Abstract
To investigate the temporal relationship between human immunodeficiency virus type 1 (HIV-1) replicative capacity and syncytium-inducing (SI) phenotype, biological and genetic characteristics of longitudinally obtained virus clones from two HIV-1-infected individuals who developed SI variants were studied. In one individual, the emergence of rapidly replicating SI and non-syncytium-inducing (NSI) variants was accompanied by a loss of the slowly replicating NSI variants. In the other subject, NSI variants were always slowly replicating, while the coexisting SI variants showed an increase in the rate of replication. Irrespective their replicative capacity, the NSI variants remained present throughout the infection in both individuals. Phylogenetic analysis of the V3 region showed early branching of the SI variants from the NSI tree. Successful SI conversion seemed a unique event since no SI variants were found among later-stage NSI variants. This was also confirmed by the increasing evolutionary distance between the two subpopulations. At any time point during the course of the infection, the variation within the coexisting SI and NSI populations did not exceed 2%, indicating continuous competition within each viral subpopulation.
Human immunodeficiency virus type 1 (HIV-1) is susceptible to genetic recombination (43) and has an error-prone reverse transcriptase enzyme, which combined with the absence of proofreading leads to a misincorporation rate of 10−4 to 10−5 per base, or approximately one misincorporation per genome per replication cycle (38, 40–42). With 1010 new viruses produced each day and a half-life of approximately 6 h (39), an HIV-1 infected-individual harbors a swarm of closely related viruses that comprise the so-called HIV-1 quasispecies. These HIV-1 variants have been shown to differ in biological properties such as replication rate, cell tropism, and syncytium-inducing (SI) capacity (2, 5, 55). Virus isolated in the early asymptomatic phase of infection is predominantly slowly replicating, macrophagetropic, and non-syncytium-inducing (NSI) in vitro (8, 44, 48, 57, 61). During progression to disease rapid replicating, more T-cell-tropic viruses appear, in about 50% of infected individuals associated with the emergence of SI variants (26, 48, 55).
Determinants which govern these biological properties have been mainly mapped to the envelope gene (19, 37, 51), especially to the variable (V) regions (9, 18, 20, 23, 52), although accessory genes like nef, vif, vpr, and vpu have also been shown to influence replication rate in certain cell types (3, 22, 24, 46, 56). Chimeric clones, constructed from an SI and an NSI molecular clone, showed that exchange of the gp120 V1-V2 fragment together with the V3 fragment was sufficient to confer SI capacity (20). Sequence analysis of the V3 fragment of a large panel of HIV-1 isolates with distinct biological phenotypes demonstrated the presence of positively charged amino acids at either one or two fixed positions in the V3 loop of SI variants (18). During the transition from NSI to SI phenotype, the hypervariable V2 region is thought to undergo increases both in length, mainly through insertion of potential N-linked glycosylation sites, and charge (17, 20).
Although replicative capacity and SI phenotype in general are coinciding biological features, their temporal relationship in the pathway of virus phenotype evolution is not known. SI variants could evolve from rapidly replicating NSI variants which are more likely to accumulate the mutations required for NSI-to-SI transition. Alternatively, SI variants could also initially replicate slowly but evolve to rapidly replicating HIV-1. When the V3 sequences of virus populations present around the time of NSI-to-SI conversion were analyzed, few V3 sequences intermediate between the NSI and SI variants were detected, in agreement with the hypothesis that less fit stages may have to be crossed in order to reach the more fit SI stage (30).
In this study, we analyzed the temporal relationship between different biological properties of HIV-1. We studied replication kinetics of biologically cloned SI and NSI variants in two individuals who developed SI variants in the course of infection. From all virus clones, V3 sequences were determined and used for phylogenetic analyses. For comparison, viruses isolated around the time of AIDS diagnosis from two individuals harboring only NSI variants throughout infection were analyzed.
MATERIALS AND METHODS
Study subjects.
Four participants of the Amsterdam Cohort Studies on HIV infection and AIDS in homosexual men were analyzed. Characteristics of the four study subjects are depicted in Table 1. Three subjects (ACH0039, ACH0208, and ACH0424) seroconverted in the course of the cohort studies and progressed to AIDS within 5 years thereafter. One subject (ACH0142) was seropositive at entry into the cohort studies and progressed to AIDS 9 years after seropositive follow-up.
TABLE 1.
Characteristics of the four subjects under studya
| Subject | CCR5 genotype | Time of:
|
AIDS defining events | ||
|---|---|---|---|---|---|
| Seroconversion or entry | First MT-2-positive sample | AIDS (time postseroconversion [mo]) | |||
| ACH0039 | wt | Oct. 1987 (S) | Mar. 1989 | Nov. 1990 (37) | Candida esophagitis |
| ACH0142 | wt | Nov. 1984 (E) | NA | Dec. 1993 (109) | Kaposi’s sarcoma |
| ACH0208 | Heterozygous | Dec. 1985 (S) | Mar. 1987 | June 1990 (54) | Pneumocystis pneumonia, Kaposi’s sarcoma |
| ACH0424 | wt | Aug. 1988 (S) | NA | Oct. 1991 (38) | Candida esophagitis |
wt, wild type; NA, not applicable; S, seroconversion; E, entry.
Limiting-dilution analysis.
