Abstract
Herpes simplex virus specifies two sets of transcripts from the UL24 gene, short transcripts (e.g., 1.4 kb), processed at the UL24 poly(A) site, and long transcripts (e.g., 5.6 kb), processed at the UL26 poly(A) site. The 1.4- and 5.6-kb transcripts initiate from the same promoter but are expressed with early and late kinetics, respectively. Measurements of transcript levels following actinomycin D treatment of infected cells revealed that the 1.4- and 5.6-kb UL24 transcripts have similar stabilities, consistent with UL24 transcript kinetics being regulated by differential polyadenylation rather than by differential stabilities. Although the UL24 poly(A) site, which gives rise to short transcripts, is encountered first during processing, long transcripts processed at the UL26 site are equally or more abundant; thus, operationally, the UL24 site is weak. Using a series of viral ICP27 mutants, we investigated whether ICP27, which has been suggested to stimulate the usage of weak poly(A) sites, stimulates 1.4-kb transcript accumulation. We found that accumulation of 1.4-kb transcripts did not require ICP27 during viral infection. Rather, ICP27 was required for full expression of 5.6-kb transcripts, and the decrease in 5.6-kb transcripts relative to 1.4-kb transcripts was not due solely to reduced DNA synthesis. Our results indicate that temporal expression of UL24 transcripts can be regulated by differential polyadenylation and that although ICP27 is not required for processing at the operationally weak UL24 poly(A) site, it does modulate 5.6-kb transcript levels at a step subsequent to transcriptional initiation.
During viral infection, transcriptional regulation plays a major role in maintaining the coordinated expression of viral genes. However, other regulatory mechanisms, such as those that exert their effects posttranscriptionally, are important in the viral life cycle. Examples include translational control (17, 25, 58, 68), RNA editing (3, 22, 65), RNA transport (8, 12), RNA stability (24, 59), splicing (12), and 3′-end formation and polyadenylation (11, 40). However, many of the cis- and trans-regulatory elements that govern posttranscriptional control during viral infection remain uncharacterized.
We have been studying differential polyadenylation of the herpes simplex virus type 1 (HSV-1) UL24 gene. UL24 transcripts possess two distinct 3′ ends due to the alternative use of two poly(A) sites (6). One site is promoter proximal, located 191 nucleotides downstream of the UL24 termination codon (nucleotide 48737, using the numbering of McGeoch et al. [34]), whereas the other is located roughly 4.0 kb downstream, 49 nucleotides 3′ of the UL26 open reading frame (ORF) (6) (Fig. 1). As three different 5′ ends have been detected for UL24 transcripts (Fig. 1) (26, 45, 46, 67), the UL24 locus gives rise to six different mRNAs. For example, the 1.4- and 5.6-kb UL24 transcripts initiate at or near nucleotide 47402 but have different 3′ termini due to processing at the UL24 and UL26 poly(A) sites, respectively (6). It is usually the rule that transcripts are processed at the promoter-proximal poly(A) site (9, 11, 29, 66) because 3′ processing is coupled with transcription and occurs on nascent transcripts (33). Only when the promoter-proximal polyadenylation signal is weak does processing occur at downstream poly(A) sites (11, 66). The observation that 5.6-kb UL24 transcripts, processed at the promoter-distal UL26 poly(A) site, are as abundant as or more abundant than 1.4-kb transcripts (6) processed at the promoter-proximal UL24 poly(A) site indicates that the UL24 poly(A) site is operationally weak. Interestingly, the patterns of temporal expression of these two transcripts are different, with the 1.4-kb transcript displaying early gene kinetics and the 5.6-kb transcript exhibiting leaky-late gene kinetics (6). One explanation for the different kinetics displayed by the two transcripts is that the 1.4-kb UL24 transcript decays more rapidly than the 5.6-kb transcript. However, an alternative explanation is that the temporal expression of these two transcripts is modulated in part by differential polyadenylation.
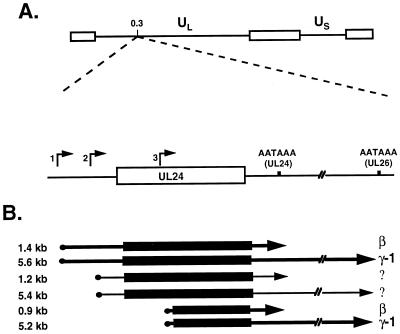
FIG. 1.
The mRNAs transcribed from the UL24 locus. (A) Schematic representation of the HSV-1 genome (prototype arrangement) indicating the location of the UL24 gene at map coordinate 0.3. The major repeat sequences are represented by the open rectangles that flank the unique long (UL) and unique short (US) regions as shown. The UL24 locus is diagrammed below the HSV-1 genome. The three UL24 promoters are indicated by arrows numbered 1, 2, and 3 and are located at nucleotides 47402, 47666, and 48076 relative to the HSV-1 strain 17 sequence (34). Positions of the UL24 ORF (open rectangle) and of the UL24 and UL26 poly(A) sites are shown. (B) Locations of the six mRNAs transcribed from the UL24 locus. Solid circles represent the 5′ cap, thin lines represent the 5′ and 3′ UTRs, the solid box represents the UL24 ORF, and the poly(A) tail is indicated by arrowheads. The thicker lines in the 1.4-, 5.6-, 0.9-, and 5.2-kb transcripts are reflective of the strengths of promoters 1 and 3, which are stronger than promoter 2. The molecular lengths of the transcripts are indicated to the left of the transcripts; the temporal gene classes for the 1.4-, 5.6-, 0.9-, and 5.2-kb transcripts are indicated at the right; the temporal gene class for the 5.4- and 1.2-kb transcripts has not been determined conclusively due to their low abundance.
