Abstract
Circadian clock performance during winter dormancy has been investigated in chestnut by using as marker genes CsTOC1 and CsLHY, which are homologous to essential components of the central circadian oscillator in Arabidopsis. During vegetative growth, mRNA levels of these two genes in chestnut seedlings and adult plants cycled daily, as expected. However, during winter dormancy, CsTOC1 and CsLHY mRNA levels were high and did not oscillate, indicating that the circadian clock was altered. A similar disruption was induced by chilling chestnut seedlings (to 4°C). Normal cycling resumed when endodormant or cold-treated plants were returned to 22°C. The behavior of CsTOC1 and CsLHY during a cold response reveals a relevant aspect of clock regulation not yet encountered in Arabidopsis.
Keywords: chilling, winter dormancy, ecodormancy, cold response
Winter dormancy is an important adaptive strategy that enables plants to persist during periods of stressful environmental conditions (1). Dormancy parameters are key determinants in woody plants in agriculture and forestry. Dormancy determines to what degree fruit crops will survive winter and early spring without shoot and flower bud damage, and, in long-lived forest species, the length of rest limits the growing season and thus affects wood production and quality. The onset of winter deep dormancy (endodormancy) is preceded by a stage of ecodormancy. Endodormancy is caused by plant endogenous factors, and, once established, no growth can be achieved until a chilling requirement has been satisfied. In order for bud break to occur, plants need to be exposed to low temperatures for a cumulative number of hours (chilling requirement). In contrast, during ecodormancy, growth is arrested by an adverse environment and resumes when conditions become favorable (1, 2). The onset of endodormancy is one of the most frequently studied photoperiodic phenomena. Some important endodormancy-related traits, such as growth cessation, bud set, and the initial stages of cold acclimation, can be induced in many tree species by a short day (SD) photoperiod (3, 4). Overexpression of the oat phytochrome A photoreceptor in hybrid aspen prevents cold acclimation in response to SDs and significantly changes the critical day length, which is defined as the longest photoperiod inducing growth cessation (5, 6). Other changes, including advanced stages of cold acclimation, leaf senescence, and abscission, are not induced by SDs alone but require exposure to low temperatures (7–9).
The molecular basis of endodormancy induction, as well as the control of most other plant photoperiodic processes, are poorly understood (2, 10). Among these processes, flowering has been most extensively studied. It has been reported recently that the circadian clock controls flowering time in Arabidopsis in response to photoperiod, in agreement with an external coincidence model (11, 12). This model proposes that day length measurement relies on a circadian oscillator that controls the level of a regulatory molecule, the activity of which is modulated by light. Transcriptional regulation of the CO (CONSTANS) gene by the circadian clock, combined with posttranscriptional regulation of the CO protein by light, has been implicated in the photoperiodic control of flowering induction (13–15).
In an attempt to identify genes that are specifically expressed during winter dormancy in a deciduous tree, we performed a cDNA subtraction on stem tissue (2-year-old branch internodes) collected around midday in June and December from adult European chestnut (Castanea sativa Mill.). Among the clones specifically expressed in winter, a cDNA fragment with high similarity to the Arabidopsis gene TOC1 was identified. This gene is considered a central element of the plant circadian clock, and its transcript levels cycle daily in Arabidopsis, peaking at dusk (16–18). The presence of TOC1 homologous transcripts during daytime in chestnut stems collected in winter were therefore anomalous and led us to analyze the circadian clock in this tissue, during both dormancy and vegetative growth.
Reciprocal interactions between TOC1 and two related MYB genes (LHY and CCA1) constitute the core mechanism of the circadian oscillator in Arabidopsis (19). LHY and CCA1 are partially redundant in function, and their transcript levels oscillate in a similar pattern, peaking soon after dawn (20–23). Circadian rhythms are based on feedback loops in which the proteins LHY and CCA1 negatively control their own synthesis by interacting with the evening element in the TOC1 promoter and inhibiting the expression of the positively regulating TOC1 transcription factor (19).
Here, we report that the chestnut circadian clock behaves as in Arabidopsis during the growing season. However, this behavior is disrupted during winter or in immediate response to low temperatures. The winter clock alteration is related to the ecodormancy state induced by cold, and it is not intrinsic to tree endodormancy. The possible roles of circadian clock disruption on dormancy physiology are discussed.
