Abstract
The nicotinic acetylcholine receptor is a well characterized ligandgated ion channel, yet a proper description of the mechanisms involved in gating, opening, closing, ligand binding, and desensitization does not exist. Until recently, atomic-resolution structural information on the protein was limited, but with the production of the x-ray crystal structure of the Lymnea stagnalis acetylcholine binding protein and the EM image of the transmembrane domain of the torpedo electric ray nicotinic channel, we were provided with a window to examine the mechanism by which this channel operates. A 15-ns all-atom simulation of a homology model of the homomeric human α7 form of the receptor was conducted in a solvated palmitoyl-2-oleoyl-sn-glycerol-phosphatidylcholine bilayer and examined in detail. The receptor was unliganded. The structure undergoes a twist-to-close motion that correlates movements of the C loop in the ligand binding domain, via the β10-strand that connects the two, with the 10° rotation and inward movement of two nonadjacent subunits. The Cys loop appears to act as a stator around which the α-helical transmembrane domain can pivot and rotate relative to the rigid β-sheet binding domain. The M2–M3 loop may have a role in controlling the extent or kinetics of these relative movements. All of this motion, along with essential dynamics analysis, is suggestive of the direction of larger motions involved in gating of the channel.
The nicotinic acetylcholine receptor is one of the best-studied ligand-gated ion channels (LGICs) (1). It has been found to have a role in a wide range of pathological conditions and diseases including epilepsy (2), schizophrenia (3), Alzheimer's (4), Parkinson's (5), neuromuscular diseases (6), inflammation (7), nicotine addiction (8), and addiction to other drugs such as alcohol (9) and cocaine (10), as well as a role in anesthetic action (11). The nicotinic acetylcholine receptor is a member of a superfamily of pentameric receptors that include the serotonin (excitory), GABA, and glycine (inhibitory) receptors that are all involved in the transmission and modification of electrical signals in excitory cells. The family is known as the Cys-loop LGIC family, because of a conserved pair of linked cysteines in the N-terminal domain of the receptor (12). Within the superfamily, an array of different nicotinic receptor subunits exists (13, 14), making different subunit assemblies possible, for different roles, in different cell types. In all of these receptors, the ligand binds at the interface between the α and other subunits of the ligand binding domain (LBD). This event begins a chain reaction of conformational changes within the receptor that transmits the message of ligand binding to the transmembrane domain (TMD), which then opens to allow the passage of ions through the channel. Although the genetics, kinetics, electrophysiology, and many topological aspects are well characterized for the nicotinic receptor (15, 16), the exact nature of the structural rearrangments involved in activation, gating, and desensitization of the channel is not known.
The neuromuscular form of the nicotinic receptor has a heteropentameric arrangement with α2βδγ subunits and only two ligands binding at the α-γ and α-δ interfaces (17). Many structural aspects of this channel were studied because of the availability of a homologue in the electric organ of the torpedo ray (18). It was from this source that the EM structure of the TMD was solved at ≈4-Å resolution (19). Before this process, a structure of a LBD homologue from another source, the Lymnea stagnalis water snail, was determined (20). This soluble acetylcholine binding protein (AChBP) was more amenable to x-ray crystallography because of the natural lack of a membrane spanning domain and therefore it was solved at higher resolution (2.7 Å). AChBP has up to 23% sequence identity with the LBD of the complete receptors, and it is most similar to that in the human α7 (hα7) subunit (21, 22). AChBP structure has subsequently been solved with ligands bound [Protein Data Bank ID codes 1UV6 (carbamylcholine), 1UW6 (nicotine), and 1UX2 (Hepes)] (23). As for AChBP, the hα7 receptor is a homopentamer (24) rather than the hetero arrangement seen in many other forms of the channel. The present study involved the production and simulation of a homology model so that it seemed prudent to use a form of the receptor that was most similar to the solved high-resolution structure. Therefore, we focus on the hα7 receptor. All of the nicotinic receptor channels are cation-selective but mostly conduct sodium at nerve synapses and neuromuscular junctions. Hα7, though, has a high calcium ion current (25) as it fulfills its role in calcium regulation in many different nerve cell types in the central and peripheral nervous systems (26).
Given a high-resolution starting structure, molecular dynamics (MD) has proven to be a valuable tool in the analysis of structure–function relationships and mechanisms. Transport processes for membrane proteins are somewhat accessible within the time-scale limitations of all-atom simulations, and MD has had much success in analyzing these, for example, in K channels (27, 28) and aquaporins (29–31), where the mechanism was first shown in a homology model (30). Previous studies of the LBD in MD have yielded valuable information about its function (32, 33). Until recently, the availability of membrane protein structures was a limiting factor in their general study, because of difficulties in solving the structures (34). But because of improving technology and the use of bacterial analogues (and other lower organisms as seen with the nicotinic receptor) of human proteins required for study, this situation is beginning to be rectified (35).
MD simulation is limited mostly to submicrosecond time scales at the present time. Therefore, long time-scale motions, such as gating, are not directly accessible with this method in its conventional form. Yet, all-atom MD provides the most complete representation of the structure of a biomolecule and its immediate environment. Here, we use this method to observe the types and direction of motions that hint at the complete motions involved in activation, gating, and desensitization.