Previous studies have suggested that the HSV regulatory protein ICP27 (also known as IE63, IE2, and UL54) stimulates 3′-end processing of genes containing nonoptimal or weak polyadenylation signals, at least in transfected cells and in in vitro polyadenylation reactions (35, 57). ICP27 is required for lytic infection and efficient viral DNA replication (32, 48, 52). It can repress the expression of several immediate-early and early genes (1, 10, 19, 38, 47, 50, 62) and can trans activate viral gene expression, as evidenced by the requirement of ICP27 for early and late gene expression (21, 32, 35, 48, 52, 64). Other roles proposed for ICP27 include inhibition of host cell pre-mRNA splicing (18, 20, 41, 43, 56, 57, 60), stimulation of late gene transcription (28) and posttranscriptional processes (60), and export of late gene mRNAs from the nucleus to the cytoplasm (42, 61).
In this study, we examined the stabilities of the UL24 transcripts and the role of ICP27 in expression of these transcripts. We hypothesized that ICP27, which has been reported to stimulate weak polyadenylation signal usage (57), might play a role in stimulating processing at the UL24 poly(A) site early in infection. We found that the 1.4- and 5.6-kb transcripts have similar stabilities in infected cells. Contrary to our expectations, we found that an ICP27 null mutant expressed higher levels of the 1.4-kb transcript than of the 5.6-kb transcript. These results provide evidence that alternative polyadenylation site usage is in part responsible for the temporal expression of the two transcripts and that although usage of the operationally weak 1.4-kb UL24 poly(A) site does not require ICP27 in infected cells, ICP27 acts at a step subsequent to transcriptional initiation, leading to an increase in the levels of the 5.6-kb transcript.
MATERIALS AND METHODS
Viruses and cells.
The wild-type (wt) HSV-1 strain used in this study was KOS1.1 (23), which was propagated in Vero cells (American Type Culture Collection, Rockville, Md.) as described previously (4). ICP27 mutant viruses d27, n504R, and n263R (48, 50) were propagated in the ICP27-complementing cell line V27 (48). Cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 5% newborn calf serum, 10 mM HEPES (pH 7.5), penicillin-streptomycin (100 U/ml), and 200 mM l-glutamine. For studies of mRNA stability, Dulbecco’s modified Eagle’s medium was replaced at 5 h postinfection with medium containing actinomycin D (5 μg/ml; Sigma Chemical, St. Louis, Mo.). Where indicated, viral DNA replication was inhibited by the addition of sodium phosphonoacetic acid (PAA; Sigma) to the medium at a concentration of 400 μg/ml at the time of virus adsorption.
RNA isolation and Northern blot analysis.
Total RNA was prepared from infected cells by using an RNAqueous kit (Ambion, Inc., Austin, Tex.) or as described previously (7). Cytoplasmic RNA was prepared as described previously (64). Polyadenylated mRNA was selected from 10 μg of total or cytoplasmic RNA, using a MicroPoly(A)Pure mRNA purification kit (Ambion) according to the manufacturer’s instructions except that the RNA pellet was washed twice with 70% ethanol, air dried briefly, and resuspended in sample buffer (10 mM morpholinepropanesulfonic acid [MOPS; pH 7.0], 4 mM sodium acetate, 0.5 mM EDTA, 50% freshly thawed formamide, 6% formaldehyde, 7% glycerol). Samples were heated at 68°C for 15 min, placed immediately on ice, and then separated by electrophoresis into a 1% denaturing agarose formaldehyde gel (SeaKem LE agarose; FMC BioProducts, Rockland, Maine) at 115 V in 10 mM MOPS (pH 7.0)–4.0 mM sodium acetate–0.5 mM EDTA (55). RNA was transferred overnight to Duralose membranes (Stratagene) in 20× SSC (3.0 M NaCl, 0.3 M sodium citrate [pH 7.0]) and was cross-linked to the membrane by using UV light. Northern blot hybridization and probe preparation were performed essentially as described previously (15). The UL24 probe, a 550-bp EcoRI-HindIII fragment that does not contain tk sequences, was isolated from pJC2. pJC2 was made by digesting pLS/ts+5/+15 (5) with EcoRI and HindIII and ligating the purified 550-bp fragment into EcoRI- and HindIII-digested pBluescriptII KS(+) (Stratagene, La Jolla, Calif.). All blots were stripped and reprobed for gB and ICP8. The probe for gB transcripts was a 900-bp PstI fragment from pSG18:SalE (44). The ICP8 probe was a 1.4-kb PstI-PstI fragment from plasmid pJC8, which was constructed by digesting pSG18Sal I-A (27) with PstI and ligating the 1.4-kb PstI-PstI fragment into PstI-digested pBluescriptII KS(+). Hybridized blots were exposed to Kodak X-ray film, and the resulting autoradiograms were scanned by using ScanWizard and Adobe Photoshop. Quantification was performed with NIH Image.
