Abstract
Background
Targeted surveillance allows public health authorities to implement testing and isolation strategies when diagnostic resources are limited, and can be implemented via the consideration of social network topologies. However, it remains unclear how to implement such surveillance and control when network data are unavailable.
Methods
We evaluated the ability of sociodemographic proxies of degree centrality to guide prioritized testing of infected individuals compared to known degree centrality. Proxies were estimated via readily available sociodemographic variables (age, gender, marital status, educational attainment, household size). We simulated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemics via a susceptible-exposed-infected-recovered individual-based model on 2 contact networks from rural Madagascar to test applicability of these findings to low-resource contexts.
Results
Targeted testing using sociodemographic proxies performed similarly to targeted testing using known degree centralities. At low testing capacity, using proxies reduced infection burden by 22%–33% while using 20% fewer tests, compared to random testing. By comparison, using known degree centrality reduced the infection burden by 31%–44% while using 26%–29% fewer tests.
Conclusions
We demonstrate that incorporating social network information into epidemic control strategies is an effective countermeasure to low testing capacity and can be implemented via sociodemographic proxies when social network data are unavailable.
Keywords: COVID-19, Madagascar, epidemic control, social network
In empirical social networks from rural Madagascar, epidemic simulations show that targeted testing guided by sociodemographic characteristics controls epidemics as effectively as targeted testing guided by known social network characteristics.
A key process of epidemic control of infectious disease is surveillance, whereby health systems test and isolate infectious individuals [1]. However, many health systems lack the resources to test all symptomatic individuals and must allocate resources accordingly. This is especially the case for emerging infectious diseases, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), where testing resources are unequally distributed across countries [2]. Limited testing capacity can be mediated by targeting surveillance of individuals or households based on their network characteristics, such as connectivity. The structure and characteristics of edges and nodes in social networks can determine population transmission dynamics [3, 4], especially for directly transmitted diseases and respiratory illnesses that require close contact for transmission [5–9]. In a social network, each individual's contribution to disease transmission is a function of their connections and location in the network, and therefore differs among individuals. In general, 20% of individuals are responsible for 80% of secondary infections [10]. By contributing disproportionately to disease transmission, these individuals represent a logical focus for targeted testing.
However, social network data are rarely available for a population, limiting authorities’ ability to use the information to guide targeted surveillance. Social network characteristics of individuals may, however, be predicted via more commonly available variables. For example, a range of sociodemographic variables have been used to predict age-specific social contact rates across 152 countries [11] and in marketing analytics to predict “central clients” that influence the purchasing behavior of others [12]. Thus, when it can be shown that sociodemographic variables predict network centrality, it may be possible to use those variables as proxies for the risk of onward transmission in targeted surveillance approaches, greatly increasing the feasibility of including network data in epidemic control.
Here, we explore the possibility of using sociodemographic proxies of network connectivity to implement targeted testing strategies in a resource-limited context. We focus on SARS-CoV-2 as a case study, given its current global relevance, the role of social contacts in driving transmission of the disease [13], and the existing past work modeling prioritized testing strategies for the disease [14–16]. We simulate SARS-CoV-2 outbreaks on 2 close-contact social networks derived from social and spatial movement data on individuals living in rural communities in Sambava district, Sava, Madagascar [17]. We then compare the effectiveness of testing strategies that test randomly, that target testing based on known social connectivity, and that target testing guided by sociodemographic proxies of social connectivity involving age, household size, marital status, and educational attainment. Effectiveness is evaluated via the time needed to control the epidemic, the total infection burden, and the number of tests needed. By comparing effectiveness, we can thus investigate whether these commonly available sociodemographic data effectively capture heterogeneity in transmission when designing testing schema.
METHODS
Social Contact Networks
We modeled SARS-CoV-2 dynamics using a susceptible-exposed-infected-recovered (SEIR) model simulated across undirected, weighted networks. Empirical contact networks contain unique network topologies that may be lacking from simulated networks, with important consequences for disease transmission [18]. To ensure our results were most relevant to settings with limited testing capacities, we simulated epidemics on 2 contact networks obtained from rural communities in Madagascar, where testing capacities for SARS-CoV-2 infection have been and remain limited. The social contact networks were constructed using survey and GPS tracker data of consenting adults (over 18 years of age) living in 2 villages in the Sava region of northeastern Madagascar, Mandena (estimated population 2700) and Sarahandrano (estimated population 900). While these villages have similar livelihood practices, Sarahandrano is closer to a larger city (Andapa). Full details on sampling and social network construction are provided in Kauffman et al [17] and the Supplementary Materials. The final networks included 120 and 318 individuals for the Mandena and Sarahandrano networks, respectively. Edge weights between individuals represented the time spent in contact during a week, with a weight of 1 corresponding to 24 hours.
