Abstract
We have recently described an evolutionarily conserved protein motif, designated the THAP domain, which defines a previously uncharacterized family of cellular factors (THAP proteins). The THAP domain exhibits similarities to the site-specific DNA-binding domain of Drosophila P element transposase, including a putative metal-coordinating C2CH signature (CX2–4CX35–53CX2H). In this article, we report a comprehensive list of ≈100 distinct THAP proteins in model animal organisms, including human nuclear proapoptotic factors THAP1 and DAP4/THAP0, transcriptional repressor THAP7, zebrafish orthologue of cell cycle regulator E2F6, and Caenorhabditis elegans chromatin-associated protein HIM-17 and cell-cycle regulators LIN-36 and LIN-15B. In addition, we demonstrate the biochemical function of the THAP domain as a zinc-dependent sequence-specific DNA-binding domain belonging to the zinc-finger superfamily. In vitro binding-site selection allowed us to identify an 11-nucleotide consensus DNA-binding sequence specifically recognized by the THAP domain of human THAP1. Mutations of single nucleotide positions in this sequence abrogated THAP-domain binding. Experiments with the zinc chelator 1,10-o-phenanthroline revealed that the THAP domain is a zinc-dependent DNA-binding domain. Site-directed mutagenesis of single cysteine or histidine residues supported a role for the C2CH motif in zinc coordination and DNA-binding activity. The four other conserved residues (P, W, F, and P), which define the THAP consensus sequence, were also found to be required for DNA binding. Together with previous genetic data obtained in C. elegans, our results suggest that cellular THAP proteins may function as zinc-dependent sequence-specific DNA-binding factors with roles in proliferation, apoptosis, cell cycle, chromosome segregation, chromatin modification, and transcriptional regulation.
Keywords: protein motif, zinc finger, Caenorhabditis elegans, cell cycle
We have recently described an evolutionarily conserved ≈90-residue protein motif, designated the THAP domain, which defines a previously uncharacterized family of cellular factors, the THAP proteins (1, 2). This motif is characterized by a putative metal-coordinating C2CH module (CX2–4CX35–53CX2H) and four additional invariant residues, P26, W36, F58, and P78, in human THAP1 (Fig. 1). The THAP domain was found to be restricted to animals and is present in both vertebrates (from zebrafish to humans) and invertebrates (e.g., fly and worm) (1). Interestingly, the THAP-motif signature was identified (1) in the site-specific DNA-binding domain of Drosophila melanogaster P element transposase (3). This finding suggested that the THAP domain may constitute an example of a DNA-binding domain shared between cellular proteins and transposases from mobile genomic parasites and that the THAP proteins may correspond to a previously uncharacterized family of cellular DNA-binding proteins (1).
Fig. 1.
THAP proteins in model animal organisms. (A) Identification of a consensus THAP domain in the zebrafish orthologue of cell-cycle transcription factor E2F6. Shown is clustalw multiple alignment of zebrafish E2F6 with human E2F6 and the THAP domain of human THAP1. Conserved residues are boxed. Black boxes indicate identical residues, whereas boxes shaded in gray show similar amino acids. The consensus THAP motif, defined by the C2CH signature and four other invariant residues (P26, W36, F58, and P78 in human THAP1), is shown above the alignment. (B) Primary structures of C. elegans THAP proteins with known functions. THAP domains are shown in black. Divergent THAP domains containing the C2CH signature are shown in gray. Known protein motifs are indicated. The transcriptional-corepressor function of CtBP has not yet been confirmed in C. elegans.
In humans, the THAP family comprises 12 distinct members, including nuclear proapoptotic factor THAP1 (2), death-associated protein DAP4/THAP0 (4, 5), transcriptional repressor THAP7 (6), and 9 other human proteins (1). Both THAP1 and DAP4/THAP0 appear to function in nuclear apoptotic pathways. THAP1 interacts and colocalizes within promyelocytic leukemia nuclear bodies with the proapoptotic leucine-zipper protein Par-4 and potentiates both serum withdrawal- and TNFα-induced apoptosis (2). DAP4/THAP0 was initially identified in a screen for genes involved in IFNγ-induced apoptosis in HeLa cells (4) and, more recently, as a nuclear partner of MST1 (5), a proapoptotic kinase that phosphorylates histone H2B during apoptosis (7). Recently, THAP7 has been shown to function as a transcriptional repressor that binds to hypoacetylated histone H4 tails and may also induce the hypoacetylation of histone H3 by recruiting corepressor NcoR and histone deacetylase HDAC3 to chromatin (6).