To isolate biological virus clones and estimate the frequency of productively infected cells, limiting-dilution cultures were performed as described previously (27, 48). Briefly, participant peripheral blood mononuclear cells (PBMC; 0.5 × 104 to 4 × 104 cells per well; 24 to 96 replicates per concentration) were cocultivated with phytohemagglutinin (PHA)-stimulated healthy donor PBMC (105 per well) in 96-well microtiter plates. Every week for 5 weeks, 65 μl of each culture supernatant was collected for detection of p24 antigen by an in-house p24 antigen capture enzyme-linked immunosorbent assay (ELISA). At the same time, half of the cells were transferred to new 96-well plates, and 105 fresh PHA-stimulated healthy donor PBMC were added to propagate the culture. The proportion of productively infected CD4+ T cells was estimated by the formula for Poisson distribution: F = −1n (F0), in which F0 is the fraction of negative cultures. PBMC from wells tested positive were transferred to 25-ml culture flasks containing 5 × 106 fresh PHA-stimulated PBMC in 5 ml of culture medium to grow virus stocks. All viruses obtained from one individual were grown on target PBMC from one seronegative blood donor. Virus containing cell-free culture supernatant was stored at −70°C until further use, cells were frozen, and approximately 106 cells were used for isolation of DNA. SI capacity of virus clones was determined by cocultivation with MT-2 cells (28).
In vitro characterization of virus replication rate.
Target PBMC from the same seronegative healthy CCR5 homozygous wild-type blood donor (determined as described previously [11]) were used in all replication experiments. All viruses obtained from one individual were analyzed within the same experiment. The titer of the virus stocks was quantified by determination of the 50% tissue culture infectious dose (TCID50) in PHA-stimulated healthy donor PBMC. From each virus clone, 102 and 103 TCID50 were added to 5 × 106 2-day PHA-stimulated PBMC derived from the same blood donor on which the titer was determined. Cells were incubated for 2 h at 37°C in a shaking water bath in 15-ml conical tubes in a 1.5-ml volume. PBMC were then washed twice, resuspended in 5 ml of culture medium containing recombinant interleukin-2 (rIL-2; 20 U/ml; Proleukin; Chiron Benelux B.V.), and cultured for 14 days in a 25-ml culture flask. After 5, 8, and 11 days, approximately 3 × 106 fresh stimulated target cells were added in 3 to 4 ml of culture medium containing rIL-2 (30 U/ml). Samples (75 μl) for determination of p24 production were harvested every day after infection for 14 days and stored at −70°C. All samples obtained for virus clones from one individual were tested for p24 production at the same time, using an in-house p24 ELISA. Cultures were considered positive for virus production when p24 antigen levels exceeded background twofold, which equaled 31 ng of p24/ml. p24 production per milliliter of supernatant was determined and corrected for the differences in volume of culture supernatants between the moments of sampling. Due to the relatively high detection limit of our p24 ELISA, the very early replication kinetics are not measured.
DNA isolation, PCR, and sequencing.
Total DNA from PBMC harboring the biological HIV-1 clones was isolated as described previously (4). Envelope V3 sequences from subject ACH0208 were amplified by PCR as described previously (53). PCR products were purified with a Geneclean kit (Bio 101, Inc., Vista, Calif.) and sequenced directly by the dideoxy-chain termination method with Sequenase (U.S. Biochemical, Cleveland, Ohio), both as instructed by the manufacturers.
Envelope V3 sequences from subject ACH0039 were amplified by PCR using primers Seq1 (5′-TACATAATGTTTGGGCCACACATGCC-3′, nucleotide positions 6417 to 6443, sense) and Seq2 (5′-TCCTTCATATCTCCTCCTCCAGGTC-3′, positions 7629 to 7653, antisense) in the presence of 3 mM MgCl2 in the first reaction and primers Seq5 (5′-GTCAACTCAACTGCTGTTAAATGGC-3′, positions 6988 to 7012, sense) and Seq6 (5′-ATCTAATTTGTCCACTGATGGGAGG-3′, positions 7532 to 7556, antisense) in the presence of 3 mM MgCl2 in the nested reaction. For amplification, the following PCR amplification cycles were used: 5 min at 95°C once; 1.5 min at 95°C, 1 min at 55°C, and 1 min at 72°C, repeated 25 times; followed by a 5-min extension at 72°C and subsequent cooling to 4°C. Nested PCR products were purified with QIAquick PCR purification kit (Qiagen, Hilden, Germany). Dye terminator cycle sequencing with AmpliTaq DNA polymerase (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.) was performed with the following amplification cycles: 30 s at 92°C, 15 s at 50°C, and 4 min at 60°C, repeated 25 times with the sense nested PCR primer Seq5. Sequence analysis was performed on ABI 373S automated sequencer according to the manufacturer’s protocol.
Phylogenetic and sequence data analysis.
Alignment of the sequences was straightforward and done with the PILEUP program, checked manually and keeping codons intact. Phylogenetic analyses were done with the neighbor-joining program (45) as implemented in the PHYLIP package (15). Bootstrap resampling was used to assess the strength of support for each branch of the phylogenetic trees (14). For bootstrapping, the SEQBOOT, DNADIST, and CONSENSE programs from this package were used. PHYLIP’s DRAWTREE program was used to produce the plots. For direct comparison of nucleotide sequences, the number of mismatches as a proportion of sequence length (Hamming distance) was used (21). The distance matrix input for the neighbor-joining analysis was generated by using Kimura’s two-parameter estimation for nucleotides (25). Estimation of the number of silent and nonsilent substitutions was done according to Nei and Gojobori’s method (36) as implemented in MEGA (31). In the statistical analysis of the data, to avoid the problem of dependence between data points, instead of the commonly used pairwise Hamming distance between clones [which results in n(n − 1)/2 data points for n sequences], we calculated the distance between all clones and their consensus sequence, which results in n data points for n sequences, thus evading the problem of correlated observations. The significance of differences between the variation of groups of clones was evaluated by using the t test.
