Abstract
Initially identified in Caenorhabditis elegans Lin-2 and Lin-7, L27 domain is a protein–protein interaction domain capable of organizing scaffold proteins into supramolecular assemblies by formation of heteromeric L27 domain complexes. L27 domain-mediated protein assemblies have been shown to play essential roles in cellular processes including asymmetric cell division, establishment and maintenance of cell polarity, and clustering of receptors and ion channels. The structural basis of L27 domain heteromeric complex assembly is controversial. We determined the high-resolution solution structure of the prototype L27 domain complex formed by mLin-2 and mLin-7 as well as the solution structure of the L27 domain complex formed by Patj and Pals1. The structures suggest that a tetrameric structure composed of two units of heterodimer is a general assembly mode for cognate pairs of L27 domains. Structural analysis of the L27 domain complex structures further showed that the central four-helix bundles mediating tetramer assembly are highly distinct between different pairs of L27 domain complexes. Biochemical studies revealed that the C-terminal α-helix responsible for the formation of the central helix bundle is a critical specificity determinant for each L27 domain in choosing its binding partner. Our results provide a unified picture for L27 domain-mediated protein–protein interactions.
Keywords: Lin-2, Lin-7, cell polarity
Polarization of cells is a fundamental process for all eukaryotes. In the metazoan, establishment of cell polarity is essential for the asymmetric cell divisions that specify different cell fates at the early stages of development. Cell polarity is also functionally indispensable after cell fate specification. Genetic and molecular studies have identified distinct groups of genes that function coordinately to establish cell polarity (see refs. 1 and 2 for recent reviews). The products of many of these genes are multidomain scaffold proteins, and they often interact with each other to form large protein complexes that mediate processes including assembly of cell junctions, organization of signal transduction complexes, regulation of cytoskeletal dynamics, and asymmetric trafficking of cell fate determinants (3).
L27 domain, initially identified in the Caenorhabditis elegans Lin-2 and Lin-7 proteins, is a previously unrecognized protein interaction module that exists in a large family of scaffold proteins (4). L27 domain-containing proteins are emerging as multidomain scaffold proteins that play critical roles in cell polarity. Formation of the evolutionarily conserved Lin-2·Lin-7·Lin-10 ternary complex requires heterodimerization of L27 domains (5, 6) and has been shown to play a central role in targeting receptor tyrosine kinase Let-23 signalsome to the basolateral surface of vulval precursor cells (7, 8). The mammalian counterparts of Lin-2, Lin-7, and Lin-10 are mLin-2/CASK, mLin-7/Velis/Mals, and mLin-10/X11α/Mint1, respectively (9–11). The L27 domain-mediated formation of the mLin-2·mLin-7·mLin-10 tripartite complex was observed in the brain (5, 9) and implicated in the targeting of NMDA receptors in neurons (12). Recently, it was shown that the assembly of another set of evolutionarily conserved cell polarity tripartite complexes, composed of Pals1·Patj·Crumbs, is also mediated by L27 domains (13–16).
The 3D structure of the tetrameric L27 domain complex formed by the L27 domain of SAP97 and the N-terminal L27 domain of mLin-2 showed that each L27 domain contains three α-helices. The two N-terminal helices (αA and αB) of each L27 domain pack together to form a tight, four-helix bundle in the heterodimer. The third helix (αC) of each L27 domain forms another four-helix bundle that assembles the two units of the heterodimer into a tetramer (17). Formation of heteromeric L27 domain complexes are highly specific (15, 17, 18). A subsequent structural study of the L27 domain complex formed by the L27 domain of Patj and the N-terminal L27 domain of Pals1 also showed that the packing of the two N-terminal helices from each L27 domain into a four-helix bundle is a general mechanism for L27 heterodimer formation (18). Despite the availability of the two pairs of L27 domain structures, the molecular basis for the formation of the cognate L27 domain complexes is still poorly defined. In addition, the mode of assembly of L27 domain heterodimers into higher-order structures is controversial (17, 18). Elucidation of the higher-order assembly mode of cognate pairs of L27 domains is critical for understanding the roles of L27 domain scaffold proteins in organizing supramolecular protein complexes.
In this work, we determined the 3D structures of the L27 domain complexes formed by mLin-7/mLin-2 and Pals1/Patj. The assembly mode of each pair of the L27 domains was further studied using biochemical and NMR approaches. Our results demonstrate that the tetrameric structure is the general assembly mode for cognate pairs of L27 domains. Additionally, we show that the αC plays critical roles in the specificity of the L27 domain complex formation.