Estimating Sociodemographic Proxies of Degree Centrality
We estimated proxies of degree centrality using sociodemographic variables to test the efficiency of prioritized testing when social network data are unavailable. While there are many indices of node connectivity (discussed further in the Supplementary Materials), we focus here on degree centrality, as it is the most intuitive index to explain to public health practitioners and is often highly correlated with other measures of centrality [19, 20]. We fit a generalized linear mixed-model to predict each individual's degree percentile in their respective network using the following sociodemographic variables: age, gender, household size, marital status (single vs cohabiting/married), and schooling level (none, primary, secondary, higher). A set of models was fit exploring all potential main effects of sociodemographic variables and interactions with gender, and the final model was an average of all models within 4 Akaike information criterion (AIC) units of the top model [21]. The full details of model fitting are described in the Supplementary Materials. Using this model, we predicted an estimated degree percentile for each individual in the 2 networks. This estimation then served as the proxy for degree centrality in the control scenarios where true network structure is unknown.
Epidemic Model Simulations
At each time step, equivalent to 1 day, an individual could become susceptible, exposed, presymptomatic, infected (symptomatic and asymptomatic), or recovered (Supplementary Figure 2.1). For each contact event (eg, an edge between a susceptible and infected node), a susceptible individual's probability of becoming exposed was a function of the transmission probability of the infected contact and the edge weight. Each simulation was initiated by randomly selecting 2 individuals to be exposed. These exposed individuals thus started the simulation on the first day of their latent period. The number of susceptible, exposed, presymptomatic, infected (asymptomatic and symptomatic), isolated, and recovered individuals were recorded at each time step. Transition rates and further model specification are described in the Supplementary Materials.
Evaluating Control Strategies
We considered 3 different testing strategies: random testing, targeted testing using known degree centrality, and targeted testing using sociodemographic proxies. Both targeted testing strategies prioritized testing of well-connected individuals, as measured by degree centrality or sociodemographic proxies of degree centrality. We focused on passive surveillance, which tests only infected, symptomatic individuals because this approach is favored in low-resource settings where diagnostic supplies are limited, particularly at the beginning of an epidemic, and is the approach used in Madagascar [22]. Infected individuals that were positively identified via testing were isolated by moving them immediately to the isolated class. We accounted for imperfect isolation by allowing for household transmission, at a reduced transmission rate, for isolated individuals. Isolated individuals remained isolated until 7 days after symptom onset, after which they moved to the recovered class.
In addition to 2 testing strategies, we considered low and high testing capacities, corresponding to monthly testing capacities of 25% and 100% of the total population. Low testing capacities corresponded to 1 test per day on the Mandena network and 3 tests per day on the Sarahandrano network. We accounted for imperfect surveillance and ascertainment by assigning a 0.75 probability of an individual being successfully identified for testing. Testing began on day 4 of all simulations, with a range of start dates explored in the sensitivity analyses. All strategies, including a control of no testing, were simulated 1000 times.
We evaluated each strategy and testing capacity combination based on how efficiently it controlled the epidemic, using 3 metrics to evaluate the outcomes: the duration of the epidemic, the cumulative number of infected individuals per capita, and the number of tests used. We assessed each strategy and testing capacity based on its ability to reduce the infection burden and the length of the epidemic while minimizing the tests needed.
Sensitivity Analysis
We assessed the robustness of our results by varying 3 categories of parameters in our simulations: intervention parameters (start date and imperfect surveillance rate), biological parameters (transmission rate), and network parameters (network size and assortativity). Further details on these methods and results are reported in the Supplementary Materials.
Data Availability Statement
All code and data needed to reproduce the simulations and analysis are located in a figshare repository (https://doi.org/10.6084/m9.figshare.19942139.v1). Individual-level sociodemographic variables are available upon request.
RESULTS
Estimating Degree Centrality Proxies
We focused on 5 sociodemographic variables as predictors of an individual's degree centrality: age, gender, household size, marital status, and education level. A model including sociodemographic variables did a poor job of predicting degree percentile across the 2 networks (R2 = 0.03). However, the model was able to loosely rank individuals by degree centrality (Mandena, Spearman ρ = 0.15, P = .09; Sarahandrano, Spearman ρ = 0.18, P = .002). The model distinguished high-degree individuals from low-degree individuals: the top 10 individuals in each network had a predicted degree that was on average twice as high as the bottom 10 individuals (Supplementary Figure 3.3). Marital status was the only variable included in all models within 4 AIC units of the top model, but all sociodemographic variables were included in the averaged model. Specifically, cohabiting individuals had lower degree centrality than single individuals. In summary, sociodemographic characteristics did not accurately predict an individual's exact degree centrality, but, across the population, successfully sorted individuals into those with higher and lower connectivity. Further details on the model are reported in the Supplementary Materials.
