Abstract
A fundamental appreciation for how biological macromolecules work requires knowledge of structure and dynamics. Molecular dynamics simulations provide powerful tools for the exploration of the conformational energy landscape accessible to these molecules, and the rapid increase in computational power coupled with improvements in methodology makes this an exciting time for the application of simulation to structural biology. In this Perspective we survey two areas, protein folding and enzymatic catalysis, in which simulations have contributed to a general understanding of mechanism. We also describe results for the F1 ATPase molecular motor and the Src family of signaling proteins as examples of applications of simulations to specific biological systems.
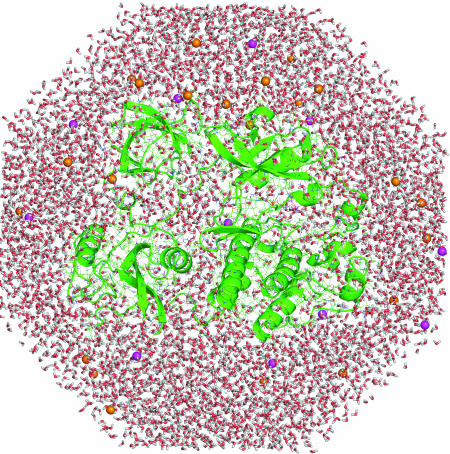
The conformational dynamics of protein molecules is encoded in their structures and is often a critical element of their function. A fundamental appreciation for how proteins work therefore requires an understanding of the connection between three-dimensional structure, obtained with increasing rapidity by x-ray crystallography and NMR, and dynamics, which is much more difficult to probe experimentally. Molecular dynamics simulations provide links between structure and dynamics by enabling the exploration of the conformational energy landscape accessible to protein molecules (1–3). The first molecular dynamics simulation of a protein was reported in 1977 and consisted of a 9.2-ps trajectory for a small protein in vacuum (4). Eleven years later, a 210-ps simulation of the same protein in water was reported (5), and the phenomenal increase in computing power since then now makes it routine to run simulations of much larger proteins that are 1,000–10,000 times as long as the original simulation (≈10–100 ns), in which the protein is surrounded by water and salt (Fig. 1). Significant improvements in the potential functions have also been achieved, making the simulations much more stable and accurate (6).
Fig. 1.
Simulation of a solvated protein. This slice through a simulation system shows a Src kinase protein (green) surrounded by ≈15,000 water molecules (oxygen atoms are red and hydrogen atoms are white). The simulation system consists of ≈50,000 atoms, including potassium and chloride ions (purple and orange spheres, respectively). A 1-ns molecular dynamics trajectory for this system can be generated in 4 days by using a cluster of four inexpensive Linux-based computers. (Courtesy of Olga Kuchment.)
The engineering principles underlying protein design are truly baroque, with evolutionary tinkering resulting in molecular machines that may be effective and efficient but whose three-dimensional structures are often so complicated as to obscure the mechanism of action. Clues to the essential dynamics of the molecule are quite often provided by the separation of the protein into domains connected by hinges and the availability of structures corresponding to different functional states, but it is not easy from visual inspection or simple calculations to deduce the workings of these molecular machines. Molecular dynamics simulations can provide the ultimate detail concerning individual atomic motions as a function of time; thus, they can be used to answer specific questions about the properties of a model system often more readily than experiments on the actual system. For many aspects of biomolecule function, these details are of interest (e.g., which of the many residues surrounding ATP in a motor protein are most important for coupling ATP binding to molecular movement).
The combination of increased computer power and improved potential functions has resulted in an ability to generate simulations that approach the point at which they can survive critical examination by the experimentalists who determine the structures of the proteins being simulated. For small proteins or protein domains that are not expected to undergo large conformational transitions (e.g., an SH2 domain bound to a phosphopeptide), a simulation lasting several nanoseconds in a fully solvated environment and using all-atom potentials without truncation of the electrostatic interactions will typically result in rms deviations from the crystal structure of ≈1 Å for the backbone atoms of the core region of the protein (7). Such simulations engender a degree of confidence that the interesting structural changes that result from the simulation of larger assemblies [e.g., intact Src kinases containing SH2 domains (8)] are meaningful, as we describe later.
Simulations are most effective when analyzed in close conjunction with experiments on protein function, which play an essential role in validating and improving the simulations (see, for example, refs. 9–12). One outcome of the increase in readily available computer power and the standardization of simulation protocols is that it is now becoming feasible for nonspecialists to reproduce and check some, if not all, of the conclusions of simulation analyses published by others. This kind of cross-validation will aid in the integration of simulations into the normal process of interpreting newly emerging structural results. A prescription for making the analysis of simulation results more rigorous and a list of possible errors has been provided (13).
In this Perspective, we illustrate the application of molecular dynamics simulations to biology by describing several selected examples, emphasizing our own work. We start with a broader survey of two areas, protein folding and enzymatic catalysis, in which simulations combined with experiment have led to a general understanding of mechanism. The explosive growth of structural results for an increasingly comprehensive range of biological processes demands simulations that address issues that are specific to particular systems. As examples, we describe simulation results for the F1 ATPase molecular motor and the Src family of signaling proteins.
Protein Folding
An understanding of how the newly synthesized polypeptide chain is able to fold to its native structure is fundamental to the description of life at a molecular level. This issue is made all the more critical because misfolding is involved in a range of diseases (14). A synergy between simulation and experiment has led to a conceptual solution for one aspect of the folding problem that is concerned with the mechanism by which the protein chain finds the native conformation despite the immensity of the potential search space. This advance has occurred despite the fact that folding is slow on the simulation time scale. (Even the fastest proteins take microseconds to fold, and the process cannot be simulated directly.) Most of the simulations of folding have used simplified models to capture the essential aspects of the process (15–17).
Key insights as to how the native structure can be found by the polypeptide chain in a reasonable time were obtained by using Monte Carlo dynamics for the folding of simplified bead models on a cubic lattice, a process fast enough so that the hundreds of trajectories needed to obtain statistically meaningful results could be calculated with the available computers (18). These lattice Monte Carlo studies showed that folding to the native state occurred on a time scale many orders of magnitude shorter than that required to sample all of the configurations (the so-called Levinthal time); e.g., with time measured in Monte Carlo steps, only ≈5 × 107 steps were required for folding a 27-bead polymer that had 1016 possible configurations. Although each folding trajectory is very different, the bias to the native state on the free energy surface is sufficient that the stochastic search samples only a small fraction of the total configurations in finding the native state. This principle is now generally accepted as the solution of the search problem in protein folding (17, 19), and the objective of present studies is concerned with the specifics of the folding of individual proteins and the ways that misfolding is avoided.