Materials and Methods
Protein Expression and Purification. Genes corresponding to the L27 domain of mouse mLin-7b (L27Lin7, residues 2–78) and the C-terminal L27 domain of mouse mLin-2 (L27Lin2C, residues 403–460) were PCR-amplified from the respective full-length cDNAs. The single-chain fusion proteins, each containing L27Lin7 and L27Lin2C connected with either a linker (Ser-Gly) or a thrombin-cleavable segment (Leu-Val-Pro-Arg-Gly-Ser-Ser-Gly), were cloned into a modified version of the pET32a vector, in which the S-tag and the thrombin recognition site were replaced by a sequence encoding a protease 3C cleavage site (Leu-Glu-Val-Leu-Phe-Gln-Gly-Pro). Similarly, the two single-chain fusion proteins containing the L27 domain of rat Patj (L27Patj, residues 1–69) and the N-terminal L27 of human Pals1 (L27Pals1N, residues 116–177) connected with a thrombin-cleavable fragment (Leu-Val-Pro-Arg-Gly-Ser-Ser-Gly) were PCR-amplified and inserted into the modified pET32a vector. Proteins were expressed at 37°C. The His-tagged, thioredoxin-containing protein was purified under native conditions by using Ni2+-nitrilotriacetic acid agarose (Qiagen, Valencia, CA) affinity chromatography. After protease 3C digestion, the L27Lin2C/L27Lin7 complex was purified by passing the digestion mixture through a DEAE-Sepharose column and a size-exclusion column. The purified L27Lin2C/L27Lin7 complex and the L27Patj/L27Pals1N complex, each connected with a protease cleavable fragment, were further digested with thrombin. After cleavage, thrombin was removed from each complex by size-exclusion chromatography.
NMR Structure Determination. NMR samples contained ≈1.5 mM L27Lin2C/L27Lin7 domain complex or L27Patj/L27Pals1N complex (calculated as the concentration of each monomeric subunit) in 100 mM potassium phosphate (pH 6.5). NMR spectra were acquired on a Varian Inova 750 MHz spectrometer. The NMR spectra for the L27Lin2C/L27Lin7 domain complex were acquired at 42°C, and the spectra for the L27Patj/L27Pals1N complex were collected at 45°C. Sequential backbone and nonaromatic, non-exchangeable side-chain resonance assignments of the protein were obtained by standard heteronuclear correlation experiments (19, 20). The side chains of the aromatics were assigned by 1H 2D total correlation spectroscopy/NOESY experiments. The stereo-specific assignments of the Val and Leu methyl groups were obtained by using a 10% 13C-labeled sample (21). Interproton distance restraints were derived from NOESY spectra (a 1H 2D NOESY and a 15N-separated- and 13C-separated NOESY). Nuclear Overhauser effects between two mLin-2/mLin-7 (or Patj/Pals1) L27 domain heterodimers were identified by using a 13C-edited (F1), 13C,15N-filtered (F3) 3D NOESY spectrum on a 13C,15N-labeled/nonlabeled single-chain fusion protein mixture (1:1) (22). Distance restraints were generated as described in ref. 17. Backbone dihedral angle restraints were derived from the secondary structure of the protein and backbone chemical shift analysis program talos (23). Structures were calculated by using the program cns (24). Figures were generated by using molmol (25), molscript (26), povscript+ (27), and grasp (28).
Construction of a Chimera L27 Domain. A chimera L27 domain containing the αA- and αB-helices of SAP97 (residues 1–41) and the αC-helix of Patj (residues 43–69) was constructed by using a PCR-based method and cloned into the modified pET32a vector. The L27 domain complex containing the chimera SAP97/Patj L27 domain (L27SP) and L27Pals1N was obtained by coexpression of these two domains. The resulting L27 domain complex was purified by using a method similar to that described for the purification of the L27Patj/L27Pals1N complex.
CD Measurement. CD spectra of isolated L27 domains, as well as their complexes, were collected on a J-720 spectropolarimeter (Jasco, Tokyo) at room temperature. The protein samples (≈10 μM) were dissolved in 100 mM phosphate buffer (pH 6.5) containing 1 mM DTT.
Mass Spectrometry. The molecular masses of the purified proteins were measured on a QSTAR-Pulsar mass spectrometer (Applied Biosystems).