Materials and Methods
Plant Material and Growth Conditions. Seeds, stem material (2-year-old branch internodes), leaves, and winter buds of European chestnut (C. sativa Mill.) were harvested from adult trees growing in Zarzalejo, Madrid (4°11′ W, 40°35′ N). Samples were collected during the months of June (22.8°C average temperature; 15 h, 5 min average day length) and December (4.9°C average temperature; 9 h, 16 min average day length). After germination, chestnut seedlings were kept in controlled-environment growth chambers for 16–24 weeks. For long day (LD) experiments, seedlings were grown at 22°C with a 16 h light/8 h dark (16:8) photoperiod (150 μE·m–2·s–1, where E = 1 mol of photons) using TLD 36W/83 (Philips, Eindhoven, The Netherlands) and GRO-LUX F36W/GRO-T8 (Sylvania Electric Products, Fall River, MA) lighting at 70% relative humidity. SD experiments were performed under the same conditions as the LD experiments, except that the photoperiod was set to 8 h light/16 h dark (8:16) during the last week. Cold treatments were carried out at 4°C under the same light regimes. Continuous light (LL) experiments were performed with plants that were grown at LD and 22°C and were subsequently transferred to LL at dawn.
To differentiate responses intrinsic to endodormancy from those that are caused by low temperatures in dormant plants, the following experiment was designed. Chestnut seeds germinated in December were grown in a greenhouse until March. Subsequently, seedlings were transferred to natural light and temperature conditions in Fuentidueña de Tajo, Madrid (3°7′ W, 40°5′ N). When plants entered endodormancy (at the stage of leaf abscission, between November 20 and December 10, 2004; 8.0°C average temperature; 9 h, 33 min average day length), they were kept for 1 week in a growth chamber at 22°C with a 16 h light/8 h dark photoperiod before sample collection. To monitor the degree of bud dormancy attained, control plants in the same growth chambers were checked daily for bud break. Twenty control plants were included for each experiment, and the experiments were repeated three times. The number of days to the first bud break was 34.3 ± 7.3, and the number of days for 50% of the plants to have at least one bud burst was 45.3 ± 6.5.
After plants were subjected to the different treatments, samples were collected at 3-h intervals. Each experiment was performed at least twice.
Isolation of cDNA Clones. Chestnut TOC1 (CsTOC1) and LHY (CsLHY) full-length cDNAs were isolated from a λ Uni-ZAP XR cDNA library following standard procedures (24). The library was constructed by using chestnut stem poly(A)+ RNA isolated from plants growing in winter. The RNA was reverse-transcribed, and the resulting cDNAs were cloned by using the ZAP-cDNA Gigapack III Gold Cloning Kit (Stratagene). The probe used for CsTOC1 was a cDNA fragment obtained by enriching for winter-specific transcripts using the PCR-Select cDNA subtraction kit (Clontech). To detect CsLHY clones, a LHY homologous probe from Arabidopsis thaliana was isolated and used under low stringency hybridization and washing conditions. Total cDNA from A. thaliana seedlings was synthesized by using the SuperScript First-Strand Synthesis System (Invitrogen). LHY fragments were amplified by PCR using 5′-TGGACATAGAAATTCCGCCTCCTCG-3′ and 5′-CTTTTGAAATTAGGAGCCAATGGC-3′ as forward and reverse primers, respectively, as described in ref. 25. To find the CsLHY full-length cDNA clone, high-stringency hybridization and washing conditions were performed. Probes were labeled with [α-32P]dATP by using a random-primed DNA labeling kit (Roche Applied Science, Indianapolis).
Gene Expression Analysis. Total RNA from chestnut stems and leaves was obtained as described in ref. 26, separated in 1.2% agarose gels with 2.2 M formaldehyde (27), and subsequently transferred to Magna nylon membranes (Osmonics, Westborough, MA). A fragment encompassing CsTOC1 nucleotides 1380–1591 and a CsLHY full-length clone were labeled as described above and used as probes to detect each transcript. The CsTOC1 fragment specifically recognizes the TOC1/PRR1 member of the pseudoresponse regulator (PRR) family gene. Northern blot hybridizations were carried out following the membrane manufacturer's recommendations. Membranes were washed at high stringency, exposed on storage phosphor screens, visualized in a TYPHOON 9400 phosphorimage scanner (Amersham Pharmacia Biotech), and quantified with quantity one software (Bio-Rad). Raw hybridization data were normalized to the rRNA value. The rRNA loading reference was estimated by staining gels with ethidium bromide. Quantified data are shown schematically in graphs as relative amounts of mRNA.