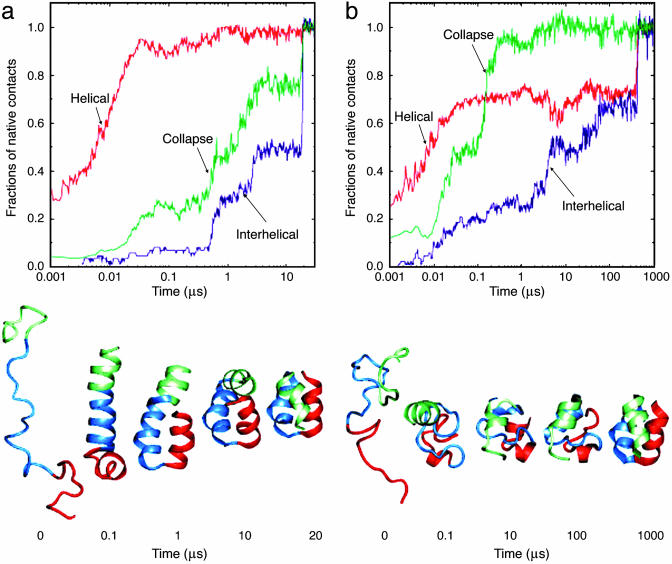
One such study of three-helix bundle proteins used a Cα model to represent the protein chain and a square well potential for the interactions between pairs of nonbonded residues. These simplifications made possible the use of discrete molecular dynamics algorithms for studying the folding process (20). The speed of the latter is such that several hundred folding trajectories could be calculated for different relative weights of native and nonnative interactions in the model potential function. Fig. 2 shows typical trajectories for a model with the native state strongly favored (Go-like potential) and a model in which nonnative interactions make a significant contribution along the folding pathway. In the former model, the helices form first and then diffuse to find the native fold [a limiting case of the diffusion–collision model (21)], whereas, in the latter model, there first is a collapse to a relatively disordered globule and the helices form simultaneously with the native tertiary structure. The model results correlate with recent folding experiments and all-atom (unfolding) simulations in explicit solvent (22). Interestingly, an extension of the discrete dynamics methodology has recently been used to simulate fibril formation from random coil peptides, illustrating a possible mechanism by which large, relatively well ordered β-sheet aggregates could appear in solution (23).
Fig. 2.
Simulations of the folding of a three-helix bundle protein. (a and b)(Upper) Semilog plots of the time dependence of the fractions of native helical and interhelical contacts and the inverse fractions of native volume (calculated from the inverse cube of the radius of gyration) for two different trajectories. (Lower) Structures of the protein molecule at selected times. [Reproduced with permission from ref. 20 (Copyright 1999, Nature Publishing Group).]
It is not yet possible to do statistically meaningful folding simulations of proteins with all-atom models. However, peptides composed of 20 or so residues with a well defined native structure are being simulated by molecular dynamics with implicit (15, 24, 25) and explicit (16) solvent. Although the peptides are small, the complexity of the simulations can be usefully interpreted in terms of disconnectivity graphical (24) and network (26) descriptions of the folding process. Given the increase in the speed of computers, particularly through the development of massively parallel machines, it is likely that within the next 10 years ab initio folding will reach the stage at which it can be used not only to determine the details of the folding mechanism but also to aid in solving the “other” folding problem, that of determining the structure of the native state directly from the sequence by molecular dynamics simulations.
Enzyme Catalysis
Enzyme catalysis can produce rate accelerations of a factor of 1019 (27). This process involves “molecular recognition” at the highest level; the catalysis of proton-transfer reactions, for example, requires the recognition of a change in a C—;H bond length of ≈0.5 Å in going from the reactant to the transition state. In 1946, before structural information was available, Linus Pauling (28) proposed that enzymes can accelerate reaction rates because they bind the transition state better than the substrate and thereby lower the activation free energy, ΔG‡. The validity of this key concept in enzyme catalysis has been confirmed by many studies (29, 30), although the dynamical contribution to the preexponential factor, A(T), in the Arrhenius expression k = A(T) exp(–ΔG‡/RT), where k is the rate constant, has to be considered as well (30).
Computer simulations are essential for understanding the lowering of the activation free energy in terms of the structure of the enzyme and its flexibility. The structure provides a “preorganized environment” (31) that enhances catalysis by a greater stabilization of the transition state than of the reactant state, with contributions from interactions with the bound substrate (32, 33) and from the enzyme itself. A certain degree of enzyme flexibility is essential for catalysis because atomic motions of the enzyme are required in all reactions (34). Moreover, larger scale motions also can be involved directly in catalysis and in providing a protected catalytic site for the reaction while permitting the substrate to enter and the product to escape. The changes in enzyme structure and vibrational modes associated with the progress along the reaction coordinate have been shown to promote catalysis most efficiently by lowering ΔG‡. This effect is distinct from the role of such motions in determining A(T).