RESULTS
Stabilities of the 1.4- and 5.6-kb UL24 transcripts.
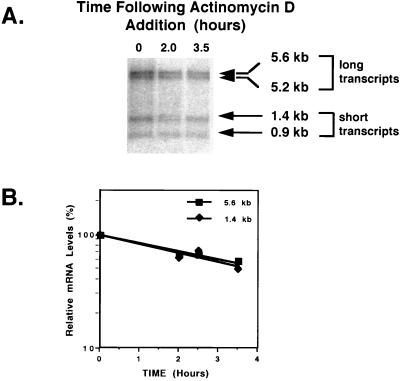
Previous data suggested that the temporal expression of UL24 transcripts was regulated by differential polyadenylation (6). Short transcripts (e.g., 1.4 kb), utilizing the UL24 poly(A) site, are expressed maximally at early times in infection and then decrease, whereas long transcripts (e.g., 5.6 kb), processed at the UL26 poly(A) site, continue to accumulate throughout infection. However, another interpretation of the data was that the shorter transcripts are inherently less stable than the longer transcripts. To address these alternative hypotheses, we assessed the stabilities of the 1.4- and 5.6-kb transcripts. Vero cells were infected with HSV-1 KOS1.1 (wt) at 10 PFU/cell. At 5 h postinfection, further RNA synthesis was inhibited by addition of actinomycin D (5 μg/ml). Total RNA was isolated at 0.5-h intervals, poly(A) selected, and analyzed by Northern blot hybridization with a UL24-specific probe. Representative data are shown in Fig. 2. Quantification of the 1.4- and 5.6-kb bands showed that the relative levels of the two transcripts were similar at all time points following actinomycin D addition, indicating that the decay rates were similar. Additionally, the relative levels of the 0.9- and 5.2-kb UL24 transcripts that initiate at or near nucleotide 48076 (6) were similar following actinomycin D treatment (Fig. 2A), indicating that the stabilities of these transcripts were not significantly different. These data are consistent with the idea that the different kinetics of expression of the short and long UL24 transcripts are due to differential usage of the UL24 and UL26 poly(A) sites and not due to different decay rates.
FIG. 2.
Stabilities of UL24 transcripts. (A) Northern blot analysis of UL24 5.6-, 5.2-, 1.4-, and 0.9-kb transcripts isolated at various times after actinomycin D treatment. KOS1.1-infected Vero cells (10 PFU/cell) were treated with actinomycin D (5 μg/ml) at 5 h postinfection. At the times indicated, polyadenylated RNA was isolated from total RNA and analyzed by Northern blotting as described in Materials and Methods. Positions of the long (5.6- and 5.2-kb) and short (1.4- and 0.9-kb) transcripts are indicated by arrows. (B) Relative amounts of 1.4- and 5.6-kb UL24 transcripts following actinomycin D treatment were determined by scanning the autoradiograms following Northern hybridization.
Infection of cells with ICP27 mutants has little effect on 1.4-kb transcript levels but causes a reduction in 5.6-kb transcript levels.
During transcription of the UL24 locus, the UL24 poly(A) site is encountered first by the cellular transcriptional/processing machinery and thus has a major advantage over the more distal UL26 poly(A) site (11, 66). However, long UL24 transcripts, which are processed at the UL26 site, are as abundant as or more abundant than those that initiate at the same promoter and utilize the UL24 poly(A) site. Thus, based on the abundance of short transcripts relative to long transcripts in virus-infected cells, the UL24 polyadenylation signal is operationally weak. Previous studies using transfected plasmids and in vitro polyadenylation reactions suggested that the essential regulatory HSV-1 gene, ICP27, stimulates usage of weak polyadenylation signals (35, 57). Thus, we hypothesized that ICP27 might stimulate the expression of the 1.4-kb UL24 transcript that is processed at the UL24 poly(A) site. This hypothesis predicted that deletion of the ICP27 gene would decrease levels of the 1.4-kb mRNA but not the larger 5.6-kb transcript.
Therefore, we infected Vero cells with three ICP27 mutants, d27, n263R, and n504R (48), and determined the relative levels of the 1.4- and 5.6-kb UL24 transcripts. Mutant d27 harbors a 1.6-kbp deletion at the ICP27 locus and is devoid of any known ICP27 function. Mutant n263R contains a nonsense codon at ICP27 codon 264 and expresses only the N-terminal half of the protein; mutant n504R has a nonsense codon at ICP27 codon 505 and lacks eight codons from the ICP27 C terminus. Additionally, both d27 and n263R accumulate reduced levels of viral DNA (6 to 23% of wt levels), whereas n504R synthesizes viral DNA levels similar to those of wt virus. The three ICP27 mutants do not express true late (γ-2) genes; n504R expresses wt levels of leaky-late (γ-1) genes, mutant n263R is able to express some γ-1 genes, and mutant d27 does not fully activate the expression of most γ-1 genes (48). All three viral mutants are unable to form plaques on Vero cells but can replicate on the complementing cell line V27 (48).