Results and Discussion
Structure of the L27 Domain Tetramer Formed by mLin-7 and mLin-2. We chose the L27 domain of mLin-7 and the C-terminal L27 domain of mLin-2 for structural studies, because these two domains represent the prototype L27 domains capable of forming a specific heteromeric complex (5, 8, 9). The purified L27Lin7/L27Lin2C complexes, both in the form of two separate chains and in the form of a single-chain fusion protein, were eluted as a single peak from an analytical gel filtration column with a molecular mass of ≈38 kDa (data not shown). The 1H,15N heteronuclear single quantum coherence (HSQC) spectra of the L27Lin7/L27Lin2C complexes in both forms are highly similar, except for the residues in the linker region of the protein (Fig. 7 a and b, which is published as supporting information on the PNAS web site), indicating that the Ser-Gly linker connecting L27Lin7 and L27Lin2C had a limited effect on the assembly and conformation of the complex. The observation of a single set of backbone resonance for both forms of the L27Lin7/L27Lin2C complexes demonstrates that the complex is symmetric in solution. The single-chain L27Lin7-L27Lin2C fusion protein was chosen for the structural determination of the complex by NMR, because the protein has superior sample stability. Additionally, the single-chain fusion protein offered another advantage in the complex structure determination, because the L27Lin7/L27Lin2C heterotetramer was simplified into a “homodimer” of two units of the L27Lin7-L27Lin2C. Intersubunit nuclear Overhauser effects between the two units of L27Lin7-L27Lin2C could be readily identified by using 13C-filtered NOESY experiments.
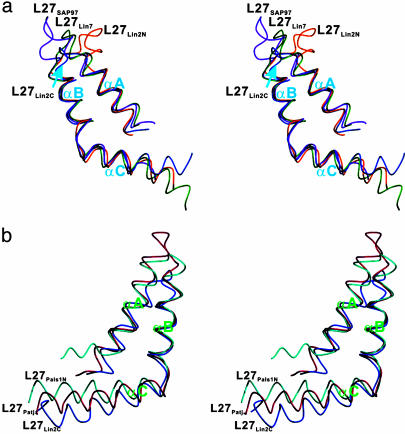
The overall structure of the L27Lin7/L27Lin2C tetramer is similar to the tetramer structure of the L27 domain complex formed by the SAP97 (L27SAP97) and mLin-2 (L27Lin2N) (17). Other than a few residues from the two termini, the structure of the L27Lin7/L27Lin2C tetrameric complex is well defined (Fig. 1a; see also Table 1, which is published as supporting information on the PNAS web site). Each L27 domain contains three α-helices (αA, αB, and αC). The N-terminal two α-helices of each L27 domain pack with each other to form a four-helix bundle. One end of this four-helix bundle is capped with the two αC-helices from each L27Lin7/L27Lin2C heterodimer unit (Fig. 1b). Each L27Lin7/L27Lin2C heterodimer is further assembled into a dimer of the heterodimer by the αC-helix from each L27 domain (Fig. 1b). The topology of the central four-helix bundle is somewhat unique. In this helix bundle, the two αC-helices from the same L27 domain form an antiparallel coiled coil. The two antiparallel coiled coils further pack with each other to form a two-layered, four-helix bundle with an intercoil angle of ≈65°. Molecular surface analysis suggests that the assembly of the L27Lin7/L27Lin2C heterodimer, as well as the dimer of the heterodimer, is primarily mediated by the extensive hydrophobic interactions inside the core of the complex (Fig. 1c). The amino acid residues involved in the hydrophobic packing are highly conserved (Fig. 2). In addition to the hydrophobic interactions, complementary charged residues on the surface contribute to the stabilization of the complex. The hydrophobic surfaces that contribute to the L27Lin7/L27Lin2C heterodimer and the dimer of heterodimer assemblies are continuous, indicating that the tetrameric structure of the L27 domain complex is likely to be an integral structural unit.
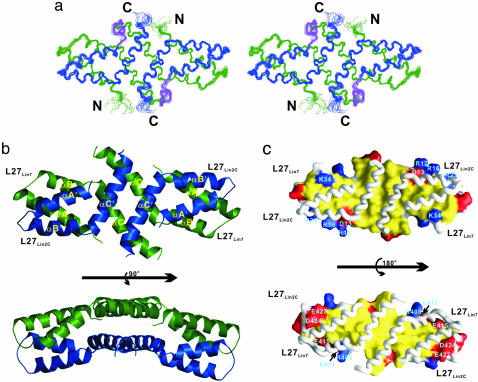
Fig. 1.
Structure of the mLin-2/mLin-7 L27 tetramer complex. (a) Stereoview showing the backbone of 20 superimposed NMR-derived structures of the tetramer complex. L27Lin7 is shown in green, L27Lin2C is drawn in blue, and the linker connecting L27Lin7 and L27Lin2C is in purple. (b) Ribbon diagram of a representative NMR structure of the tetramer complex, colored as in a. A prime after each secondary element (e.g., αA′) in L27Lin2C is used to indicate the same secondary structure as in L27Lin7.(c) Surface representation showing the packing interface of L27Lin7/L27Lin2C.In Upper, L27Lin7 is in the surface representation, and L27Lin2C is in the worm model. In Lower, L27Lin2C is in the surface model, and L27Lin7 is shown in the worm model. The hydrophobic residues are in yellow, the positively charged residues are in blue, the negatively charged residues are in red, and the polar residues are in white.