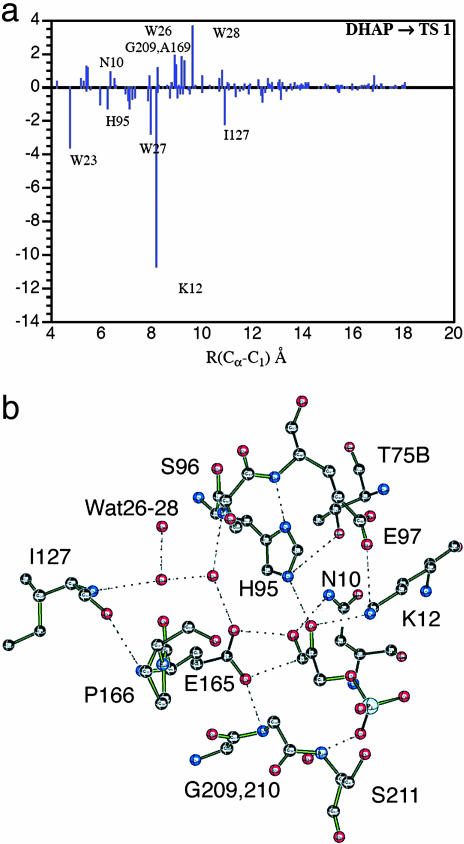
One of the enzymes that has been studied in detail experimentally and by simulations is triosephosphate isomerase (TIM), which catalyzes the conversion of dihydroxyacetone phosphate (DHAP) to (R)-glyceraldehyde 3-phosphate. The apparent barrier for the reaction in the enzyme has been calculated to be 11–13 kcal/mol (1 cal = 4.18 J) lower than that for the reaction in aqueous solution (35, 36). The rate-determining step is the transfer of a proton from DHAP to Glu-165 and the contributions obtained from a perturbation model (35) of individual residues to lowering the activation energy of this step are shown in Fig. 3A; the positions of important residues in the active site are shown in Fig. 3B. The charged residue Lys-12 makes the most important contribution, but the neutral His-95 side chain as well as certain main chain NH groups also contribute. This type of decomposition, which can be validated in part by experiment, also provides a basis for determining the evolutionary variation in ΔG‡ for the large number of available TIM sequences (M.K., unpublished data). Because water can lead to a side reaction, protection of the active site is achieved by a “lid” motion, which makes the active site of TIM accessible to the substrate but closes it off for catalysis (37, 38).
Fig. 3.
Mechanism of TIM. (a) Electrostatic contribution of individual residues (in kcal/mol on the ordinate) to the lowering of the activation energy barrier (TS1) of the reaction of the DHAP substrate to form the enolate intermediate. This is the rate-determining step of the overall chemical reaction. The residues are plotted on the abscissa as a function of the distance from the Cα carbon of the residue [or the oxygen of a water molecule (W)] to C1 of the substrate. Negative values correspond to the lowering of the barrier. (b) Active-site structure at transition state showing important residues and water molecules. [Reproduced with permission from ref. 30 (Copyright 2004, AAAS).]
Another enzyme that has been studied extensively is dihydrofolate reductase (DHFR), which catalyzes hydride transfer between nicotinamide adenine dinucleotide phosphate and 7,8-dihydrofolate. Experiments (39, 40) and simulations (41, 42) point to the fact that the reaction coordinate is complex, involving motions of parts of the enzyme distant from the active site. Although enzyme motion is involved, the catalysis is primarily due to the lowering of the activation barriers.
Dynamic effects associated with the preexponential factor, A(T), are normally very difficult to isolate experimentally. One contribution concerns the Eyring-type transmission coefficient, which can be less than unity because of the recrossing of the barrier. Simulations have shown that, although the recrossing factor is quantitatively interesting in terms of a full understanding of the reaction, the magnitude of its effect is generally small (no more than a factor of 2 or 3) as compared with other contributions (30). By contrast, tunneling can be more important in enzymatic reactions, particularly those involving the transfer of hydrogen (hydrogen atom, proton, or hydride ion) (43–45). Because inferences concerning this effect from isotope studies are indirect (46, 47), simulations have been important for a direct determination of the magnitude of the tunneling. At ambient temperatures, the calculated rate enhancements due to tunneling range from a factor of 1.5 for the intermolecular proton transfer in TIM (43) to 780 for soybean lipoxygenase (48). These rate accelerations are equivalent to thermodynamic free energy effects of 0.2–3.9 kcal/mol, a significant contribution but one that is still small compared with the lowering of the activation free energy by 10 kcal/mol or more. Detailed calculations of the tunneling contribution in DHFR are providing additional insights concerning its role in enzyme catalysis (49).
Molecular Dynamics Analyses of F1 ATPase
The enzyme Fo–F1 ATP synthase is perhaps the most remarkable of the molecular machines that have been studied at atomic resolution (Fig. 4A). This enzyme is central to life because it synthesizes ATP by harnessing the chemical potential of proton gradients across cell membranes. One component of Fo–F1 ATP synthase (Fo) is mainly within the membrane and is responsible for converting the chemical energy of the proton gradient into rotatory motion of a drive shaft that is located within the second component, which is known as F1 ATPase (50). The rotational motion of the drive shaft is converted by F1 ATPase into the production of ATP from ADP and Pi (inorganic phosphate, H2PO4–). The landmark crystal structures of F1 ATPase (51, 52), along with the results of single molecule studies (53) and more conventional biochemical experiments (54), have provided the basis for a series of simulations aimed at understanding how F1 ATPase works. Of particular interest is the demonstration that F1 ATPase, which by itself normally hydrolyzes ATP, can also synthesize ATP if the central shaft is rotated by external forces (55). High-resolution structures for the Fo component are not available at present, but this experiment demonstrates that it is reasonable to hope to understand how F1 ATPase synthesizes ATP without including the Fo component in simulations (but see ref. 56 for a simulation analysis of how Fo functions).
Fig. 4.
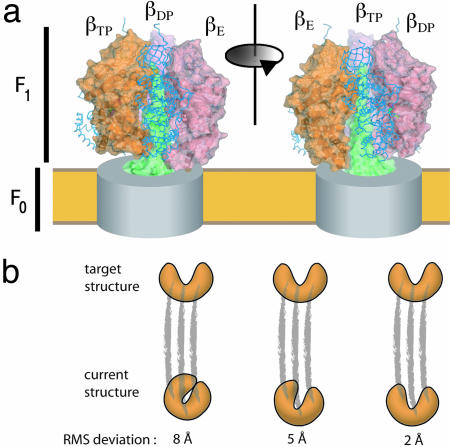
F1 ATPase and targeted molecular dynamics. (a) Structure of Fo–F1 ATP synthase based on crystal structures of the F1 ATPase (51, 52). The Fo component is indicated as a gray cylinder within the membrane (yellow), and the γ-subunit shaft is shown in green. The three α-subunits are indicated by blue backbone traces, and the molecular surfaces of the three β-subunits are shown. The γ-subunit rotates in a clockwise direction during ATP synthesis, as viewed from the membrane, and the effects of a 120° rotation of the γ-subunit are manifested as a change in the conformation of the β-subunits. (b) Schematic representation of targeted molecular dynamics. The simulated structure is restrained to be at a specified rms distance from the target structure, and this distance is gradually decreased during the simulation. Because the restraint is applied in an overall rms sense, the intermediate structures are not specified explicitly and different parts of the structure can relax toward the final structures at different rates.
