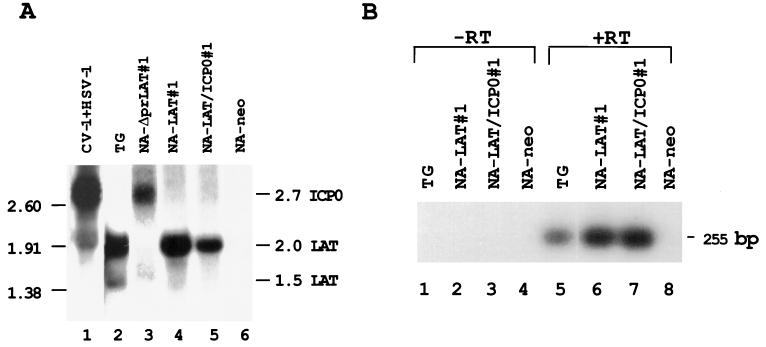
The neuronal cell clones were examined for expression of the 2.0- and 1.5-kb LATs by Northern blot analysis (Fig. 2A) and by RT-PCR (Fig. 2B). While the 2.0-kb LAT was present at amounts high enough to be easily visualized by Northern blot analysis in NA-LAT#1 and NA-LAT/ICP0#1 cells and latently infected mouse TG (Fig. 2A, lanes 4, 5, and 2, respectively) and was absent from NA-ΔprLAT#1 and NA-neo cells (lanes 3 and 6, respectively), the 1.5-kb LAT was produced at low levels and was observed on Northern blots only as a faint band and after overexposure of the film (data not shown). This transcript was clearly identified in RNA from latently infected mouse TG (lane 2). However, a DNA band with an expected size of 255 bp, representative of the 1.5-kb LAT (Fig. 1F), was obtained by RT-PCR analysis of RNA obtained from NA-LAT#1 and NA-LAT/ICP0#1 clones (Fig. 2B, lanes 6 and 7, respectively) and of RNA from TG of latently infected mice (Fig. 2B, lane 5). Appropriate controls without RT enzyme in the reaction mixture gave no DNA band (Fig. 2B, lanes 1 to 4).
FIG. 2.
Transcription of HSV-1 LATs and ICP0 in stably transfected neuronal cell clones. (A) Northern blot analysis of total RNAs obtained from NA-ΔprLAT#1, NA-LAT#1, NA-LAT/ICP0#1, and NA-neo cell clones. Lane 1, RNA from CV-1 cells infected with HSV-1 (MOI of 5) and harvested 5 h postinfection; lane 2, RNA from latently infected TG; lane 3, RNA from NA-ΔprLAT#1 cells; lane 4, RNA from NA-LAT#1 cells; lane 5, RNA from NA-LAT/ICP0#1 cells; lane 6, RNA from NA-neo cells. Ten micrograms of RNA was loaded in each lane, except lanes 1 and 2, which were loaded with 5 μg of RNA. The DNA probe used was BstEII-BamHI. The positions of RNA markers (in kilobases) are indicated on the left. Sizes of the relevant HSV-1 transcripts (in kilobases) are indicated on the right. (B) Southern blot analysis of RT-PCR products from RNAs obtained from NA-LAT#1, NA-LAT/ICP0#1, and NA-neo cells. Lanes 1 and 5, latently infected TG; lanes 2 and 6, NA-LAT#1; lanes 3 and 7, NA-LAT/ICP0#1; lanes 4 and 8, NA-neo. Lanes 1 to 4: control PCR without RT enzyme in the reaction mixture. The 255-bp DNA fragment is representative of the 1.5-kb LATs (Fig. 1F). The DNA probe used was BstEII-BstEII (Fig. 1). RT was performed with primer P3, and PCR was performed with primers P3 and P2.
By Northern blot analysis, clone NA-ΔprLAT#1 was found to produce the ICP0 transcript (Fig. 2A, lane 3). As expected, both the 2.7-kb ICP0 transcript and the 2.0-kb LAT were observed in HSV-1-infected CV-1 cells (Fig. 2A, lane 1).
HSV-1 replication is suppressed in neuronal cell clones that express the LATs.
In order to study the effect of the gene(s) expressed during HSV-1 latency on viral replication in neuronal cells, we infected the three neuronal cell clones, NA-LAT/ICP0#1, NA-ΔprLAT#1, and NA-LAT#1, with HSV-1 (strain F). NA-neo cells were used as a control (Table 1). Twenty-four hours postinfection, plates were stored at −70°C. Prior to titration on CV-1 cells, the infected cells together with the medium were sonicated. All experiments were performed in triplicate. Presented are the averaged results of a representative experiment. To ensure that the MOI would be accurate, control plates were subjected to cell quantification by the MTT assay.
TABLE 1.
LAT effect on HSV-1 replication in the stably transfected neuronal clonesa
| MOI | Value (PFU/ml) for clone:
|
Fold of inhibition for NA-neo vs:
|
||||
|---|---|---|---|---|---|---|
| NA-neo | NA-ΔprLAT#1 | NA-LAT/ICP0#1 | NA-LAT#1 | NA-LAT#1 | NA-LAT/ICP0#1 | |
| 10 | 3.8 × 104 | 2.0 × 104 | 1.6 × 104 | 1.4 × 104 | 2.6 | 2.4 |
| 1 | 3.3 × 103 | 3.5 × 103 | 10 | 15 | 220 | 330 |
| 0.1 | 3.0 × 101 | 3.0 × 101 | <4 | <4 | ||
HSV-1, produced by the LAT-expressing NA-LAT/ICP0#1 and NA-LAT#1 clones and by the control NA-ΔprLAT#1 and NA-neo clones, was quantitated on CV-1 cells. Since titration was performed on 0.25 ml of a total volume of 1 ml, lack of plaques in the titration assay is represented as <4. All experiments were performed in triplicate, and the results represent the average calculations.