Although orthologous relationships with the human THAP proteins were not obvious, analysis of the D. melanogaster (1) and Caenorhabditis elegans (8) THAP families revealed several interesting features. Two of the predicted Drosophila THAP proteins were found to contain more than one THAP domain, the double-THAP protein CG14860 and the multi-THAP protein CG10631, which is predicted to contain 27 THAP domains occurring as internal repeats (1). A third multi-THAP protein, designated HIM-17, has recently been described in C. elegans and plays a critical role in chromosome segregation during meiosis by linking chromatin modification and competence for initiation of meiotic recombination by double-strand breaks (8). A substantial fraction of HIM-17 was found to comprise six internal repeats of the THAP domain (Fig. 1), including two divergent C2CH modules lacking the invariant W, F, or P residues. Similar divergent or consensus THAP domains were also identified in other C. elegans proteins (8), including LIN-36, LIN-15A, and LIN-15B, three proteins initially characterized for their role in vulval development (9–12). Remarkably, LIN-36, LIN-15A, LIN-15B, and HIM-17 have all been found to interact genetically with LIN-35/Rb, the sole C. elegans retinoblastoma homolog (8, 9, 12, 13). In addition, LIN-36 and LIN-15B have been found to function as inhibitors of the G1-to-S-phase cell-cycle transition (13), and LIN-36 has also been shown to function redundantly with FZR-1, the C. elegans homolog of APC regulator Cdh1, in the global control of cell proliferation (14).
Although the THAP motif has been well defined and several THAP proteins have been functionally characterized, the biochemical role of the THAP domain in cellular THAP proteins has not yet been described. In this study, using the THAP domain of human THAP1 as a prototype, we show that the THAP domain is a zinc-dependent sequence-specific DNA-binding module, and we demonstrate that its DNA-binding activity absolutely requires the C2CH signature and the four other conserved residues (P, W, F, and P) of the THAP motif.
Materials and Methods
Plasmid Constructions. The THAP domain of human THAP1 (amino acids 1–90) was amplified by PCR using as a template pGADT7-THAP1 (2) with primers 5′-GCGCATATGGTGCAGTCCTGCTCCGCCTACGGC-3′ and 5′-GCGCTCGAGTTTCTTGTCATGTGGCTCAGTACAAAG-3′. The PCR product was digested with NdeI and XhoI and cloned in-frame with a carboxyl-terminal His tag into plasmid pET-21c (Novagen). Similarly, the THAP1 ORF was amplified by PCR to generate pCDNA3-THAP1 or pCDNA3.1-THAP1 (THAP1-Myc). The THAP1-C5A, THAP1-C10A, THAP1-C54A, T H A P1-H57A, T H A P1-P26A, T H A P1-W36A, THAP1-F58A, and THAP1-P78A single-point mutants were obtained by PCR using specific primers containing the corresponding mutations and cloned as EcoRI–XbaI fragments in pCDNA3 expression vector.
Protein Expression and Purification. pET-21c-THAP-domain recombinant protein was produced in Escherichia coli strain BL-21 pLysS according to the supplier's instructions (Novagen). The cells were lysed by sonication in buffer A [50 mM sodium phosphate (pH 7.5)/300 mM NaCl/0.1% 2-mercaptoethanol/10 mM imidazole], and the lysate was cleared by centrifugation. The supernatant was loaded onto a Ni nitrilotriacetate (NTA) agarose column (Amersham Pharmacia Biotech) equilibrated in buffer A. After washing, the protein was eluted with a linear gradient of imidazole in buffer A. Fractions containing the THAP domain were pooled, concentrated with YM-3 filter devices (Amicon), and applied to a Superdex 75 gel-filtration column (Amersham Pharmacia Biotech) equilibrated in buffer B [50 mM Tris·HCl (pH 7.5)/150 mM NaCl/1 mM DTT). Fractions containing the THAP domain were pooled and stored at 4°C or frozen at –80°C in 20% glycerol. The purity of the sample was assessed by SDS/PAGE, and the protein concentration was determined with Bradford protein assay (Bio-Rad).
Systematic Evolution of Ligands by Exponential Enrichment (SELEX) Assay. DNA-binding specificity of the THAP domain from human THAP1 was determined by SELEX, essentially as described in ref. 15. The following 62-bp oligonucleotide was synthesized: 5′-TGGGCACTATTTATATCAACN25AATGTCGTTGGTGGC CC-3′ (where N is any nucleotide) along with primers complementary to each end, F-HindIII 5′-ACCGCAAGCTTGGGCACTATTTATATCAAC-3′ and R-XbaI 5′-GGTCTAGAGGGCCACCAACGCATT-3′. A pool of double-stranded 80-bp degenerate oligonucleotides was amplified by PCR using the F-HindIII and R-XbaI primers. Recombinant THAP domain (≈250 ng) was incubated with Ni-NTA magnetic beads (Qiagen) in NT2 buffer [20 mM Tris·HCl (pH 7.5)/100 mM NaCl/0.05% Nonidet P-40] for 30 min at 4°C, and the beads were washed twice with 500 μl of NT2 buffer. The immobilized THAP domain was incubated with the random pool of oligonucleotides (2–5 μg) in 100 μl of binding buffer [20 mM Tris·HCl (pH 7.50)/100 mM NaCl/0.05% Nonidet P-40/0.5 mM EDTA/100 μg/ml BSA/20–50 μg of poly(dI-dC)] for 10 min at room temperature. The beads were then washed six times with 500 μl of NT2 buffer, the protein–DNA complexes were extracted with phenol/chloroform, and DNA was precipitated with ethanol with glycogen as a carrier. About 20% of the recovered DNA was amplified by PCR (15–20 cycles) and used for the next round of selection. After 12 rounds of selection by the THAP domain, selected double-stranded oligonucleotides were digested with XbaI and HindIII, cloned into the pBluescript II KS E. coli vector (Stratagene), and sequenced.