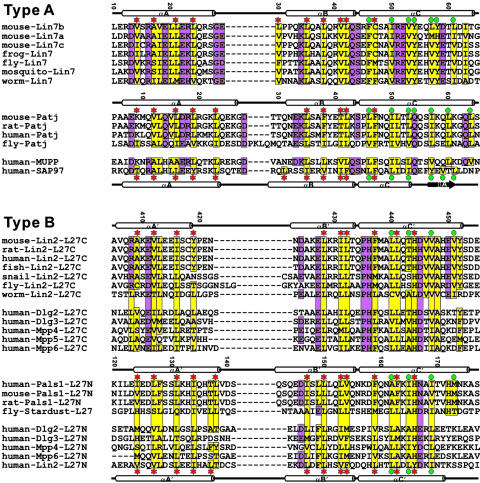
Fig. 2.
Amino acid sequence alignment of selected L27 domains derived from the 3D structures of three different complexes. In this alignment, the secondary structures of L27 domains with known 3D structures are shown at the top of each sequence. The highly conserved hydrophobic residues are highlighted in yellow. The other conserved residues are shown in purple. The amino acid residues involved in the L27SAP97/L27Lin2N heterodimer packing are indicated with red asterisks, and the amino acid residues involved in the packing of the central four-helix bundle of the tetramer are highlighted with turquoise dots. One may note from this structure-based sequence alignment that the structural role of the αC-helix in the type A and type B L27 domains (see ref. 17 for domain classification) is significantly different. In the type A L27 domains, the αC-helix is primarily involved in the packing of the central four-helix bundle. With the exception of one residue in the N terminus of the helix, the rest of the helix makes very little contact with the αA- and αB-helices. In contrast, in addition to an involvement in the formation of the central four-helix bundle, the αC-helix in each type B L27 domain also packs intimately with the αA- and αB-helices from the same domain.
Solution Structure of the Patj and Pals1 L27 Domain Complex. The unexpected assembly mode suggested by the x-ray crystal structure of the Patj and Pals1 L27 domain complex prompted us to reinvestigate the structure of this pair of the L27 domain complex (18). Structure-based amino acid sequence analysis of L27Patj and L27Pals1N suggested that the construct lengths of the two L27 domains used in this study, as well as in the work by Li et al. (18), are sufficient to cover the entire L27 domains (Fig. 2). We have again prepared L27Patj/L27Pals1N complexes in two forms: one in the form of separate L27Patj and L27Pals1N chains and the other in the form of L27Patj-L27Pals1N single-chain fusion protein. Analytical gel filtration analysis of the L27Patj/L27Pals1N complexes in both forms indicated a tetrameric structure of the complexes (data not shown). The 1H,15N HSQC spectra of the L27Patj/L27Pals1N complexes in both forms display a single set of backbone peaks for the tetramer, and the two spectra are nearly identical (except the linker region) (Fig. 7 c and d). We conclude that the L27Patj/L27Pals1N complex exists as a symmetric tetramer in solution. Because the covalent linking of the two L27 domains does not alter the structure and assembly of the complex, we determined the high-resolution structure of the L27Patj/L27Pals1N complex, using the single-chain fusion protein, by NMR spectroscopy.
The overall structure of the L27Patj/L27Pals1N tetramer is similar to the structures of the SAP97/mLin-2 and mLin-7/mLin-2 L27 domain complexes (ref. 17 and Figs. 1 and 3). In particular, the conformations of the four-helix bundles formed by the αA- and αB-helices from each pair of cognate L27 heterodimers are particularly similar. The conformation of the four-helix bundle formed by the αA- and αB-helices in our structure is essentially the same as the conformation of the corresponding four-helix bundle in the crystal structure (Fig. 3 b and c). However, the assembly of the central helix bundle of the L27Patj/L27Pals1N tetramer in solution is significantly different from the central helix bundles of the SAP97/mLin-2 and mLin-7/mLin-2 L27 domain complexes (Fig. 4). In the central helix bundle of the L27Patj/L27Pals1N tetramer, the two αC-helices from each heterodimer unit form an antiparallel helix–helix interface using the N-terminal half of each helix. The assembly of the two heterodimer units into a tetramer is mediated by two distinct parallel helix–helix interfaces involving the C-terminal half of each αC-helix (Fig. 4b). The interactions between the two heterodimers are primarily hydrophobic in nature (Fig. 8, which is published as supporting information on the PNAS web site). The topology of the L27Patj/L27Pals1N tetramer central helix bundle is highly similar to that of the four-helix bundle formed by the tetramerization domain of p53 (29–31).