Methods
Protein Modeling. Homology modeling was conducted with modeller 4.0 (36) (www.salilab.org/modeller/modeller.html). The 2.7-Å resolution x-ray structure of the AChBP (Protein Data Bank ID 1I9B) (20) and the EM structure of the TMD (Protein Data Bank ID 1OED) (19) were combined (at residue T231) to form the template from which the homology model was created. The modeled hα7 sequence did not include 23 residues at the N terminus, as these were not in the aligned template structure (AChBP), and it did not include the 142 residues between M3 and M4, which correspond to the cytoplasmic vestibule domain, as these were not included in the TMD structure. Four residues were not included at the C terminus for the same reason. The model was built by bringing these templates together, using an overlaid all-α-subunit makeup for the TMD, and then modeling the full pentameric receptor. Subunits (named A, B, C, D, and E) were arranged with a counterclockwise adjacency, looking from the extracellular side, as in the AChBP structure. Five-fold symmetry was imposed when modeling the pentamer structure but no α-helical or β-sheet restraints were applied to actively retain secondary structure. Two disulphide bridges in each subunit were restrained in the modeling process between residues 127 and 141 (Cys loop) and between residues 189 and 190 (C loop). Sequence alignments for input to modeling were generated by using clustalw in jalview (www.ebi.ac.uk:80/jalview/index.html). Structures and models were checked and compared by using procheck (37) and whatcheck (38). Before running simulations, side-chain ionization states were adjusted to match the results of pKa calculations, performed as described (39), using uhbd (40) to calculate free energy differences between ionized and unionized side chains of the protein. Protonation states were then checked manually to assure no inappropriate results caused by the approximations of the method (41), and the H-bonding network was not optimized before the calculation.
System Set-Up and Treatment. An initial 110 × 110-Å palmitoyl-2-oleoyl-sn-glycerol-phosphatidylcholine (POPC) bilayer slab, with water molecules positioned to hydrate the headgroups, was created by using the membrane package in vmd (42). This preliminary bilayer (consisting of 326 POPC molecules, 163 in each leaflet, and 5,529 water molecules) was then relaxed by steepest descent minimization for 100 steps and equilibrated in MD for 500 ps to optimize the lipid–lipid and lipid–water packing.
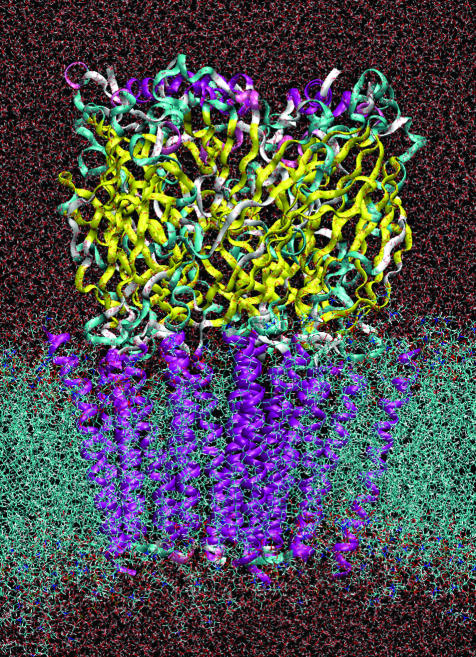
The hα7 receptor model was then inserted into the center of the bilayer, and all overlapping lipids and waters were removed (the system then contained 218 palmitoyl-2-oleoyl-sn-glycerol-phosphatidylcholine and 3,698 water molecules). The system was then fully solvated to a box size of 110 Å × 110 Å × 140 Å by using solvate (43). Counterions were added to neutralize the charge on the protein plus enough extra Na+ and Cl– ions to give a 0.1 M solution (Fig. 1). Disulphide bonds were retained throughout the simulation as described for the modeling. The system was then relaxed for 100 steps followed by an equilibration MD run, with the positions of the heavy atoms of the protein restrained to their initial positions throughout. During the equilibration, the temperature was raised from 10 to 310 K under constant volume conditions. This restrained run allowed the water and lipid to relax around the protein. System topology set-up was conducted by using the psfgen program that is part of the namd package (44). Images were prepared by using vmd and raster3d (45).
Fig. 1.
The set-up of the system with the pentameric hα7 nicotinic receptor model (ribbon) inserted into the palmitoyl-2-oleoyl-sn-glycerol-phosphatidylcholine lipid bilayer (cyan strings) and fully hydrated with transferable intermolecular potential 3-point waters (red background). The colors on the protein indicate secondary structure: purple is α-helix, yellow is β-strand, and cyan is random coil.
namd Simulation Details. MD simulation was run by using the charmm forcefield (46) (version 27) in namd (44) with a non-bonded van der Waals cut-off of 9 Å. The restrained equilibration (500 ps) and production runs (15 ns) were conducted at 310 K, although they were initially restrained at 10 K, and then maintained at 310 K by using a Langevin temperature piston. Production simulations (those after minimization and equilibration) were conducted at constant pressure. Particle mesh Ewald electrostatics settings were applied to each system. All of the simulations were run with shake (47) with a 2-fs time step. Transferable intermolecular potential 3-point water was used. All of the MD runs were conducted on the Blue Horizon machine at the San Diego Supercomputer Center. Preparation, visualization, and analysis were performed on a Dell Dual Xeon processor, desktop machine, under Red Hat Linux 9. Essential dynamics was performed by using John Mongan's (University of California, San Diego) ied 2.0 add-on for vmd (http://mccammon.ucsd.edu/ied) (48).