Control Efficiency
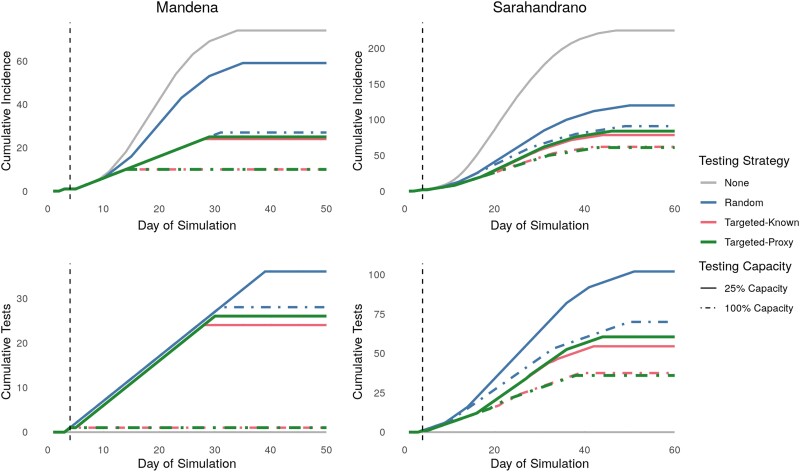
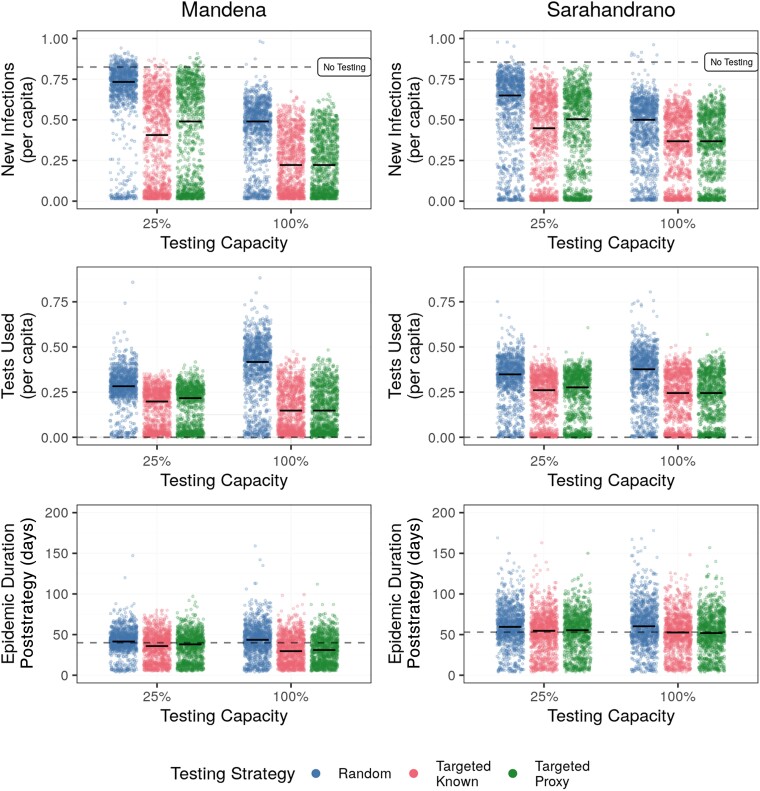
While a poor proxy for the absolute degree centrality itself, sociodemographic proxies resulted in similar epidemic dynamics as degree centrality when used to guide prioritized testing. The targeted strategies resulted in the lowest daily incidence rates and number of tests required, particularly at the lowest testing capacity (Figure 1 and Figure 2). At low testing capacities, targeting known highly connected individuals reduced infections by 31%–44% compared to random testing (Table 1). Targeted testing using sociodemographic proxies reduced infection burdens compared to random testing, but resulted in infection burdens 13%–19% higher compared to targeted testing when the social network was known (Table 1 and Figure 2). The infection burden decreased with increasing testing capacity for all control strategies, with no difference between using known or proxy degree centralities at a testing capacity of 100% (Table 1 and Figure 2). Notably, targeted testing was more effective on the Mandena network, where it reduced per capita infections by over 32%, compared to 21% on the Sarahandrano network.
Figure 1.
Targeted testing using either known social network data or an estimated proxy reduces daily incidence while requiring fewer tests than random testing. Cumulative daily incidence (top row) and cumulative tests required (bottom row) for the 3 testing strategies across 2 testing capacities on the Mandena and Sarahandrano networks. Testing capacities refer to ability to test a percentage of the total population monthly. The vertical dashed line represents the start of the control strategies at day 4. Lines represent median values from 1000 simulations.
Figure 2.
Targeted testing using proxies performs better than random at low testing capacity and similar to using known degree centralities at high testing capacities. Testing capacities correspond to monthly testing capacities equal to testing 25% and 100% of the total population. The dashed black line represents median values from simulations with no testing. Raw data are represented by points and median values per strategy are represented by bold horizontal lines. The figure displays results from 1000 simulations for each combination of testing capacity and control strategy.
Table 1.