Nucleotide sequence accession numbers.
All newly generated sequences have been deposited in GenBank under the following accession numbers: for ACH0039, AF022257 to AF022302; for ACH0208, AF021477, AF021478, AF021494, AF021495, AF021499, AF0214503, AF021505, AF021508, AF021510, AF021513, AF021514, AF021516, AF021517, AF021518, AF021521, AF021522, AF021523, AF021524, AF021532, AF021533, AF021536, AF021537, AF021540, AF021542, AF021607, AF021608, AF021610, AF021611, AF021612, AF021613, AF021614, AF021616, AF021617, AF021618, AF021620, AF021622, AF021646, AF021647, AF021650, AF021651, and AF021652.
RESULTS
Replicative capacity of longitudinally obtained biological virus clones.
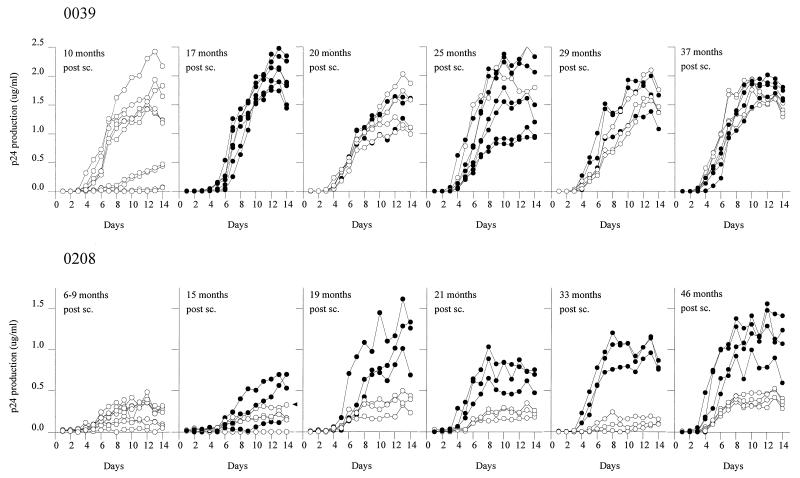
From two participants (ACH0039 and ACH0208) from the Amsterdam cohort, biological HIV-1 clones were isolated at several time points between seroconversion and AIDS diagnosis. Individual clones were compared for SI/NSI phenotype and analyzed for replication kinetics. It is important to note that not all virus clones detected in the limiting-dilution experiment were used in the replication experiments, but only a random sample of 6 to 10 virus clones from each time point. The NSI clones detected in the 17 months sample from subject ACH0039 were lost due to technical difficulties. In each experiment, the viral replication kinetics after infections with 100 TCID50 confirmed the observations after infection with 1,000 TCID50 (data not shown). Therefore, only the results for the inoculation with 1,000 TCID50 are shown in the figures. Four slowly replicating NSI variants were isolated 10 months after seroconversion of subject ACH0039, but the majority of the isolated NSI virus clones already had rapid replication kinetics (Fig. 1, top left panels). The first and rapidly replicating, SI variants were obtained 17 months after seroconversion. Throughout follow-up, coexisting SI and NSI variants that had similar rapid replication kinetics were isolated (Fig. 1, top right panels).
FIG. 1.
Replication kinetics of SI (•) and NSI (○) HIV-1 clones during the course of HIV-1 infection in two subjects harboring both SI and NSI virus variants. From each virus clone, 103 TCID50 was added to 5 × 106 2-day PHA-stimulated PBMC derived from the same donor on which the titer was determined. After 5, 8, and 11 days, approximately 3 × 106 fresh stimulated target cells were added. Samples (75 μl) for determination of p24 production were harvested every day after infection for 14 days. Each line represents the results obtained for one virus clone; ▸ indicates NSI clone 12B3 with arginines at positions 10 and 25. AIDS diagnosis for ACH0039 and ACH0208 was at 37 and 54 months respectively, postseroconversion (post sc.).
In both the early and late phases of infection, different replication patterns were observed in subject ACH0208 (Fig. 1, bottom panels). During the first 9 months after seroconversion of subject ACH0208, only slowly replicating NSI viruses were obtained (Fig. 1, bottom left panels). After 15 months the first, slowly replicating SI variants were detected; these were replaced within 4 months by rapidly replicating SI variants. The SI variants obtained from ACH0208 gained increasing replication kinetics over time, while the coexisting NSI variants maintained slow replication kinetics (Fig. 1, bottom right panels). At the time of AIDS diagnosis, faster-replicating NSI variants were isolated, although replication was still markedly slower than that of the coexisting SI variants.
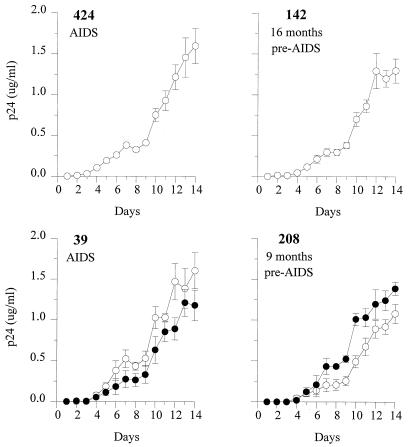
The viruses isolated around the time of AIDS diagnosis from these two patients were compared with those obtained from two individuals harboring only NSI variants throughout infection, ACH0142 and ACH0424. In general, viruses obtained from all four individuals had rapid replication kinetics around the time of AIDS diagnosis, irrespective of the presence of SI variants (Fig. 2).