The major structural element of F1 ATPase is a hexameric assembly of three α-subunits and three β-subunits surrounding the γ-subunit, which has a globular base and an extended coiled-coil domain (51) (see Fig. 4A). All six of the α- and β-subunits bind nucleotides, but only the three β-subunits are catalytically active. The crystal structures of F1 ATPase have proven to be enormously informative regarding the mechanism of the motor because each crystallographic snapshot provides views of three distinct states of the catalytic β-subunits (51, 52, 57). The centrally located and asymmetric γ-subunit forms a shaft, and its orientation determines the conformation of the β-subunits. The original crystal structure of F1 ATPase led to the identification of three conformations of the β-subunit: βE (empty), βTP (ATP bound), and βDP (ADP bound) (51). The open conformation of the βE-subunit is different from that of the βTP- and βDP-subunits, which are both closed and very similar to each other.
The prescient analysis of kinetic data by Paul Boyer (58) led to his proposal (in advance of structural information) of a remarkable and unprecedented “binding change mechanism” that described the action of F1 ATPase. In terms of this mechanism, ATP synthesis proceeds by the cyclical conversion of the β-subunit from an “open” state that binds ATP only weakly to a “loose” state that has higher affinity for ATP to a “tight” state that has the highest affinity for ATP. The crystal structure (51), with its three distinct conformations of the β-subunits and asymmetric disposition of the γ-subunit shaft, clearly supported the binding change mechanism proposed by Boyer (58).
Molecular dynamics simulations have made contributions to our understanding of two aspects of the mechanism of F1 ATPase. As is usually the case in crystallography, the structures are silent about the nature of the transitions from one state to the other and the forces involved. Simulations have been useful in piecing together at least some of what must happen as the motor moves through its duty cycle, and we discuss these studies first (59–61). The second issue concerns the identification of particular conformations of the β-subunit with the open, loose, and tight states of the binding change mechanism. It is apparent that the empty conformation (βE) is the low-affinity state of the β-subunit, but the βDP and βTP states are very similar to each other, and it has not been possible on the basis of experimental data alone to identify which of the two is the high-affinity binding site for ATP. Free energy difference simulations of nucleotide binding to the β-subunits have resolved these ambiguities (62), allowing the large body of experimental data on the complex to be integrated into a detailed kinetic scheme that models the catalysis of ATP hydrolysis by F1 ATPase in the absence of the Fo component (63).
Conformational Transitions in F1 ATPase. The time scale for one rotation of the γ shaft is in the microsecond to millisecond range (53) and is therefore not accessible to the submicrosecond time scales probed by standard molecular dynamics simulations. Because the beginning and end states corresponding to a 120° rotation of the γ-subunit are known (51), the challenge is essentially one of mapping a low-energy path that connects one stable conformation of the system to another, a general problem that is receiving considerable attention at present (64). The earliest attempt to characterize the intermediate structures in F1 ATPase used an interpolation method in a cyclindrical coordinate system defined by the rotation axis of the γ-subunit (65). These interpolated structures were recently used after local relaxation by molecular dynamics to analyze the breaking of interactions between the ATP and the protein as the system is driven from the βTP state to the βE state (59). More details concerning the conformational transitions in F1 ATPase were obtained by molecular dynamics simulations in the presence of biasing forces that were applied to the γ-subunit alone (60, 61) or to the entire structure in a procedure that drives the system from one state to the other without explicitly constraining the nature of the transition path (61). The spirit in which these simulations are done entails “pushing” on the molecular assembly (e.g., by forcing the γ-subunit to rotate) and analyzing how the rest of the structure responds (Fig. 4B). Because the time scale of the forced rotational transition of the γ-subunit is orders of magnitude faster than the actual rotation rate, the implicit assumption in such studies is that meaningful information concerning the mechanism can be still be obtained.
Both of these studies demonstrated that the rotation of the γ-shaft triggers the opening of the nucleotide-bound βTP-subunit and the closure of the open βE-subunit (60, 61). The importance of a track of ionic residues on the γ-shaft and on the inward-facing surfaces of the α- and β-subunits has been highlighted (61). These ionic residues provide a mechanism for smooth rotation of the γ-subunit by enabling the sequential handover of ion pairing interactions. The functional difference between the α- and β-subunits is demonstrated by smooth bypass of the α-subunits by the rotating γ-subunit. In contrast, interlocking elements from the γ- and β-subunits generate responses in the β-subunits as the γ-subunits move.
One interesting phenomenon that emerged from both sets of simulations (60, 61) is the rapid relaxation of the βE-subunit once the steric block imposed by the γ-subunit is removed by rotation. Although solution NMR data for the isolated βE-subunit indicate that the equilibrium is shifted toward the open conformation until the nucleotide binds, the rapid closure of the βE-subunit seen in the simulations with (61) and without (60) nucleotides is consistent with the results of normal mode calculations (66), which show that one of the lowest frequency (i.e., most readily excitable) normal modes of the β-subunit accounts for a significant fraction of the conformational difference between the open and closed forms of this subunit.
Free Energy Simulations of F1 ATPase. The simulations described so far do not address the energetics of ATP binding to the various states. The essential difficulty here arises from the striking similarity in structure between the βDP and βTP states (52, 57). A recently determined high-resolution structure of F1 ATPase has the ATP analog ADP.AlF4 bound at the βDP and the βTP sites, with tight coordination of the nucleotide in both cases (52). Which site features high-affinity ATP binding and which site more closely resembles the structure of the subunit when the chemical catalysis step occurs? Answers to these questions have been provided by free energy difference simulations in which ATP.H2O is converted computationally to ADP.Pi (62). These calculations, which are often referred to as “computational alchemy,” provide estimates of the standard free energy change, ΔGo, for the conversion of ATP into ADP at each of the occupied binding sites. This free energy change is calculated to be –9 kcal/mol in the βDP site (i.e., ADP is strongly favored over ATP at this site) and 1.5 kcal/mole at the βTP site (i.e., ATP and ADP have almost the same chemical potential at this site).