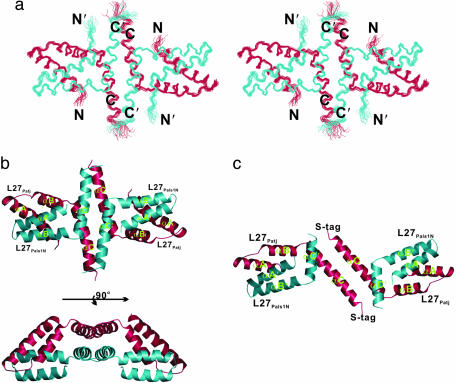
Fig. 3.
Structure of the Patj/Pals1 L27 tetramer complex. (a) Stereoview showing the backbone of 20 superimposed NMR-derived structures of the tetramer complex. L27Patj and L27Pals1N are shown in red and turquoise, respectively. The flexible linker is omitted for clarity. (b) Ribbon diagram of a representative NMR structure of the tetramer complex, colored as in a.(c) Ribbon diagram of the Patj/Pals1 L27 tetramer structure solved by x-ray crystallography (18).
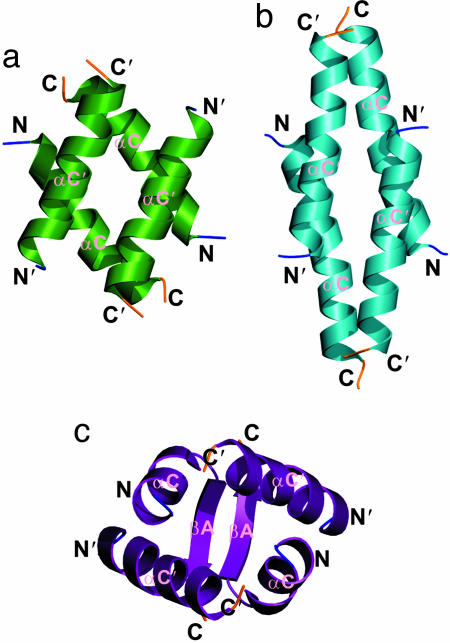
Fig. 4.
Comparison of the central four helix bundles. The topologies of the central four helix bundles of the L27Lin2C/L27Lin7 (a), L27Patj/L27Pals1N (b), and L27SAP97/L27Lin2N (c) complexes were drawn by using ribbon diagrams.
The structure of the central helix bundle in the solution structure of the L27Patj/L27Pals1N tetramer is radically different in a number of aspects from that in the crystal structure (Fig. 3 b and c). In the crystal structure of the L27Patj/L27Pals1N complex, the αC-helix of the Patj L27 domain is significantly longer than any of the αC-helices in the L27 domains with known structures (Fig. 3c). The packing of the two L27Patj/L27Pals1N heterodimers is mediated solely by the antiparallel coiled coil formed by the elongated αC-helix of L27Patj. The two αC-helices from L27Pals1N are too far apart to make any direct contact. Therefore, the interheterodimer packing is much less extensive when compared with our NMR structure. A number of interheterodimer nuclear Overhauser effects observed from the 13C half-filtered NOESY spectrum of the L27Patj/L27Pals1N complex in solution clearly argued against the assembly mode seen in the crystal structure (Fig. 9, which is published as supporting information on the PNAS web site). The crystal structure of L27Patj/L27Pals1N further suggested that the two heterodimer units in the tetramer were asymmetric, an observation contradicted by our NMR observations of the symmetric dimer of the heterodimer of the L27Patj/L27Pals1N complex (as well as in two other pairs of L27 tetramers). These large differences between the x-ray structure and our NMR structure of the L27Patj/L27Pals1N complex could result from a 17-residue helix-promoting S-tag fused to the αC-helix of L27Patj used in the crystal structure determination (32). With no artificial tag sequence attached, we found that the αC-helix of the Patj L27 domain stops at Ser-65 (instead of two more additional helical turns extending into the linker sequence in the crystal structure). The discrepancy between the NMR and crystal structures could also derive from the crystal packing of the four elongated αC-helices from L27Patj between two asymmetric units (18).