FIG. 2.
Replication kinetics of SI (•) and NSI (○) HIV-1 clones in four subjects at the time of AIDS diagnosis. From each virus clone, 103 TCID50 was added to 5 × 106 2-day PHA-stimulated PBMC derived from the same donor on which the titer was determined. After 5, 8, and 11 days, approximately 3 × 106 fresh stimulated target cells were added. Samples (75 μl) for determination of p24 production were harvested every day after infection for 14 days. Means and standard errors of the results for four to five virus clones per phenotype are shown.
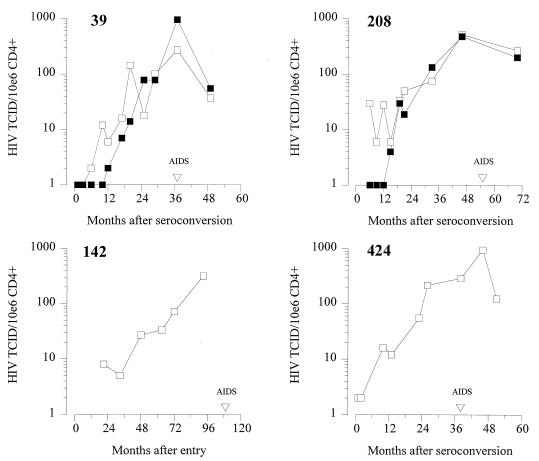
Contribution of phenotypically different HIV-1 clones to virus load.
To analyze the significance of slowly replicating coexisting NSI variants, we calculated the contribution of NSI and SI virus clones to the total cellular infectious load in the limiting-dilution analysis. For each time point analyzed, the contributions of SI and NSI HIV-1 variants to the total infectious cellular load are shown in Fig. 3. SI variants were first detected 17 and 15 months after seroconversion in ACH0039 and ACH0208, respectively. The equally rapidly replicating coexisting SI and NSI variants in subject ACH0039 contributed equally to the infectious cellular load (Fig. 3, top left). However, also in subject ACH0208 the NSI variants constituted 50% of the total virus population throughout follow-up despite their slow replication rates (Fig. 3, top right). The two NSI-harboring individuals, ACH0142 and ACH0424, also showed a gradual load increase over time, reaching viral loads similar to the two SI- and NSI-harboring individuals around the moment of AIDS diagnosis.
FIG. 3.
Changes in infectious cellular SI (▪) and NSI (□) HIV-1 load in relation to time after seroconversion or seropositive entry. The frequency of cells productively infected with NSI or SI variants was determined by limiting-dilution analysis of patient PBMC on PHA-stimulated target cells.
Phylogenetic analysis.
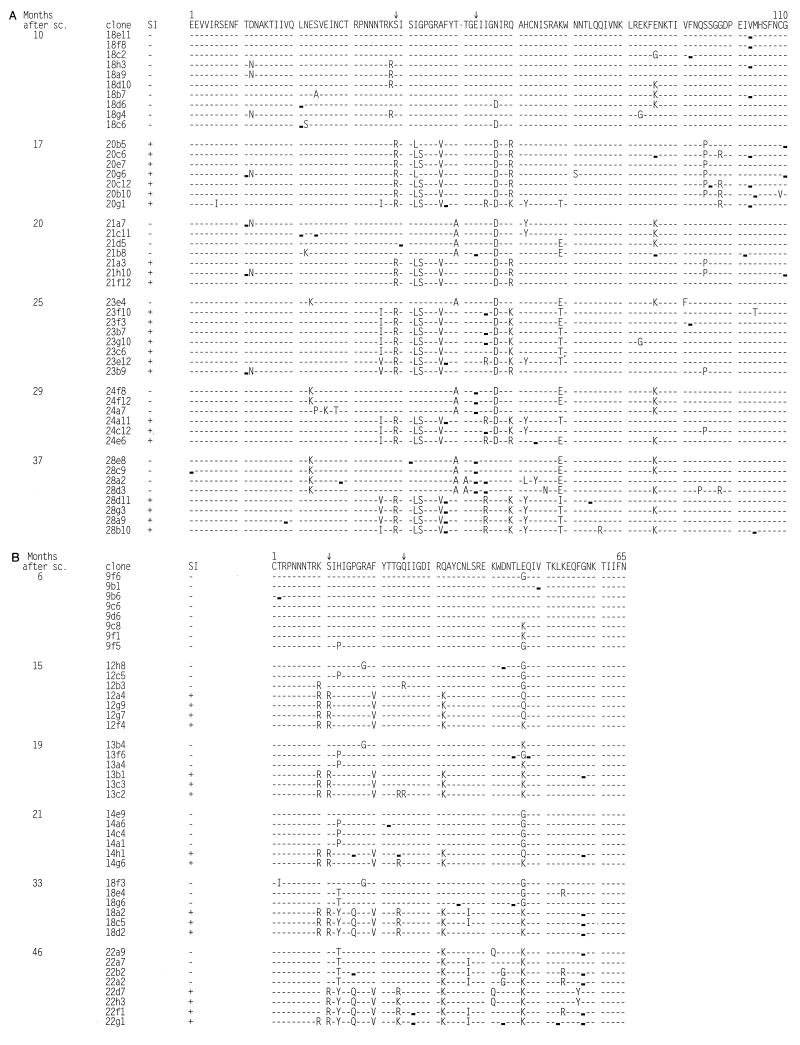
To understand the phylogenetic relationship between the coexisting SI and NSI variants in the two SI-harboring subjects, a region encompassing the V3 domain of the gp120 envelope molecule was sequenced, 330 nucleotides for ACH0039 and 195 nucleotides for ACH0208. When comparing the deduced 65 amino acids sequenced for both subjects, the V3 sequences were distinct, grouping separately into two groups, with a mean Hamming distance between the two groups of 15%. Bootstrap resampling supported the distinction of two separate groups, 100 of 100 bootstraps. The deduced amino acid sequences for the two subjects are shown in Fig. 4.