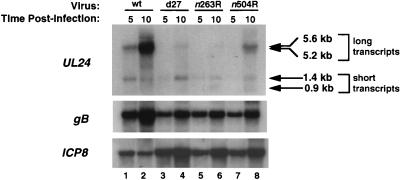
RNA was isolated from Vero cells infected with wt, d27, n263R, or n504R at 5 and 10 h postinfection and analyzed by Northern blot hybridization (Fig. 3). The data were quantified by scanning the resulting autoradiograms. At 5 h h-postinfection, the levels of both short and long UL24 transcripts were lower in mutant-infected cells than in wt-infected cells. However, by 10 h postinfection in the mutant-infected cells, the levels of the short transcripts were similar to or exceeded those in the wt-infected cells at any time postinfection (Fig. 3). In contrast, at 10 h postinfection, the levels of long UL24 transcripts were reduced in n504R-infected cells and were even more dramatically reduced in cells infected with d27 or n263R compared to wt-infected cells (Fig. 3). Accordingly, at 10 h postinfection, the molar ratios of the short to long transcripts in n504R-, d27-, and n263R-infected cells were 0.7, 9.2, and >10.0, respectively. The molar ratios for d27 and n263R were much higher than that of the wt virus (0.1).
FIG. 3.
Northern blot hybridization of total RNA isolated from cells infected with wt virus or UL24 mutant viruses d27, n263R, and n504R at 5 or 10 h postinfection. The blot was hybridized with probes specific for UL24 (top), gB (middle), and ICP8 (bottom) as described in Materials and Methods. Positions of the long (5.6- and 5.2-kb) and short (0.9- and 1.4-kb) transcripts are indicated by arrows.
Previous results have shown that ICP27 is required for γ-2 gene expression and can stimulate the expression of certain γ-1 genes (32, 48, 49, 51, 52). The 5.6-kb transcript is expressed with γ-1 kinetics (6). To determine if the substantially decreased 5.6-kb transcript levels in the three ICP27 mutants resulted solely from a general inability of the mutants to express γ-1 genes, the Northern blot in Fig. 3 was stripped and reprobed for the HSV γ-1 gene gB. At 10 h postinfection, there was somewhat less gB mRNA in cells infected with the three ICP27 mutants than in wt-infected cells (Fig. 3, middle). Next, we reprobed the blot in Fig. 3 for ICP8, an HSV β gene. Consistent with previous results (32, 64), ICP8 expression was increased or unaffected in ICP27 mutant-infected cells. In particular, there was ample expression of gB and ICP8 transcripts at 5 h postinfection; thus, the low levels of UL24 transcripts detected at this time were not due to trivial causes such as poor RNA recovery. Taken together, these results indicate that in ICP27 mutant-infected cells, the reduced expression of the long UL24 transcripts at 10 h postinfection was a bona fide property of the ICP27 mutants.
The ICP27-complementing cell line V27 restores the expression of the 5.6-kb transcript by d27.
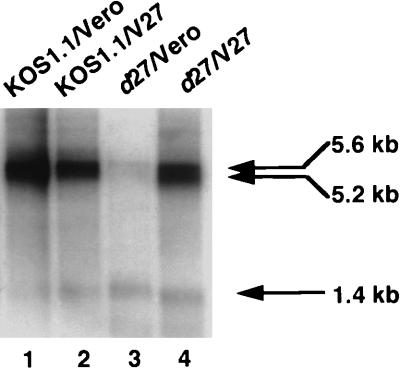
To determine if the observed decrease in 5.6-kb transcripts relative to 1.4-kb transcripts in the d27-infected cells was due to absence of ICP27, we asked whether the wt ratio of 1.4-kb transcript to 5.6-kb transcript could be restored by d27 infection of V27 cells, an ICP27-complementing cell line (48). Vero or V27 cells were infected with wt virus or mutant d27, and total RNA was isolated, poly(A) selected, and analyzed by Northern blot hybridization. At 10 h postinfection, the levels of the 1.4-kb transcript were similar in all cases; however, the levels of the 5.6-kb transcript were drastically reduced in d27-infected Vero cells. In this experiment, d27 infection of Vero cells yielded a ratio of 1.4-kb transcripts to 5.6-kb transcript of 1.9 (Fig. 4, lane 3). However, when the ICP27-complementing cell line V27 was infected with d27, the ratio of 1.4-kb transcript to 5.6-kb transcript was similar to that of wt-infected V27 cells (Fig. 4, lanes 2 and 4). Similar ratios were observed in wt- and d27-infected V27 cells at 7.5 h postinfection (16). Thus, the reduced levels of 5.6-kb UL24 transcripts observed in d27-infected Vero cells can be attributed to the absence of ICP27.
FIG. 4.
Northern blot analysis of UL24 5.6- and 1.4-kb transcripts isolated from Vero or V27 cells infected with wt virus or the ICP27 deletion mutant virus d27. Cells were infected at 10 PFU/ml, and polyadenylated mRNA was isolated from total RNA isolated at 10 h postinfection as described in Materials and Methods. Position of the 5.6-, 5.2-, and 1.4-kb transcripts are indicated by arrows.