Median and 95% Confidence Interval of Efficiency Metrics for 3 Control Strategies on 2 Empirical Social Contact Networks From Rural Madagascar
| Efficiency Metric | Testing Capacity, % | Mandena Network | Sarahandrano Network | ||||
|---|---|---|---|---|---|---|---|
| Random Testing | Targeted Testing, Known | Targeted Testing, Proxy |
Random Testing | Targeted Testing, Known |
Targeted Testing, Proxy | ||
| Epidemic duration | 0 | 44 (13–67) |
44 (13–67) |
44 (13–67) |
57 (19–85) |
57 (19–85) |
57 (19–85) |
| 25 | 46 (11–73) |
40 (10.75–71) |
42 (11–71) |
64 (11–116) |
59 (11–106) |
59.5 (11–103) |
|
| 100 | 48 (11–88) |
34 (11–71) |
35 (11–69) |
58 (39–91) |
57 (16–89) |
56 (11–102) |
|
| Infections per capita | 0 | 0.82 (.03–.87) |
0.82 (.03–.87) |
0.82 (.03–.87) |
0.86 (.02–.9) |
0.86 (.02–.9) |
0.86 (.02–.9) |
| 25 | 0.73 (.02–.87) |
0.41 (.02–.78) |
0.49 (.02–.82) |
0.65 (.01–.83) |
0.45 (.01–.75) |
0.51 (.01–.78) |
|
| 100 | 0.49 (.02–.69) |
0.22 (.02–.57) |
0.22 (.02–.55) |
0.5 (.01–.71) |
0.37 (.01–.62) |
0.37 (.01–.64) |
|
| Tests per capita | 25 | 0.28 (0–.45) |
0.20 (.01–.32) |
0.22 (.01–.34) |
0.35 (.01–.51) |
0.26 (0–.4) |
0.28 (0–.41) |
| 100 | 0.42 (.02–.64) |
0.15 (.01–.39) |
0.15 (.01–.38) |
0.38 (.01–.56) |
0.25 (0–.42) |
0.25 (0–.43) |
|
Represents median and confidence intervals from 1000 simulations. Testing capacity corresponds to monthly testing capacity, with 100% equal to the ability to test the full population monthly. Note that efficiency at 0% testing capacity is the same for all strategies because it represents the control strategy of no testing.
Testing was more efficient when targeting highly connected individuals using known or proxy degree centralities, requiring less than three-quarters of the number of tests needed when testing randomly at 25% testing capacity (Table 1). For example, on the Mandena network, 34 tests were required when testing randomly, 24 tests when using targeted-known testing, and 26 tests when using targeted-proxy testing. Targeted testing only shortened the epidemic length on the Mandena network at high levels of testing (Table 1 and Figure 2), where it was able to stop transmission chains earlier in the epidemic than random testing (Figure 1 and Figure 3). On the Sarahandrano network, all control strategies flattened the epidemic curve by reducing the number of infections, with the targeted testing strategies only slightly reducing epidemic length at a testing capacity of 25% (Figure 1 and Figure 2). While flattening the epidemic curve lengthens epidemics, it also reduces the daily incidence of cases to prevent overwhelming the health system [23]. Therefore, a targeted testing strategy that flattens the curve, rather than shortening the epidemic, can also be an effective form of epidemic control.
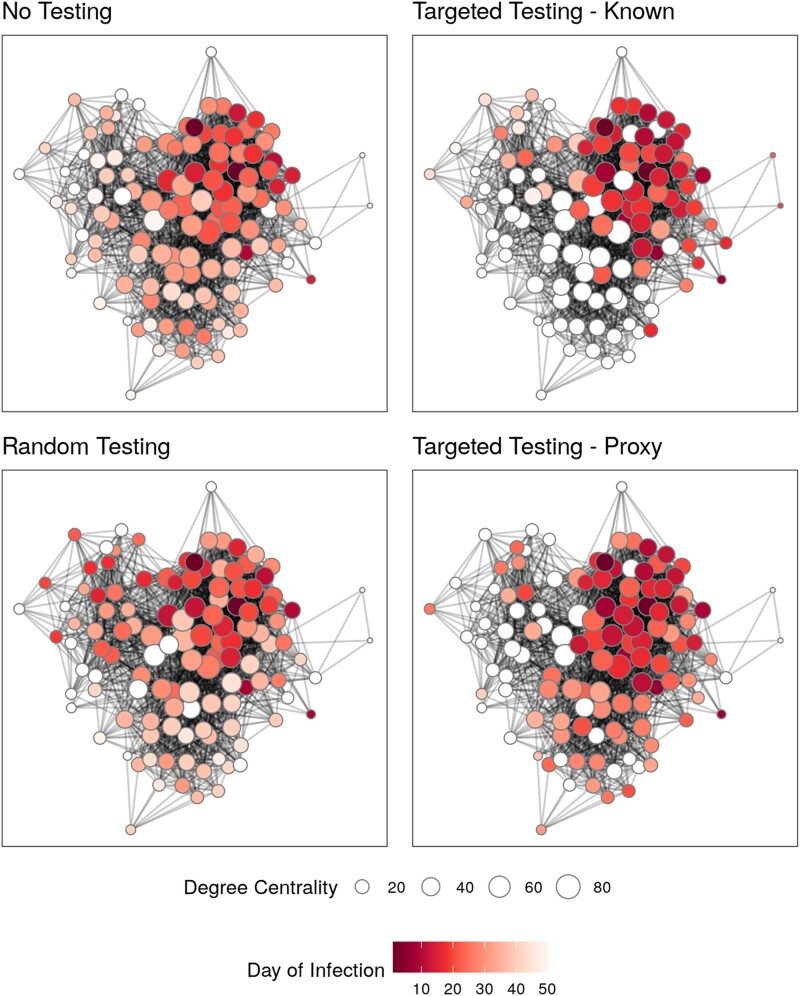
Figure 3.