FIG. 4.
Deduced amino acid sequences of the V3 region. The sequences are aligned with the consensus sequence of the variants present in the first sample for each patient. Amino acid positions involved in SI capacity are marked (↓). Dashes indicate identity with the reference sequence. , silent mutation compared with reference sequence. (A) Alignment of V3 sequences from virus clones obtained during the course of infection of participant ACH0039. Position 1 corresponds to amino acid 268 of the HXB2 envelope protein. (B) Alignment of V3 sequences from virus clones obtained during the course of infection of participant ACH0208. Position 1 corresponds to amino acid 296 of the HXB2 envelope protein.
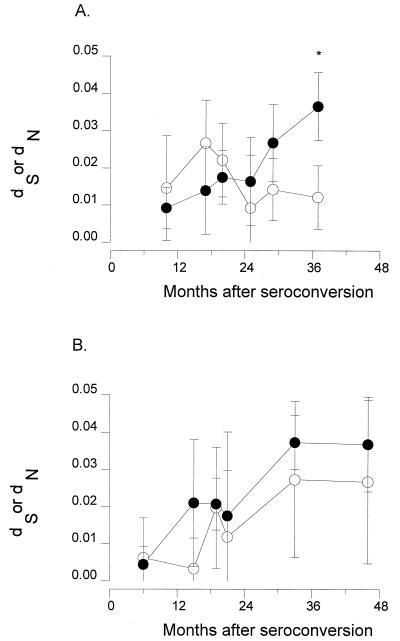
The mean synonymous substitution rate (dS) in the V3 region was 0.025 (±0.015) for ACH0039 and 0.032 (±0.030) for ACH0208 over 27 and 40 months of follow-up, respectively. The mean rates of nonsynonymous substitution (dN) were 0.036 (±0.018) and 0.040 (±0.023), respectively, which in both cases seems higher than the rate of synonymous substitutions but not significantly different due to the large standard deviations. When comparing dS and dN for each time point separately, only at the last time point (37 months) for subject ACH0039 did we observe a significant difference (Student’s t test, P = 0.0040). Figure 5 shows the changes in dS and dN detected during follow-up. Variants in subject ACH0039 had a stable rate of synonymous substitutions (time effect analysis of covariance [ANCOVA], P = 0.2227), whereas the rate of nonsynonymous substitutions increased sharply during follow-up (P < 0.0001), resulting in a decreasing dS/dN ratio. In subject ACH0208, both the rate of silent substitutions and the rate of nonsynonymous substitutions increased over time (time effect, P = 0.005 and P < 0.0001, respectively).
FIG. 5.
Plots of synonymous (dS) and nonsynonymous (dN) substitution rates in the V3 region of the virus clones obtained in the course of HIV-1 infection from two subjects, ACH0039 (A) and ACH0208 (B). Estimation of the number of silent and nonsilent substitutions between all sequences from one time point and the consensus sequence of that time point was done according to Nei and Gojobori’s method (36) as implemented in MEGA (31). Numbers of synonymous (○) and nonsynonymous (•) substitutions from the same time point are shown. ∗, P = 0.0040.
