Abstract
Vaccinia virus encodes a 37-kDa palmitylated protein (p37) that is required for envelopment, translocation, and cell-to-cell spread of virions. We have analyzed the biological significance of the palmitate modification by constructing a recombinant vaccinia virus that expresses a nonpalmitylated p37 and comparing its biological activity to that of the wild-type virus. The mutant virus is inefficient at cell-to-cell spread and does not produce or release enveloped virions, although it produces normal amounts of nonenveloped virions. Furthermore, the mutant virus is not able to nucleate actin to propel itself through and out of the cell, a function requiring the indirect participation of p37. The deficiency in protein function appears to result from a lack of appropriate targeting to the membranes of the trans-Golgi network (TGN) which leaves p37 soluble in the cytoplasm. We conclude that the palmitate moiety is necessary for targeting or anchoring p37 to the TGN membrane, where, along with other vaccinia virus-encoded proteins, p37 is involved in the complex process of virion envelopment and release.
Vaccinia virus (VV) is a member of the Poxviridae, a family of large, complex DNA viruses that replicate in the cytoplasm of infected cells (32). Its nearly 200-kbp genome has been completely sequenced and appears to encode more than 200 gene products (16). While expressing many of the enzymes and cofactors required for nucleotide metabolism, including the replication and expression of its genome, the virus is dependent on the host cell translational machinery for protein production. Proteins are expressed in discrete temporal classes starting with the early class as soon as 15 min after entry into the cell. The intermediate class of genes is expressed from about 2 to 4 h postentry, followed by the late class, which is expressed from 4 h postentry until cell death. Virion assembly coincides with late protein production.
The first immature virions appear (by electron microscopy) as hollow membrane crescents that loosely encapsidate the core components. The first infectious virions, which are referred to as intracellular mature virus (IMV), are formed as the core, which is composed of the genome, packaged enzymes and cofactors, and numerous structural proteins, condenses. The envelope surrounding the IMV particle, most likely derived from the intermediate compartment between the endoplasmic reticulum and the Golgi apparatus (45), also contains virus-encoded proteins. In tissue culture systems, IMV represents the majority of virions produced, but the in vivo significance of this virion form remains to be established. Depending on the strain of virus and host cell, 25 to 40% of the total IMV produced are enveloped with additional membranes and released from the cell (36). These multiply enveloped forms of the virus are likely of greater significance in vivo, since they have been implicated in the cell-to-cell spread and long-range dissemination of the virus (37).
The envelopment of IMV is a complex process that involves targeting of the IMV particle to the trans-Golgi network (TGN), in which, by budding through the compartment, two additional surrounding membranes are acquired, forming intracellular enveloped virus (IEV) (20). The IEV particle uses actin polymerization to propel itself through and out of the cell, sometimes into neighboring cells without exposure to the extracellular environment (2, 7, 19, 21, 47). At the plasma membrane, the outermost envelope of the IEV particle fuses with the cell membrane, releasing virions to the outside of the cell with the loss of the outermost membrane. When the virion remains attached (or reattaches) to the cell, it is referred to as cell-associated enveloped virus (CEV), and when it is free floating in the extracellular medium, it is referred to as extracellular enveloped virus (EEV).
The formation of infectious enveloped virions is dependent on numerous VV-encoded proteins, including the products of the A27L (40), A33R (42), A34R (4, 30, 55), A36R (33), B5R (12, 54), and F13L (encoding p37) (1) open reading frames (ORF) of VV. All of these, except for the product of A27L, a 14-kDa IMV-associated protein, are specific for the enveloped forms of the virus and are located in the outermost envelope of EEV. The A34R gene product is glycosylated (10), while the A33R and B5R gene products are both glycosylated and palmitylated. The p37 protein is palmitylated but is not glycosylated (35). The A36R protein has not been extensively characterized with regard to protein modification.