Results
Homology Model Analysis. A description of the homology model's structure is worthwhile for later discussions about structural rearrangement and functional relevance, even though the model is essentially the same topologically as the x-ray (AChBP) and EM (TMD) structures from which it was templated (Fig. 5 Left, which is published as supporting information on the PNAS web site). This model can be compared with the human nicotinic acetylcholine receptor α7 sequence (Fig. 5 Right). The LBD consists mainly of β-sheet (β1to β10), but with a short helix (H1) at the N terminus. The loop between β9 and β10, called the C loop, encloses the ligand binding site that exists between adjacent subunits. At the base of the LBD, inserted in between the tops of the four TM helices, is the conserved Cys loop from which the superfamily was named. A disulphide bridge exists between two cysteines in this loop. In α7, a disulphide is also present between two adjacent residues in the C loop, but it is not a well conserved feature across the nicotinic subfamily. The loop between β8 and β9 is long and relatively unstructured. It is the region of most sequence variation in the LBD. Both N and C termini are extracellular with the protein passing through the membrane four times as α-helices (M1 to M4). M2 is the pore-lining helix. Because of the lack of a structure for the vestibule domain that exists between M3 and M4, on the cytoplasmic side of the membrane, M4 is not covalently connected to the rest of the protein.
The quality of the homology was checked by using procheck (36) and whatcheck (38). The overall procheck score of 93% in the most favorable region is actually better than the AChBP structure. For both whatcheck packing and χ1/χ2 analysis, the model compares favorably with the original LBD (AChBP) and TMD templates, as well as other membrane protein x-ray and EM structures. There is a small distortion of χ1/χ2 angles in the residues at the interface between the LBD and TMD. There is also a loss of perfect β-sheet arrangement between a couple of residues at the center of strands β6 and β9 of the binding domain during the homology modeling process. This change could have enabled some extra flexibility in these regions but as the analysis of the simulation shows no real flexibility was actually observed there.
The AChBP from which the LBD was modeled is thought to be in the active/desensitized state (20, 23, 49). The TMD is probably in the closed/desensitized state (19). Added to this factor, there is no ligand present in the active-state LBD in this simulation. This possible contradiction of states is unavoidable but relaxation of this obvious imperfection in the complete homology model may accelerate motions and make relevant transition motions more observable.
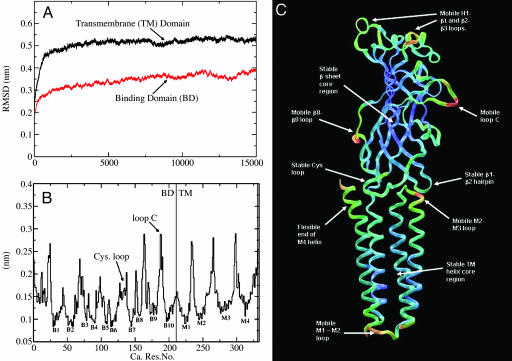
Structural Fluctuations. The rms deviations of the Cα atoms in the LBD and TMD are displayed as functions of time in Fig. 2A. After the first few ns, both domains have stabilized. The deviations from the starting structure are not unexpected for a system of this size. Overall, this analysis shows that the model is stable in simulation but as we might expect for any protein undergoes some motion. It is important to analyze this motion in more detail later and extract from it important information regarding the structure–function relationship.
Fig. 2.
Protein motion. (A and B) These plots illustrate the structural motions that take place in the simulation, in terms of rms deviation (RMSD), separately in the LBD and TM region (A), and rms fluctuations for the complete structure, averaged over the five subunits (B). (C) These average rms fluctuation values are displayed as red-green-blue values on a single subunit of the structure to better illustrate the areas of interest.
We would expect proteins to be less flexible in the areas of secondary structure, which appears to be the case in both the LBD and TMD. The rms fluctuations of the Cα atoms, averaged over the five subunits (Fig. 2B), indicate that the secondary structure (both β-sheet in the binding domain and α-helix in the TMD) is much less mobile relative to the loops in the structure. It includes loops that may be functionally important, such as the Cys loop and the C loop. These average fluctuations in structure are plotted as a red-green-blue scale onto a single subunit structure so that the areas of motion can be more easily seen (Fig. 2C).
A closer look at the motions is necessary to extract biologically relevant information. Much motion is seen in the C loop, which in two of subunits (B and D) moves away from the binding pocket. The C loop (residues 180–197 in each subunit) encloses the pocket in which the ligand would bind. This finding is consistent with previous studies (50, 51) that indicate that the C loop is mobile when no ligand is present and that contact of the C loop with a bound ligand, as in the liganded AChBP structure (23), helps stabilize the binding pocket and the C loop in a more closed state. Therefore an open/mobile C loop, in at least two subunits, seems to correlate with a closed state of the TM channel.
The radius of gyration relative to the central axis of the receptor for the two domains of each subunit is plotted (Fig. 6 A and B, which is published as supporting information on the PNAS web site). Fig. 4 A and B indicates which parts of the receptor move in and which move out. This movement is dominated by loop motion in the LBD and twisting of the helices in the TMD. There is some expansion of the subunits in the LBD. In two of the subunits (B and D), this expansion is attributable to the opening of the C loop. Another two subunits have an increase in radius (A and C) from the center of the pore, resulting from the movement of the H1–β1 and β2–β3 loops.