At an MOI of 0.1, no HSV-1 replication could be detected in NA-LAT/ICP0#1 and NA-LAT#1 cells (Table 1). Since titration was performed in 0.25 ml of a total volume of 1 ml, lack of plaques on titration assay is represented as <4. HSV-1 replication was detectable in NA-neo and NA-ΔprLAT#1 cell clones. At an MOI of 1, HSV-1 replication was markedly suppressed in LAT/ICP0#1 and NA-LAT#1 cells compared to that in NA-neo cells by similar orders of magnitude of about 330- and 220-fold, respectively. The NA-ΔprLAT#1 cells were unable to suppress HSV-1 replication. At an MOI of 10, the suppressive effect of the LATs upon HSV-1 replication was almost completely abolished. Similar results were obtained in repeated experiments, with the same cell clones, and in experiments with three additional cell clones randomly selected from the NA-LAT/ICP0, NA-ΔprLAT, and NA-LAT cell populations (data not shown).
To validate these results, similar infection experiments were carried out with the three neuronal cell populations as well. The parental neuronal NA cells were used as the control. As expected, HSV-1 replication was repressed in the LAT-expressing populations (NA-LAT/ICP0 and NA-LAT) compared to that in the NA-ΔprLAT and NA populations, and the suppressive effect was MOI dependent (data not shown).
Thus, HSV-1 replication was suppressed in NA-LAT/ICP0#1 cells, which contain the entire DNA sequence transcriptionally active during latency, including the coding sequences for the LATs, mLAT, ICP0, γ34.5, ORF P, and ORF O. A similar effect was demonstrated with NA-LAT#1 cells, which are capable of expressing only the LATs, suggesting that the observed inhibition of HSV-1 replication is mediated by the LATs and not by the ICP0, γ34.5, ORF P, or ORF O gene. Indeed, the repressive effect upon HSV-1 replication was not present in NA-ΔprLAT#1 cells, which are incapable of expressing the LATs but express ICP0 and also contain the three genes γ34.5, ORF P, and ORF O.
The suppressive effect of the LATs upon HSV-1 replication was MOI dependent (Table 1). At an MOI of 0.1, no viral replication could be detected in NA-LAT/ICP0#1 and NA-LAT#1 cells, compared to that in NA-neo cells, while at an MOI of 1 the differences were 330- and 220-fold, respectively. At an MOI of 10, the suppressive effect was almost completely abolished.
Replication of HSV-1 LAT-negative mutant is suppressed in a neuronal cell clone that expresses the LATs.
In order to further substantiate the findings that HSV-1 LATs suppress viral replication in neuronal cell clones, we repeated these experiments in parallel and under similar conditions, with a LAT-negative HSV-1 mutant. We used the FS1001K mutant, in which a 1.1-kb fragment starting from the 5′ end is deleted from the 2.0-kb LAT coding sequence to abolish all LAT expression (15).
Twenty-four hours postinfection, virus yield in NA-LAT#1 and NA-neo cells was determined by titration on CV-1 indicator cells. This experiment was performed in triplicate, and the average results are presented in Table 2. HSV-1 LAT-negative mutant replication was suppressed in NA-LAT#1 cells compared to that in the control NA-neo cells. The effect was again MOI dependent, i.e., more pronounced at a low MOI of 0.1, where the inhibition was 160-fold. Similar results were obtained in repeated experiments. These results substantiate the findings that the LATs suppress HSV-1 replication in neuronal cells and indicate that the LAT effect has a trans-acting component.
TABLE 2.
LAT effect on HSV-1 LAT-negative mutant replication in the stably transfected neuronal clonesa
| MOI | Value (PFU/ml) for clone:
|
Fold of inhibition for NA-neo vs NA-LAT#1 | |
|---|---|---|---|
| NA-neo | NA-LAT#1 | ||
| 10 | 2.3 × 105 | 7.0 × 104 | 3.3 |
| 1 | 3.9 × 105 | 1.6 × 104 | 24.4 |
| 0.1 | 4.8 × 102 | 3.0 | 160 |
FS1001K LAT-negative mutant, produced by the LAT-expressing NA-LAT#1 and the control NA-neo clones, was quantitated on CV-1 cells. All experiments were performed in triplicate, and the results represent the average calculations.
ICP0 mRNA levels are suppressed in a neuronal cell line that expresses the LATs.
Since HSV-1 replication is dependent on the expression of the viral IE genes, we hypothesized that the LATs may reduce HSV-1 replication in neuronal cells by down-regulating expression of the IE genes. We chose initially to examine the influence of the LATs on ICP0 expression for the following reasons. (i) ICP0 is the first IE gene to be expressed at very early times postinfection (36). (ii) This IE protein is a transactivator which enhances HSV-1 replication during acute infection as well as reactivation from latency (9) and is capable of activating all three kinetic classes of HSV genes (10). (iii) The sequences of ICP0 mRNA and of the LATs overlap at their 3′ end (and therefore an antisense mechanism for LAT action has been proposed elsewhere [64]). (iv) The pLAT/ICP0 vector used in this work (Fig. 1C) includes the entire DNA sequences required for ICP0 expression (46). (v) The LATs have been shown elsewhere to inhibit ICP0 protein synthesis (20) and ICP0-mediated transactivation (16).
(i) The levels of ICP0 mRNA produced by an HSV-1 LAT-negative mutant are suppressed in a neuronal cell line that expresses the LATs.