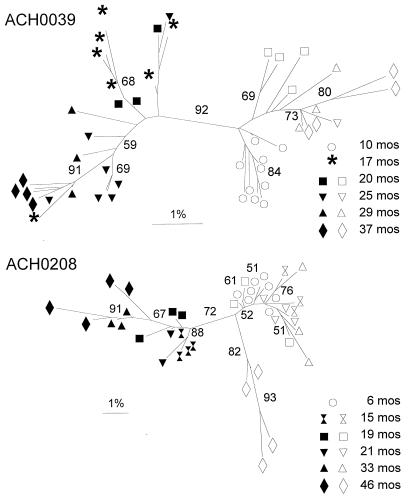
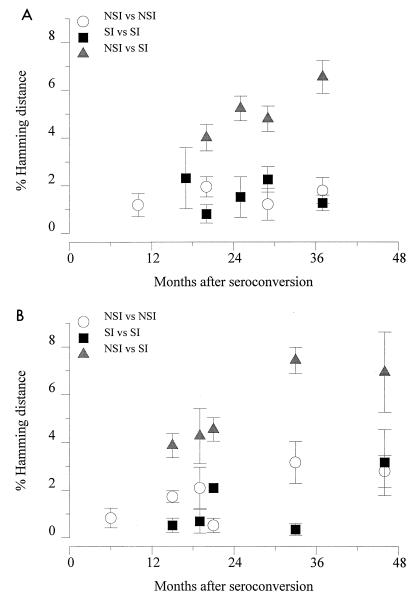
Early after seroconversion both subjects harbored a rather homogeneous group of NSI V3 sequences (mean Hamming distances of 1.2 and 0.8%, respectively). For both individuals, the first SI variants isolated were different from the coexisting NSI variants located at least 4 amino acid positions of the 35 amino acids in the V3 loop (mean Hamming distances were 3.8 and 3.9%, respectively at the first time point of detection of SI variants) (Fig. 4). To strengthen this observation, neighbor-joining trees with bootstrap resampling were constructed (Fig. 6). Indeed, in both patients the SI and NSI variants from all time points were separated but not with very high bootstrap values (92 and 72%, respectively). Total Hamming distances between the variants present at each time point increased over time in both patients (Fig. 7). Variation within each phenotypic group at each time point was relatively stable (mean Hamming distance of approximately 1.5%). However, the evolutionary distance between the two groups of variants increased steadily over time (mean Hamming distances were 6.7 and 7.0%, respectively, at the last time point available). Accordingly, Hamming distances were 3.2 and 4.9% between the late NSI variants and the early NSI variants respectively, and were 3.9 and 4.5% between the late SI and the early SI variants, respectively, for both patients. To test if the course of the variation over time differs between SI and NSI variants, an analysis of covariance was done with the variation (distance of each clone to the consensus) as the dependent variable, the time since seroconversion as a continuous and SI/NSI as a dichotomous predictor. The time effect was significant in both patients for the comparison between SI sequences and NSI consensus or between NSI sequences and SI consensus (ACH0039, P = 0.0009 and P = 0.0001, respectively; ACH0208, P < 0.0001 and P < 0.0001 respectively), but only in patient ACH0208 was a significant interaction effect observed, indicating that the NSI and SI variation developed differently over time (ACH0039, NSI P = 0.5760 and SI P = 0.6952; ACH0208, NSI P = 0.0001 and SI P = 0.0009).
FIG. 6.
Results of phylogenetic analysis of the V3 region (neighbor-joining method, unrooted tree) from virus clones obtained during the course of infection of participants ACH0039 and ACH0208. Bootstrap values indicate the percentages of trees showing the observed specific groupings. Filled symbols, SI sequences; open symbols, NSI sequences.
FIG. 7.
Plots of Hamming distances for the V3 region between the virus clones obtained in the course of HIV-1 infection from two subjects, ACH0039 (A) and ACH0208 (B). Distances were calculated by using DNADIST as implemented in the PHYLIP program. Comparisons between NSI variants, between SI variants, and between SI and NSI variants from the same time point are shown.
One of the NSI variants (12B3) isolated 15 months after seroconversion from subject ACH0208 had arginines at positions 10 and 25, two amino acids positions that are positively charged in the coexisting SI variants but not in the other NSI variants (Fig. 4). Interestingly, this NSI variant had the highest replication kinetics of the NSI variants present at that time point (Fig. 1, second bottom panel, line marked with ◂). The NSI variants present in this individual at 46 months after seroconversion had certain amino acid changes that were only detected in SI variants before that time (for example, the lysines at positions 32 and 47, the isoleucine at position 37, and the glutamine at position 41) (Fig. 4). When the four NSI variants were omitted from the data set the bootstrap value separating the SI and NSI branches of the phylogenetic tree increased from 72 to 91% (data not shown).
DISCUSSION
In this study, we analyzed the temporal relationship between changes in virus replication characteristics and the evolution of SI and NSI virus populations during the natural course of HIV disease. Interestingly, rapidly replicating SI virus clones had emerged more than 3 years before AIDS diagnosis in two individuals (7). After the emergence of, and compared to, SI variants, the coexisting NSI variants had slower replication kinetics in one subject and similar replication kinetics in the other. The similar replication kinetics of the coexisting SI and NSI variants with very different V3 regions in this latter subject again illustrate that regions other than V3 are important for replication rate (19, 20).
When SI variants are present in patient PBMC, they always outgrow the coexisting NSI variants in in vitro bulk coculture of the patient PBMC with PHA-stimulated target PBMC (unpublished observations). In vivo, however, irrespective their replicative capacity, NSI and SI HIV-1 clones each constituted 50% of the total infectious cellular load, in agreement with our previous observations (27, 48). The selective growth advantage for SI HIV-1 as observed in vitro is apparently not present in vivo. It is possible that the NSI variants detected in the periphery originate solely from compartments where virus production from macrophages predominates. Most primary SI variants are not able to infect macrophages in vitro (49), probably because their entry depends on much higher concentrations of CD4 on the cell surface than is expressed on macrophages (29). Therefore, it seems likely that virus production from macrophage compartments is predominantly of the NSI phenotype. The SI variants are apparently unable to compete with the direct spread of NSI variants from the macrophages to CD4+ T cells which may occur during the close cell-to-cell contact during antigen presentation.
Alternatively, the persistence of slowly replicating NSI variants may be explained by the existence of different target T-cell populations for NSI and SI variants. Recently, several members of the CC and CXC chemokine receptor families have been shown to function as the coreceptors for HIV entry. NSI variants primarily use the β-chemokine receptor CCR5, while primary SI variants were shown in vitro to use both CCR5 and the α-chemokine receptor CXCR4 (1, 6, 10, 12, 13, 16, 54, 60). The equal contribution to the infectious cellular load indicates the ability of the slow NSI variants to compete with the more rapid SI variants for the same CCR5-expressing targets. This may be mediated by increased affinity for the CCR5 and/or CD4 receptor, allowing replication in cells with lower expression of these molecules.