Solution measurements have shown that, under “unisite” hydrolysis conditions (i.e., when the ATP concentration is so low that only the strongest binding site is involved), the reaction free energy is near zero; the experimental value is 0.4 kcal/mol for the mitochondrial enzyme, whose structure has been determined, and –0.6 kcal/mol for the Escherichia coli enzyme (63). Given the free energy simulation results just cited, it is possible to identify the βTP as the strong binding site for ATP and the βDP site as the intermediate binding site. This “missing link” between the solution measurements and the crystal structure has made possible a complete assignment of the measured binding constants not only for ATP but also for ADP.Pi (62). Interestingly, it was also shown that the βDP site is the strong binding site for ADP.Pi (63). These results indicate that there are two major contributions to the driving force (actually a biasing free energy) that rotates the γ-subunit when ATP is hydrolysed by F1 ATPase. The first contribution is the increase in the binding free energy of ATP in going from the βE to the βTP state; this result is in agreement with the original model of Wang and Oster (65). However, the second contribution is a new result; it arises from the fact that once hydrolysis has taken place and ADP.Pi occupies the binding site, the conformation is biased toward βDP. A detailed kinetic model for ATP hydrolysis by F1 ATPase has been developed with these results and solution kinetic constants identified with the different β-subunits (63).
Dynamics of the Src Tyrosine Kinases
The Src tyrosine kinases are a family of closely related proteins that transmit signals initiated by growth factor receptors in human cells (67). A mutant and constitutively activated form of Src is encoded by the first oncogene to be discovered (v-Src; so named for the sarcomas caused by the expression of this retroviral gene). The Src proteins contain a tyrosine kinase domain that catalyzes the transfer of phosphate from ATP to tyrosine residues on substrate proteins. The inappropriate activation of tyrosine kinases such as Src can have deadly consequences in terms of the onset of cancers because phosphorylated tyrosines serve as targeting signals for SH2 domains in signaling proteins that control cell growth and differentiation (68).
The Src kinases themselves contain an SH2 domain and another peptide-binding module known as the SH3 domain, both of which are important for the maintenance of the inactive state of these proteins. Crystal structures of inactive Src kinases unexpectedly revealed that the SH2 and SH3 domains bind to the distal face of the kinase domain, rather than near the active site (69, 70). How do the SH2 and SH3 domains affect the catalytic activity of the kinase domain without being located near the active site? One possibility is that these domains affect the global dynamics of the protein, making it more difficult for the kinase domain to undergo the change in conformation from the observed inactive state (69, 70) to the structurally different active form (71).
Molecular dynamics simulations of the Src kinases c-Src and Hck have revealed an unexpected feature of the SH2 and SH3 domains that may be a key to the regulation of the Src kinases (8). Before the simulation analysis, the SH2 and SH3 domains were considered to be flexibly linked and independently functioning modules (72, 73). Thus, it came as a surprise that the simulations showed the SH2–SH3 unit of the assembled Src proteins to be coupled together relatively rigidly. Unbiased simulations extending for several nanoseconds revealed that the SH2 and SH3 domains tended to move together as a unit, with the linker between them playing a particularly important role in clamping these domains to the kinase domain. In contrast, NMR studies of isolated SH2–SH3 units of Src kinases had shown that these domains are linked together flexibly (73). Indeed, simulations of isolated SH2–SH3 domains do reveal them to be extremely flexible in terms of their relative orientation (8), suggesting that the linker between them functions as an “inducible snap lock,” a term introduced by Wright and coworkers (74) to describe flexible connections between certain zinc finger modules that snap into rigidity when the zinc fingers bind to their cognate DNA recognition elements. In the Src proteins, the linker between the SH2 and SH3 domains is stabilized in the assembled and inactive state by several hydrogen bonds. When released from the interactions with the kinase domain, simulations show that these hydrogen bonds are broken by water molecules and the linker becomes flexible (8).
Targeted molecular dynamics simulations in which an “activation loop” that is located near the active site of the kinase domain is driven from the inactive conformation to the active conformation while leaving the SH2 and SH3 domains unrestrained showed that movements at the active site of the kinase reverberate through to the SH2 domain, even though it is located >40 Å away (8). The coupling between the SH2 and SH3 domains is the key to this intersite communication in Src and underlies the importance of the phosphotyrosine–SH2 linkage at the base of the distal surface of the kinase domain. If this linkage is broken, as in the form of the protein produced by the v-Src oncogene, the kinase activity of Src is turned on constitutively (67). The molecular dynamics simulations show that the residues in the SH2 domain that interact with the phosphotyrosine residue in the tail move significantly when the activation loop in the kinase domain is displaced. The SH2–phosphotyrosine linkage would be expected to resist these movements of the activation loop and, thereby, to impede the activation process.
The importance of rigidity in the SH2–SH3 linker was tested experimentally and computationally (8). Simulations in which linker residues were replaced by glycine caused the SH2 and SH3 domains to become less tightly coupled. Introduction of corresponding mutations into Src proteins in cell-based assays led to their constitutive activation, bypassing the inhibitory action of the distal SH2–phosphotyrosine linkage (8). We note that these mutations were designed only as a consequence of the molecular dynamics simulations, despite years of prior experimental investigation into Src function.
The Abelson (Abl) Kinase and the Specificity of Imatinib Mesylate (Gleevec)
An interesting corroboration of these results from molecular dynamics was provided by a subsequent structural analysis of the Abl tyrosine kinase (75). A close relative of the Src kinases, Abl contains SH2 and SH3 domains that are similar in sequence to those of Src, and, although it lacks the tyrosine residue that anchors the SH2 domain to the kinase in Src when phosphorylated, the overall structure of autoinhibited Abl is similar to that of Src (75). Molecular dynamics simulations of Abl carried out as for Src suggested that these domains form a similar snap lock in Abl (75). As in Src, mutations in the SH2–SH3 linker activate the Abl kinase (76, 77). The difference is that the SH2 domain in Abl is anchored to the kinase domain not by a phosphotyrosine-mediated linkage, which keeps the SH2 and kinase domains apart in Src, but through an intimate and direct interface that forms between the SH2 domain and the kinase domain.