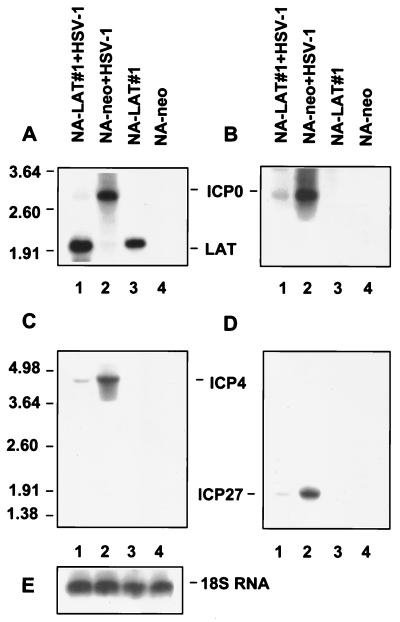
The LAT effect upon ICP0 expression was first examined by infecting LAT-expressing NA-LAT#1 and NA-neo control cells with the LAT-negative mutant at an MOI of 1. RNA was extracted 6 h postinfection, resolved by gel electrophoresis, Northern blotted, and hybridized with the BstEII-BamHI DNA probe specific to both ICP0 mRNA and LATs (Fig. 1B). ICP0 mRNA was barely expressed in NA-LAT#1 cells (Fig. 3A, lane 1) and was readily identified in NA-neo cells (lane 2). Bio-Imager quantification of ICP0 mRNA suggested a 16-fold inhibition.
FIG. 3.
Expression of ICP0, ICP4, and ICP27 genes by an HSV-1 LAT-negative mutant following infection of a LAT-expressing neuronal cell line. Results of Northern blot analysis of total RNAs (10 μg) obtained from NA-LAT#1 and NA-neo cells following infection (MOI of 1) with HSV-1 LAT-negative mutant are shown. RNAs were harvested 6 h postinfection. The order of the lanes is identical in all panels. Lane 1, RNA from infected NA-LAT#1 cells; lane 2, RNA from infected NA-neo cells; lane 3, RNA from control NA-LAT#1 cells; lane 4, RNA from control NA-neo cells. Each of the four identical filters was hybridized with a different probe (Fig. 1B and Materials and Methods): BstEII-BamHI probe for the LATs and ICP0 (A), single-stranded DNA probe specific for ICP0 mRNA (B), AflIII-BamHI probe for ICP4 (C), and PCR product specific for ICP27 (D). The positions of RNA markers (in kilobases) are indicated on the left. The relevant HSV-1 transcripts are indicated in the center. (E) 18S rRNA hybridized with a specific single-stranded DNA probe (see Materials and Methods).
To confirm that the identified band was indeed ICP0 mRNA, we repeated this experiment with an ICP0-specific single-stranded DNA probe (Fig. 3B). As expected, a 2.7-kb RNA band, of the size of ICP0 mRNA, was identified in RNA obtained from NA-neo cells infected with HSV-1 (lane 2) and not in control RNA from uninfected cells (lanes 3 and 4). The ICP0 mRNA was barely visualized in the HSV-1-infected LAT-expressing NA-LAT#1 cells (lane 1). To validate the finding that equal amounts of RNA were loaded in each lane, we also hybridized the blot with a single-stranded DNA probe specific for the 18S rRNA (Fig. 3E). Measurement of rRNA levels by Bio-Imager showed a 1.1-fold difference in the amounts of RNA from NA-LAT#1-infected cells compared to that from NA-neo-infected control cells (Fig. 3E, lanes 1 and 2, respectively).
These results indicate that expression of HSV-1 LATs in a neuronal cell clone reduces the ICP0 mRNA levels. It also shows that this effect can be mediated in trans.
(ii) LATs reduce ICP0 mRNA levels following transfection of pLAT/ICP0 into NA cells.
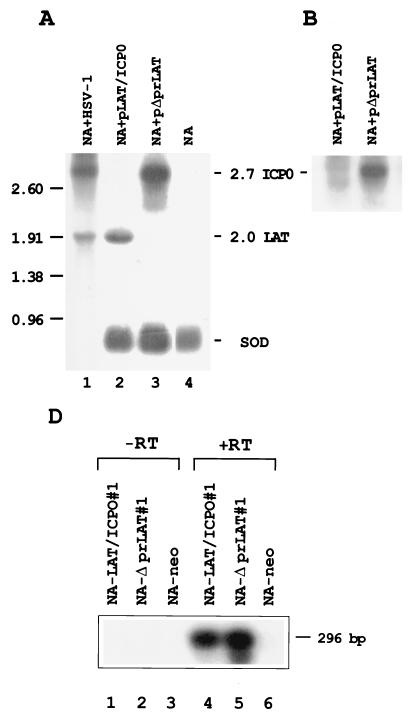
To rule out the possibility that the LATs interfere with HSV-1 entry into cells, with subsequent reduction of ICP0 mRNA, and to examine whether the LAT effect upon ICP0 mRNA is a direct one, we used a second approach. NA cells were transfected with either pLAT/ICP0, which contains the LAT and ICP0 genes (Fig. 1C), or with pΔprLAT, which has deletions of the CMV promoter and of LAP2 (Fig. 1D) and is therefore unable to express the LATs but still expresses ICP0 (Fig. 2).
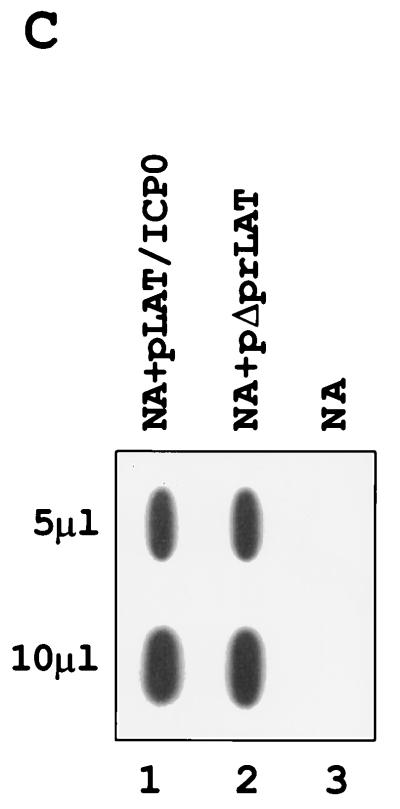
RNAs were extracted from NA cells 48 h posttransfection, resolved by gel electrophoresis, Northern blotted, and hybridized with a BstEII-BamHI probe specific to both ICP0 mRNA and the LATs (Fig. 1B). While ICP0 mRNA was visualized in NA cells transfected with pΔprLAT (Fig. 4A, lane 3), no ICP0 mRNA was observed in pLAT/ICP0-transfected NA cells (lane 2). Bio-Imager quantification suggested a difference of at least 58-fold in ICP0 mRNA levels between the NA cells transfected with each of these vectors. As described previously, single-stranded ICP0-specific probe confirmed the identity of the ICP0 mRNA (Fig. 4B), following rehybridization of the same blot shown in Fig. 4A. To ensure that equal amounts of RNA were loaded in each lane, we also hybridized the blot with the SOD probe (2) (Fig. 4A, bottom). Measurement of SOD mRNA levels by Bio-Imager showed a 1.4-fold difference in the amounts of RNA from pΔprLAT-transfected cells compared to that from pLAT/ICP0-transfected cells (Fig. 4A, lanes 3 and 2, respectively). To quantitate the efficiency of transfection in the NA cells, a Hirt procedure followed by DNA dot blot hybridization was performed. It indicated that similar amounts of pΔprLAT and pLAT/ICP0 DNAs entered the NA cells (Fig. 4C).