Results and Discussion
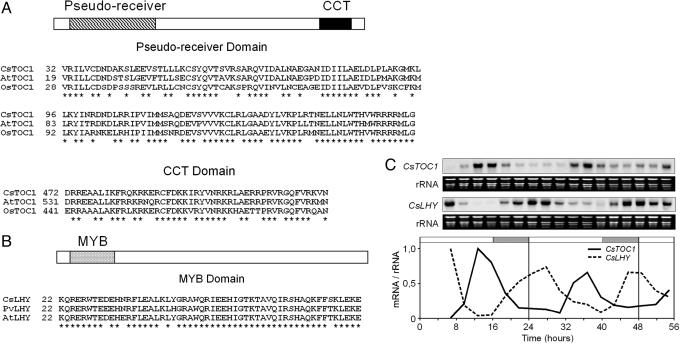
Isolation and Characterization of Chestnut TOC1 and LHY cDNA Clones. To characterize the circadian clock in chestnut, we isolated two full-length cDNA clones from a C. sativa stem library that were homologous to genes TOC1 and LHY from Arabidopsis. The chestnut TOC1 cDNA clone (CsTOC1) encoded a polypeptide containing two domains characteristic of the PRR protein family. Arabidopsis TOC1 belongs to this family and is also known as APRR1 (16, 17, 28). We confirmed that the similarity between CsTOC1 and TOC1/APRR1 (52% identity) was greater than that between CsTOC1 and the other members of the Arabidopsis PRR family. The CsTOC1 protein (545 aa residues) was smaller than its Arabidopsis counterpart TOC1 (618 residues) and greater than a protein deduced from a putative ortholog from Oryza sativa (518 residues). The pseudoreceiver domain and the CCT (CONSTANS, CONSTANS-LIKE, and TOC1) motif of TOC1 proteins were highly conserved among the three species (Fig. 1A). Similar to the Arabidopsis protein, the chestnut CsTOC1 pseudoreceiver domain lacked two invariant aspartic residues required for normal response in true response regulator proteins (16, 17). The chestnut CsLHY sequence encoded a polypeptide of 768 aa with an MYB-like domain and exhibited significantly higher similarity to LHY from Arabidopsis and Phaseolus vulgaris than to CCA1 from Arabidopsis (20, 21, 29). A comparison of the MYB-like domains of LHY proteins from the three plant species is shown in Fig. 1B.
Fig. 1.
Characterization of central genes from the chestnut circadian clock. (A) Representation of the structure of PRRs containing two types of common domains (the pseudoreceiver and the CCT motif) and comparison of the pseudoreceiver and CCT domains of TOC1 from chestnut (GenBank accession no. AY611028), A. thaliana (AF272039), and rice (AK111828). (B) LHY protein structure showing the position of the MYB domain, and comparison of LHY's MYB domain from chestnut (AY611029), P. vulgaris (AJ420902), and A. thaliana (AJ006404). Asterisks indicate identical residues. (C) CsTOC1 and CsLHY gene expression rhythms in chestnut leaves from 16- to 24-week-old seedlings grown under standard conditions (LD, 22°C) and subsequently transferred to LL and 22°C. CsTOC1 and CsLHY RNA blot analysis and mRNA abundance are shown. Samples were collected at 3-h intervals. The experiment was performed at least twice with similar results; one representative data set is shown. Raw hybridization data obtained by using a phosphoimage scanner was quantified by normalizing it to the rRNA value. The rRNA loading reference was detected by staining gels with ethidium bromide. Quantified data are shown schematically in graphs as relative amounts of mRNA. The open and shaded bars above the graphs represent the subjective day and night lengths, respectively.
To test the circadian behavior of CsTOC1 and CsLHY genes, we analyzed their expression in chestnut leaves collected from 16- to 24-week-old seedlings under LL conditions at 22°C. A clear profile of sequential circadian waves of the mRNA levels of both genes in LL was observed (Fig. 1C).