EMSA. EMSAs were performed with purified recombinant THAP domain of human THAP1, in vitro translated THAP1, or THAP1-Myc proteins synthesized in rabbit reticulocyte lysate (RRL) with the TNT-T7 kit (Promega). The THAP-domain-binding sequence (THABS) probes, 25-bp (5′-AGCAAGTAAGGGCAACTACTTCAT-3′) and 36-bp (5′-TATCAACTGTGGGCAAACTACGGGCAACAGGTAATG-3′), were used in the assays. After annealing of the complementary oligonucleotides, double-stranded probes were purified on 12% polyacrylamide gels, 32P-end-labeled, and quantified by Cerenkov counting. Purified THAP domain (≈20 ng) was incubated with 30,000 cpm of the appropriate probe (≈2 ng). Binding reactions were carried out for 10 min at room temperature in 20 μl of binding buffer [20 mM Tris·HCl (pH 7.5)/100 mM KCl/0.1% Nonidet P-40/100 μg/ml BSA/2.5 mM DTT/5% glycerol/10 μg/ml poly(dI-dC)]. For in vitro translated proteins, 3 μl of RRLs expressing THAP1 or THAP1-Myc were incubated in 20 μl of binding buffer containing 150 mM KCl, 50 μg/ml of poly(dI-dC), and 50 μg/ml salmon sperm DNA. Electrophoresis was performed on 6% or 8% (29:1) polyacrylamide gels containing 5% glycerol. The gels were run in 0.25× TBE [1× TBE = 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA (pH 8.3)] at 150 V at 4°C, dried, and exposed on a PhosphorImager screen (Molecular Dynamics) or autoradiographed. For competitive EMSA, unlabeled oligonucleotides were added to the reaction mixture just before the addition of the probe. Supershift experiments were performed by using 1 μg of anti-Myc (Sigma) or isotype-control mouse monoclonal antibodies. For metal-chelation experiments, the proteins were preincubated with EDTA or 1,10-o-phenanthroline (Sigma) for 20 min at room temperature. Metal salts (Sigma), as indicated, were added at 100 μM final concentration. The reactions were allowed to equilibrate for 10 min at room temperature before the addition of the probe.
In Silico Sequence Analysis. GenBank nucleotide, protein, EST, and genome databases at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/) were searched with both the nucleotide and amino acid sequences of human THAP proteins (1), by using the programs blastn, tblastn, and blastp (16). clustalw (17) was used to carry out the alignment of zebrafish E2F6 (locus CAH68904) with human E2F6 (locus NP_001943) and the THAP domain of human THAP1 (locus NP_060575). The oligonucleotide sequences recovered by SELEX were analyzed by using the motif discovery program meme (Multiple Em for Motif Elicitation; http://meme.sdsc.edu/meme/website/meme.html), and the logo representation of the THABS consensus motif was generated with weblogo (18).
Results
A Comprehensive List of THAP Proteins in Model Organisms. We performed extensive searches of sequence databases in an effort to extend our previous work (1) and build up a comprehensive list of proteins containing THAP domains in model organisms. We identified ≈100 distinct THAP-protein sequences in model animal organisms Homo sapiens, Mus musculus, Gallus gallus (chicken), Xenopus laevis, Danio rerio (zebrafish), D. melanogaster, and C. elegans (Table 1 and see Table 2 and Fig. 6, which are published as supporting information on the PNAS web site). Surprisingly, we found that five of the human THAP genes (THAP5, THAP6, THAP8, THAP9, and THAP10) did not appear to have orthologues in the M. musculus genome, which encoded only seven distinct THAP proteins. In contrast, we identified 23 distinct THAP family members in the tetraploid frog X. laevis, including two multi-THAP proteins predicted to contain two THAP domains at their amino termini. We also found >30 distinct THAP family members (32 distinct genes) in the zebrafish D. rerio, likely because of the large-scale gene duplication that occurred in this species (19). Interestingly, one of these THAP proteins (locus CAH68904) was found to correspond to the zebrafish orthologue of cell-cycle transcription factor E2F6 (Fig. 1A). The THAP–E2F6 fusion gene was validated by several overlapping ESTs (GenBank accession nos. CK029739, CF348112, and CO813444), and a similar THAP-E2F6 fusion protein was also identified in other fish species, including Tetraodon nigroviridis (accession no. CAG01476) and Gasterosteus aculaetus (accession no. CD497753, partial cDNA).
Table 1. The THAP family in model animal organisms.
| Model organism | THAP proteins | Putative orthologues |
|---|---|---|
| H. sapiens | 12 | THAP 0-THAP 11 |
| M. musculus | 7 | THAP 0, 1, 2, 3, 4, 7, and 11 |
| G. gallus (chicken) | 6 | THAP 0, 1, 4, 5, and 7 |
| X. laevis | 23 | THAP 1, 4, 7, and 11 |
| D. rerio (zebrafish) | 32 | THAP 0, 1, 7, 9, and 11 |
| D. melanogaster | 9 | |
| C. elegans | 8 |
See Table 2 and Fig. 6 for complete THAP-domain sequences, alignment, and accession numbers.