FIG. 4.
Transcription of ICP0 and LATs in a transfected neuronal cell line. (A) Northern blot analysis of total RNAs (10 μg) obtained from NA cells following transient transfection with vectors pLAT/ICP0 and pΔprLAT. Lane 1, RNA from NA cells infected with HSV-1 (MOI of 5) and harvested 5 h postinfection; lane 2, RNA from NA cells transfected with pLAT/ICP0 vector; lane 3, RNA from NA cells transfected with pΔprLAT vector; lane 4, RNA from NA control cells. The probe used is the BstEII-BamHI DNA fragment (Fig. 1). The positions of RNA markers (in kilobases) are shown on the left. Sizes (in kilobases) of the relevant HSV-1 transcripts are indicated on the right. At the bottom is shown the mRNA of the CuZn SOD gene hybridized with an SOD-specific probe (see Materials and Methods). (B) Rehybridization of the same blot as in panel A with a single-stranded DNA probe specific for ICP0 mRNA. (C) Dot blot analysis of plasmid DNA extracted from transfected NA cells by the Hirt procedure. Lane 1, NA cells transfected with pLAT/ICP0 vector; lane 2, NA cells transfected with pΔprLAT; lane 3, control NA cells. The amount of the DNA in each row is indicated in microliters. The probe used is the BstEII-BamHI DNA fragment (Fig. 1). (D) Southern blot analysis of RT-PCR products from total RNAs obtained from NA-LAT/ICP0#1, NA-ΔprLAT#1, and NA-neo cells. Lanes 1 and 4, NA-LAT/ICP0#1; lanes 2 and 5, NA-ΔprLAT#1; lanes 3 and 6, NA-neo; lanes 1 to 3, control PCR without RT enzyme in the reaction. RT was performed with primer P11, and PCR was performed with P11 and P10. The probe used is the BstEII-BamHI DNA fragment. The 296-bp DNA is representative of the spliced ICP0 mRNA (Fig. 1F).
To investigate whether in the presence of the LATs small amounts of ICP0 mRNA may still be detected, we have applied the much more sensitive technique of RT-PCR followed by Southern blot analysis. The 296-bp DNA fragment representative of the spliced ICP0 mRNA (Fig. 1F) was present in the two respective preparations (Fig. 4D, lanes 4 and 5). Appropriate controls without RT enzyme in the reaction mixture identified no DNA band (lanes 1 to 3). This would indicate that, while the LATs suppress ICP0 mRNA, residual levels may still be detected.
In order to confirm that the results of the transfection experiments were not due to interference by the strong CMV promoter in pLAT/ICP0 with the native ICP0 promoter, we cotransfected 2.5 μg of pIE-0-CAT (a vector containing the CAT gene under the control of the HSV-1 ICP0 promoter) with increasing amounts (0.5 to 2.5 μg) of the CMV promoter cloned in the expression vector pCH2N (37). No effect upon CAT expression from the presence of the CMV promoter was noted in the cotransfected NA cells (data not shown).
Taken together, these results (i) indicate that HSV-1 LATs directly reduce ICP0 mRNA levels in neuronal cells and (ii) rule out the possibility that the findings are the outcome of entry of different viral DNA amounts into the two neuronal cell lines or are an experimental in vitro artifact due to the influence of the CMV promoter upon the ICP0 promoter.
LATs do not reduce ICP0 mRNA levels via repression of the ICP0 promoter.
Since HSV-1 LATs substantially reduce the steady-state levels of ICP0 mRNA, we went on to examine their direct effect upon the ICP0 promoter. pIE-0-CAT (19) was transfected into NA-LAT#1 and into NA-neo control cells. CAT activity was determined in duplicate in crude cell extracts (Table 3). All experiments included a control plasmid, pRSV-CAT, in which the Rous sarcoma virus promoter drives the expression of CAT (23), to correct for differences in transfection efficiency. This control vector was transfected in duplicate in each experiment, and the reported results represent the average.
TABLE 3.
LAT effect upon ICP0 promotera
| Vector | Extract (μl) | % for clone:
|
NA-LAT#1 vs NA-neo | |
|---|---|---|---|---|
| NA-neo | NA-LAT#1 | |||
| pIE-0-CAT | 2 | 9.1 | 36.2 | 3.9 |
| 4 | 18.7 | 69.6 | 3.7 | |
| pRSV-CAT | 2.5 | 10.3 | 37.9 | 3.6 |
| 5.0 | 19.9 | 68.0 | 3.4 | |
Quantification of results was performed with the Bio-Imager analyzer. Values indicate the percentages of chloramphenicol acetylated in lysates prepared from the transfected cells. All transfections included a control plasmid (pRSV-CAT) to correct for differences in transfection efficiency. The control plasmid was transfected in duplicate in each experiment, and the reported results represent the averages.