Targeted testing most efficiently reduces disease spread across social networks, whether using known degree centralities or proxies. Final epidemic spread of 1 simulation on the Mandena network for 3 different control strategies and 1 strategy of no control. Nodes are represented by points, colored based on day of infection, and sized according to their degree centrality. Nodes that were never infected are white. All control strategies used a monthly testing capacity of 25%.
Visualizing a simulated SEIR epidemic on the Mandena network illustrates how each strategy works (Figure 3). The strategy of no control allowed for the highest infection burden, including individuals with low centrality who were infected later in the epidemic than in other strategies. Testing randomly resulted in rapid, early spread, with the highest daily incidence of 7 cases on day 32 of the epidemic. In contrast, targeted testing slowed transmission by halting transmission chains that would result in a high number of secondary infections (Figure 3); daily incidence never rose above 5 cases with using known degree centralities or 6 cases with proxies. Notably, while the 2 targeted strategies had similar overall incidence, different proportions of the population were infected.
DISCUSSION
In the face of global diagnostic and vaccine inequity, many countries are tasked with developing novel public health interventions that optimize limited diagnostic capacities to control epidemics. Considering social network topologies is an effective way to guide testing strategies when testing capacity is limited, but social networks are rarely known. We compared the performance of control strategies that prioritized testing using sociodemographic proxies of individuals’ degree centralities to those using known social network data, simulating SARS-CoV-2 epidemics in rural Madagascar as a case study. We found that strategies that target well-connected, infected individuals are the most effective, reducing overall infection burden while requiring fewer tests. In simulations on empirical social contact networks from rural Madagascar, targeted testing reduced the infection burden and shortened the epidemic even at a testing capacity of only 1 test per day, equivalent to a monthly testing capacity of 25% of the population. These strategies were robust even when targeting was imperfect because the true social network was not known and prioritized testing was based solely on sociodemographic variables. Importantly, the use of sociodemographic proxies highlights one way to implement an otherwise theoretical network-based approach when social network data are not available. Our findings therefore demonstrate the benefits of considering social networks in data-driven epidemic control strategies even when social network data are incomplete or unavailable.
We find that strategies that prioritize testing highly connected individuals using either known or proxy measures of connectivity offer the most benefit in contexts with low testing capacities. In our simulations, this is achieved by controlling the epidemic before it reaches the point at which limited testing capacity cannot contain it. However, even when the start of testing is delayed by 24 days, the targeted strategy can avoid on average 9 infections on the Mandena network, or 0.075 infections per capita (Supplementary Figure 4.1). Early, aggressive testing has been used to successfully control SARS-CoV-2 in several countries [24, 25], and a similar mechanism explains why strategies that target highly connected individuals are so efficient in our simulations. In addition to delayed testing, high transmission rates can result in epidemics that targeted testing is unable to control at limited testing capacities. Indeed, other mathematical models of SARS-CoV-2 have shown that the effectiveness of testing to control epidemics becomes limited at increasing transmission rates [15, 16]. This was seen in our sensitivity analyses (Supplementary Figure 4.3) and on the Sarahandrano network, where higher average edge weights resulted in higher community transmission than on the Mandena network. Future work that incorporates a range of diseases and their associated epidemiological parameters could identify the conditions that impact the effectiveness of targeted testing with sociodemographic proxies.