Analysis of the C. elegans THAP family (Fig. 1B) gave additional interesting clues to the biological roles of THAP proteins in vivo. In addition to the recently described chromatin-associated protein HIM-17 and cell-cycle regulators LIN-15B and LIN-36 (8, 13), we identified five other proteins exhibiting consensus THAP motifs in C. elegans, including CDC-14B, an isoform of cell-cycle regulator tyrosine phosphatase CDC-14 (20), which contains both consensus and divergent THAP motifs at its carboxyl terminus. The C. elegans THAP family also included three previously uncharacterized proteins (Table 2 and Fig. 6) as well as the orthologue of CtBP, a well conserved transcriptional corepressor for homeodomain, nuclear hormone receptor, and C2H2 zinc-finger proteins (21), which does not exhibit a THAP domain in other animal species. Finally, LIN-15A and seven other predicted C. elegans proteins that contained divergent THAP domains similar to those found in HIM-17, LIN-15B and CDC-14B, were also identified (Table 2 and Fig. 1).
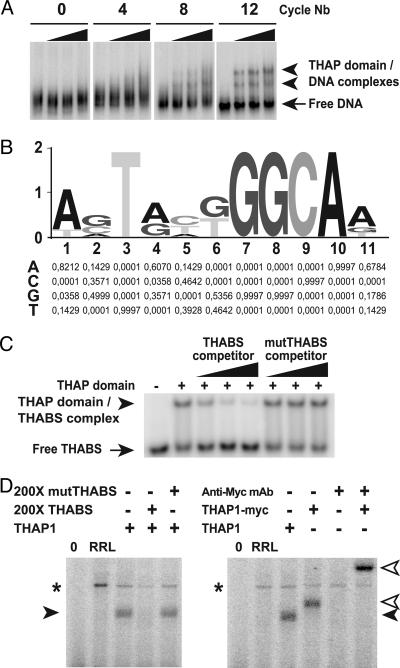
The THAP Domain Is a Sequence-Specific DNA-Binding Domain. To determine whether the THAP domain possesses sequence-specific DNA-binding activity and to identify a consensus target-binding site, we used a PCR-based approach, SELEX (15). For that purpose, we used as a prototype the THAP domain of human THAP1, recently characterized in our laboratory (2). We first expressed and purified the THAP domain in E. coli, then used this protein to select a preferential binding site from a degenerate pool of 25-bp oligonucleotides flanked by conserved sequences to facilitate amplification and cloning. After 4, 8, and 12 cycles, we evaluated the DNA–protein interaction by EMSA (Fig. 2A). High-affinity binding sequences selected after 12 cycles were cloned and sequenced. Analysis of 25 selected sequences with meme identified an 11-bp consensus motif (Fig. 2B) that was found twice in most selected sequences, as either direct or inverted repeats. The consensus sequence, which we refer to as THABS, comprised a core “GGCA” motif. Competitive EMSA experiments with unlabeled THABS or mutant THABS oligonucleotides demonstrated that purified recombinant THAP domain binds the THABS target sequence specifically (Fig. 2C). These results were confirmed and further extended by using in vitro translated full-length THAP1 protein, which provided an independent source of THAP domain. Recognition of the THABS by the THAP domain was observed in the context of the whole THAP1 protein (Fig. 2D). This binding was specific, and the DNA–protein complex formed between the THABS and a Myc-tagged THAP1 protein was supershifted by an anti-Myc antibody (Fig. 2D) but not by an unrelated control antibody (data not shown).
Fig. 2.
The THAP domain is a sequence-specific DNA-binding domain. (A) Identification of THAP-domain DNA-target sequence by SELEX. DNA recovered after 0, 4, 8, and 12 rounds of selection was labeled and incubated with increasing amounts of recombinant THAP domain (1, 6, and 60 ng, respectively). Resulting protein–DNA complexes were analyzed in EMSA. (B) Identification of a consensus THABS. The oligonucleotide sequences recovered after 12 rounds of selection were analyzed by using the motif-discovery program meme. The position-specific probability matrix returned by meme is given. (C) Specificity of THAP-domain–THABS interaction. Recombinant THAP domain was incubated with THABS probe in the absence or presence of increasing amounts (50-, 150-, and 250-fold molar excess) of either THABS (AGTAAGGGCAA) or mutTHABS (AGTAATTTCAA) unlabeled competitor. The protein–DNA complexes were analyzed in EMSA. (D) Binding of full-length THAP1 to the THABS. In vitro translated THAP1 or THAP1-Myc was incubated with labeled THABS probe in the absence or presence of a 200-fold molar excess of either THABS or mutTHABS unlabeled competitor, and protein–DNA complexes were analyzed in EMSA. For the supershift experiment, THAP1-Myc was incubated with the THABS probe in the presence of anti-Myc mAb. RRL, THABS probe incubated with unprogrammed RRL; black arrowhead, THAP1–THABS DNA complex; white arrowhead, THAP1-Myc–THABS DNA complex; *, nonspecific complex.