Fatty acylation has been demonstrated to be an important modification for VV-encoded proteins, contributing to more than one stage of virion assembly. In addition to numerous myristylproteins (13), VV also encodes at least six palmitylated proteins of 14, 17, 23-28 (a single protein), 37, 42, and 92 kDa (6). The identities of all but the 14- and 17-kDa proteins are known. The 23- to 28-kDa protein is the glycosylated product of the A33R gene previously thought to be encoded by the A34R gene (43). The 37-kDa protein is p37 (22), the product of the F13L gene and the object of this study. The 42-kDa protein is the glycosylated product of the B5R gene (25), which is also known as gp42. The 92-kDa protein is the A-type inclusion protein and represents the only known example of a VV protein modified by both myristic and palmitic acids (29). Only the site(s) of modification for p37, which is palmitylated within a hydrophobic domain on cysteines 185 and 186 of the 372-amino-acid protein is known.
p37 is expressed from 4 h postinfection (hpi) until cell death. Its electrophoretic mobility indicates a relative mass of 37 kDa, but its predicted mass based on amino acid content is 41 kDa. Within infected cells, it is targeted to the TGN (20) and, when it is virion associated, is specific for the enveloped forms of the virus (18, 34). Hydrophobicity and membrane topology predictions suggest that it is a transmembrane protein based on a hydrophobic alpha-helical region in the central part of the protein. Recent biochemical studies argue against the predictions and indicate that the protein is only peripherally associated with membranes, albeit very tightly (44). The significance of the fatty acyl moiety in anchoring the protein to membranes has been demonstrated by two methods. The protein normally fractionates with the membranous components of infected cells and cannot be stripped from membranes by salt or carbonate extraction. If the membranes are treated with neutral hydroxylamine, which cleaves the thioester bonded palmitate moiety from the protein, the protein is released from the membranes (44). Indirect immunofluorescence analysis of cells in which p37 is transiently expressed demonstrates that the wild-type protein is present in discrete cytoplasmic foci, while a nonpalmitylated mutant is evenly diffuse throughout the cytoplasm (17).
Recent evidence suggests that p37 is a member of the phospholipase D superfamily, all of which contain a motif (HXKXXXXD with the single-letter amino acid code) also present in p37 (48). Mutation of this motif in p37 or other phospholipase D superfamily members results in loss of function. Baek et al. have demonstrated that p37 exhibits phospholipase A and C activity rather than phospholipase D activity. The in vivo substrates for p37 are unknown. It has been suggested that the products of the lipase reactions may be involved in membrane fusion.
While these studies have contributed to our understanding of p37 function and protein-membrane biochemistry, they have not answered the fundamental question of why p37 is palmitylated. In this study, we have attempted to answer that question by the construction and analysis of a mutant virus that expresses a nonpalmitylated p37 rather than the wild-type fatty acylated protein. Our findings indicate that the biological significance of the acyl moiety is in targeting p37 to intracellular membranes whereby the protein is functional in cooperatively enveloping IMV.
MATERIALS AND METHODS
Cells.
BSC40 (African green monkey kidney) and RK13 (rabbit kidney) cell lines were cultured in Eagle’s minimal essential medium supplemented with 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, and 10 μg of gentamicin sulfate per ml (MEM-10 LG/GS) at 37°C in a 5% CO2 humidified atmosphere. Upon infection with VV, the cells were then cultured in Eagle’s minimal essential medium supplemented with 2 mM l-glutamine and 10 μg of gentamicin sulfate (MEM LG/GS) per ml at 37°C in a 5% CO2 humidified atmosphere.
Viruses.
The IHD-J strain of VV was routinely propagated, and titers on BSC40 cells were determined as previously described (11). It has not been genetically altered and encodes the wild-type p37. The mutant virus vRB10 has been previously described (1). In vRB10, 93% of the F13L ORF encoding p37 has been deleted by insertion of the mycophenolic acid resistance gene gpt under transcriptional control by the VV 7.5-kDa promoter. It was propagated by low-multiplicity passage through BSC40 cells in the presence of mycophenolic acid, xanthine, and hypoxanthine. Plaque titrations were performed by inoculating serial dilutions of the virus onto BSC40 monolayers, followed by transfection of a plasmid-borne rescuing copy of the F13L gene behind a VV promoter. Plaques were visualized by crystal violet staining at 72 hpi.
Construction of recombinant VV.