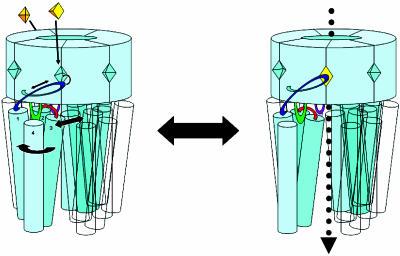
Fig. 4.
The binding of only two ligands is required for action, binding to two nonadjacent subunits. This cartoon illustrates the gating mechanism proposed here. Ligand binding pulls the C loop onto the binding site. This conformational shift is transmitted through the β10 sheet to the TMD. The helices in two nonadjacent subunits undergo small clockwise rotation and outward motions. Once open, ions can flow through the pore. Throughout this process, the Cys loop, connected to the rigid inner β-sheet LBD, acts like a stator around which the TMD can move and rotate. The interaction of the M2–M3 loop and the β1–β2 loop may limit TMD motion and may be the target of gating modification by several allosteric effectors such as anesthetics.
The rotation measured in the LBD (Fig. 6C) does not appear to be consistent or stable. The twisting motion in most of the subunits seems to be minimal and fluctuates to an extent that it cannot be correlated with the other motions observed. Because the bulk of the LBD is essentially rigid and the only substantial motions are those of the loops, these rotations seem to represent vibrational motions.
One of the most mobile regions in the LBD, besides the C loop, is the long, relatively unstructured loop that connects β8 and β9. It sits at the interface between subunits. This linker loop moves inward toward its subunit. This motion occurs in all of the subunits, but it occurs to the greatest degree in the subunits adjacent to those whose C loops move out the most, i.e., in subunits C and E. It represents motion in and around the same ligand binding sites in subunits B and D.
TMD and Pore in Simulation: Correlated Motion. The major observation of the simulation is the closure of the pore along with the outward motion of the C loop. It closes via a counterclockwise twisting motion of the helices in subunits B and D (Figs. 3C and 6D). The pore is pinched off at a ring of hydrophobic residues, protruding from the center of the pore-lining M2 helices, mostly from the M2 helices of subunit B and D. The main residues involved are L278, V274, and the so-called conserved leucine, L270. This leucine actually forms the narrowest point in the channel even at the beginning of the simulation. This point is not quite the narrowest in the EM structure, which has a slightly dissimilar pore profile because of small differences in side-chain conformations. The differences in side-chain orientation and initial pore profile between the homology model and the EM structure are eliminated by just a few steps of energy minimization of the structures (data not shown).
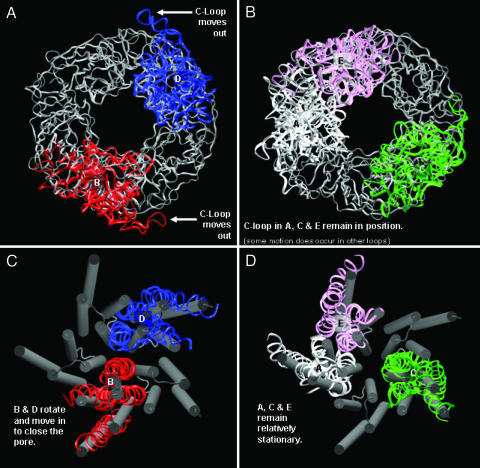
Fig. 3.
These pictures illustrate the important structural transitions that take place during the simulation. (A) The outward motion of the LBD C loops in subunits B and D is shown. (B) This lack of motion can be compared with the motion seen in A and shows how the C loops in subunits A, C, and E do not move out. (C) The closing of the pore by the M2 helices of subunits B and D is demonstrated. (D) Shown are how similar motions do not occur in subunits A, C, and E. Pictures were taken from the same snapshot at 14 ns.
Rotation in the TMDs (Figs. 3 C and D and 6D) occurs in two ways: rotation of whole subunits relative to the other subunits and rotation of the helices in each subunit relative to the other helices in the subunit. Rotation in either sense does not occur equally in each subunit but in fact occurs mostly in two of the subunits (B and D), which see a total average rotation of 10° counterclockwise, to close the pore. This finding compares to a 15° clockwise rotation for channel opening previously seen (52, 53). Helices also twist relative to each other in these subunits with M2 and M3 moving toward the pore. These are the same subunits for which outward C-loop motion is observed. These motions also appear to occur in the same time interval, over the first 10 ns of the simulation. Similar helix twisting motions are not seen in TMD subunits A, C, and E. Subunit E TM helices actually appear to tilt slightly outward as the M2 helices from B and D move in. The subunit E LBD is the only one that moves in. It appears in most of the subunits that the axial movements of the two domains are anticorrelated (at least considering the C loop motion in the LBDs), with an inward motion of the LBD producing an outward motion of the attached TMD and vice versa.
All of the TM helices retain their secondary structure. As stated earlier, the TM helices undergo less change in structure than their connecting loops. This even includes M4, which is not covalently attached to the rest of the structure, because of the lack of the intervening vestibule domain (18). The M4 helix undergoes the most structural change in a helix of the TMD. This finding is perhaps not surprising because the M4 helix has the fewest contacts with other parts of the protein. The M4 bends outward at the top (C-terminal/extracellular end) in all five subunits.
It has been proposed that M2 kinking may be part of the gating mechanism of the channel (54–57). In this simulation, M2 does not kink, although the time scale of the simulation may preclude the possibility of this happening. The M2 helix is somewhat curved outward, away from the pore center as the M2 comes in contact with the binding domain, above the membrane, a factor that is present in the starting structure. This simulation suggests a mode of gating involving rotation and inward/outward motion rather than kinking of M2.