Tetramer Structure as a General Assembly Mode of Cognate L27 Domains. The three pairs of cognate L27 domain structures that we have determined all adopt symmetric tetrameric assembly. Each specific L27 domain complex is formed by two different L27 domains, one from a protein containing a single L27 domain and the other from a protein containing two L27 domains connected in tandem. Formation of tetrameric L27 complexes provides a mechanistic basis for the polymerization of L27 domain scaffold proteins (6, 15, 17). We further showed that mutations leading to the disruption of the central helix bundle also destroyed L27 heterodimer formation in SAP97 and mLin-2. This biochemical observation is consistent with continuous hydrophobic packing mediating the assembly of both the heterodimer and dimer of the heterodimer in all three pairs of L27 domain complexes. Dissociation of the L27 tetramer into two heterodimers would leave a large patch of hydrophobic surface exposed to the solvent. However, Li et al. (18) argued that the L27Patj/L27Pals1N heterodimer can stably exist in solution and acts as a biologically relevant unit. We used NMR spectroscopy to study whether the two pairs of L27 tetramers would dissociate into heterodimers in solution when the concentration of the complexes was lowered to micromolar range. We reasoned that, upon dissociation of the L27 tetramer into two heterodimers, the disruption of the extensive packing between the αC-helices in the central helix bundle would induce significant chemical shift changes to residues in each of the αC-helices even if the heterodimer still retained its fold. We compared the 1H,15N HSQC spectra of the L27Patj/L27Pals1N tetramer (i.e., the two L27 domains that are not covalently linked) at protein concentrations of 1.0 mM and 10 μM (expressed as each L27Patj/L27Pals1N heterodimer concentration). Lowering the sample concentration did not result in significant chemical shift and line-width changes to the spectra (Fig. 10, which is published as supporting information on the PNAS web site). This result demonstrated that the L27Patj/L27Pals1N tetramer has a Kd value significantly <5 μM. We have also compared the HSQC spectra of the L27Lin7/L27Lin2C complex at concentrations of 1.0 mM and 10 μM (heterodimer unit concentration). Again, the complete overlap of the two spectra demonstrated that no detectable amount of tetramer-to-heterodimer dissociation could be observed at a tetramer concentration of 5 μM (data not shown). Taken together, the NMR data presented in this study and our earlier mutagenesis analysis of the SAP97 mLin-2 L27 domain complex suggest that the tetrameric structure is a general assembly mode for cognate pairs of L27 domains. It remains to be established whether the dimer-of-heterodimer assembly mode of the L27 domains observed in our in vitro studies represents functional complex organization in vivo.
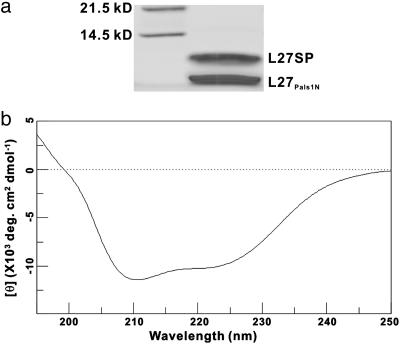
The Central Helix Bundle as a Key Specificity Determinant for the L27 Heteromeric Complexes. Having determined the structures of three distinct pairs of L27 domain complexes, we next asked what might be the molecular basis underlying the specificity for the formation of cognate pairs of L27 domains. Structural analysis showed that the conformations of six different L27 domains with known structure are highly similar (Fig. 5). The structural similarity is particularly high for the L27 domains from SAP97, mLin-2, and mLin-7 (Fig. 5a). The orientations of the αC-helix of L27 domains from Patj and Pals1 show some variation (Fig. 5b). Additionally, both the overall topology and the detailed packing of the helix bundles formed by the αA- and αB-helices in each pair of L27 domain complexes are similar. The most striking conformational differences among the three pairs of L27 tetramers are in the central four helix bundles formed by the αC-helices (Fig. 4). This observation prompted us to test the idea that the αC-helix might be a critical specificity determinant for the interaction of cognate pairs of L27 domains. We used two different approaches to test this hypothesis. In this first case, we mutated a pair of contacting hydrophobic residues in the interface of the two L27Patj/L27Pals1N heterodimer units (Ile-57 in L27Patj and Ile-169 in L27Pals1N) (Fig. 11a, which is published as supporting information on the PNAS web site). Consistent with the structural-based prediction, mutation of these two Ile residues to Ser disrupted dimer-of-dimer assembly (Fig. 11 b and c). The same mutations are not predicted to have a major impact on the tetramer assembly if the complex assumes the structure shown in the crystal study. Similarly, mutation