Many theoretical studies have shown the effectiveness of incorporating network data and topology into epidemic control strategies [3, 26], but the feasibility of doing so has been questioned because the true social network is almost never known. One alternative is occupationally targeted strategies that target high contact rates or high-risk environments (eg, health care or food service workers) [7]. However, in rural communities such as Mandena and Sarahandrano, there is little variation in occupation: nearly all community members are agriculturalists. To overcome this obstacle, we considered sociodemographic predictors of network centrality to guide targeted testing rather than the true values of network centrality or occupationally based targeting. While demographic predictors did not accurately rank individuals by degree centrality, they were able to distinguish between individuals with high and low contact rates, and they performed as well as “true” degree centrality when used to prioritize testing schemes in our simulated epidemics. Despite these predictors being poor proxies of degree centrality itself, they were accurate enough to guide targeted testing, suggesting that even imperfect targeted testing strategies can be effective. Health authorities can implement targeted control strategies by taking into account easily acquired individual characteristics, such as age, gender, household size, and marital status, many of which are available in health care and governmental records, or can be quickly generated through surveys. The exact sociodemographic variables to include will vary depending on local demographics and cultural practices and will require input from local experts. This is particularly important to ensure prioritized testing strategies do not target already vulnerable groups nor lead to stigmatization.
Our study advances previous work on disease control strategies configured by social networks [27–31]. In particular, our study is one of the few that explicitly considers limited testing capacities on par with those in low-income countries and pairs simulations with social network data from such a context. Madagascar tested 26 425 individuals (less than 0.01% of the total population) for SARS-CoV-2 between March and September 2020 [32], which is a fraction of the testing capacity of mass-testing campaigns that have been implemented elsewhere [33]. This is further complicated by the relative remoteness of some communities, with more than 50% of the population living further than 2 hours from a hospital [34]. In both Mandena and Sarahandrano, for example, no SARS-CoV-2 testing has been available to date. While cost and physical access to testing are significant barriers to disease control in Madagascar [22], our findings suggest that, if and when testing is available to rural communities, targeted testing can mitigate the negative impact of limited testing capacity on epidemic control. For example, antigen-based rapid diagnostic testing could be implemented at a local scale via outreach teams of skilled health workers [35]. However, for prioritized testing schemes such as this to be possible, the global inequality in access to diagnostics must first be overcome [36].
Our study had several limitations. First, our social networks represented realistic, but necessarily simplified, versions of true social networks. They assumed social contacts were static, only included individuals over 18 years old, were relatively small, and did not record all social contacts in the community. Both social networks were constructed based on GPS tracker data, with missing links between GPS-wearers imputed based on overlapping movement patterns, a social survey with limited response categories, and demographic data on the individuals. While still small (n = 120 and 318), the resulting close-contact network is over 10 times as dense as the network based solely on survey data [17]. In addition, these network sizes are not an outlier in the context of other social network studies, where less than 100 nodes are common [19], and are necessarily limited due to the small size of communities in rural Madagascar. Our sensitivity analyses found that the targeted strategy was most effective for a variety of network sizes and assortativity values, evidence that our results are not simply an artifact of our empirical social networks. However, our networks may underestimate the rate of spread of simulated epidemics compared to a more complete network. Second, we focused on only one characteristic of a social network, centrality, and only one measure of centrality, degree centrality. Other indices that consider indirect, higher-degree connections, such as betweenness or closeness centrality, may contain more information relevant for onward transmission and could be even more effective at controlling epidemics. Third, we only considered the testing of symptomatic individuals because that is most relevant in extremely resource-limited contexts. However, active surveillance strategies are common in high-income settings and future research should evaluate whether sociodemographic proxies perform similarly for these strategies. Finally, as with any targeted public health intervention, there is a risk that those targeted by the intervention will be stigmatized. Local experts should assess the risk of stigma and how this could exacerbate existing inequalities before implementing prioritized testing strategies. In certain contexts, this could represent a significant obstacle to the implementation of targeted testing.
By using empirical social contact networks, we included realistic social networks that more accurately represent exposure risk in rural Madagascar than simulated networks or networks based on studies from the Global North, which is the source of the majority of social networks used in infectious disease simulations. A recent meta-analysis found only 4 social contact studies, less than 7% of those included in the meta-analysis, incorporated data from sub-Saharan Africa [37]. Expanding social network data collection outside of the Global North would allow for more realistic and context-specific estimates of disease dynamics on social networks globally.
Incorporating social network data into testing strategies greatly increases their efficiency under limiting testing capacities. Prioritized control strategies were effective even when individuals’ true degree centralities were unknown and testing was prioritized using only common sociodemographic variables. This theoretical study focused on testing to control epidemics at the population level, and assumed that diagnostics were not tied to treatment. In practice, control strategies should also consider factors such as the severity of disease in different age groups to ensure equitable distribution of diagnostics. As social network data becomes more widely available, considering social network information and structure is a promising method for allocating limited resources during public health crises. We demonstrate data-driven control strategies are effective even when social network data are missing, overcoming one of the major barriers to implementing this currently theoretical approach.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Michelle V Evans, Maladies Infectieuses et Vecteurs : Écologie, Génétique, Évolution et Contrôle, Université Montpellier, CNRS, IRD, Montpellier, France.
Tanjona Ramiadantsoa, Maladies Infectieuses et Vecteurs : Écologie, Génétique, Évolution et Contrôle, Université Montpellier, CNRS, IRD, Montpellier, France.