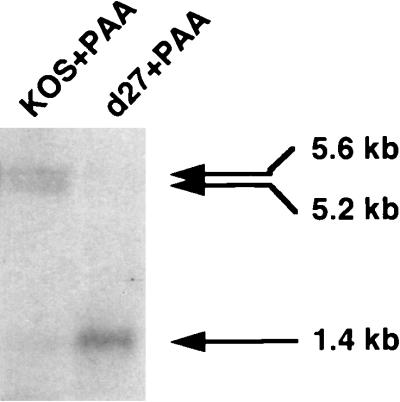
The reduced levels of the 5.6-kb UL24 transcript are not due solely to inhibition of DNA replication.
The results in Fig. 3 showed that cells infected with all ICP27 mutants accumulated reduced amounts of UL24 transcripts compared to wt-infected cells. Only when cells were infected with mutant n504R were levels of the 5.6-kb transcript readily detectable at 10 h postinfection (Fig. 3, lane 8). As mutant n504R replicates viral DNA efficiently (48), it was conceivable that the low levels of the 5.6-kb transcript in d27- and n263R-infected cells resulted solely from reduced DNA replication and not the absence of ICP27. To determine if inhibition of viral DNA synthesis was responsible for the low 5.6-kb transcript levels, we infected Vero cells with either wt or d27 in the presence of the viral DNA synthesis inhibitor PAA. Northern blot hybridization of poly(A)-selected RNA isolated at 5.5 h postinfection (Fig. 5) showed that in the presence of PAA, the 5.6-kb transcript was evident in wt-infected cells, with a 1.4-kb/5.6-kb transcript ratio of 0.2, whereas in d27-infected cells the 5.6-kb transcript was not detected and the 1.4-kb/5.6-kb transcript ratio was >10.0. Similar amounts of gB transcripts were observed for the two viruses when the blot shown in Fig. 5 was stripped and reprobed with a gB-specific probe (16). Thus, the decrease in the 5.6-kb transcript in d27-infected cells was not due solely to reduced DNA synthesis.
FIG. 5.
Northern blot analysis of polyadenylated UL24 5.6- and 1.4-kb transcripts isolated from wt- or d27-infected Vero or V27 cells at 5.5 h postinfection in the presence of PAA (400 μg/ml). In this experiment, polyadenylated mRNA was selected from cytoplasmic RNA. Cells were infected with virus at 20 PFU/cell as described in Materials and Methods. Positions of the 5.6-, 5.2-, and 1.4-kb transcripts are indicated by arrows.
DISCUSSION
In this study, we have investigated further the temporal regulation of two UL24 transcripts that are transcribed from the same promoter. Our data argue that during viral infection, the temporal expression of the 1.4- and 5.6-kb UL24 transcripts is not due to altered mRNA stabilities. This finding supports the hypothesis that the temporal regulation of the 1.4- and 5.6-kb UL24 transcripts is governed, at least in part, at the level of differential polyadenylation. The promoter-proximal UL24 poly(A) site, which generates the 1.4-kb transcript, should be more efficiently used than the downstream UL26 poly(A) site because it is encountered first by the processing machinery (11, 66). Instead, except at very early times postinfection, 5.6-kb transcripts processed at the UL26 poly(A) site are more abundant than 1.4-kb transcripts processed at the UL24 poly(A) site; this finding indicates that during viral infection, the UL24 site is operationally weak. Using a series of ICP27 mutants, we found that ICP27, which has been suggested to stimulate usage of weak polyadenylation sites, was not required for 3′ processing of the 1.4-kb UL24 transcript. We did find, however, that the accumulation of the 5.6-kb transcript required ICP27. Taken together, our results suggest that ICP27 does not stimulate usage of the operationally weak UL24 poly(A) site but may instead play a role in increasing 5.6-kb transcript levels.
ICP27 does not stimulate 3′ processing at the UL24 poly(A) site.
Our results showing that ICP27 is not required for utilization of the operationally weak UL24 polyadenylation site appear to differ from results of transient transfection and cell-free polyadenylation assays (35, 57) which suggested that ICP27 stimulates the expression of genes containing nonoptimal or inefficiently utilized polyadenylation signals (35, 57). One interpretation of this apparent difference is that transient transfection and in vitro assays may not accurately reflect the environment within HSV-infected cells. For example, during adenovirus infection, a switch from early to late polyadenylation site usage requires a cis change in the viral genome (11); thus, viral DNA alterations in cis may be needed for bona fide regulation by ICP27. It is also possible that the UL24 poly(A) site is not representative of other inefficiently utilized polyadenylation sites in that it does not require ICP27 for 3′ processing.
It is thought that cleavage-stimulatory factor is the rate-limiting factor in determining polyadenylation site usage (13, 14, 63). This multisubunit protein binds GU- or U-rich tracts 3′ to the poly(A) site signal, AAUAAA (30, 66). Both the UL24 and UL26 poly(A) sites contain a GU-rich element downstream of the hexanucleotide AAUAAA but not other previously identified consensus elements (31, 53). In vitro polyadenylation assays have shown that four U’s are sufficient for binding of the cleavage-stimulatory factor 64-kDa subunit and that the distance of this site from the AAUAAA motif can influence the polyadenylation site strength (30). This is of interest since in the UL24 3′ untranslated region (UTR) the U4 element is 46 nucleotides downstream of the AAUAAA whereas in the UL26 3′ UTR the distance between the hexanucleotide AAUAAA and the U tract is shorter, with U7 35 nucleotides downstream. Whether this is responsible for the weak utilization of the UL24 poly(A) site remains to be determined.