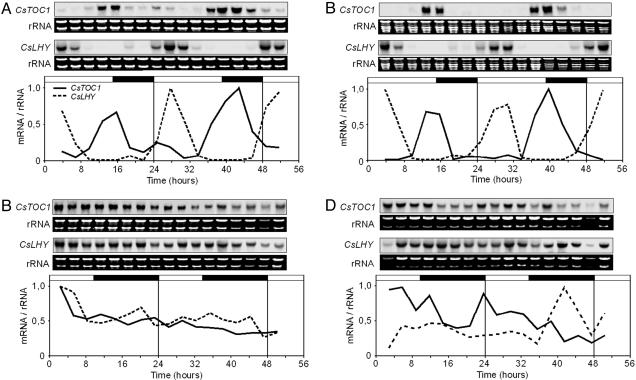
Seasonal Differences in Circadian Clock Performance in Adult Chestnut. We investigated the performance of the circadian clock in adult chestnuts grown under natural light and temperature conditions in Zarzalejo, Madrid (4°11′ W, 40°35′ N) during the month of June, when vegetative growth takes place, and during the month of December, when trees undergo winter rest. The expression dynamics of CsTOC1 and CsLHY were studied by Northern blot analysis, using specific probes derived from cDNA clones. Similar to what has been reported in Arabidopsis (18), during June, the mRNA levels of these two genes cycled robustly according to the light–dark regime in both stems and leaves (Figs. 2 A and B). However, in stems and winter buds undergoing dormancy during December, TOC1 and LHY mRNA levels were relatively high and did not follow the typical daily cycles (Fig. 2 C and D). Therefore, these results indicate that the circadian clock oscillator did not function during winter dormancy, probably due to disruption of the transcriptional regulatory mechanisms, although other possibilities should not be ruled out. In Drosophila, expression of clock genes requires regulation at the transcriptional, posttranscriptional, and posttranslational levels for normal rhythmicity (30, 31). We attempted to correlate the observed disruption with factors that determine the onset of tree endodormancy, namely low temperature and SD photoperiod.
Fig. 2.
CsTOC1 and CsLHY gene expression rhythms in adult chestnut under different seasonal conditions. (A and B) Stem material (second-year branch internodes) and leaves, respectively, collected in June. (C and D) Stem material and winter buds, respectively, collected in December. Samples were collected at 3-h intervals. Each experiment was performed at least twice with similar results; one representative data set is shown. The quantification was as described in the legend of Fig. 1. The open and filled bars above each graph represent the natural day and night lengths, respectively, as provided by the National Institute of Meteorology in Madrid.
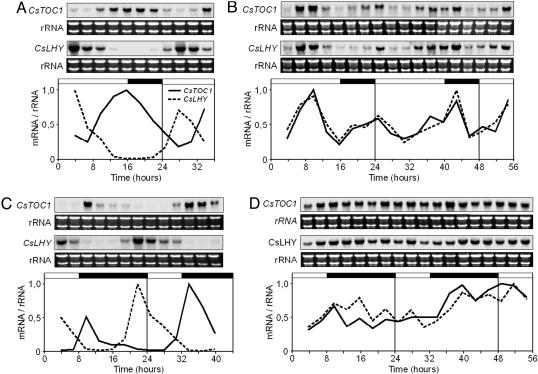
Circadian Clock Responses in Chestnut Seedlings to Low Temperature and SD Photoperiod. Changes throughout daily cycles in the mRNA levels of the CsTOC1 and CsLHY genes in chestnut stems and leaves collected from 16- to 24-week-old seedlings grown under normal conditions [16 h light/8 h dark (16:8 LD) and 22°C] were compared with those in seedlings that had been subjected to cold treatment for 1 week (16:8 LD and 4°C). Normal cycling of CsTOC1 and CsLHY mRNA levels in the stems of plants growing under standard conditions was evident, whereas in the cold-treated seedlings, the mechanism of the circadian oscillator was disrupted (Fig. 3 A and B), and the apparent expression pattern of the CsTOC1 gene was similar to that of the CsLHY gene, instead of the normal opposite phase expression. We also compared the behavior of the oscillator in SDs (8:16 SD) at 22°C and after 1 week at 4°C. At 22°C, the onset of CsLHY expression took place a few hours earlier for the SD than for the LD treatment, indicating that transcription begins in the dark (Fig. 3C). Although the beginning of CsTOC1 transcript synthesis was not significantly affected in the SD treatment, its expression was shifted toward the dark period. Similar variations have been described in Arabidopsis (28, 32). The performance of the circadian clock was disrupted in cold-treated plants exposed to SD and low temperature, and the CsTOC1 and CsLHY mRNAs were also simultaneously accumulated (Fig. 3D). Expression patterns in leaves were essentially similar to those observed in stems (Fig. 6, which is published as supporting information on the PNAS web site). It could not be excluded that synchrony of rhythms among the different tissues at 22°C was due to simultaneous but independent resetting of the corresponding circadian clocks (33).
Fig. 3.