While the vector pIE-0-CAT expressed CAT in NA-LAT#1 cells at about 3.8-fold-higher levels relative to that in NA-neo cells, the control vector, pRSV-CAT, also expressed about 3.5-fold-higher amounts of CAT in NA-LAT#1 cells relative to that in NA-neo cells. Thus, the difference in the levels of activity of the ICP0 promoter between these two cell lines originated from the difference in transfection efficiencies rather than from any LAT effect on the ICP0 promoter.
The levels of ICP4 and ICP27 mRNA produced by HSV-1 LAT-negative mutant are suppressed in a neuronal cell line that expresses the LATs.
Following the observation that ICP0 mRNA was reduced by the LATs, we went on to examine LAT effects upon ICP4 and ICP27 mRNA levels, because these two IE genes are essential for viral replication. Since it was proposed that the LATs affect ICP0 via an antisense mechanism, the effect of the LATs on the expression of other IE genes might shed light upon the LAT action mechanisms.
Measurements of ICP4 and ICP27 mRNA levels were performed in parallel with those of ICP0 mRNA levels, on the RNA samples from NA-LAT#1 and NA-neo cells following infection with HSV-1 LAT-negative mutant at an MOI of 1. RNAs were resolved by gel electrophoresis, Northern blotted, and hybridized with an AflIII-BamHI DNA probe specific for ICP4 mRNA (Fig. 1B) or with the PCR product-specific probe for ICP27 mRNA (see Materials and Methods). Both ICP4 and ICP27 mRNA levels were reduced in the NA-LAT#1 cells compared to the control NA-neo cells (Fig. 3C, lanes 1 and 2, and 3D, lanes 1 and 2, respectively). Bio-Imager quantification indicated a difference of about fourfold in ICP4 mRNA levels and about threefold in ICP27 mRNA levels in NA-neo cells compared to that in NA-LAT#1 cells, after correction for different amounts of RNA loaded from NA-LAT#1 and from NA-neo cells (1.1-fold).
These results indicate that the LATs also reduce ICP4 and ICP27 mRNA levels in the neuronal cell line.
DISCUSSION
The LATs suppress viral replication in neuronal cell lines.
Our aim in this study was to examine the role of the latency-associated gene(s) in HSV-1 replication and to map the viral genomic region responsible for any observed effect. Any approach to addressing this question must take into account the fact that, within the sequence(s) of the HSV-1 latency-associated gene(s), other genes such as γ34.5, ICP0, ORF P, and ORF O and transcripts designated L/STs (69) are located. Therefore, we constructed three different vectors which enabled us to separate the expression of LAT from that of the other genes expressed from this viral genomic region.
Our findings demonstrate that, at an MOI of 0.1, no HSV-1 replication could be detected in the NA-LAT/ICP0#1 neuronal cell clone and that, at an MOI of 1, it was reduced up to 330-fold compared to that in the control NA-neo cells (Table 1). However, the other genes located within pLAT/ICP0 may have also affected HSV-1 replication (8, 10, 44, 48, 55). In order to delineate the viral locus responsible for replication inhibition, we used the two other cell clones, NA-LAT#1 and NA-ΔprLAT#1. Our findings indicate that the suppressive effect upon HSV-1 replication may be mediated by the LATs alone and not by the other genes located downstream from the LAT gene (Table 1). These results were substantiated by a complementary approach, in which a virus having the LAT gene deleted was applied. We showed that a LAT-negative HSV-1 mutant replicated better in the control NA-neo cells than in NA-LAT#1, the latter cell line complementing in trans the lack of LATs (Table 2). In this experiment, we conclusively indicate that the LATs may act via a trans mechanism.
In comparing replication abilities of the LAT-negative mutant and HSV-1 in the various NA cell clones, two observations should be noted: (i) the LAT-negative mutant replicated better than HSV-1 (in 10- to 100-fold-higher titers) in NA-neo cells (Tables 1 and 2), and (ii) the suppressive effect upon viral replication was less pronounced with the LAT-negative mutant. These findings may suggest that the suppressive effect upon HSV-1 replication is dependent upon the cellular amounts of the LATs and that it has also a cis-acting component.
The impact of the LATs upon HSV-1 replication was MOI dependent, being more pronounced at lower MOIs. This probably reflects a biologically important mechanism, since the viral titer that is obtained during acute infection in mice TG suggests a low viral load (reference 61 and our unpublished data). A similar MOI-dependent effect was demonstrated for the neuronal transcription factors Oct-2 (35) and N-Oct-3 (25), which suppressed viral replication in neuronal cells. It seems, therefore, that cellular and viral factors, which act to reduce HSV-1 replication in neuronal cells, are most effective at low viral loads, which are indeed present during the in vivo infection.
The LATs reduce ICP0, ICP4, and ICP27 mRNA levels in neuronal cell clones.
To study the mechanism responsible for viral replication suppression by the LATs, we chose first to examine the influence of LATs on ICP0 gene expression. Using an HSV-1 LAT-negative mutant for infection of LAT-expressing neuronal cell lines, we demonstrated that the LATs reduce ICP0 mRNA steady-state levels at an MOI of 1 (Fig. 3A), an HSV-1 MOI that also showed effective suppression of viral replication in these cells (Tables 1 and 2). These findings are in accordance with studies indicating that ICP0-negative mutants grow poorly at low MOIs (9, 10, 51). A transfection experiment with a plasmid coding for both ICP0 and the LATs gave identical results (Fig. 4A), indicating that the LAT effect upon ICP0 steady-state mRNA levels was most probably mediated directly and not through other viral gene products. The LAT effect upon ICP0 mRNA steady-state levels is not the result of ICP0 promoter suppression. This conclusion is based on the observation that the LATs were unable to reduce CAT expression driven by the ICP0 promoter, following transfection of pIE-0-CAT into NA-LAT#1, the LAT-expressing neuronal cell line (Table 3).