Possible mechanisms of ICP27 in modulating the 5.6-kb transcript accumulation.
Our data show that ICP27 is required for efficient accumulation of the 5.6-kb transcript. Because the 1.4- and 5.6-kb transcripts are transcribed from the same promoter (6), it is apparent that ICP27 does not selectively stimulate transcriptional initiation of the 5.6-kb transcript over the 1.4-kb transcript. Rather, ICP27 must exert its selective effects on the 5.6-kb transcript at a step subsequent to transcriptional initiation. We can envision several mechanisms by which ICP27 may stimulate the expression of the UL24 5.6-kb transcript at a step subsequent to transcriptional initiation: by promoting transcriptional elongation and thereby extending the transcripts to the UL26 poly(A) signal, by stimulating poly(A) site selection at the UL26 site, by altering mRNA transport, by stabilizing the 5.6-kb transcript, or by a combination of two or more of these processes. (It is, of course, possible that ICP27 acts both at one of these steps and during transcriptional initiation and that these two processes are linked; however, the ensuing discussion will focus on each of these promoter-distal steps.)
Although there is no direct evidence that ICP27 can stimulate transcriptional elongation, this could explain why expression of the longer 5.6-kb UL24 transcript requires ICP27. Evidence suggests that ICP27 may be required for an HSV-induced factor called LPF (36, 37). This factor has been shown to promote 3′ processing at promoter distal poly(A) sites at late times postinfection (37). Ordinarily, the efficiency of 3′ processing is reduced by increasing the distance between the 5′ end of the mRNA and the polyadenylation site; LPF abrogates this effect (37). If ICP27 is indeed a component of LPF, it may act by increasing transcriptional elongation, permitting efficient utilization of the UL26 poly(A) site, located roughly 4,000 nucleotides downstream of the UL24 poly(A) site. A role for ICP27 in increasing transcriptional elongation could also explain why Uprichard and Knipe found that of two differentially polyadenylated UL52 mRNAs, accumulation of the larger transcript was especially dependent on ICP27 expression (64). It is also possible that ICP27 is ordinarily required for suppression of polyadenylation late in infection and that this permits the synthesis of longer transcripts.
The possibility that ICP27 promotes usage of the UL26 poly(A) site is consistent with previous observations that ICP27 regulation of certain genes is 3′-end dependent (57) and that in vitro, ICP27 stimulates processing at certain late polyadenylation sites (35, 36). Recently, it has been shown that the carboxy-terminal domain of RNA polymerase II can specifically bind 3′-end processing factors (33). Thus, it is possible that ICP27 interacts simultaneously with the elongating polymerase II complex and 3′-end processing apparatus. Alternatively, direct binding of ICP27 to nascent transcripts could also promote use of the UL26 poly(A) site.
ICP27 has been implicated in nuclear and cytoplasmic shuttling (38a, 42, 55a, 61). It has been proposed that ICP27 may stimulate late gene expression by specifically transporting late gene mRNAs to the cytoplasm (38a, 61). Thus, mRNA regulatory elements present within the 5.6-kb but not the 1.4-kb transcript may be required for export and expression of the 5.6-kb transcript at late times postinfection. It has also been proposed that ICP27 stabilizes mRNAs (2, 39, 60). In the three ICP27 mutants tested, the peak levels of the 5.6-kb transcript, but not the 1.4-kb transcript, were specifically decreased compared to wt-infected cells. Thus, although the 1.4- and 5.6-kb transcripts exhibited similar stabilities in the presence of ICP27, we cannot rule out the possibility that the stability of the 5.6-kb transcript is decreased in the absence of functional ICP27. The extremely low amounts of long transcripts present in d27- and n263R-infected cells make it difficult to address this issue directly. It has been suggested that the ability of ICP27 to stabilize labile mRNAs resides in its extreme C terminus (2). This observation is interesting in light of the lower levels of long transcript observed in n504R-infected cells relative to wt-infected cells, since the ICP27 synthesized by n504R lacks the C-terminal eight amino acids.
Studies of ICP27 have been complicated by the multifunctional nature of the protein. The effect of ICP27 mutants on UL24 transcript levels should prove useful in deciphering the role of ICP27. Since distinct UL24 transcripts are generated from the same promoter but utilize different polyadenylation sites, the apparent effects of ICP27 on transcriptional initiation (54) can be separated from regulation occurring subsequently. Thus, the UL24 gene should provide a useful model for elucidating ICP27 function.
ACKNOWLEDGMENTS
This work was supported by NIH postdoctoral fellowships to L.E.H. (AI09525) and W.J.C. (AI08940) and research grants to D.M.K. (AI20530) and D.M.C. (AI26126). S.L.U. was supported by Public Health Service training grant AI07245.