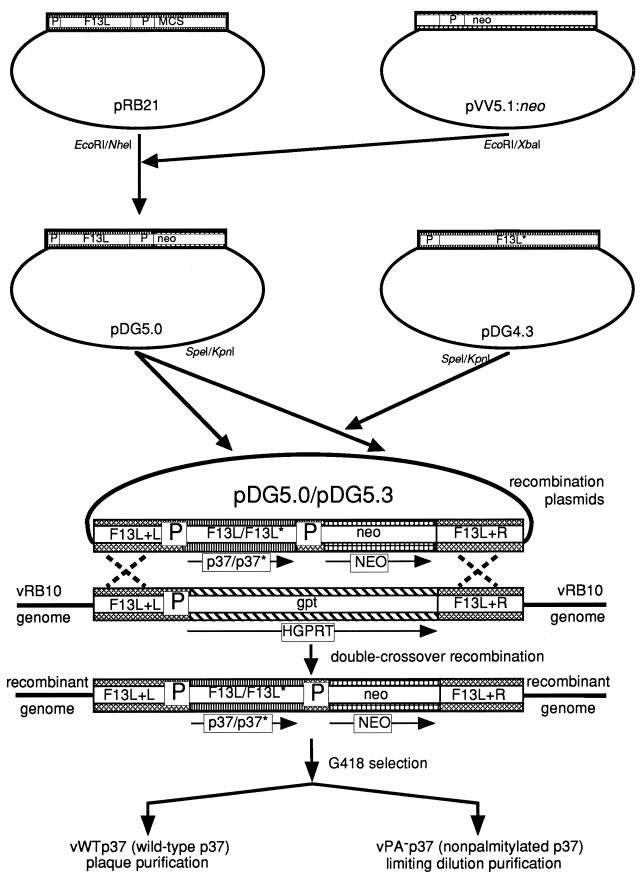
The plasmid pVV5.1:neo (14) was restricted with EcoRI and XbaI to release a 1,192-bp fragment containing the entire bacterial neomycin phosphotransferase II (neo) gene (see Fig. 1). The neo gene was ligated into EcoRI-NheI-restricted pRB21 (3), placing the gene downstream from a VV synthetic early-late promoter. The resulting plasmid was named pDG5.0. The plasmid pDG4.3, which was previously demonstrated to contain an engineered mutation in the F13L ORF resulting in the expression of a nonpalmitylated form of p37 (17), was restricted with SpeI and KpnI to release a 672-bp internal fragment of the F13L ORF. This fragment was ligated into SpeI-KpnI-restricted pDG5.0, replacing the wild-type F13L ORF with the mutated copy. This plasmid was named pDG5.3. In this plasmid, the codons for the normally palmitylated cysteines (cysteines 185 and 186) were mutated to code for serine instead. The presence of the mutated copy of the F13L ORF was confirmed by restriction digestion with HindIII, since there is an additional site in the mutated gene not found in the wild-type sequence and by DNA sequencing.
FIG. 1.
Diagram of the construction of recombinant viruses vWTp37 and vPA−p37. Plasmids used or constructed in this study were pRB21, pVV5.1:neo, pDG5.0, pDG4.3, and pDG5.3. P, VV promoters. Protein coding regions for each plasmid are indicated. F13L encodes p37, neo encodes neomycin phosphotransferase II, and gpt encodes hypoxanthine-xanthine guanosine phosphoribosyltransferase. The nucleotide sequences flanking the F13L ORF in the VV genome are indicated by F13L+L and F13L+R. The mutated F13L gene sequence and the nonpalmitylated p37 protein that it encodes are indicated by an asterisk.
The plasmids were transfected into BSC40 cells simultaneously infected by vRB10 as follows. Ten micrograms of plasmid DNA (either pDG5.0 or pDG5.3) was added to 1 ml of MEM LG/GS in polystyrene tubes. Thirty microliters of the liposome transfection reagent DMRIE-C (Life Technologies, Inc.) was added to each tube, followed by incubation at room temperature for 15 min. After the DNA-liposome complexes had formed, 106 infectious U of vRB10 was added to the mixture. The virus-DNA-liposome mixture was inoculated onto 106 BSC40 cells in 35-mm well of a six-well tissue culture plate and incubated for 48 h at 37°C in a 5% CO2 humidified atmosphere. At 48 hpi, the infected cells were washed free of the tissue culture plate and suspended in the culture medium. Cells were pelleted from the suspension by centrifugation at 700 × g and 4°C for 10 min. The cells were resuspended in 100 μl of phosphate-buffered saline (PBS [137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4]) and freeze-thawed three times to lyse cells and release virus. A 10-μl volume of the infected cell lysates was used as an inoculum to infect 107 BSC40 cells on 100-mm plates that had been pretreated with G418 at a concentration of 200 μg/ml in the culture medium for 24 h prior to infection. Upon infection, the G418 concentration was increased to 400 μg/ml in the culture medium (MEM LG/GS). The infection continued for 48 h at 37°C in a 5% CO2 humidified atmosphere. The cells were then scraped free from the plate and resuspended in the culture medium. The cells were pelleted from the culture medium by centrifugation at 700 × g and 4°C for 10 min. The cells were resuspended in 1 ml of PBS and freeze-thawed three times to lyse cells and release virus. A 100-μl volume of the infected cell lysates was used as an inoculum to infect BSC40 cells. The process of passing virus through G418-pretreated BSC40 cells in the presence of G418 was repeated three times to enrich for recombinant viruses expressing NEO.