CsTOC1 and CsLHY gene expression rhythms in chestnut stems from 16- to 24-week-old seedlings grown under different conditions of temperature and photoperiod. (A) Stems from seedlings grown at LD [16 h light/8 h dark (16:8)] and 22°C. (B) Stems from seedlings grown for 1 week at LD and 4°C. (C) Stems from seedlings grown at SD (8:16) and 22°C. (D) Stems from seedlings grown for 1 week at SD (8:16) and 4°C. Samples were collected at 3-h intervals. Each experiment was performed at least twice with similar results; one representative data set is shown. The quantification was as described in the legend of Fig. 1. The open and filled bars above each graph represent the lights being on and off, respectively.
The effects of low temperature on plant circadian oscillator components have not been previously investigated, although some observations have been reported concerning downstream genes under circadian control. Cycling of mRNA levels corresponding to cold-circadian rhythm RNA-binding protein (CCR) and the chlorophyll a/b (CAB) binding proteins is observed in Arabidopsis seedlings grown for 5 days at 4°C under a light/dark photoperiod (34). However, 4°C cold pulses for 12 or 20 h under LL causes a phase delay of 4 and 12 h, respectively, in the rhythms of CAB and CCR2 RNA levels (34). Additionally, low-temperature treatment (4°C for 16 h) of chilling-sensitive tomato plants interrupts circadian regulation of transcriptional activity of certain genes, including those encoding CAB protein, Rubisco activase, and nitrate reductase (35, 36). Upon rewarming, the circadian rhythm of transcriptional activity is restored but is out of phase with the actual time of day by the amount of time that the tomato plants are at low temperature. In tomato experiments, extended time periods have not been investigated (35, 36).
We analyzed the circadian clock response in chestnut during the first 3 days under low temperatures. These experiments were carried out at 4°C by using an LD photoperiod. Treatments began either in the morning (CsLHY mRNA high; CsTOC1 mRNA low) or in the evening (CsLHY mRNA low; CsTOC1 mRNA high). In both instances, we observed that the expression of CsTOC1 and CsLHY changed from the beginning of the experiments and that their cycling patterns were interrupted, with their mRNA levels remaining at the initial values (Fig. 4 A and B; see also Fig. 7, which is published as supporting information on the PNAS web site). After 24 h, the transcript level, which was low in each case, began to increase, and, in both experiments, the mRNA level of the two genes reached a plateau after 36 h and remained high until the end of the experiment, a status that was similar to that observed after 1 week at 4°C (Fig. 3B). These data show that at low temperatures, the normal function of clock components is disrupted, because the increase of CsLHY mRNA did not result in an inhibition of CsTOC1 expression. These observations point to the existence of clock regulatory components in chestnut not yet identified in Arabidopsis, where LHY mRNA negatively correlates with TOC1 mRNA (19).
Fig. 4.
CsTOC1 and CsLHY gene expression rhythms in chestnut stems from 16- to 24-week-old seedlings exposed to continuous cold treatment (4°C). Seedlings were grown at LD [16 h light/8 h dark (16:8)] and 22°C and subsequently transferred to 4°C for 52 h. Plants were transferred either in the morning (3 h after the lights were turned on), when LHY mRNA levels are high (A) or at dusk (1 h before lights went off), when TOC1 mRNA levels are high (B). Samples were collected at 3-h intervals. Each experiment was performed at least twice with similar results; one representative data set is shown. The quantification was as described in the legend of Fig. 1.
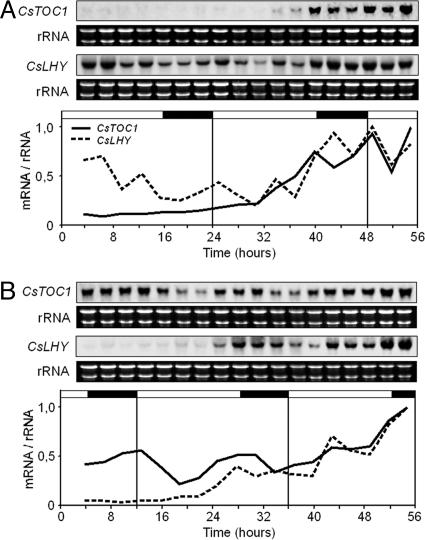
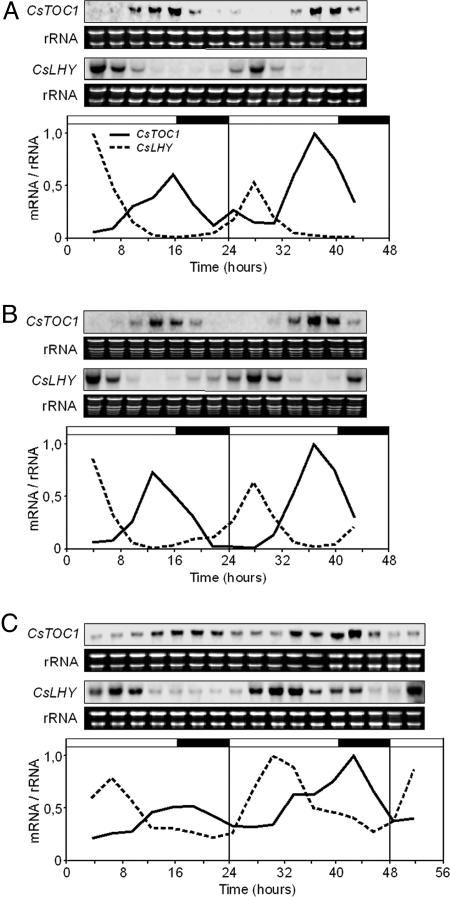