REFERENCES
- 1.Block T, Jordan R. Herpes simplex virus type 1 alpha gene containing plasmids can inhibit expression regulated from an alpha promoter in CV-1 but not HeLa cells. Virus Res. 1988;11:269–279. doi: 10.1016/0168-1702(88)90001-9. [DOI] [PubMed] [Google Scholar]
- 2.Brown C R, Nakamura M S, Mosca J D, Hayward G S. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J Virol. 1995;69:7187–7195. doi: 10.1128/jvi.69.11.7187-7195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casey J L, Gerin J L. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J Virol. 1995;69:7593–7600. doi: 10.1128/jvi.69.12.7593-7600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coen D M, Fleming H E, Jr, Leslie L K, Retondo M J. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug sensitivity mutations to the DNA polymerase locus. J Virol. 1985;53:477–488. doi: 10.1128/jvi.53.2.477-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coen D M, Weinheimer S P, McKnight S L. A genetic approach to promoter recognition during trans induction of viral gene expression. Science. 1986;234:53–59. doi: 10.1126/science.3018926. [DOI] [PubMed] [Google Scholar]
- 6.Cook W J, Coen D M. Temporal regulation of herpes simplex virus type 1 UL24 mRNA expression via differential polyadenylation. Virology. 1996;218:204–213. doi: 10.1006/viro.1996.0180. [DOI] [PubMed] [Google Scholar]
- 7.Cook W J, Gu B, DeLuca N A, Moynihan E B, Coen D M. Induction of transcription by a viral regulatory protein depends on the relative strengths of functional TATA boxes. Mol Cell Biol. 1995;15:4998–5006. doi: 10.1128/mcb.15.9.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen B R, Greene W C. Regulatory pathways governing HIV-1 replication. Cell. 1989;58:423–426. doi: 10.1016/0092-8674(89)90420-0. [DOI] [PubMed] [Google Scholar]
- 9.Denome R M, Cole C N. Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol Cell Biol. 1988;8:4829–4839. doi: 10.1128/mcb.8.11.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett R D. The products of herpes simplex virus type 1 (HSV-1) immediate early genes 1, 2, and 3 can activate HSV-1 gene expression in trans. J Gen Virol. 1986;67:2507–2513. doi: 10.1099/0022-1317-67-11-2507. [DOI] [PubMed] [Google Scholar]
- 11.Falck-Pedersen E, Logan J. Regulation of poly(A) site selection in adenovirus. J Virol. 1989;63:1–14. doi: 10.1128/jvi.63.2.532-541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmartin G M, Nevins J R. Molecular analysis of two poly(A) site-processing factors that determine recognition and efficiency of cleavage of the pre-mRNA. Mol Cell Biol. 1991;11:2432–2438. doi: 10.1128/mcb.11.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmartin G M, Nevins J R. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3:2180–2189. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 15.Hann L E, Gehrke L. mRNAs containing the unstructured 5′ leader sequence of alfalfa mosaic virus RNA 4 translate inefficiently in lysates from poliovirus-infected HeLa cells. J Virol. 1995;69:4986–4993. doi: 10.1128/jvi.69.8.4986-4993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hann, L. E., D. M. Knipe, and D. M. Coen. Unpublished results.
- 17.Hann L E, Webb A, Cai J M, Gehrke L. Identification of a competitive translation determinant in the 3′ untranslated region of alfalfa mosaic virus coat protein mRNA. Mol Cell Biol. 1997;17:2005–2013. doi: 10.1128/mcb.17.4.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardwicke M A, Vaughn P J, Sekulovich R E, O’Connor R E, Sandri-Goldin R M. The regions important for the activator and repressor functions of herpes simplex virus type 1 α protein ICP27 map to the C-terminal half of the molecule. J Virol. 1989;63:4590–4602. doi: 10.1128/jvi.63.11.4590-4602.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hibbard M K, Sandri-Goldin R M. Arginine-rich regions succeeding the nuclear localization region of the herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J Virol. 1995;69:4656–4667. doi: 10.1128/jvi.69.8.4656-4667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horikami S, Moyer S. Synthesis of leader RNA and editing of the P mRNA during transcription by purified measles virus. J Virol. 1991;65:5342–5347. doi: 10.1128/jvi.65.10.5342-5347.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes R G, Munyon W H. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J Virol. 1975;16:275–283. doi: 10.1128/jvi.16.2.275-283.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy I M, Haddow J K, Clements J B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J Virol. 1991;65:2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerry J A, Priddy M A, Kohler C P, Staley T L, Weber D, Jones T R, Stenberg R M. Translational regulation of the human cytomegalovirus pp28 (UL99) late gene. J Virol. 1997;71:981–987. doi: 10.1128/jvi.71.2.981-987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kibler P K, Duncan J, Keith B D, Hupel T, Smiley J R. Regulation of herpes simplex virus true late gene expression: sequences downstream from the US11 TATA box inhibit expression from an unreplicated template. J Virol. 1991;65:6749–6760. doi: 10.1128/jvi.65.12.6749-6760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C K, Knipe D M. Thermolabile in vivo DNA-binding activity associated with a protein encoded by mutants of herpes simplex virus type 1. J Virol. 1983;46:909–911. doi: 10.1128/jvi.46.3.909-919.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin, J. K., and D. M. Knipe. Unpublished data.