The M2–M3 loop is “docked” into the underside of the binding domain and links the pore lining M2 and the LBD, as it is in contact with both the Cys loop and a β-hairpin [formed between strands β1 and β2 (sometimes called loop 2)]. There is significant motion of the M2–M3 loop, yet the β1–β2 hairpin and the Cys loop both remain relatively static. Therefore, the motion of these loops does not appear to be coupled, at least in this simulation.
Essential dynamics is a useful tool for deconstructing the dynamics present in a protein simulation (58, 59). It confirms what is observable by watching the trajectory as well as other analyses that have already been described. It can also reduce the dimensionality of the trajectory data, and in the process, highlight important dynamical features. The first two eigenvectors in the simulation involve the twisting/closing movement of the pore domain and opening of the C loop. These motions then appear to be correlated. This motion accounts for ≈60% of motion seen here. The next largest eigenvector describes the motion of the M2–M3 loop, which does not feature equally in all of the subunits. The loops undergo a large amount of curling into the hydrophobic cavity that exists in that region of the structure.
Conclusions and Discussion
One of the major findings of this study is a relationship between motions in the binding domain and those in the pore domain that close the channel in the unliganded state. The various forms of analysis indicate how the motion of the single subunit domains (LBD to TMD) is more correlated than that between the adjacent subunits in the separate domains. This finding is consistent with the lack of conservation in residues at the subunit interface in the LGIC family, which suggests that the precise interactions at the interface are not important for conserving the gating activity of the pentamer (60). The LBD and TMD actually have three major points of contact: the direct peptide connection of the C loop to M1 via β10, the Cys loop that sits in between the tops of the four TM helices, and the β1–β2 hairpin loop that associates with the M2–M3 loop. The Cys loop serves like a stator or pivot point around which the motion of the binding domain and the TMD occurs.
The rigid β-sheet region seems to be important to this mechanism. Without it, the loop motion around the binding site would not be transmitted to the TMD, it could simply be absorbed by the LBD. Previously, fitting of the activated nicotinic receptor LBD to AChBP showed movement of β9 and β10 on activation in the α-subunits (51). This finding is consistent with the motion seen in these simulations and the concept that the “outer” β-strands (β9 and β10) actually form a lever that connects the events at the binding site, involving the C loop, with the motions in the TMD. This rigidity extends down to the Cys loop, around which the TMD can rotate. The lack of motion in the Cys loop is somewhat contrary to previous observations seen in LBD-only simulations (32), and this difference likely represents the influence of the associated TMD. Movements of the C loop, similar to those seen here, have been observed (50, 51), and the opposite behavior, with the C loop being held over the binding site by a ligand, also has been noted (50).
The M2–M3 loop has been shown to be an important linker between the LBD and the TMD in several Cys-loop LGIC, especially during channel opening (61–64). It is of potential importance because it connects the M2 and M3 helices, which, for two nonadjacent subunits, move into the pore during the simulation. Several different allosteric affectors may also interact with the M2–M3 region in the Cys-loop receptors, especially anesthetics. Although not likely to bind anesthetics directly, the M2–M3 loop plays a critical role in transmitting the effect to the channel gate (65, 66). As a mobile contact point between the LBD and TMD, the M2–M3 loop would have an important role in conformational transmission and gating, and this is not the first time that this has been suggested for Cys-loop receptors (67). In the simulation, the loop is seen to curl down into the hydrophobic pocket in the TMD, between M1, M2, and M3 where anesthetics are thought to bind to the receptor (68), and in other Cys-loop receptors such as the GABA receptor (69, 70). For nicotinic channels, a view that anesthetics bind solely to M2 (71, 72) seems to prevail. The disorder of the M2–M3 loop seen here may be an important indicator of the mechanisms involved in controlling gating, and previous work has shown the importance of this segment of the structure in other Cys-loop LGICs (63). The loop is not obviously dynamically correlated with the nearby LBD β1–β2 loop of the binding domain and is seemingly in a mobile/disordered state, although the interaction may still be of importance (67). The disorder of the loop may be characteristic of the closed state of this receptor, whereas a more ordered loop may be part of the stabilization of the open state. This stabilization of either state, and differences in the structure/sequence of the M2–M3 loop between Cys-loop family members, may be an important factor in the response to anesthetic binding because these molecules have opposing allosteric affects on very similar members of the family (73), e.g., opening the hyperpolarizing/inhibiting receptors (glycine and GABA) and favoring the closed state of polarizing/excitable members (nicotinic ACh and polarizing 5-HT3).
The pore domain closure was not completely unexpected because evidence suggests that the channel, in this model (from the similar template), is already in a resting/desensitized state (19, 74). The gate in the nicotinic channel and other Cys-loop LGICs undoubtedly exists in the M2 helix (75–79). Much evidence also suggests that the gate is in different conformations in the closed and desensitized states (80, 81). That is, that the gate may be more extensively closed in the desensitized state than in the resting state (82). The motions seen here are suggestive of the differences between these two states.