Recovery of Standard Performance of the Circadian Clock in Ecodormant and Endodormant Chestnut Seedlings. Standard performance of the circadian clock, which was interrupted by low temperatures in stems and leaves of chestnut seedlings (Figs. 3 and 6), was recovered in both tissues once the plants were returned to normal conditions, LD and 22°C, for 1 week (Fig. 5 A and B). In a similar experiment carried out with endodormant plants grown under natural conditions that had not yet met the chilling requirement (see Materials and Methods), the circadian clock also recovered its standard performance after 1 week under normal conditions in a controlled-environment growth chambers (Fig. 5C). This behavior indicated that clock disruption is not intrinsic to the endodormancy state but is a direct consequence of the response to cold. However, a role for clock disruption in the onset of endodormancy cannot be ruled out. Once the short photoperiod induces winter dormancy processes, perhaps through a mechanism of external coincidence such as that described for flowering control in Arabidopsis (11, 12), disruption of the circadian clock could be necessary to reach advanced dormancy stages that would also require low temperatures (7–9). Analysis of quantitative trait loci associated with dormancy in hybrid poplars indicates that genetic differences in photoperiodic responses play a modest role in explaining genetic differences in bud set timing under natural field conditions, thus suggesting that responses to other environmental factors, such as temperature, could be relevant (37, 38). Additionally, it has been recently reported (39) that, in birch, preexposure to SD followed by low-temperature treatment resulted in a remarkable increase in expression of a C repeat-binding factor-controlled dehydrin gene when compared with low-temperature-treated plants grown at LD photoperiod. Transgenic Arabidopsis experiments indicated that this potentiation could be birch- or tree-specific (39).
Fig. 5.
Recovery of standard performance of the circadian clock in ecodormant and endodormant chestnut seedlings. (A and B) CsTOC1 and CsLHY gene expression rhythms in chestnut stems (A) and leaves (B) from 16- to 24-week-old seedlings grown under standard conditions (LD, 22°C) and subsequently transferred to 4°C for 7 days and placed back on standard growth conditions for 1 week. (C) CsTOC1 and CsLHY gene expression rhythms in chestnut stems from 11-month-old endodormant plants (see Material and Methods) grown under natural conditions in Madrid and subsequently transferred to controlled-environment growth chambers at standard conditions for 1 week. Samples were collected at 3-h intervals. Each experiment was performed at least twice with similar results; one representative data set is shown. The quantification was as described in the legend of Fig. 1.
Clock disruption results in increased expression of both CsTOC1 and CsLHY, rather than none of the clock components, suggesting a positive role of these components during dormancy. Indeed, it has been proposed (40) that both ABA and the transcriptional factor ABI3, whose expression occurs after perception of the critical day length, are essential components for bud set in poplar, which in turn is a precondition for the establishment of winter dormancy (40). The interaction between ABI3 and TOC1 in Arabidopsis shown by the yeast two-hybrid system (41) suggests that altered expression of TOC1 by low temperatures could induce changes in ABI3 behavior.
The expression of at least 10% of the Arabidopsis genes is under circadian clock control. The circadian clock is involved in the coordination and proper functioning of major metabolic pathways, as well as in the control of developmental processes (42–44). Therefore, the clock disruption observed in chestnut during winter could control physiological changes that take place in woody perennials during dormancy.
The effect of cold temperatures on endodormancy release and on promoting flowering through vernalization are well known (4, 45, 46), and considerable progress has been made in understanding the molecular basis of vernalization in Arabidopsis (47, 48). In both processes, the effect of cold requires a prolonged period of exposure, so that plants have the ability to measure a complete winter season. The fast response of the chestnut circadian clock to low temperatures reported here points out that cold effects on dormancy induction are of a different nature. Additionally, the different behavior of Arabidopsis and chestnut circadian clocks under cold temperatures indicates that cold acclimation in temperate woody plants may have significant specific features compared with annual herbaceous plants (49, 50). In fact, Fowler et al. (51) have recently shown that induction by low temperature of C repeat-binding factor genes in Arabidopsis is gated by the circadian clock.