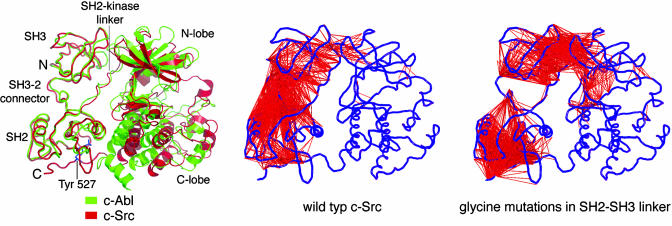
The Abl kinase domain is the target of the successful cancer drug Gleevec, which works by blocking the catalytic activity of a form of the Abl protein that is activated in chronic myelogenous leukemia (78). Gleevec binds with high affinity to the inactive Abl kinase domain but not to that of Src, even though all of the residues that make contact with the drug in Abl are conserved in Src (79, 80). The striking feature of the structures of the inactive forms of Abl and Src is that it is the conformation of the SH2–SH3 unit and not that of the kinase domain that is conserved between the two proteins (Fig. 5). It appears that the rigidity of the SH2–SH3 unit, which is a feature of the Src and Abl simulations (8, 75), is manifested as a conservation of the structure of the SH2–SH3 unit in the inactive states of both proteins. Because the SH2 domain in Abl is closer to the kinase domain than in Src, the SH3 domain in the rigid SH2–SH3 unit swings away, and the kinase domain adjusts its conformation so as to preserve the interaction with the SH3 domain. As a result, the kinase domain of Abl is more open, and it is this difference in conformation to which Gleevec is sensitive.
Fig. 5.
Structure and dynamics of the Src and Abl kinases. (Left) The structures of c-Abl (green) and c-Src (red) are shown superimposed on their SH2 and SH3 domains (69, 70, 75). Note the dissimilarity in the conformation of the kinase domains. (Center and Right) The results of unbiased molecular dynamics simulations of c-Src. Residues in different domains that move in a correlated manner in the simulation are linked by a red line. These correlations were calculated by superimposing each instantaneous structure in the simulation on the C-terminal lobe of the kinase domain, and motions that are correlated to the C-terminal lobe are removed by this procedure. (Right) The mutation of residues in the SH2–SH3 linker to glycine reduces the correlation in the dynamics of these domains. Similar results were obtained for c-Abl. (Modified from refs. 8 and 75).
Future Prospects
It is now often the case that experimental structures are available for more than one functional state of a molecular assembly, so that the problem of finding transition paths between low-energy regions of a complex conformational space is becoming increasingly important. Despite the difficulties inherent in the difference between the experimental and simulation time scales, it is likely that creative solutions to the problem will be found, as illustrated by the recent application of a transition path sampling method (64) to the problem of fidelity checking by DNA polymerases (81). It will be particularly interesting to be able to calculate the free energy barriers to conformational transitions, which has not as yet been widely done for protein conformational changes because of the difficulties in specifying the reaction coordinate for the transition. A prominent exception occurs in the study of ion channels, for which the reaction coordinate is essentially defined by the system (82). Alternatively, the generation of multiple targeted molecular dynamics trajectories could be used to extract the free energy directly (83).
It is to be hoped that experimental structural biologists, who know their systems better than anyone else, will make increasing use of molecular dynamics simulations for obtaining a deeper understanding of particular biological systems. When molecular dynamics simulations are a routine part of structural biology, it will become clearer what refinements and extensions of the methodology are most needed to improve the results and to perfect the constructive interplay between the simulations and experiment. These conclusions will, in turn, provide challenges for the simulation experts and catalyze new developments in the field. In addition to reaction path search methods, improved treatments of solvent by implicit methods (84) are likely to play a role here. Although simulations with explicit water molecules are the gold standard, implicit solvent models make possible the testing of hypotheses by repeated simulations (e.g., with different mutants and/or with different constraints) for larger systems. Given the availability of several molecular dynamics programs, large amounts of computer time, and examples for which molecular dynamics has really played a role in furthering our understanding of protein functions, we look forward to a wide field of biological applications for molecular dynamics in the future.
Acknowledgments
We thank the National Energy Research Scientific Computing Center for supercomputing resources and Lore Leighton for preparing Fig. 4. J.K. thanks Matthew Young for many stimulating discussions regarding molecular dynamics. M.K. thanks M. Viloca-Garcia, J. Gao, and D. G. Truhlar for helpful discussions and collaboration on ref. 30, which forms the basis of the enzyme kinetics text.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Karplus, M. & McCammon, J. A. (2002) Nat. Struct. Biol. 9, 646–652. [DOI] [PubMed] [Google Scholar]