- 29.Levitt N, Briggs A, Gil A, Proudfoot N J. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald C C, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLauchan J, Gaffney D, Whitton J L, Clements J B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 1985;13:1347–1367. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 34.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 35.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLauchlan J, Phelan A, Loney C, Sandri-Goldin R M, Clements J B. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate virus mRNA 3′ processing. J Virol. 1992;66:6939–6945. doi: 10.1128/jvi.66.12.6939-6945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLauchlan J, Simpson S, Clements J B. Herpes simplex virus induces a processing factor that stimulates poly(A) site usage. Cell. 1989;59:1093–1105. doi: 10.1016/0092-8674(89)90765-4. [DOI] [PubMed] [Google Scholar]
- 38.McMahan L, Schaffer P A. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J Virol. 1990;1990:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Mears W E, Rice S A. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 39.Mosca J D, Pitha P M, Hayward G S. Herpes simplex virus infection selectively stimulates accumulation of beta interferon reporter gene mRNA by a posttranscriptional mechanism. J Virol. 1992;66:3811–3822. doi: 10.1128/jvi.66.6.3811-3822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevins J R, Wilson M C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981;290:113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- 41.Phelan A, Carmo-Fonseca M, McLauchlan J, Lamond A L, Clements J B. A herpes simplex virus type 1 immediate-early gene product IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phelan A, Clements J B. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol. 1997;78:3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 43.Phelan A, Dunlop J, Clements J B. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J Virol. 1996;70:5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rafield L F, Knipe D M. Characterization of the major mRNA transcribed from genes for glycoprotein B and the DNA-binding protein ICP8 of herpes simplex type 1. J Virol. 1984;49:960–969. doi: 10.1128/jvi.49.3.960-969.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Read G S, Sharp J A, Summers W C. In vitro and in vivo transcription initiation sites on the tk-encoding BamH1 Q fragment of HSV-1 DNA. Virology. 1984;138:368–372. doi: 10.1016/0042-6822(84)90363-5. [DOI] [PubMed] [Google Scholar]
- 46.Read G S, Summers W C. In vitro transcription of the thymidine kinase gene of herpes simplex virus. Proc Natl Acad Sci USA. 1982;79:5215–5219. doi: 10.1073/pnas.79.17.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice S A, Knipe D M. Gene-specific transactivation by herpes simplex virus type 1 α protein ICP27. J Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice S A, Knipe D M. Genetic evidence for two distinct transactivation functions of the herpes simplex virus α protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice S A, Lam V. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J Virol. 1994;68:823–833. doi: 10.1128/jvi.68.2.823-833.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice S A, Su L, Knipe D M. Herpes simplex virus α protein ICP27 possesses separable positive and negative regulatory activities. J Virol. 1989;63:3399–3407. doi: 10.1128/jvi.63.8.3399-3407.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice S A, Lam V, Knipe D M. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J Virol. 1993;67:1778–1787. doi: 10.1128/jvi.67.4.1778-1787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sacks W R, Greene C C, Aschman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadofsky M, Alwine J C. Sequences on the 3′ side of the hexanucleotide AAUAAA effect efficiency of cleavage at the polyadenylation site. Mol Cell Biol. 1984;4:1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samaniego L A, Webb A L, DeLuca N A. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J Virol. 1995;69:5705–5715. doi: 10.1128/jvi.69.9.5705-5715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55a.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandri-Goldin R M, Mendoza G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 58.Schmechel S, Chute M, Skinner P, Anderson R, Schiff L. Preferential translation of reovirus mRNA by a sigma 3-dependent mechanism. Virology. 1997;232:62–73. doi: 10.1006/viro.1997.8531. [DOI] [PubMed] [Google Scholar]
- 59.Scott J, Imperiale M. Promoter-proximal poly(A) sites are processed efficiently, but the RNA products are unstable in the nucleus. Mol Cell Biol. 1997;17:2127–2135. doi: 10.1128/mcb.17.4.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith I L, Hardwicke M A, Sandri-Goldin R M. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 61.Soliman T M, Sandri-Goldin R M, Silverstein S J. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su L, Knipe D M. Herpes simplex virus alpha protein ICP27 can inhibit or augment viral gene transactivation. Virology. 1989;170:496–504. doi: 10.1016/0042-6822(89)90441-8. [DOI] [PubMed] [Google Scholar]
- 63.Takagaki Y J, Manley C C, MacDonald C C, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 64.Uprichard S L, Knipe D M. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J Virol. 1996;70:1969–1980. doi: 10.1128/jvi.70.3.1969-1980.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volchkov V, Becker V, Volchkowa V, Ternovoj V, Kotov A, Netesov S, Klenk H. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and T7 and vaccinia virus polymerase. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 66.Weiss E A, Gilmartin G M, Nevins J R. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991;10:215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilkie N M, Eglin R P, Sanders P G, Clements J B. The association of herpes simplex virus with squamous carcinoma cells of the cervix, and studies of the virus thymidine kinase gene. Proc R Soc Lond Biol Sci. 1980;210:411–421. doi: 10.1098/rspb.1980.0143. [DOI] [PubMed] [Google Scholar]
- 68.Yager D, Coen D M. Translational regulation of herpes simplex virus DNA polymerase. J Virol. 1990;64:2217–2225. doi: 10.1128/jvi.64.5.2217-2225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]