The fact that in this simulation, and previous simulations of the LBD (32, 50), only two nonadjacent subunits seem to undergo substantial displacements is of both functional and perhaps evolutionary importance. It seems characteristic of these pentameric receptors that only two subunits are necessary to convey the activation/gating action of the channel, because the maximum Hill coefficient observed is two (83). Such asymmetry has been demonstrated before in the TMD (84) and the binding domain (32). For other members of the nicotinic receptor family, this two-site asymmetry seems to be “hardwired” into the available functional oligmers because in these forms, such as muscular α2β2δγ and neuronal α4β2, only two active agonist binding sites exist, at the α-β interfaces. In the homomeric α7 receptor, this two-site asymmetry appears also to be present because of the spatial restriction that only two TMD subunits can move into the pore as observed. Of course, in this receptor, each subunit is equivalent, but the simulation is not long enough to examine all of the possible states. It is merely consistent with the idea that only two subunits/interfaces/binding sites need to be activated to convey functionality of the channel.
So a mechanism that links all of these components together can be seen operating, on a reduced scale, in this simulation. Data from here, previous work, and other sources can be brought together to propose an overall mechanism for gating, as demonstrated in Fig. 4. The reverse situation to that seen in this simulation would enable channel opening. The C loop encloses the binding site and is the handle of a lever. As the lever is moved by the binding of an agonist, its direct link to the TM domain forces a rotation and slight inward/outward motion of that domain. The rigid β-sheet binding domain acts as a stationary point for the lever to push against. The Cys loop, which extends down from the LBD into the center of the TM domain, acts like a pivot point around which the TM domain can rotate. The M2–M3 loop's contact with the β1–β2 hairpin either acts as a second contact point for transmission of motion from the LBD to the TMD or as a latch that limits and/or allows modification of that of the lever-rotor mechanism. An antagonist such as d-tubocurarine is a large molecule that when bound in the active site of the LBD not only blocks the active site from agonists but also pushes the C loop into a more open position. In the lever-rotor mechanism this open lever would rotate two of the subunits in the TMD into a “closed” conformation, much like that seen in this simulation, as would be the case with no ligand bound.
Supplementary Material
Acknowledgments
We thank the J.A.M. group for all of its useful discussions. This work was supported in part by the National Science Foundation, the National Institutes of Health, Howard Hughes Medical Institute, the San Diego Supercomputer Center, the National Biomedical Computation Resource, Accelrys, the Keck Foundation, and the National Science Foundation Center for Theoretical Biological Physics.
Author contributions: R.J.L. designed research; R.J.L. performed research; R.J.L. and R.H.H. contributed new reagents/analytic tools; R.J.L. analyzed data; and R.J.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LGIC, ligand-gated ion channel; MD, molecular dynamics; LBD, ligand binding domain; TM, transmembrane; TMD, TM domain; AChBP, acetylcholine binding protein; hα7, human α7.
References
- 1.Karlin, A. (2002) Nat. Rev. Neurobiol. 3, 102–114. [DOI] [PubMed] [Google Scholar]
- 2.Hogg, R. & Bertrand, D. (2003) Drug News Perspect. 16, 261–266. [DOI] [PubMed] [Google Scholar]
- 3.Freedman, R. C. H., Myles-Worsley, M., Orr-Urtreger, A., Olincy, A., Davis, A., Polymeropoulos, M., Holik, J., Hopkins, J., Hoff, M., Rosenthal, J., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis, P., Palmer, A., Snape, M. & Wilcok, G. (1999) J. Neurol. Neurosurg. Psychiatric Res. 66, 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fodale, V., Practico, C. & Santamaria, L. (2004) Parkinsonism Related Dis. 10, 189–190. [DOI] [PubMed] [Google Scholar]
- 6.Lindstrom, J. (2003) Ann. N.Y. Acad. Sci. 998, 41–52. [DOI] [PubMed] [Google Scholar]
- 7.Wang, H. Y., Ochani, M., Amella, C. A., Tanovic, M., Susaria, S., Li, J. H., Wang, H., Yang, H., Ulloa, L., Al-Abed, Y., et al. (2003) Nature 421, 384–388. [DOI] [PubMed] [Google Scholar]