- 2.Wang, W., Donini, O., Reyes, C. M. & Kollman, P. A. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 211–243. [DOI] [PubMed] [Google Scholar]
- 3.Hansson, T., Oostenbrink, C. & van Gunsteren, W. (2002) Curr. Opin. Struct. Biol. 12, 190–196. [DOI] [PubMed] [Google Scholar]
- 4.McCammon, J. A., Gelin, B. R. & Karplus, M. (1977) Nature 267, 585–590. [DOI] [PubMed] [Google Scholar]
- 5.Levitt, M. & Sharon, R. (1988) Proc. Natl. Acad. Sci. USA 85, 7557–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackerell, A. D., Jr. (2004) J. Comput. Chem. 25, 1584–1604. [DOI] [PubMed] [Google Scholar]
- 7.Price, D. J. & Brooks, C. L., III (2002) J. Comput. Chem. 23, 1045–1057. [DOI] [PubMed] [Google Scholar]
- 8.Young, M. A., Gonfloni, S., Superti-Furga, G., Roux, B. & Kuriyan, J. (2001) Cell 105, 115–126. [DOI] [PubMed] [Google Scholar]
- 9.Case, D. A. (2002) Acc. Chem. Res. 35, 325–331. [DOI] [PubMed] [Google Scholar]
- 10.Soares, T. A., Daura, X., Oostenbrink, C., Smith, L. J. & van Gunsteren, W. F. (2004) J. Biomol. NMR 30, 407–422. [DOI] [PubMed] [Google Scholar]
- 11.Krieger, E., Darden, T., Nabuurs, S. B., Finkelstein, A. & Vriend, G. (2004) Proteins 57, 678–683. [DOI] [PubMed] [Google Scholar]
- 12.Horita, D. A., Zhang, W., Smithgall, T. E., Gmeiner, W. H. & Byrd, R. A. (2000) Protein Sci. 9, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Gunsteren, W. F. & Mark, A. E. (1998) J. Chem. Phys. 108, 6109–6116. [Google Scholar]
- 14.Dobson, C. M. (2003) Nature 426, 884–890. [DOI] [PubMed] [Google Scholar]
- 15.Cavalli, A., Ferrara, P. & Caflisch, A. (2002) Proteins 47, 305–314. [DOI] [PubMed] [Google Scholar]
- 16.Simmerling, C., Strockbine, B. & Roitberg, A. E. (2002) J. Am. Chem. Soc. 124, 11258–11259. [DOI] [PubMed] [Google Scholar]
- 17.Wolynes, P. G. (2005) Philos. Trans. R. Soc. London A 363, 453–467. [DOI] [PubMed] [Google Scholar]
- 18.Dobson, C. M., Sali, A. & Karplus, M. (1998) Angew. Chem. 37, 868–893. [DOI] [PubMed] [Google Scholar]
- 19.Karplus, M. (1997) Fold Des. 2, S69–S75. [DOI] [PubMed] [Google Scholar]
- 20.Zhou, Y. & Karplus, M. (1999) Nature 401, 400–403. [DOI] [PubMed] [Google Scholar]
- 21.Islam, S. A., Karplus, M. & Weaver, D. L. (2002) J. Mol. Biol. 318, 199–215. [DOI] [PubMed] [Google Scholar]
- 22.Gianni, S., Guydosh, N. R., Khan, F., Caldas, T. D., Mayor, U., White, G. W., DeMarco, M. L., Daggett, V. & Fersht, A. R. (2003) Proc. Natl. Acad. Sci. USA 100, 13286–13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen, H. D. & Hall, C. K. (2004) Proc. Natl. Acad. Sci. USA 101, 16180–16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krivov, S. V. & Karplus, M. (2004) Proc. Natl. Acad. Sci. USA 101, 14766–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, R. (2003) Proteins 53, 148–161. [DOI] [PubMed] [Google Scholar]
- 26.Rao, F. & Caflisch, A. (2004) J. Mol. Biol. 342, 299–306. [DOI] [PubMed] [Google Scholar]
- 27.Wolfenden, R. & Snider, M. J. (2001) Acc. Chem. Res. 34, 938–945. [DOI] [PubMed] [Google Scholar]
- 28.Pauling, L. (1946) Chem. Eng. News 24, 1375–1377. [Google Scholar]
- 29.Schowen, R. L. (1978) in Transition States of Biochemical Processes, eds. Gandour, R. D. & Schowen, R. L. (Plenum, New York), pp. 77–114.
- 30.Garcia-Viloca, M., Gao, J., Karplus, M. & Truhlar, D. G. (2004) Science 303, 186–195. [DOI] [PubMed] [Google Scholar]
- 31.Villa, J. & Warshel, A. (2001) J. Phys. Chem. B 105, 7887–7907. [Google Scholar]
- 32.Dinner, A. R., Blackburn, G. M. & Karplus, M. (2001) Nature 413, 752–755. [DOI] [PubMed] [Google Scholar]
- 33.Fromme, J. C., Bruner, S. D., Yang, W., Karplus, M. & Verdine, G. L. (2003) Nat. Struct. Biol. 10, 204–211. [DOI] [PubMed] [Google Scholar]
- 34.Brooks, C. L., Karplus, M. & Pettitt, B. M. (1988) Proteins: A Theoretical Perspective of Dynamics, Structure and Thermodynamics (Wiley, New York).
- 35.Cui, Q. & Karplus, M. (2002) J. Phys. Chem. B 106, 1768–1798. [Google Scholar]
- 36.Feierberg, I. & Åqvist, J. (2002) Theor. Chem. Acc. 108, 71–84. [Google Scholar]
- 37.Joseph, D., Petsko, G. A. & Karplus, M. (1990) Science 249, 1425–1428. [DOI] [PubMed] [Google Scholar]
- 38.Kursula, I., Salin, M., Sun, J., Norledge, B. V., Haapalainen, A. M., Sampson, N. S. & Wierenga, R. K. (2004) Protein Eng., Des. Sel. 17, 375–382. [DOI] [PubMed] [Google Scholar]
- 39.Schnell, J. R., Dyson, H. J. & Wright, P. E. (2004) Annu. Rev. Biophys. Biomol. Struct. 33, 119–140. [DOI] [PubMed] [Google Scholar]
- 40.Sawaya, M. R. & Kraut, J. (1997) Biochemistry 36, 586–603. [DOI] [PubMed] [Google Scholar]
- 41.Rod, T. H., Radkiewicz, J. L. & Brooks, C. L., III (2003) Proc. Natl. Acad. Sci. USA 100, 6980–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watney, J. B., Agarwal, P. K. & Hammes-Schiffer, S. (2003) J. Am. Chem. Soc. 125, 3745–3750. [DOI] [PubMed] [Google Scholar]
- 43.Cui, Q. & Karplus, M. (2002) J. Am. Chem. Soc. 124, 3093–3124. [DOI] [PubMed] [Google Scholar]
- 44.Billeter, S. R., Webb, S. P., Iordanov, T., Agarwal, P. K. & Hammes-Schiffer, S. (2001) J. Chem. Phys. 114, 341–349. [DOI] [PubMed] [Google Scholar]