Supplementary Material
Acknowledgments
We thank F. Garcia-Olmedo, S. Herrero, J. Paz-Ares, and R. Sederoff for critical reading of the manuscript and J. M. Malpica for helpful comments. This work was supported by a grant (BIO02-805) and Predoctoral Fellowship from the Spanish Ministerio de Ciencia y Tecnología (to A.R.) and a grant (07M/0047/2000) and Postdoctoral Fellowship from the Comunidad Autónoma de Madrid (to E.P.-S.). C.I. is a Predoctoral Fellow of the Chilean Government-Banco Interamericano de Desarrollo.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SD, short day; LD, long day; LL, continuous light; PRR, pseudoresponse regulator.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY611028 (CsTOC1) and AY611029 (CsLHY)].
References
- 1.Lang, G. A. (1987) HortScience 22, 817–820. [Google Scholar]
- 2.Rhode, A., Howe, G. T., Olsen, J. E., Moritz, T., Van Montagu, M., Junttila, O. & Boerjan, W. (2000) in Molecular Biology of Woody Plants, eds. Jain, S. M. & Minocha, S. C. (Kluwer, Dordrecht, The Netherlands), Vol. 1, pp. 89–134. [Google Scholar]
- 3.Wareing, P. F. (1956) Annu. Rev. Plant Physiol. 7, 191–214. [Google Scholar]
- 4.Thomas, B. & Vince-Prue, D. (1997) in Photoperiodism in Plants (Academic, London), 2nd Ed.
- 5.Olsen, J. E., Junttila, O., Nilsen, J., Eriksson, M. E., Martinussen, I., Olsson, O., Sandberg, G. & Moritz, T. (1997) Plant J. 12, 1339–1350. [Google Scholar]
- 6.Welling, A., Moritz, T., Palva, T. & Junttila, O. (2002) Plant Physiol. 129, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiser, C. J. (1970) Science 169, 1269–1278. [DOI] [PubMed] [Google Scholar]
- 8.Howe, G. T., Davis, J., Jeknic, Z., Chen, T. H. H., Frewen, B., Bradshaw, H. D. & Saruul, P. (1999) HortScience 34, 1174–1184. [Google Scholar]
- 9.Arora, R., Rowland, L. J. & Tanino, K. (2003) HortScience 38, 911–921. [Google Scholar]
- 10.Horvath, D. P., Anderson, J. V., Chao, W. S. & Foley, M. E. (2003) Trends Plant Sci. 8, 534–540. [DOI] [PubMed] [Google Scholar]
- 11.Yanovsky, M. J. & Kay, S. (2003) Nat. Rev. Mol. Cell. Biol. 4, 265–275. [DOI] [PubMed] [Google Scholar]
- 12.Hayama, R. & Coupland, G. (2003) Curr. Opin. Plant Biol. 6, 13–19. [DOI] [PubMed] [Google Scholar]
- 13.Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F. & Coupland, G. (2001) Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- 14.Yanovsky, M. J. & Kay, S. A. (2002) Nature 419, 308–312. [DOI] [PubMed] [Google Scholar]
- 15.Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A. & Coupland, G. (2004) Nature 303, 1003–1006. [DOI] [PubMed] [Google Scholar]
- 16.Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Más, P., Panda, S., Kreps, J. A. & Kay, S. A. (2000) Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- 17.Makino, S., Kiba, T., Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Ueguchi, C., Sugiyama, T. & Mizuno, T. (2000) Plant Cell Physiol. 41, 791–803. [DOI] [PubMed] [Google Scholar]
- 18.Yanovsky, M. J. & Kay, S. (2001) Curr. Opin. Plant Biol. 4, 429–435. [DOI] [PubMed] [Google Scholar]
- 19.Alabadí, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Más, P. & Kay, S. A. (2001) Science 293, 880–883. [DOI] [PubMed] [Google Scholar]
- 20.Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I. A. & Coupland, G. (1998) Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- 21.Wang, Z. Y. & Tobin, E. M. (1998) Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- 22.Alabadí, D., Yanovsky, M. J., Más, P., Harmer, S. L. & Kay, S. A. (2002) Curr. Biol. 12, 757–761. [DOI] [PubMed] [Google Scholar]
- 23.Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H-R., Carré, I. A. & Coupland, G. (2002) Dev. Cell 2, 629–641. [DOI] [PubMed] [Google Scholar]
- 24.Strauss, W. M. (1987) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A. & Struhl, K. (Wiley, New York), pp. 6.3.1–6.3.6.