Kayla Kauffman, Department of Evolutionary Anthropology, Duke University, Durham, North Carolina, USA; Duke Global Health Institute, Durham, North Carolina, USA; Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, California, USA.
James Moody, Department of Sociology, Duke University, Durham, North Carolina, USA.
Charles L Nunn, Department of Evolutionary Anthropology, Duke University, Durham, North Carolina, USA; Duke Global Health Institute, Durham, North Carolina, USA.
Jean Yves Rabezara, Department of Science and Technology, University of Antsiranana, Antsiranana, Madagascar.
Prisca Raharimalala, Andapa, Madagascar.
Toky M Randriamoria, Association Vahatra, Antananarivo, Madagascar; Zoologie et Biodiversité Animale, Domaine Sciences et Technologies, Université d’Antananarivo, Antananarivo, Madagascar.
Voahangy Soarimalala, Association Vahatra, Antananarivo, Madagascar; Institut des Sciences et Techniques de l’Environnement, Université de Fianarantsoa, Fianarantsoa, Madagascar.
Georgia Titcomb, Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, California, USA; Marine Science Institute, University of California, Santa Barbara, California, USA; Department of Fish, Wildlife, and Conservation Biology, Colorado State University, Fort Collins, Colorado, USA.
Andres Garchitorena, Maladies Infectieuses et Vecteurs : Écologie, Génétique, Évolution et Contrôle, Université Montpellier, CNRS, IRD, Montpellier, France; Pivot, Ifanadiana, Madagascar.
Benjamin Roche, Maladies Infectieuses et Vecteurs : Écologie, Génétique, Évolution et Contrôle, Université Montpellier, CNRS, IRD, Montpellier, France.
Notes
Acknowledgments. We thank the communities of Mandena and Sarahandrano for participating in this research. We also thank Jessica Metcalf for her feedback.
Financial support. This work was supported by the Agence Nationale de la Recherce (to M. V. E. and A. G.); National Institues of Health-National Science Foundation-National Institute of Food and Agriculture Ecology and Evolution of Infectious Disease Award (grant number R01-TW011493-01 to K. K., J. M., C. L. N., J. Y. R., P. R., T. M. R., V. S., and G. T.); and a Duke University Provost's Collaboratory Grant.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Peeling RW, Heymann DL, Teo Y-Y, Garcia PJ. Diagnostics for COVID-19: moving from pandemic response to control. Lancet 2022; 399:757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eyawo O, Viens AM. Rethinking the central role of equity in the global governance of pandemic response. J Bioethical Inq 2020; 17:549–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pellis L, Ball F, Bansal S, et al. Eight challenges for network epidemic models. Epidemics 2015; 10:58–62. [DOI] [PubMed] [Google Scholar]
- 4. Rothenberg RB, Potterat JJ, Woodhouse DE, Muth SQ, Darrow WW, Klovdahl AS. Social network dynamics and HIV transmission. AIDS 1998; 12:1529. [DOI] [PubMed] [Google Scholar]
- 5. Cauchemez S, Bhattarai A, Marchbanks TL, et al. Role of social networks in shaping disease transmission during a community outbreak of 2009 H1N1 pandemic influenza. Proc Natl Acad Sci U S A 2011; 108:2825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meyers LA, Pourbohloul B, Newman MEJ, Skowronski DM, Brunham RC. Network theory and SARS: predicting outbreak diversity. J Theor Biol 2005; 232:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bansal S, Pourbohloul B, Meyers LA. A comparative analysis of influenza vaccination programs. PLoS Med 2006; 3:e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klovdahl AS. Social networks and the spread of infectious diseases: the AIDS example. Soc Sci Med 1985; 21:1203–16. [DOI] [PubMed] [Google Scholar]
- 9. Anderson RM, Gupta S, Ng W. The significance of sexual partner contact networks for the transmission dynamics of HIV. J Acquir Immune Defic Syndr (1988) 1990; 3:417. [PubMed] [Google Scholar]
- 10. Woolhouse MEJ, Dye C, Etard J-F, et al. Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proc Natl Acad Sci U S A 1997; 94:338–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prem K, Cook AR, Jit M. Projecting social contact matrices in 152 countries using contact surveys and demographic data. PLoS Comput Biol 2017; 13:e1005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernandes A, Gonçalves PCT, Campos P, Delgado C. Centrality and community detection: a co-marketing multilayer network. J Bus Ind Mark 2019; 34:1749–62. [Google Scholar]
- 13. Susswein Z, Bansal S. Characterizing superspreading of SARS-CoV-2: from mechanism to measurement. medRxiv, 10.1101/2020.12.08.20246082, 11 December 2020, preprint: not peer reviewed. [DOI] [Google Scholar]
- 14. Firth JA, Hellewell J, Klepac P, Kissler S, Kucharski AJ, Spurgin LG. Using a real-world network to model localized COVID-19 control strategies. Nat Med 2020; 26:1616–22. [DOI] [PubMed] [Google Scholar]