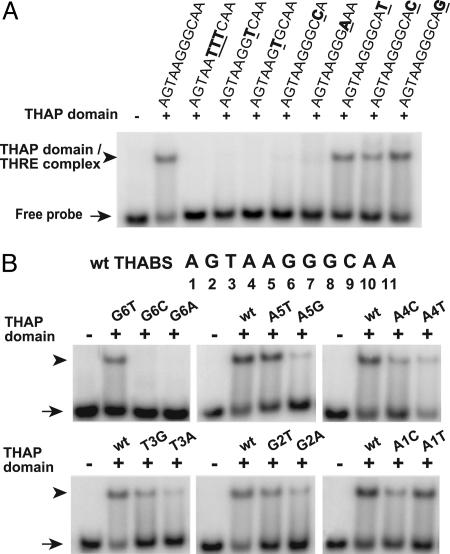
DNA-Binding-Site Specificity of the THAP Domain of Human THAP1. To further characterize the DNA-binding-site specificity of the THAP domain, we performed scanning mutagenesis of a THABS consensus oligonucleotide obtained from the SELEX experiment. Labeled oligonucleotides bearing substitutions in each of the bases comprising the THABS were incubated with recombinant THAP domain. These analyses revealed that the core GGCA motif of the THABS is essential for recognition by the THAP domain (Fig. 3A). In addition, a G or a T at position 6 was also strictly required (Fig. 3B). In contrast, single base substitutions at other nucleotide positions upstream of the core motif did not abrogate DNA-binding but, rather, were found to modulate the strength and affinity of THAP-domain–THABS interaction (Fig. 3B). Scanning mutagenesis results were generally in good agreement with the percentage occurrence of each base at each position in the sequences identified by SELEX (Fig. 2B). For instance, very low binding was observed with mutant oligonucleotides bearing T or G in positions 4 or 5 of the THABS, respectively (Fig. 3B), and these bases never occurred at these positions in the SELEX sequences (Fig. 2B). A notable exception was the T at position 3, which was found in all of the oligonucleotides obtained from the SELEX experiments but did not appear to be absolutely required for THAP-domain binding.
Fig. 3.
Mutations of single nucleotide positions in the THABS abrogate recognition by the THAP domain of human THAP1. (A) The GGCA core motif of the THABS is required for THAP-domain–THABS interaction. EMSAs were performed by using recombinant THAP domain and labeled oligonucleotides bearing mutations in the GGCA core motif of the THABS. (B) Scanning mutagenesis of the THABS sequence reveals that bases upstream of the GGCA core motif modulate the strength and affinity of THAP-domain–THABS interaction. EMSAs were performed by using recombinant THAP-domain and DNA targets bearing single-point mutations in the THABS sequence.
The THAP Domain Is a Zinc-Dependent DNA-Binding Domain. A notable feature of the THAP domain is the C2CH motif (CX2–4CX35–53CX2H), which may constitute a metal-coordinating module, as in previously described zinc fingers. However, to our knowledge, a large C2CH module such as the one found in the THAP domain (up to 53 residues of spacing between the C2 and CH coordinating residues) has not previously been described in the zinc-finger superfamily. Therefore, we investigated whether the THAP domain is a zinc-dependent DNA-binding domain. EMSA was performed after incubation of the THAP domain with metal-chelating agents 1,10-o-phenanthroline and EDTA. Increasing amounts of 1,10-o-phenanthroline (up to 5 mM) and, to a lesser extent, EDTA (up to 50 mM) gradually inhibited DNA-binding activity (Fig. 4A), indicating that the THAP domain requires divalent metal ions for its functional activity. To examine the role of zinc in THAP-domain activity, we added back zinc, calcium, magnesium, or iron to binding reactions after preincubation of the THAP domain with 5 mM 1,10-o-phenanthroline. Significant THAP-domain DNA-binding activity was restored by the addition of zinc but not calcium, magnesium, or iron (Fig. 4B), demonstrating that the THAP domain possesses zinc-dependent DNA-binding activity.
Fig. 4.
The THAP domain is a zinc-dependent DNA-binding domain. (A) Inhibition of THAP-domain DNA-binding activity by metal-chelating agents EDTA and 1,10-o-phenanthroline. THAP-domain–THABS DNA complexes were analyzed by EMSA. Lane 0, THABS probe alone; UT, THABS probe incubated with untreated THAP domain; MeOH, methanol vehicle alone. (B) Role of zinc in THAP-domain DNA-binding activity. Recombinant THAP domain was treated with 5 mM 1,10-o-phenanthroline, and the chloride of Zn2+, Ca2+, Mg2+, or Fe2+ was subsequently added to the binding reactions before analysis of THAP-domain–DNA complexes (arrowhead) by EMSA.
The C2CH Motif and the Four Other Conserved Residues of the THAP Domain Are Required for Sequence-Specific DNA Binding. To investigate the role of the putative metal-coordinating residues of the C2CH module in THAP-domain functional activity, we generated four single-point mutants in the C2CH signature. The three cysteines and the single histidine residues were replaced by alanines to generate THAP1 mutants C5A, C10A, C54A, and H57A, respectively. Mutagenesis of the putative metal-coordinating residues of the C2CH module did not appear to affect translation or stability of the mutant proteins because in vitro translation of the four mutant proteins in RRL revealed expression levels similar to those of the wild-type THAP1 protein (Fig. 5A). However, the four THAP1 mutants lost DNA-binding activity and failed to recognize the THABS in EMSAs (Fig. 5B). Mutagenesis of single cysteine or histidine residues in the C2CH motif had the same effect as treatment of the wild-type THAP1 protein with the zinc chelator 1,10-o-phenanthroline (Fig. 5B), supporting an important role for the C2CH motif in zinc coordination and DNA-binding activity of the THAP domain. The absolute conservation in all THAP domains of four additional residues (P26, W36, F58, and P78 in human THAP1) suggested a possible requirement for these residues in DNA-binding function. Four additional THAP1 mutants (P26A, W36A, F58A, and P78A) were therefore generated and expressed in RRL (Fig. 5A). These mutants lost DNA-binding activity and failed to recognize the THABS in EMSAs (Fig. 5C). These results favor an important role for both the C2CH motif and the four other conserved residues of the THAP domain in DNA-binding activity.