Interestingly, the subject that harbored rapid SI and slow NSI variants was heterozygous for the 32-bp deletion (Δ32) in the CCR5 gene (32, 47). It is likely that the NSI viruses in this individual have adapted to entry of target cells expressing lower levels of CCR5, for example, by using other coreceptors or developing higher affinity for CCR5 itself. Indeed, we have recently found evidence for adaptation of viruses from CCR5 Δ32 heterozygous individuals for growth in cells from CCR5 Δ32 heterozygous blood donors (3a). As the NSI virus variants from subject ACH0208 have slower replication kinetics than the NSI variants from subject ACH0039, this adaptation may be associated with decreased replication kinetics.
The fact that the evolutionary distance between coexisting NSI and SI populations increases over time may indeed indicate that in general there is only little interaction between these two compartments of HIV-1 replication. The two compartments evolved away from the common ancestral sequence with no sign of saturation, which may indicate that SI variants are newly generated in an individual rather than transmitted together with the NSI variant, as the latter scenario would result in two highly distinct coexisting SI and NSI populations already at the very first time point of detection of SI variants, of which we find no evidence in these patients or in several other cohort participants studied for this phenomenon (data not shown). Only in one case, where SI variants were detected immediately after infection of a new individual, did we find coexisting SI and NSI populations that were more than 5% apart, reflecting transmission of both coexisting variants from the donor (57).
After the first emergence of the SI variants in both subjects, no new SI variants generated from late NSI variants were detected. Once SI variants are present, it may be more difficult for newly arising switch variants to compete. Alternatively, there may only be a certain type of NSI variants from which SI variants can be generated. When this stage in the evolution of NSI variants has been passed, no new SI variants will emerge or the new SI variants may not be able to compete with the late NSI variants. Indeed, at the time of AIDS diagnosis, high virus loads and rapid virus replication kinetics were found in all individuals, including the two individuals harboring only NSI variants throughout the course of infection. Apparently, the presence of rapid NSI variants was sufficient to establish progression to AIDS without the generation of SI variants. Possibly, these late rapid NSI variants were as cytopathic as the late rapid SI variants in the other two individuals (50). Subject ACH0039, however, harbored rapid NSI variants 10 months after seroconversion. Still, in this subject SI variants emerged. This may suggest that the increased target cell repertoire of SI variants in some individuals provides enough selective growth advantage for SI variants to emerge in the presence of rapidly replicating NSI variants.
In both subjects, the coexisting SI and NSI populations each are surprisingly homogeneous throughout the study period. In subject ACH0039, the average Hamming distance rarely exceeded 2%, and in subject ACH0208 only after more than 30 months of infection did the average Hamming distance per phenotype reach the 5% level. The homogeneity per phenotypic population may indicate fierce competition within the NSI and SI HIV-1 populations, where only the fittest variant in both groups evolves further, generating the new virus population detected at the next time point. The possible recombination observed in subject ACH0208 suggests, however, that interaction between the coexisting SI and NSI variants may occur, which would imply at least some overlap of target cell populations, in agreement with the reported CCR5 usage of primary SI variants (54). Despite the homogeneity of each phenotypic virus population at a particular time point, there is a continued evolution toward new variants reflected in the increasing Hamming distance between the NSI and SI virus populations and between each population and the populations present at earlier time points.
Overall, sequence variation increased with time in the two subjects, due to increases in both the number of synonymous and nonsynonymous substitutions in ACH0208 and due to increases in the number of nonsynonymous substitutions only in ACH0039. Nonsynonymous virus variation in both p17 and V3 has been shown to be correlated with immune selective pressures (33–35, 53, 58, 59). The HIV-specific cytotoxic T-lymphocyte reactivity against autologous virus epitopes in the V3 region in these two subjects is currently under study.
ACKNOWLEDGMENTS
This study was performed as part of the Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Municipal Health Service, The Academic Medical Centre, and the Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, Amsterdam, The Netherlands. Proleukin (rIL-2) was kindly provided by R. Rombouts, Chiron Benelux B.V., Amsterdam, The Netherlands. We are greatly indebted to all cohort participants for their continuous participation, to Marijke Roos and colleagues for excellent technical assistance, to Ana-Maria de Roda-Husman for providing data on CCR5 genotype, to Catherine Macken, Andrew Leigh-Brown, and David Krakauer for discussions and advice, and to Frank van Engelenburg, Maarten Koot, and Frank Miedema for critical reading of the manuscript.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Åsjö B, Albert J, Karlsson A, Morfeldt-Månson L, Biberfeld G, Lidman K, Fenyö E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 3.Balliet J W, Kolson D L, Eiger G, Kim F M, MacGann K A, Srinivasan A, Collman R. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 3a.Blaak, H. Unpublished observations.