of two hydrophobic residues in the dimer-of-heterodimer interface of the L27Lin7/L27Lin2C complex disrupted the tetramer assembly (Fig. 11 d–f). This disruption approach indicates that the αC-helix-mediated bundle formation is important for cognate L27 domain complex formation. In the second approach, we swapped the αC-helix of L27SAP97 with the corresponding αC-helix of L27Patj and assayed whether the resulting SAP97/Patj L27 chimera (referred to as L27SP) could form a stable complex with L27Pals1N. The thioredoxin-fused L27SP and L27Pals1N were coexpressed as two separate chains. After Ni2+-nitrilotriacetic acid affinity purification, the thioredoxin fusion tag was cleaved by protease 3C. After two additional steps of chromatographic purification (gel filtration and anion exchange), we obtained a protein complex that appears as two bands with ≈1:1 stoichiometry on SDS/PAGE (Fig. 6a). This protein complex was eluted as a single peak on an analytical gel filtration chromatography with an elution volume corresponding to a molecular mass of ≈30 kDa. The molecular masses of the two bands in Fig. 6a measured by mass spectrometry were 8,285.2 Da (Upper) and 7,440.2 Da (Lower), corresponding to the calculated molecular masses for L27SP (8,286.5 Da) and L27Pals1N (7,441.4 Da), respectively. The CD spectrum indicated a well folded, helix-rich structure of the purified protein L27SP/L27Pals1N complex (Fig. 6b). We conclude that the L27SP and L27Pals1N can form a stable tetrameric complex. As a control, we also coexpressed thioredoxin-fused L27SAP97 and L27Pals1N single-chain fusion protein. Protease 3C digestion of the coexpressed Trx-L27SAP97 and Trx-L27Pals1N resulted in complete degradation of both L27SAP97 and L27Pals1N, suggesting that these two L27 domains do not form a stable complex (data not shown and refs. 17 and 33). This observation is consistent with the earlier observation that L27SAP97 does not bind to the L27 domains from Pals1 (6, 18). Taken together, our biochemical data demonstrated that the αC-helix in each L27 domain is a critical specificity determinant for the domain to recognize its binding partner.
Fig. 5.
Comparison of the conformations of individual L27 domains. (a) Stereoview of the four L27 domains with very similar conformations. In these four L27 domains, all three helices share very similar conformation and can be superimposed with each other. (b) Stereoview showing that the orientations of the αC-helices of L27Patj and L27Pals1N are noticeably different from that of the L27 domain shown in a.
Fig. 6.
Characterization of the L27 domain tetramer formed by L27Pals1N and a chimera L27 domain containing the αA- and αB-helices of L27SAP97 and the αC-helix of L27Patj (L27SP). (a) SDS/PAGE gel showing the purified L27Pals1N/L27SP complex with ≈1:1 stoichiometry. (b) CD spectrum showing the well folded, helix-rich structure of the L27Pals1N/L27SP complex.
In summary, the structures of the prototype L27 domain complex formed by L27 domains from mLin-2 and mLin-7, together with the other two pairs of L27 domain complexes, showed that the tetrameric structure is a general assembly mode for cognate pairs of L27 domain complexes. The carboxyl α-helix of each L27 domain that mediates the tetramer formation plays a critical role in recognizing its binding partner. The tetramer assembly of cognate L27 domains provides a molecular basis for polymerization of L27 domain-containing scaffold proteins. Such L27 domain-mediated multimeric scaffolds provide nucleation platforms for organizing supramolecular protein complexes that have been implicated in a number of cellular processes, including asymmetric cell divisions, cell polarity control, and signal transduction regulations.
Supplementary Material
Acknowledgments
We thank Dr. Robert Qi for mass spectrometric analysis, Miss Jing Yan for technical help, and Dr. James Hackett for reading the manuscript. This work was supported by Competitive Earmarked Research Grants and an Area of Excellence grant from the Research Grants Council of Hong Kong (to M.Z.). The NMR spectrometer used in this work was purchased with funds donated to the Biotechnology Research Institute by the Hong Kong Jockey Club. M.Z. is a Croucher Foundation Senior Research Fellow.
Author contributions: W.F., J.-f.L., and M.Z. designed research; W.F. and J.-f.L. performed research; W.F., J.-f.L., and M.Z. analyzed data; and M.Z. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: HSQC, heteronuclear single quantum coherence.
Data deposition: The atomic coordinates of the mLin-2/mLin-7 and Patj/Pals1 L27 domain complexes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1Y74 and 1Y76, respectively).