HSV-1 infection experiments have also shown that the LATs reduce the steady-state mRNA levels of two other IE genes, ICP4 and ICP27 (Fig. 3C and D). However, while the LATs suppressed ICP0 mRNA by 16-fold, they reduced ICP4 and ICP27 mRNA by only 3- to 4-fold.
How do the LATs act upon HSV-1 IE genes? The LAT sequence overlaps the 3′ end of the ICP0 transcript and therefore may regulate its turnover by an antisense mechanism (16, 64). Nevertheless, an antisense mechanism for ICP0 inhibition was not supported by a genetic approach that used an HSV mutant with deletion of the LAT-ICP0 overlapping sequences (43). Our present finding that the LATs act also upon ICP4 and ICP27 does not resolve this question. It might be that the LATs act directly and in a similar fashion upon all three IE genes, making the antisense hypothesis for LAT action unlikely. Alternatively, the reduced ICP4 and ICP27 mRNA levels in the presence of the LATs might be a consequence of reduced ICP0 mRNA levels. Indeed, it was shown that, in the absence of structural viral proteins such as Vmw65 (10) and at a low MOI, ICP0 can activate the expression of ICP4 and ICP27 genes (10, 30, 36). Further experiments are in progress to address the mechanism of action of the LATs upon ICP0, ICP4, and ICP27.
The possible mechanism(s) of LAT action upon HSV-1 reactivation.
In general, the consistent phenotype of LAT-negative mutants is defective reactivation (4, 6, 26, 42, 53, 61, 67). However, no evidence to prove a direct role for LATs in reactivation has so far been presented. The other possibility is that this gene(s) functions to promote the ability of HSV-1 to establish latent infection in neurons, increasing the pool of latently infected cells and indirectly improving reactivation ability (3, 53). This is supported by the recent observation that HSV-1 reactivation efficacy correlated with its ability to establish latent infection in a high percentage of neurons (66).
Our findings indicate that HSV-1 LATs suppress viral replication in neuronal cell lines in parallel with reduction of ICP0, ICP4, and ICP27 mRNA levels. By reducing these IE gene mRNA levels, the LATs may keep HSV-1 replication below levels which culminate in neuronal cell destruction and thus facilitate the establishment of viral latent infection. This hypothesis is in accordance with the observations derived from studies with latently infected mouse DRG tissues of increased numbers (i) of latently infected cells with a LAT-positive virus compared to those with LAT-negative mutants (38, 52, 53, 66) and (ii) of neurons expressing ICP4 and ICP27 during LAT-negative mutant infection compared to wild-type virus (11, 18).
Why would the LATs suppress IE genes and hence replication during primary infection but not during reactivation? Partial explanations may be based on the findings that the cellular amounts of LATs decrease during reactivation (57) and/or that, following a reactivation trigger, early genes are expressed first and prior to IE gene expression, bypassing the LAT blocking effect upon IE genes (65).
However, the LATs may not be the only factors involved in the establishment of latency, as viruses lacking LATs are still capable of establishing latent infection (28, 29, 54, 61). ORF P and ORF O together can block the expression of regulatory proteins essential for viral replication (8, 48), and at least two neuronal transcription factors may promote the establishment of latency via blockage of the transactivating action of Vmw65 (25, 35).
Further investigation of the mode of LAT action upon HSV-1 IE genes may now be performed in the cell lines constructed and characterized in this work.
ACKNOWLEDGMENTS
We thank O. Abramsky for continuous interest in our work. We are grateful to Haya Falk and Itamar Goren for technical assistance and to A. Honigman and H. Rosen for critical readings of the manuscript.
This work was supported in part by grants from the Chief Scientist, Ministry of Health, State of Israel; from the Hermann J. Abs Research Program of the Deutsche Bank; from the German-Israel Foundation (GIF) for Research; from the Israeli Ministry of Science; and from the DKFZ, Heidelberg, Germany. D.G. was supported in part by a grant from the Center of Absorption of Scientists, Ministry of Absorption, State of Israel.
REFERENCES
- 1.Augusti-Tocco G, Sato G. Establishment of functional clonal lines of neurons from mouse neuroblastoma. Proc Natl Acad Sci USA. 1969;64:311–315. doi: 10.1073/pnas.64.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benedetto M T, Anzai Y, Gordon J W. Isolation and analysis of the mouse genomic sequence encoding Cu2+-Zn2+ superoxide dismutase. Gene. 1991;99:191–195. doi: 10.1016/0378-1119(91)90126-v. [DOI] [PubMed] [Google Scholar]
- 3.Birmanns B, Reibstein I, Steiner I. Characterization of an in vivo reactivation model of herpes simplex virus from mice trigeminal ganglia. J Gen Virol. 1993;74:2487–2491. doi: 10.1099/0022-1317-74-11-2487. [DOI] [PubMed] [Google Scholar]
- 4.Block T M, Deshmane S, Masonis J, Maggioncalda J, Valyi-Nagi T, Fraser N W. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology. 1993;192:618–630. doi: 10.1006/viro.1993.1078. [DOI] [PubMed] [Google Scholar]
- 5.Block T M, Spivack J G, Steiner I, Deshmane S, McIntosh M T, Lirette R P, Fraser N W. A herpes simplex virus type 1 latency-associated transcript mutant reactivates with normal kinetics from latent infection. J Virol. 1990;64:3417–3426. doi: 10.1128/jvi.64.7.3417-3426.1990. . (Erratum, 64:4603.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom D C, Hill J M, Devi-Rao G, Wagner E K, Feldman L T, Stevens J G. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol. 1996;70:2449–2459. doi: 10.1128/jvi.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Bruni R, Roizman B. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc Natl Acad Sci USA. 1996;93:10423–10427. doi: 10.1073/pnas.93.19.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai W, Astor T L, Liptak L M, Cho C, Coen D M, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol. 1992;66:2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S-H, Kramer M F, Schaffer P A, Coen D M. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71:5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou J, Roizman B. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+ J Virol. 1990;64:1014–1020. doi: 10.1128/jvi.64.3.1014-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croen K D, Ostrove J M, Dragovic L J, Smialek J E, Straus S E. Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate early gene “anti-sense” transcript by in situ hybridization. N Engl J Med. 1987;317:1427–1432. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- 14.Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 15.Fareed M U, Spivack J G. Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J Virol. 1994;68:8071–8081. doi: 10.1128/jvi.68.12.8071-8081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrell M J, Dobson A T, Feldman L T. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci USA. 1991;88:790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser N W, Block T M, Spivack J G. The latency-associated transcripts of herpes simplex virus: RNA in search of function. Virology. 1992;191:1–8. doi: 10.1016/0042-6822(92)90160-q. [DOI] [PubMed] [Google Scholar]
- 18.Garber D A, Schaffer P A, Knipe D M. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol. 1997;71:5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelman I H, Silverstein S. Herpes simplex virus immediate-early promoters are responsive to virus and cell trans-acting factors. J Virol. 1987;61:2286–2296. doi: 10.1128/jvi.61.7.2286-2296.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glorioso J C, Deluca N A, Fink D J. Development and application of HSV-1 vectors for human gene therapy. Annu Rev Microbiol. 1995;49:675–710. doi: 10.1146/annurev.mi.49.100195.003331. [DOI] [PubMed] [Google Scholar]
- 21.Goins W F, Sternberg L R, Croen K D, Krause P R, Hendricks R L, Fink D J, Straus S E, Levine M, Glorioso J C. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J Virol. 1994;68:2239–2252. doi: 10.1128/jvi.68.4.2239-2252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldenberg D, Mador N, Ball M J, Panet A, Steiner I. The abundant latency-associated transcripts of herpes simplex virus type 1 are bound to polyribosomes in cultured neuronal cells and during latent infection in mouse trigeminal ganglia. J Virol. 1997;71:2897–2904. doi: 10.1128/jvi.71.4.2897-2904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorman C M, Merlino G T, Willingham M C, Pastan I, Howard B. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci USA. 1982;79:6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagmann M, Georgiev O, Schaffner W, Douville P. Transcription factors interacting with herpes simplex virus α gene promoters in sensory neurons. Nucleic Acids Res. 1995;23:4978–4985. doi: 10.1093/nar/23.24.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill J M, Sedarati F, Javier R T, Wagner E K, Stevens J G. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology. 1990;174:117–125. doi: 10.1016/0042-6822(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 27.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 28.Ho D Y, Mocarski E S. Herpes simplex virus latent RNA (LAT) is not required for latent infection in the mouse. Proc Natl Acad Sci USA. 1989;86:7596–7600. doi: 10.1073/pnas.86.19.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Javier R T, Stevens J G, Dissette V B, Wagner E K. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology. 1988;166:254–257. doi: 10.1016/0042-6822(88)90169-9. [DOI] [PubMed] [Google Scholar]
- 30.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp L M, Dent C L, Latchman D S. Octamer motif mediates transcriptional repression of HSV immediate early genes and octamer-containing cellular promoters in neuronal cells. Neuron. 1990;4:215–222. doi: 10.1016/0896-6273(90)90096-x. [DOI] [PubMed] [Google Scholar]
- 32.Lagunoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the γ134.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J Virol. 1994;68:6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leib D A, Bogard C L, Kosz-Vnenchak M, Hicks K A, Coen D M, Knipe D M, Schaffer P A. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989;63:2893–2900. doi: 10.1128/jvi.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lillycrop K A, Estridge J K, Latchman D S. The octamer binding protein Oct-2 inhibits transactivation of the herpes simplex virus immediate-early genes by the virion protein Vmw65. Virology. 1993;196:888–891. doi: 10.1006/viro.1993.1552. [DOI] [PubMed] [Google Scholar]
- 35.Lillycrop K A, Howard M K, Estridge J K, Latchman D S. Inhibition of herpes simplex virus infection by ectopic expression of neuronal splice variants of the Oct-2 transcription factor. Nucleic Acids Res. 1994;22:815–820. doi: 10.1093/nar/22.5.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lium E K, Silverstein S. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J Virol. 1997;71:8602–8614. doi: 10.1128/jvi.71.11.8602-8614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mador N, Panet A, Latchman D, Steiner I. Expression and splicing of the latency-associated transcripts of herpes simplex virus type 1 in neuronal and non-neuronal cell lines. J Biochem. 1995;117:1288–1297. doi: 10.1093/oxfordjournals.jbchem.a124857. [DOI] [PubMed] [Google Scholar]
- 38.Maggioncalda J, Mehta A, Su Y H, Fraser N W, Block T M. Correlation between herpes simplex virus type 1 rate of reactivation from latent infection and the number of infected neurons in trigeminal ganglia. Virology. 1996;225:72–81. doi: 10.1006/viro.1996.0576. [DOI] [PubMed] [Google Scholar]
- 39.McMorris F A, Ruddle F H. Expression of neuronal phenotypes in neuroblastoma cell hybrids. Dev Biol. 1974;39:226–246. doi: 10.1016/0012-1606(74)90237-1. [DOI] [PubMed] [Google Scholar]
- 40.Miller R R, McDevitt C A. A quantitative microwell assay for chondrocyte cell adhesion. Anal Biochem. 1991;192:380–383. doi: 10.1016/0003-2697(91)90552-5. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell W J, Lirette R P, Fraser N W. Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J Gen Virol. 1990;71:125–132. doi: 10.1099/0022-1317-71-1-125. [DOI] [PubMed] [Google Scholar]