- 4.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, Van der Noordaa J. A rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1991;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 6.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 7.Connor R I, Ho D D. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor R I, Mohri H, Cao Y, Ho D D. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Jong J J, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Suttons R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of the major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 11.De Roda Husman A M, Koot M, Cornelissen M T E, Brouwer M, Broersen S M, Bakker M, Roos M T L, Prins M, De Wolf F, Coutinho R A, Miedema F, Goudsmit J, Schuitemaker H. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann Intern Med. 1997;127:882–890. doi: 10.7326/0003-4819-127-10-199711150-00004. [DOI] [PubMed] [Google Scholar]
- 12.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 13.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP manual version 3.5. Berkeley, Calif: University Herbarium, University of California at Berkeley; 1993. [Google Scholar]
- 16.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 17.Fouchier R A M, Broersen S M, Brouwer M, Tersmette M, van ’t Wout A B, Groenink M, Schuitemaker H. Temporal relationship between elongation of the HIV-1 gp120 V2 domain and the conversion towards a syncytium inducing phenotype. AIDS Res Hum Retroviruses. 1995;11:1473–1478. doi: 10.1089/aid.1995.11.1473. [DOI] [PubMed] [Google Scholar]
- 18.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman J G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groenink M, Andeweg A C, Fouchier R A M, Broersen S M, Van der Jagt R C M, Schuitemaker H, De Goede R E Y, Bosch M L, Huisman J G, Tersmette M. Phenotype-associated env gene variation among eight related human immunodeficiency virus type 1 clones: evidence for in vivo recombination and determinants of cytotropism outside the V3 domain. J Virol. 1992;66:6175–6180. doi: 10.1128/jvi.66.10.6175-6180.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groenink M, Fouchier R A M, Broersen S M, Baker C H, Koot M, van ’t Wout A B, Huisman J G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 21.Hamming R W. Coding and information theory. Englewood Cliffs, N.J: Prentice Hall; 1986. [Google Scholar]
- 22.Hattori N, Michaels F, Fargnoli K, Marcon L, Gallo R C, Franchini G. The human immunodeficiency virus type 2 vpr gene is essential for productive infection of human macrophages. Proc Natl Acad Sci USA. 1990;87:8080–8084. doi: 10.1073/pnas.87.20.8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 24.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 25.Kimura M. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 26.Koot M, Keet I P M, Vos A H V, De Goede R E Y, Roos M T L, Coutinho R A, Miedema F, Schellekens P T A, Tersmette M. Prognostic value of human immunodeficiency virus type 1 biological phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 27.Koot M, van ’t Wout A B, Kootstra N A, De Goede R E Y, Tersmette M, Schuitemaker H. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J Infect Dis. 1996;173:349–354. doi: 10.1093/infdis/173.2.349. [DOI] [PubMed] [Google Scholar]
- 28.Koot M, Vos A H V, Keet I P M, De Goede R E Y, Dercksen W, Terpstra F G, Coutinho R A, Miedema F, Tersmette M. HIV-1 biological phenotype in long term infected individuals, evaluated with an MT-2 cocultivation assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 29.Kozak S L, Platt E J, Madani N, Ferro F E, Peden K, Kabat D. CD4, CXCR-4, and CCR5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuiken C L, De Jong J J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. CABIOS. 1994;10:189–192. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 32.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:1–20. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 33.Lukashov V V, Kuiken C L, Goudsmit J. Intrahost human immunodeficiency virus type 1 evolution is related to the length of the immunocompetent period. J Virol. 1995;69:6911–6916. doi: 10.1128/jvi.69.11.6911-6916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald R A, Mayers D L, Chung R C-Y, Wagner K F, Ratto-Kim S, Birx D L, Michael N L. Evolution of human immunodeficiency virus type 1 env sequence variation in patients with diverse rates of disease progression and T-cell function. J Virol. 1997;71:1871–1879. doi: 10.1128/jvi.71.3.1871-1879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A J, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 37.O’Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 38.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 40.Preston B D, Poiesz B J, Loeb L A. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 41.Ricchetti M, Buc H. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 1990;9:1583–1593. doi: 10.1002/j.1460-2075.1990.tb08278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 43.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 44.Roos M T L, Lange J M A, De Goede R E Y, Coutinho R A, Schellekens P T A, Miedema F, Tersmette M. Viral phenotype and immune response in primary human immunodeficiency virus type 1 (HIV-1) infection. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Sakai K, Ma X, Gordienko I, Volsky D J. Recombinational analysis of a natural noncytopathic human immunodeficiency virus type 1 (HIV-1) isolate: role of the vif gene in HIV-1 infection kinetics and cytopathicity. J Virol. 1991;65:5765–5773. doi: 10.1128/jvi.65.11.5765-5773.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C, Saragosti S, Lapouméroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 48.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, De Goede R E Y, Van Steenwijk R P, Lange J M A, Eeftinck Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuitemaker H, Kootstra N A, De Goede R E Y, De Wolf F, Miedema F, Tersmette M. Monocytotropic human immunodeficiency virus 1 (HIV-1) variants detectable in all stages of HIV infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheppard H W, Krowka J, Ascher M, Cuevas B, Dondero D, Lu E. Abstracts of the XI International Conference on AIDS. Vancouver, British Columbia, Canada. 1996. Determinants of long-term non-progression: the relative contribution of viral burden and strain variation, abstr. A390. [Google Scholar]
- 51.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 52.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Brown A J L. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either LESTR or CCR5 as co-receptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tersmette M, Gruters R A, De Wolf F, De Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman J G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terwilliger E F, Cohen E A, Lu Y, Sodroski J G, Haseltine W A. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van ’t Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolinsky S M, Korber B T M, Neumann A U, Daniels M, Kunstman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T, Koup R A. Adaptive evolution of human immunodeficiency virus type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Huang Y, He T, Cao Y, Ho D D. HIV-1 subtype and second-receptor use. Nature. 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 61.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]