Fig. 5.
The C2CH signature and the four other invariant residues of the THAP domain are essential for DNA binding. (A) Generation of THAP1 mutants in the consensus THAP motif. Wild-type THAP1 (wt) and single-point mutants THAP1-C5A, THAP1-C10A, THAP1-C54A, THAP1-H57A, THAP1-P26A, THAP1-W36A, THAP1-F58A, and THAP1-P78A were translated in vitro in RRL in the presence of 35S-labeled methionine and analyzed by SDS/PAGE and autoradiography. Molecular mass markers are shown on the left (kDa). (B) Mutation of single cysteine or histidine residues of the C2CH signature abrogates THAP-domain DNA-binding activity. EMSAs were performed with the THABS probe and THAP1 wild-type (wt) or mutant proteins (C5A, C10A, C54A, and H57A). For comparison, wild-type THAP1 was incubated with 5 mM 1,10-o-phenanthroline or methanol vehicle alone (MeOH). RRL, unprogrammed RRL; arrowhead, THAP1–THABS DNA complex; *, nonspecific complexes. (C) Mutation of the four other conserved residues of the THAP domain (P, W, F, and P) abrogates DNA binding. EMSAs were performed with the THABS probe and THAP1 wild-type (wt) or mutant proteins (P26A, W36A, F58A, and P78A).
Discussion
The THAP domain is an evolutionarily conserved protein motif restricted to animals (1), which defines a previously uncharacterized family of cellular factors, the THAP proteins, with >100 distinct members in the animal kingdom (Tables 1 and 2 and Fig. 6). In this article, we demonstrate the biochemical function of the THAP domain in cellular THAP proteins as a zinc-dependent sequence-specific DNA-binding domain belonging to the zinc-finger superfamily. We reported in ref. 1 that the site-specific DNA-binding domain of Drosophila P element transposase (3) corresponded to a consensus THAP domain. Our present data show that, although the THAP domains of human THAP1 and P element transposase share <25% sequence identity, the biochemical function of the domain, sequence-specific DNA binding, has been conserved between the two proteins. These results emphasize the evolutionary and functional relationships between cellular THAP proteins and P element transposase and are in agreement with the previous observation that one of the human THAP proteins, THAP9, appears to be an ancient descendant of P element transposase (1, 22). The THAP domain is therefore another example of a DNA-binding domain shared between cellular proteins and transposases from mobile genomic parasites. Previous examples include the DNA-binding domain of centromere protein CENP-B (23), which is homologous to that of Drosophila pogo transposase, human tigger pogo-like transposases, and the BED finger (24), an atypical zinc-finger DNA-binding domain found in both cellular chromatin-boundary element-binding proteins BEAF/DREF and AC1/Hobo-like transposases from animals, plants, and fungi.
We demonstrate that the DNA-binding activity of the THAP domain of human THAP1 is zinc-dependent and that the four putative metal-coordinating residues of the C2CH module are essential for functional activity. This DNA binding suggests that the THAP domain belongs to the zinc-finger superfamily. Classical zinc fingers have been defined as small, functional, independently folded domains that require coordination of one or more zinc ions to stabilize their structure (25, 26). Indeed, the C2H2-type zinc finger, which defines the most abundant class of DNA-binding proteins in the human genome, is a compact, ≈30-aa DNA-binding domain repeated in multiple copies in the proteins (25, 26). Similarly, the zinc-coordinating module of the C4-type zinc finger found in the GATA family of transcription factors covers only the first 30 residues of the 60-aa DNA-binding domain (27). Therefore, the size of the THAP domain (≈90 aa) and the spacing between the C2 and CH residues of the large C2CH module (up to 53 aa) are atypical in the zinc-finger superfamily. In addition, the sizes of the DNA target sequences recognized by the THAP domain, 11 nucleotides for human THAP1 (Figs. 2 and 3) and 10 nucleotides for Drosophila P element transposase (28), are considerably larger than those recognized by classical C2H2 zinc fingers, which typically recognize only 3–4 nucleotides (25). Despite these differences, we believe that the THAP domain, which is a functional, independently folded domain that requires coordination of zinc for its DNA-binding activity, should be classified as a zinc-coordinating DNA-binding domain belonging to the zinc-finger superfamily. With ≈100 distinct THAP-domain protein sequences identified in model animal organisms (Tables 1 and 2 and Fig. 6), including 12 human proteins (1), the THAP domain may define one of the most abundant class of zinc-coordinating DNA-binding proteins in the animal kingdom, after the C2H2 zinc-finger proteins and the nuclear hormone receptors.