- 25.Makino, S., Matsushika, A., Kojima, M., Yamashino, T. & Mizuno, T. (2002) Plant Cell Physiol. 43, 58–69. [DOI] [PubMed] [Google Scholar]
- 26.Chang, S., Puryear, J. & Cairney, J. (1993) Plant Mol. Biol. Rep. 11, 113–116. [Google Scholar]
- 27.Allona, I., Quinn, M., Shoop, E., Swope, K., St. Cyr, S., Carlis, J., Riedl, J., Retzel, E., Campbell, M. M., Sederoff, R. & Whetten, R. W. (1998) Proc. Natl. Acad. Sci. USA 95, 9693–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushika, A., Makino, S., Kojima, M. & Mizuno, T. (2000) Plant Cell Physiol. 41, 1002–1012. [DOI] [PubMed] [Google Scholar]
- 29.Kaldis, A. D., Kousidis, P., Kesanopoulus, K. & Prombona, A. (2003) Plant Mol. Biol. 52, 981–997. [DOI] [PubMed] [Google Scholar]
- 30.So, W. V. & Rosbash, M. (1997) EMBO J. 16, 7146–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanewsky, R., Lynch, K. S., Brandes, C. & Hall, J. C. (2002) J. Biol. Rhythms 17, 293–306. [DOI] [PubMed] [Google Scholar]
- 32.Roden, L. C., Song, H. R., Jackson, S., Morris, K. & Carré, I. A. (2002) Proc. Natl. Acad. Sci. USA 99, 13313–13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thain, S. C., Hall, A. & Millar, A. J. (2000) Curr. Biol. 10, 951–956. [DOI] [PubMed] [Google Scholar]
- 34.Kreps, J. A. & Simon, A. E. (1997) Plant Cell 9, 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martino-Catt, S. & Ort, D. R. (1992) Proc. Natl. Acad. Sci. USA 89, 3731–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, T. L., Tucker, D. E. & Ort, D. R. (1998) Plant Physiol. 118, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howe, G. T., Saruul, P., Davis, J. & Chen, T. H. H. (2000) Theor. Appl. Genet. 101, 632–642. [Google Scholar]
- 38.Frewen, B. E., Chen, T. H., Howe, G. T., Davis, J., Rohde, A., Boerjan, W. & Bradshaw, H. D. (2000) Genetics 154, 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puhakainen, T., Li, C., Boije-Malm, M., Kangasjärvi, J., Heino, P. & Palva, E. T. (2004) Plant Physiol. 136, 4299–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhode, A., Prinsen, E., De Rycke, R., Engler, G., Van Montagu, M. & Boerjan, W. (2002) Plant Cell 14, 1885–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurup, S., Jones, H. D. & Holdsworth, M. J. (2000) Plant J. 21, 143–155. [DOI] [PubMed] [Google Scholar]
- 42.Harmer, S. L., Hogenesch, J. B., Straume, M., Chang, H. S., Han, B., Zhu, T., Wang, X., Kreps, J. A. & Kay, S. A. (2000) Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- 43.Schaffer, R., Landgraf, J., Accerbi, M., Simon, V., Larson, M. & Wisman, E. (2001) Plant Cell 13, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michael, T. P. & McClung, C. R. (2003) Plant Physiol. 132, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry, T. O. (1971) Science 171, 29–36. [DOI] [PubMed] [Google Scholar]
- 46.Chouard, P. (1960) Annu. Rev. Plant Physiol. 11, 191–238. [Google Scholar]
- 47.Sung, S. & Amasino, R. M. (2004) Curr. Opin. Plant Biol. 7, 4–10. [DOI] [PubMed] [Google Scholar]
- 48.Henderson, I. R., Shindo, C. & Dean, C. (2003) Annu. Rev. Genet. 37, 371–392. [DOI] [PubMed] [Google Scholar]
- 49.Thomashow, M. F. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- 50.Shinozaki, K., Yamaguchi-Shinozaki, K. & Seki, M. (2003) Curr. Opin. Plant Biol. 6, 410–417. [DOI] [PubMed] [Google Scholar]
- 51.Fowler, S. G., Cook, D. & Thomashow, M. F. (2005) Plant Physiol. 137, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.