- 42.Perng G-C, Dunkel E C, Geary P A, Slanina S M, Ghiasi H, Kaiwar R, Nesburn A B, Wechsler S L. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol. 1994;68:8045–8055. doi: 10.1128/jvi.68.12.8045-8055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perng G-C, Ghiasi H, Slanina S M, Nesburn A B, Wechsler S L. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol. 1996;70:976–984. doi: 10.1128/jvi.70.2.976-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perng G-C, Thompson R L, Sawtell N M, Taylor W E, Slanina S M, Ghiasi H, Kaiwar R, Nesburn A B, Wechsler S L. An avirulent ICP34.5 deletion mutant of herpes simplex virus type 1 is capable of in vivo spontaneous reactivation. J Virol. 1995;69:3033–3041. doi: 10.1128/jvi.69.5.3033-3041.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry L J, McGeoch D J. The DNA sequences of the long repeat region and adjoining parts of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:2831–2846. doi: 10.1099/0022-1317-69-11-2831. [DOI] [PubMed] [Google Scholar]
- 46.Perry L J, Rixon F J, Everett R D, Frame M G, McGeoch D J. Characterization of the IE110 gene of herpes simplex virus type 1. J Gen Virol. 1986;67:2365–2380. doi: 10.1099/0022-1317-67-11-2365. [DOI] [PubMed] [Google Scholar]
- 47.Post L E, Conley A J, Mocarski E S, Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc Natl Acad Sci USA. 1980;77:4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Randale G, Lagunoff M, Roizman B. The product of ORF O located within the domain of herpes simplex virus 1 genome transcribed during latent infection binds to and inhibits in vitro binding of infected cell protein 4 to its cognate DNA site. Proc Natl Acad Sci USA. 1997;94:10379–10384. doi: 10.1073/pnas.94.19.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rock D L, Nesburn A B, Ghiasi H, Ong J, Lewis T L, Lokensgard J R, Wechsler S L. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987;61:3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roizman B, Sears A E. An inquiry into the mechanisms of herpes simplex virus latency. Annu Rev Microbiol. 1987;41:543–571. doi: 10.1146/annurev.mi.41.100187.002551. [DOI] [PubMed] [Google Scholar]
- 51.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawtell N M. Comprehensive quantification of herpes simplex virus latency at the single cell level. J Virol. 1997;71:5423–5431. doi: 10.1128/jvi.71.7.5423-5431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawtell N M, Thompson R L. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol. 1992;66:2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sedarati F, Izumi K M, Wagner E K, Stevens J G. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine sensory neurons. J Virol. 1989;63:4455–4458. doi: 10.1128/jvi.63.10.4455-4458.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spivack J G, Fareed M U, Valyi-Nagy T, Nash T C, O’Keefe J S, Gesser R M, McKie E A, MacLean A R, Fraser N W, Brown S M. Replication, establishment of latent infection, expression of the latency-associated transcripts and explant reactivation of herpes simplex virus type 1 γ34.5 mutants in a mouse eye model. J Gen Virol. 1995;76:321–332. doi: 10.1099/0022-1317-76-2-321. [DOI] [PubMed] [Google Scholar]
- 56.Spivack J G, Fraser N W. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987;61:3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spivack J G, Fraser N W. Expression of herpes simplex virus type 1 latency-associated transcripts in the trigeminal ganglia of mice during acute infection and reactivation of latent infection. J Virol. 1988;62:1479–1485. doi: 10.1128/jvi.62.5.1479-1485.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steiner I, Kennedy P G E. Herpes simplex virus latency in the nervous system—a new model. Neuropathol Appl Neurobiol. 1991;17:433–440. doi: 10.1111/j.1365-2990.1991.tb00747.x. [DOI] [PubMed] [Google Scholar]
- 59.Steiner I, Kennedy P G E. Herpes simplex virus latent infection in the nervous system. J Neurovirol. 1995;1:19–29. doi: 10.3109/13550289509111007. [DOI] [PubMed] [Google Scholar]
- 60.Steiner I, Spivack J G, Deshmane S L, Ace C I, Preston C M, Fraser N W. A herpes simplex virus type 1 mutant containing a nontransinducing Vmw65 protein establishes latent infection in vivo in the absence of viral replication and reactivates efficiently from explanted trigeminal ganglia. J Virol. 1990;64:1630–1638. doi: 10.1128/jvi.64.4.1630-1638.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steiner I, Spivack J G, Lirette R P, Brown S M, MacLean A R, Subak-Sharpe J H, Fraser N W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989;8:505–511. doi: 10.1002/j.1460-2075.1989.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steiner I, Spivack J G, O’Boyle II D R, Lavi E, Fraser N W. Latent herpes simplex virus type 1 transcription in human trigeminal ganglia. J Virol. 1988;62:3493–3496. doi: 10.1128/jvi.62.9.3493-3496.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stevens J G. Human herpesviruses: a consideration of the latent state. Microbiol Rev. 1989;53:318–332. doi: 10.1128/mr.53.3.318-332.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stevens J G, Wagner E K, Devi-Rao G B, Cook M L, Feldman L T. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 65.Tal-Singer R, Lasner T M, Podrzucki W, Skokotas A, Leary J J, Berger S L, Fraser N W. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson R L, Sawtell N M. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J Virol. 1997;71:5432–5440. doi: 10.1128/jvi.71.7.5432-5440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trousdale M D, Steiner I, Spivack J G, Deshmane S L, Brown S M, MacLean A R, Subak-Sharpe J H, Fraser N W. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J Virol. 1991;65:6989–6993. doi: 10.1128/jvi.65.12.6989-6993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsumoto H. Autoradiography of new era replacing traditional x-ray film. Cell Tech. 1990;9:456–462. [Google Scholar]
- 69.Yeh L, Schaffer P A. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J Virol. 1993;67:7373–7382. doi: 10.1128/jvi.67.12.7373-7382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zwaagstra J C, Ghiasi H, Slanina S M, Nesburn A B, Wheatley S C, Lillycrop K, Wood J, Latchman D S, Patel K, Wechsler S L. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J Virol. 1990;64:5019–5028. doi: 10.1128/jvi.64.10.5019-5028.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]