The four other residues strictly conserved in all THAP-domain sequences were also found to be required for DNA binding (Fig. 5C). Therefore, mutation of any of the eight residues that define the THAP motif abrogates DNA-binding activity. These results are important because they link our in vitro data on THAP-domain–DNA interactions to in vivo genetic data previously obtained in C. elegans, because equivalent mutations have been identified in lin-36 and him-17: Of seven single-point mutations identified in the LIN-36 protein, four mutations were found in the THAP domain, including two independent mutations in the last P residue of the THAP motif (12); two single-point mutations were identified in the HIM-17 protein, and both were found in the THAP domains, including mutation of the second C of the C2CH motif in THAP domain 6 (8). The identification of single-point mutants in the THAP domains of LIN-36 and HIM-17 supports the possibility that these THAP domains are functional and likely to exhibit DNA-binding activity in vivo.
The consensus THABS motif recognized by the THAP domain of human THAP1 (Fig. 2B) does not share significant homology with the A+T-rich motif recognized by P element transposase (28). Together with the observation that distinct THAP-domain sequences within a single species exhibit <50% identity between each other, this finding suggests that each THAP domain may possess its own specific DNA-binding site. However, we cannot exclude, at this stage, the possibility that some THAP domains may lack sequence specificity. Similarly, the divergent THAP motifs found in HIM-17 and other C. elegans proteins (8) may have lost DNA-binding activity and function instead as protein–protein-interaction modules, as shown for some C2H2 zinc fingers in ref. 29. Finally, it remains possible that a single THAP domain may function in both protein–protein and protein–DNA interactions, as suggested in ref. 6 for human THAP7.
Genetic data obtained in C. elegans indicate that cellular THAP proteins may be involved in chromatin modification. The multi-THAP C. elegans protein HIM-17 has been shown to be associated with chromatin and required for proper accumulation of histone H3 methylation at lysine-9 on meiotic prophase chromosomes (8), suggesting that HIM-17 recruits chromatin-modifying and/or -remodeling complexes essential for chromatin modification during meiosis. The link between THAP proteins and chromatin modification/remodeling is further reinforced by the observation that five distinct C. elegans proteins containing consensus and/or divergent THAP domains (LIN-36, LIN-15B, HIM-17, CDC-14B, and LIN-15A) have been shown to interact genetically with LIN-35/Rb (8, 9, 13, 20), a known component of chromatin-remodeling complexes in mammalian cells (30). Interestingly, human THAP7 has recently been found to associate with chromatin and to function as a histone-tail-binding protein that represses transcription through recruitment of corepressor NcoR and histone deacetylase HDAC3 (6). Similarly, nuclear proapoptotic factors THAP1 and THAP0 may also function in chromatin modification and/or transcriptional repression.
The genetic interactions of C. elegans THAP proteins with LIN-35/Rb (8, 9, 13, 20) and our observation that the zebrafish orthologue of cell-cycle transcription factor E2F6 contains a THAP domain (Fig. 1) suggest important roles for THAP proteins in cell proliferation and/or cell-cycle progression. Evidence for such a role has already been provided for LIN-36 (14) and LIN-15B, which have been shown to negatively regulate G1 progression (13). A third C. elegans THAP protein, the CDC-14B isoform of the cell-cycle regulator tyrosine phosphatase CDC-14, may also function with LIN-35/Rb in G1 regulation because C. elegans CDC-14 has been shown to inhibit G1–S transition by modulating nuclear levels of CDK inhibitor CKI-1 (20). Alternatively, the CDC-14B isoform, which contains the DNA-binding THAP motif, could mediate the effects of CDC-14 in the protection of the C. elegans genome against DNA damage (31). CDC-14 isoforms containing THAP domains have not yet been described in other animal species. However, it is well known that two proteins, which have homologues in another organism fused into a single protein chain, often show interaction between each other (32). Therefore, in other organisms, CDC-14 could be targeted to DNA and/or chromatin by direct interaction with a cellular THAP protein. A similar scenario may apply to C. elegans CtBP and zebrafish E2F6, which contain consensus THAP domains at their amino termini (Fig. 1) that are not observed in homologues from other animal species.
In summary, the results presented in this article show that the THAP domain of human THAP1 is a zinc-dependent sequence-specific DNA-binding domain. Together with previous data obtained in humans and C. elegans, these findings suggest that cellular THAP proteins may function as sequence-specific DNA-binding factors with roles in cell proliferation, apoptosis, cell cycle, chromosome segregation, chromatin modification, and transcriptional regulation.
Supplementary Material
Acknowledgments
We thank Laurence Nieto [Institut de Pharmacologie et de Biologie Structurale (IPBS)–Centre National de la Recherche Scientifique (CNRS)] and Sophia Kossida (Endocube) for help with EMSAs and initial characterization of THAP-family protein sequences, respectively, and members of the Laboratory of Vascular Biology (IPBS–CNRS) for stimulating discussions. We are grateful to Corinne Cayrol (IPBS–CNRS) for the identification of THAP-E2F6 fusion proteins. This work was supported by grants from Ligue Nationale Contre le Cancer (Equipe Labellisée “La Ligue 2003”) and Ministère de la Recherche Actions Concertées Incitatives “Jeunes Chercheurs.”