- 45.Truhlar, D. G., Gao, J., Alhambra, C., Garcia-Viloca, M., Corchado, J., Sanchez, M. L. & Villa, J. (2002) Acc. Chem. Res. 35, 341–349. [DOI] [PubMed] [Google Scholar]
- 46.Jonsson, T., Glickman, M. H., Sun, S. J. & Klinman, J. P. (1996) J. Am. Chem. Soc. 118, 10319–10320. [Google Scholar]
- 47.Basran, J., Sutcliffe, M. J. & Scrutton, N. S. (1999) Biochemistry 38, 3218–3222. [DOI] [PubMed] [Google Scholar]
- 48.Tresadern, G., McNamara, J. P., Mohr, M., Wang, H., Burton, N. A. & Hillier, I. A. (2002) Chem. Phys. Lett. 358, 489–494. [Google Scholar]
- 49.Hammes-Schiffer, S. (2004) Curr. Opin. Struct. Biol. 14, 192–201. [DOI] [PubMed] [Google Scholar]
- 50.Boyer, P. D. (1997) Annu. Rev. Biochem. 66, 717–749. [DOI] [PubMed] [Google Scholar]
- 51.Abrahams, J. P., Leslie, A. G., Lutter, R. & Walker, J. E. (1994) Nature 370, 621–628. [DOI] [PubMed] [Google Scholar]
- 52.Menz, R. I., Walker, J. E. & Leslie, A. G. (2001) Cell 106, 331–341. [DOI] [PubMed] [Google Scholar]
- 53.Kinosita, K., Jr., Adachi, K. & Itoh, H. (2004) Annu. Rev. Biophys. Biomol. Struct. 33, 245–268. [DOI] [PubMed] [Google Scholar]
- 54.Weber, J. & Senior, A. E. (2003) FEBS Lett. 545, 61–70. [DOI] [PubMed] [Google Scholar]
- 55.Itoh, H., Takahashi, A., Adachi, K., Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K. (2004) Nature 427, 465–468. [DOI] [PubMed] [Google Scholar]
- 56.Aksimentiev, A., Balabin, I. A., Fillingame, R. H. & Schulten, K. (2004) Biophys. J. 86, 1332–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kagawa, R., Montgomery, M. G., Braig, K., Leslie, A. G. & Walker, J. E. (2004) EMBO J. 23, 2734–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyer, P. D. (1993) Biochim. Biophys. Acta 1140, 215–250. [DOI] [PubMed] [Google Scholar]
- 59.Antes, I., Chandler, D., Wang, H. & Oster, G. (2003) Biophys. J. 85, 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bockmann, R. A. & Grubmuller, H. (2002) Nat. Struct. Biol. 9, 198–202. [DOI] [PubMed] [Google Scholar]
- 61.Ma, J., Flynn, T. C., Cui, Q., Leslie, A. G., Walker, J. E. & Karplus, M. (2002) Structure (Cambridge, MA, U. S.) 10, 921–931. [DOI] [PubMed] [Google Scholar]
- 62.Yang, W., Gao, Y. Q., Cui, Q., Ma, J. & Karplus, M. (2003) Proc. Natl. Acad. Sci. USA 100, 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao, Y. Q., Yang, W., Marcus, R. A. & Karplus, M. (2003) Proc. Natl. Acad. Sci. USA 100, 11339–11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolhuis, P. G., Chandler, D., Dellago, C. & Geissler, P. L. (2002) Annu. Rev. Phys. Chem. 53, 291–318. [DOI] [PubMed] [Google Scholar]
- 65.Wang, H. & Oster, G. (1998) Nature 396, 279–282. [DOI] [PubMed] [Google Scholar]
- 66.Cui, Q., Li, G., Ma, J. & Karplus, M. (2004) J. Mol. Biol. 340, 345–372. [DOI] [PubMed] [Google Scholar]
- 67.Martin, G. S. (2004) Oncogene 23, 7910–7917. [DOI] [PubMed] [Google Scholar]
- 68.Pawson, T. & Nash, P. (2003) Science 300, 445–452. [DOI] [PubMed] [Google Scholar]
- 69.Sicheri, F., Moarefi, I. & Kuriyan, J. (1997) Nature 385, 602–609. [DOI] [PubMed] [Google Scholar]
- 70.Xu, W., Harrison, S. C. & Eck, M. J. (1997) Nature 385, 595–602. [DOI] [PubMed] [Google Scholar]
- 71.Yamaguchi, H. & Hendrickson, W. A. (1996) Nature 384, 484–489. [DOI] [PubMed] [Google Scholar]
- 72.Engen, J. R., Smithgall, T. E., Gmeiner, W. H. & Smith, D. L. (1999) J. Mol. Biol. 287, 645–656. [DOI] [PubMed] [Google Scholar]
- 73.Arold, S. T., Ulmer, T. S., Mulhern, T. D., Werner, J. M., Ladbury, J. E., Campbell, I. D. & Noble, M. E. (2001) J. Biol. Chem. 276, 17199–17205. [DOI] [PubMed] [Google Scholar]
- 74.Laity, J. H., Dyson, H. J. & Wright, P. E. (2000) J. Mol. Biol. 295, 719–727. [DOI] [PubMed] [Google Scholar]
- 75.Nagar, B., Hantschel, O., Young, M. A., Scheffzek, K., Veach, D., Bornmann, W., Clarkson, B., Superti-Furga, G. & Kuriyan, J. (2003) Cell 112, 859–871. [DOI] [PubMed] [Google Scholar]
- 76.Hantschel, O., Nagar, B., Guettler, S., Kretzschmar, J., Dorey, K., Kuriyan, J. & Superti-Furga, G. (2003) Cell 112, 845–857. [DOI] [PubMed] [Google Scholar]
- 77.Brasher, B. B., Roumiantsev, S. & Van Etten, R. A. (2001) Oncogene 20, 7744–7752. [DOI] [PubMed] [Google Scholar]
- 78.Druker, B. J. & Lydon, N. B. (2000) J. Clin. Invest. 105, 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schindler, T., Bornmann, W., Pellicena, P., Miller, W. T., Clarkson, B. & Kuriyan, J. (2000) Science 289, 1938–1942. [DOI] [PubMed] [Google Scholar]
- 80.Nagar, B., Bornmann, W. G., Pellicena, P., Schindler, T., Veach, D. R., Miller, W. T., Clarkson, B. & Kuriyan, J. (2002) Cancer Res. 62, 4236–4243. [PubMed] [Google Scholar]
- 81.Radhakrishnan, R. & Schlick, T. (2004) Proc. Natl. Acad. Sci. USA 101, 5970–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berneche, S. & Roux, B. (2001) Nature 414, 73–77. [DOI] [PubMed] [Google Scholar]
- 83.Hummer, G. & Szabo, A. (2001) Proc. Natl. Acad. Sci. USA 98, 3658–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feig, M. & Brooks, C. L., III (2004) Curr. Opin. Struct. Biol. 14, 217–224. [DOI] [PubMed] [Google Scholar]