- 8.Jones, S. & Yakel, J. (1997) J. Physiol. (London) 504, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narahashi, T., Aistrup, G., Marszalec, W. & Nagata, K. (1999) Neurochem. Int. 35, 131–141. [DOI] [PubMed] [Google Scholar]
- 10.Francis, M. M., Cheng, E. Y., Weiland, G. A. & Oswald, R. E. (2001) Am. Chem. Soc. Pharmacol. Exp. Ther. 60, 71–79. [DOI] [PubMed] [Google Scholar]
- 11.Arias, H. R. (1999) Neurosci. Biohehav. Rev. 23, 817–843. [DOI] [PubMed] [Google Scholar]
- 12.Novere, N. & Changeux, J. (1999) Nucleic Acid Res. 27, 340–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd, G. & Williams, M. (2000) J. Pharmacol. Exp. Ther. 292, 461–467. [PubMed] [Google Scholar]
- 14.Changeux, J., Bertrand, D., Corringer, P., Dehaene, S., Edelstein, S., Lena, C., Novere, N., Marbubio, L., Picciotto, M. & Zoli, M. (1998) Brain Res. Rev. 26, 198–216. [DOI] [PubMed] [Google Scholar]
- 15.Corringer, P., Le Novere, N. & Changeux, J. (2000) Annu. Rev. Pharmacol. 40, 431–458. [DOI] [PubMed] [Google Scholar]
- 16.Corringer, P.-J., Bertrand, S., Bohler, S., Edelstein, S. J., Changeux, J.-P. & Bertrand, D. (1998) J. Neurosci. 18, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blount, P. & Merlie, J. P. (1989) Neuron 3, 349–357. [DOI] [PubMed] [Google Scholar]
- 18.Unwin, N. (2000) Philos. Trans. R. Soc. London B 355, 1813–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazawa, A., Fujiyoshi, Y. & Unwin, N. (2003) Nature 423, 949–955. [DOI] [PubMed] [Google Scholar]
- 20.Brejc, K., van Dijk, W. J. & Klaassen, R. (2001) Nature 411, 269–276. [DOI] [PubMed] [Google Scholar]
- 21.Sine, S. M. (2002) J. Neurobiol. 53, 431–446. [DOI] [PubMed] [Google Scholar]
- 22.Smit, A., Syed, N., Schaap D., Van Minnen, J., Klumperman, J., Kits, K. S., Lodder, H., Van der Schors, R., Van Elk, R., Sorgedrager, B., et al. (2001) Nature 411, 261–268. [DOI] [PubMed] [Google Scholar]
- 23.Celie, P., Van Rossum-Fikkert, S. E., Van Dijk, W., Brejc, K., Smit, A. B. & Sixma, T. (2004) Neuron 41, 907–914. [DOI] [PubMed] [Google Scholar]
- 24.Doucette-Stamm, L. M., Donnelly-Roberts, D., Wang, M. T., Lee, J., Tian, J. & Giordano, T. (1993) Drug Dev. Res. 30, 252–256. [Google Scholar]
- 25.Dajas-Bailador, F. & Wonnacott, S. (2004) Trends Pharmacol. Sci. 25, 317–324. [DOI] [PubMed] [Google Scholar]
- 26.Hogg, R., Raggenbass, M. & Bertrand, D. (2003) Rev. Physiol. Biochem. Pharmacol. 147, 1–46. [DOI] [PubMed] [Google Scholar]
- 27.Bernèche, S. & Roux, B. (2001) Nature 414, 73–77. [DOI] [PubMed] [Google Scholar]
- 28.Shrivastava, I. H. & Sansom, M. S. P. (2000) Biophys. J. 78, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law, R. J. & Sansom, M. S. P. (2004) Eur. Biophys. J 33, 477–489. [DOI] [PubMed] [Google Scholar]
- 30.de Groot, B. & Grubmuller, H. (2001) Science 294, 2353–2356. [DOI] [PubMed] [Google Scholar]
- 31.Jensen, M., Park, S., Tajkorshid, E. & Schulten, K. (2002) Proc. Natl. Acad. Sci. USA 99, 6731–6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henchman, R., Wang, H., Sine, S. M., Taylor, P. & McCammon, J. (2003) Biophys. J. 85, 1–12.12829458 [Google Scholar]
- 33.Henchman, R. H., Wang, H., Sine, S. M., Taylor, P. & McCammon, J. A. (2005) Biophys. J. 88, 2564–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenbusch, J. (2001) J. Struct. Biol. 136, 144–157. [DOI] [PubMed] [Google Scholar]
- 35.White, S. (2004) Protein Sci. 13, 1948–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sali, A. & Blundell, T. L. (1993) J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 38.Rodriguez, R., Chinea, G., Lopez, N., Pons, T. & Vriend, G. (1998) Comput. Appl. Biosci. 14, 523–528. [DOI] [PubMed] [Google Scholar]
- 39.Adcock, C., Smith, G. R. & Sansom, M. S. P. (1998) Biophys. J. 75, 1211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis, M. E., Madura, J. D., Luty, B. A. & McCammon, J. A. (1991) Comput. Phys. Comm. 62, 187–197. [Google Scholar]
- 41.Nielsen, J. & Mccammon, J. (2003) Protein Sci. 12, 1894–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphrey, W., Dalke, A. & Schulten, K. (1996) J. Mol. Graphics 14, 33–38. [DOI] [PubMed] [Google Scholar]
- 43.Grubmuller, H., Heymann, B. & Tavan, P. (1996) Science 271, 997–999. [DOI] [PubMed] [Google Scholar]
- 44.Kalé, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K. & Schulten, K. (1999) J. Comp. Phys. 151, 283–312. [Google Scholar]
- 45.Merritt, E. A. & Bacon, J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 46.MacKerrell, A. D., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., et al. (1998) J. Phys. Chem. B 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- 47.Ryckaert, J. P., Ciccotti, G. & Berendsen, H. J. C. (1977) J. Comput. Phys. 23, 327–341. [Google Scholar]