References
- 1.Macara, I. G. (2004) Curr. Biol. 14, R160–R162. [PubMed] [Google Scholar]
- 2.Kemphues, K. (2000) Cell 101, 345–348. [DOI] [PubMed] [Google Scholar]
- 3.Pawson, T. & Nash, P. (2003) Science 300, 445–452. [DOI] [PubMed] [Google Scholar]
- 4.Doerks, T., Bork, P., Kamberov, E., Makarova, O., Muecke, S. & Margolis, B. (2000) Trends Biochem. Sci. 25, 317–318. [DOI] [PubMed] [Google Scholar]
- 5.Butz, S., Okamoto, M. & Sudhof, T. C. (1998) Cell 94, 773–782. [DOI] [PubMed] [Google Scholar]
- 6.Lee, S., Fan, S., Makarova, O., Straight, S. & Margolis, B. (2002) Mol. Cell. Biol. 22, 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simske, J. S., Kaech, S. M., Harp, S. A. & Kim, S. K. (1996) Cell 85, 195–204. [DOI] [PubMed] [Google Scholar]
- 8.Kaech, S. M., Whitfield, C. W. & Kim, S. K. (1998) Cell 94, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borg, J. P., Straight, S. W., Kaech, S. M., de Taddeo-Borg, M., Kroon, D. E., Karnak, D., Turner, R. S., Kim, S. K. & Margolis, B. (1998) J. Biol. Chem. 273, 31633–31636. [DOI] [PubMed] [Google Scholar]
- 10.Hata, Y., Butz, S. & Sudhof, T. C. (1996) J. Neurosci. 16, 2488–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irie, M., Hata, Y., Deguchi, M., Ide, N., Hirao, K., Yao, I., Nishioka, H. & Takai, Y. (1999) Oncogene 18, 2811–2817. [DOI] [PubMed] [Google Scholar]
- 12.Setou, M., Nakagawa, T., Seog, D. H. & Hirokawa, N. (2000) Science 288, 1796–1802. [DOI] [PubMed] [Google Scholar]
- 13.Bachmann, A., Schneider, M., Theilenberg, E., Grawe, F. & Knust, E. (2001) Nature 414, 638–643. [DOI] [PubMed] [Google Scholar]
- 14.Hong, Y., Stronach, B., Perrimon, N., Jan, L. Y. & Jan, Y. N. (2001) Nature 414, 634–638. [DOI] [PubMed] [Google Scholar]
- 15.Roh, M. H., Makarova, O., Liu, C. J., Shin, K., Lee, S., Laurinec, S., Goyal, M., Wiggins, R. & Margolis, B. (2002) J. Cell Biol. 157, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemmers, C., Medina, E., Delgrossi, M. H., Michel, D., Arsanto, J. P. & Le Bivic, A. (2002) J. Biol. Chem. 277, 25408–25415. [DOI] [PubMed] [Google Scholar]
- 17.Feng, W., Long, J. F., Fan, J. S., Suetake, T. & Zhang, M. (2004) Nat. Struct. Mol. Biol. 11, 475–480. [DOI] [PubMed] [Google Scholar]
- 18.Li, Y., Karnak, D., Demeler, B., Margolis, B. & Lavie, A. (2004) EMBO J. 23, 2723–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bax, A. & Grzesiek, S. (1993) Acc. Chem. Res. 26, 131–138. [Google Scholar]
- 20.Kay, L. E. & Gardner, K. H. (1997) Curr. Opin. Struct. Biol. 7, 722–731. [DOI] [PubMed] [Google Scholar]
- 21.Neri, D., Szyperski, T., Otting, G., Senn, H. & Wuthrich, K. (1989) Biochemistry 28, 7510–7516. [DOI] [PubMed] [Google Scholar]
- 22.Zwahlen, C., Legault, P., Vincent, S. J. F., Greenblatt, J., Konrat, R. & Kay, L. E. (1997) J. Am. Chem. Soc. 119, 6711–6721. [Google Scholar]
- 23.Cornilescu, G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- 24.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 25.Koradi, R., Billeter, M. & Wuthrich, K. (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
- 26.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 27.Fenn, T. D., Ringe, D. & Petsko, G. A. (2003) J. Appl. Crystallogr. 36, 944–947. [Google Scholar]
- 28.Nicholls, A. (1992) grasp, Graphical Representation and Analysis of Surface Properties (Columbia Univ., New York).
- 29.Lee, W., Harvey, T. S., Yin, Y., Yau, P., Litchfield, D. & Arrowsmith, C. H. (1994) Nat. Struct. Biol. 1, 877–890. [DOI] [PubMed] [Google Scholar]
- 30.Clore, G. M., Ernst, J., Clubb, R., Omichinski, J. G., Kennedy, W. M., Sakaguchi, K., Appella, E. & Gronenborn, A. M. (1995) Nat. Struct. Biol. 2, 321–333. [DOI] [PubMed] [Google Scholar]
- 31.Jeffrey, P. D., Gorina, S. & Pavletich, N. P. (1995) Science 267, 1498–1502. [DOI] [PubMed] [Google Scholar]
- 32.Bierzynski, A., Kim, P. S. & Baldwin, R. L. (1982) Proc. Natl. Acad. Sci. USA 79, 2470–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris, B. Z., Venkatasubrahmanyam, S. & Lim, W. A. (2002) J. Biol. Chem. 277, 34902–34908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.