- 15. Cui Y, Ni S, Shen S. A network-based model to explore the role of testing in the epidemiological control of the COVID-19 pandemic. BMC Infect Dis 2021; 21:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Voigt A, Martyushenko N, Karlsen E, et al. Containing pandemics through targeted testing of households. BMC Infect Dis 2021; 21:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kauffman K, Werner CS, Titcomb G, et al. Comparing transmission potential networks based on social network surveys, close contacts and environmental overlap in rural Madagascar. J R Soc Interface 2021; 19:20210690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aleta A, de Arruda GF, Moreno Y. Data-driven contact structures: from homogeneous mixing to multilayer networks. PLOS Comput Biol 2020; 16:e1008035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valente TW, Coronges K, Lakon C, Costenbader E. How correlated are network centrality measures? Connect (Tor) 2008; 28:16–26. [PMC free article] [PubMed] [Google Scholar]
- 20. Shao C, Cui P, Xun P, Peng Y, Jiang X. Rank correlation between centrality metrics in complex networks: an empirical study. Open Phys 2018; 16:1009–23. [Google Scholar]
- 21. Symonds MRE, Moussalli A. A brief guide to model selection, multimodel inference and model averaging in behavioural ecology using Akaike's information criterion. Behav Ecol Sociobiol 2011; 65:13–21. [Google Scholar]
- 22. Rakotonanahary RJL, Andriambolamanana H, Razafinjato B, et al. Integrating health systems and science to respond to COVID-19 in a model district of rural Madagascar. Front Public Health 2021; 9:654299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matrajt L, Leung T. Evaluating the effectiveness of social distancing interventions to delay or flatten the epidemic curve of coronavirus disease. Emerg Infect Dis 2020; 26:1740–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee W, Hwang S-S, Song I, et al. COVID-19 in South Korea: epidemiological and spatiotemporal patterns of the spread and the role of aggressive diagnostic tests in the early phase. Int J Epidemiol 2020; 49:1106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jefferies S, French N, Gilkison C, et al. COVID-19 in New Zealand and the impact of the national response: a descriptive epidemiological study. Lancet Public Health 2020; 5:e612–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christley RM, Pinchbeck GL, Bowers RG, et al. Infection in social networks: using network analysis to identify high-risk individuals. Am J Epidemiol 2005; 162:1024–31. [DOI] [PubMed] [Google Scholar]
- 27. Christakis NA, Fowler JH. Social network sensors for early detection of contagious outbreaks. PLoS One 2010; 5:e12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sneppen K, Nielsen BF, Taylor RJ, Simonsen L. Overdispersion in COVID-19 increases the effectiveness of limiting nonrepetitive contacts for transmission control. Proc Natl Acad Sci 2021; 118:e2016623118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nielsen BF, Simonsen L, Sneppen K. COVID-19 superspreading suggests mitigation by social network modulation. Phys Rev Lett 2021; 126:118301. [DOI] [PubMed] [Google Scholar]
- 30. Brantley M, Schumacher C, Fields EL, et al. The network structure of sex partner meeting places reported by HIV-infected MSM: opportunities for HIV targeted control. Soc Sci Med 2017; 182:20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Metzig C, Surey J, Francis M, Conneely J, Abubakar I, White PJ. Impact of hepatitis C treatment as prevention for people who inject drugs is sensitive to contact network structure. Sci Rep 2017; 7:1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Randremanana RV, Andriamandimby S, Rakotondramanga JM, et al. The COVID-19 epidemic in Madagascar: clinical description and laboratory results of the first wave, March-September 2020. Influenza Other Respir Viruses 2021; 15:457–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pavelka M, Van-Zandvoort K, Abbott S, et al. The impact of population-wide rapid antigen testing on SARS-CoV-2 prevalence in Slovakia. Science 2021; 372:635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ouma PO, Maina J, Thuranira PN, et al. Access to emergency hospital care provided by the public sector in sub-Saharan Africa in 2015: a geocoded inventory and spatial analysis. Lancet Glob Health 2018; 6:e342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jacobs J, Kühne V, Lunguya O, Affolabi D, Hardy L, Vandenberg O. Implementing COVID-19 (SARS-CoV-2) rapid diagnostic tests in sub-Saharan Africa: a review. Front Med 2020; 7:557797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jensen N, Kelly AH, Avendano M. The COVID-19 pandemic underscores the need for an equity-focused global health agenda. Humanit Soc Sci Commun 2021; 8:1–6. [Google Scholar]
- 37. Hoang T, Coletti P, Melegaro A, et al. A systematic review of social contact surveys to inform transmission models of close-contact infections. Epidemiology 2019; 30:723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code and data needed to reproduce the simulations and analysis are located in a figshare repository (https://doi.org/10.6084/m9.figshare.19942139.v1). Individual-level sociodemographic variables are available upon request.