- 48.Mongan, J. & McCammon, J. A. (2004) J. Comp. Chem. 25, 2038–2048. [DOI] [PubMed] [Google Scholar]
- 49.Grutter, T. & Changeux, J. (2001) Trends Biochem. Sci. 26, 459–463. [DOI] [PubMed] [Google Scholar]
- 50.Gao, F., Bren, N., Burghardt, T., Hansen, S., Henchman, R., Taylor, P., Mccammon, J. & Sine, S. M. (2004) Neuron (2005) J. Biol. Chem. 280, 8443–8451. [DOI] [PubMed] [Google Scholar]
- 51.Unwin, N., Miyazawa, A., Li, J. & Fujiyoshi, Y. (2002) J. Mol. Biol. 319, 1165–1176. [DOI] [PubMed] [Google Scholar]
- 52.Lester, H., Dibas, M., Dahan, D., Leite, J. & Dougherty, D. (2004) Trends Neurosci. 27, 329–336. [DOI] [PubMed] [Google Scholar]
- 53.Unwin, N. (2003) FEBS Lett. 555, 91–95. [DOI] [PubMed] [Google Scholar]
- 54.Cymes, G., Grosman, C. & Auerbach, A. (2002) Biochemistry 41, 5548–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Law, R. J., Tieleman, D. P. & Sansom, M. S. P. (2003) Biophys. J. 84, 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unwin, N. (1998) J. Struct. Biol. 121, 181–190. [DOI] [PubMed] [Google Scholar]
- 57.Tikhonov, D. B. & Zhorov, B. S. (1998) Biophys. J. 74, 242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amadei, A., Linssen, A. B. M. & Berendsen, H. J. C. (1993) Proteins Struct. Funct. Genet. 17, 412–425. [DOI] [PubMed] [Google Scholar]
- 59.Garcia, A. (1992) Phys. Rev. Lett. 68, 2696–2699. [DOI] [PubMed] [Google Scholar]
- 60.Smit, A., Brej, K., Syed, N. & Sixma, T. (2003) Ann. N.Y. Acad. Sci. 998, 81–92. [DOI] [PubMed] [Google Scholar]
- 61.Bouzat, C., Gumilar, F., Spitzmaul, G., Wang, H., Rayes, D., Hansen, S., Taylor, P. & Sine, S. M. (2004) Nature 430, 896–900. [DOI] [PubMed] [Google Scholar]
- 62.Absalom, N., Lewis, T. & Schofield, P. (2004) Exp. Physiol. 89, 145–153. [DOI] [PubMed] [Google Scholar]
- 63.Bera, A., Chatav, M. & Akabas, M. (2002) J. Biol. Chem. 277, 43002–43010. [DOI] [PubMed] [Google Scholar]
- 64.Grosman, C., Zhou, M. & Auerbach, A. (2000) Nature 403, 773–776. [DOI] [PubMed] [Google Scholar]
- 65.Downie, D. L., Vicente-Agullo, F., Campos-Caro, A., Bushell, T. J., Lieb, W. R. & Franks, N. P. (2002) J. Biol. Chem. 277, 10367–10373. [DOI] [PubMed] [Google Scholar]
- 66.Yakamura, T., Borghese, C. & Harris, R. A. (2000) J. Biol. Chem. 275, 40879–40886. [DOI] [PubMed] [Google Scholar]
- 67.Kash, T., Jenkins, A., Kelley, J., Trudell, J. & Harrison, N. (2003) Nature 421, 272–275. [DOI] [PubMed] [Google Scholar]
- 68.Chiara, D., Dangott, L., Eckenhoff, R. G. & Cohen, J. (2003) Biochemistry 42, 13457–13467. [DOI] [PubMed] [Google Scholar]
- 69.Bali, M. & Akabas, M. (2004) Mol. Pharmacol. 65, 68–76. [DOI] [PubMed] [Google Scholar]
- 70.Mascia, M. P., Trudell, J. & Harris, R. A. (2001) Proc. Natl. Acad. Sci. USA 97, 9305–9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jozwiak, K., Ravichandran, S., Collins, J. & Wainer, I. (2004) J. Med. Chem. 47, 4008–4021. [DOI] [PubMed] [Google Scholar]
- 72.Campagna, J., Millar, K. & Forman, S. (2003) N. Engl. J. Med. 348, 2110–2124. [DOI] [PubMed] [Google Scholar]
- 73.Eckenhoff, R. G. & Johansson, J. S. (1997) Am. Chem. Soc. Pharmacol. Exp. Ther. 49, 343–364. [Google Scholar]
- 74.Corry, B. (2004) Biochim. Biophys. Acta 1663, 2–5. [DOI] [PubMed] [Google Scholar]
- 75.Wilson, G. G. & Karlin, A. (1998) Neuron 20, 1269–1281. [DOI] [PubMed] [Google Scholar]
- 76.Montal, M. O., Iwamoto, T., Tomich, J. M. & Montal, M. (1993) FEBS Lett. 320, 261–266. [DOI] [PubMed] [Google Scholar]
- 77.Hucho, F. & Hilgenfeld, R. (1989) FEBS Lett. 257, 17–23. [DOI] [PubMed] [Google Scholar]
- 78.Unwin, N. (1989) Neuron 3, 665–676. [DOI] [PubMed] [Google Scholar]
- 79.Imoto, K., Busch, C., Sakmann, B., Mishina, M., Konno, T., Nakai, J., Buyo, H., Mori, Y., Kukuda, K. & Numa, S. (1988) Nature 335, 645–648. [DOI] [PubMed] [Google Scholar]
- 80.Auerbach, A. & Akk, G. (1998) J. Gen. Physiol. 112, 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim, S., Chamberlain, A. & Bowie, J. (2004) Biophys. J. 87, 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilson, G. G. & Karlin, A. (2001) Proc. Natl. Acad. Sci. USA 98, 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corringer, P., Galzi, J., Eisele, J., Bertrand, S., Changeux, J. & Bertrand, D. (1995) J. Biol. Chem. 270, 11749–11752. [DOI] [PubMed] [Google Scholar]
- 84.Grosman, C. & Auerbach, A. (2000) J. Gen. Physiol. 115, 637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.