Author contributions: C.M. and J.-P.G. designed research; T.C., M.R., V.E., and C.M. performed research; T.C., F.A., and J.-P.G. analyzed data; and J.-P.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RRL, rabbit reticulocyte lysate; SELEX, systematic evolution of ligands by exponential enrichment; THABS, THAP-domain-binding sequence.
References
- 1.Roussigne, M., Kossida, S., Lavigne, A. C., Clouaire, T., Ecochard, V., Glories, A., Amalric, F. & Girard, J. P. (2003) Trends Biochem. Sci. 28, 66–69. [DOI] [PubMed] [Google Scholar]
- 2.Roussigne, M., Cayrol, C., Clouaire, T., Amalric, F. & Girard, J. P. (2003) Oncogene 22, 2432–2442. [DOI] [PubMed] [Google Scholar]
- 3.Lee, C. C., Beall, E. L. & Rio, D. C. (1998) EMBO J. 17, 4166–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deiss, L. P., Feinstein, E., Berissi, H., Cohen, O. & Kimchi, A. (1995) Genes Dev. 9, 15–30. [DOI] [PubMed] [Google Scholar]
- 5.Lin, Y., Khokhlatchev, A., Figeys, D. & Avruch, J. (2002) J. Biol. Chem. 277, 47991–48001. [DOI] [PubMed] [Google Scholar]
- 6.Macfarlan, T., Kutney, S., Altman, B., Montross, R., Yu, J. & Chakravarti, D. (2005) J. Biol. Chem. 280, 7346–7358. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, W. L., Ajiro, K., Samejima, K., Kloc, M., Cheung, P., Mizzen, C. A., Beeser, A., Etkin, L. D., Chernoff, J., Earnshaw, W. C. & Allis, C. D. (2003) Cell 113, 507–517. [DOI] [PubMed] [Google Scholar]
- 8.Reddy, K. C. & Villeneuve, A. M. (2004) Cell 118, 439–452. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, E. L. & Horvitz, H. R. (1989) Genetics 123, 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, S. G., Lu, X. & Horvitz, H. R. (1994) Genetics 137, 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, L. S., Tzou, P. & Sternberg, P. W. (1994) Mol. Biol. Cell 5, 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas, J. H. & Horvitz, H. R. (1999) Development (Cambridge, U.K.) 126, 3449–3459. [DOI] [PubMed] [Google Scholar]
- 13.Boxem, M. & van den Heuvel, S. (2002) Curr. Biol. 12, 906–911. [DOI] [PubMed] [Google Scholar]
- 14.Fay, D. S., Keenan, S. & Han, M. (2002) Genes Dev. 16, 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouvet, P. (2001) Methods Mol. Biol. 148, 603–610. [DOI] [PubMed] [Google Scholar]
- 16.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, D. G., Thompson, J. D. & Gibson, T. J. (1996) Methods Enzymol. 266, 383–402. [DOI] [PubMed] [Google Scholar]
- 18.Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. (2004) Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor, J. S., Braasch, I., Frickey, T., Meyer, A. & Van de Peer, Y. (2003) Genome Res. 13, 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito, R. M., Perreault, A., Peach, B., Satterlee, J. S. & van den Heuvel, S. (2004) Nat. Cell Biol. 6, 777–783. [DOI] [PubMed] [Google Scholar]
- 21.Chinnadurai, G. (2002) Mol. Cell 9, 213–224. [DOI] [PubMed] [Google Scholar]
- 22.Hammer, S. E., Strehl, S. & Hagemann, S. (2005) Mol. Biol. Evol. 22, 833–844. [DOI] [PubMed] [Google Scholar]
- 23.Smit, A. F. & Riggs, A. D. (1996) Proc. Natl. Acad. Sci. USA 93, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravind, L. (2000) Trends Biochem. Sci. 25, 421–423. [DOI] [PubMed] [Google Scholar]
- 25.Pavletich, N. P. & Pabo, C. O. (1991) Science 252, 809–817. [DOI] [PubMed] [Google Scholar]
- 26.Laity, J. H., Lee, B. M. & Wright, P. E. (2001) Curr. Opin. Struct. Biol. 11, 39–46. [DOI] [PubMed] [Google Scholar]
- 27.Omichinski, J. G., Clore, G. M., Schaad, O., Felsenfeld, G., Trainor, C., Appella, E., Stahl, S. J. & Gronenborn, A. M. (1993) Science 261, 438–446. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman, P. D., Doll, R. F. & Rio, D. C. (1989) Cell 59, 359–371. [DOI] [PubMed] [Google Scholar]
- 29.Sun, L., Liu, A. & Georgopoulos, K. (1996) EMBO J. 15, 5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, H. S., Gavin, M., Dahiya, A., Postigo, A. A., Ma, D., Luo, R. X., Harbour, J. W. & Dean, D. C. (2000) Cell 101, 79–89. [DOI] [PubMed] [Google Scholar]
- 31.Pothof, J., van Haaften, G., Thijssen, K., Kamath, R. S., Fraser, A. G., Ahringer, J., Plasterk, R. H. & Tijsterman, M. (2003) Genes Dev. 17, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcotte, E. M., Pellegrini, M., Ng, H. L., Rice, D. W., Yeates, T. O. & Eisenberg, D. (1999) Science 285, 751–753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.