After the third passage to enrich for recombinants, individual virions were amplified to obtain pure virus stocks. This was accomplished by two separate techniques. The recombinant virus expressing the wild-type p37, which was code named vWTp37, was purified by screening virus stocks grown from individual plaques formed in BSC40 monolayers overlaid with agar-containing medium plus 200 μg of G418 per ml. The expression of p37 by this virus was confirmed as described below. The recombinant virus expressing the mutated and presumably nonpalmitylated form of p37, which was code named vPA−p37, did not form plaques of normal size (see Results), and it could not be plaque purified as described for vWTp37. Instead, individual viruses were cultured by inoculating serial dilutions of the G418-passaged stocks onto BSC40 cells in 96-well tissue culture plates and by treating with 200 μg of G418 per ml in the culture medium as generally outlined by Blasco and Moss (1). At 72 hpi, each well was examined microscopically to determine if microplaques were forming. Wells which had no plaques or more than one microplaque were disregarded. The cells from individual wells with only one detectable microplaque were scraped free from the plate with a pipette tip, resuspended in the culture supernatant, transferred to microcentrifuge tubes, and freeze-thawed three times. The entire cell lysate was used as an inoculum to infect 106 BSC40 cells in six-well tissue culture plates. The cells were pretreated with 200 μg of G418 per ml for 24 h prior to infection, and the infection was in the presence of 200 μg of G418 per ml. At 24 hpi, the cells were harvested and lysates were prepared by freeze-thawing as described above. The lysates were screened for p37 production as described below, and only lysates that contained p37 were purified further. The p37-containing cell lysates were serially diluted and inoculated onto BSC40 cells in 96-well plates that had been pretreated with G418. Individual microplaques were observed microscopically at 72 hpi, harvested, and amplified in the presence of 200 μg of G418 per ml as described above. The limiting dilution purification of vPA−p37 was performed a total of three times. Purified stocks of vWTp37 and vPA−p37 were prepared as previously described (11) and stored at −70°C.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot detection of proteins.
Proteins from cell lysates or from purified virion samples were boiled in reducing sample buffer and resolved by discontinuous gel electrophoresis as described by Laemmli (27). All gels used were 12.5% polyacrylamide. The proteins were transferred from gels to nitrocellulose (49) and sequentially probed with antibodies directed against VV antigens, followed by enzyme-conjugated secondary antibodies. The blots were developed by addition of chemiluminescent substrates and exposure to film. All primary antibodies were from polyclonal antisera produced in rabbits and have been described previously. Polyclonal antiserum directed against p37 (α-p37) (44) was used at a dilution of 1:10,000, while antibodies directed against the 25-kDa VV core protein (α-25K) (53) and the IMV membrane protein encoded by the L1R ORF (α-L1R) (15) were used at a dilution of 1:1,000. The secondary antibody was a goat-anti-rabbit immunoglobulin G (IgG) polyclonal antibody conjugated to horseradish peroxidase (GαR-HRP) diluted 1:40,000 for use. The antibody-protein complexes were detected by incubating the blots with SuperSignal chemiluminescent substrate solutions (Pierce) followed by exposure to BioMax MR-2 film (Kodak).
Tritiated palmitate labeling of virion proteins and immunoprecipitation of p37.
BSC40 cells were grown to confluence (106 cells) in 35-mm wells of six-well tissue culture plates. The cells were infected with IHD-J, vRB10, vWTp37, or vPA−p37 at a multiplicity of infection (MOI) of 10. At 4 hpi, the media were aspirated and replaced with MEM LG/GS containing 200 μCi/ml of [9,10-3H]palmitic acid ([3H]PA; New England Nuclear). The infections continued until 24 hpi at 37°C in a 5% CO2 humidified atmosphere. The cells were suspended in the culture medium by repeatedly pipetting the medium over the cell monolayer. The cell suspensions were subjected to centrifugation at 12,000 × g and 4°C for 30 min to pellet cells and virus particles. The pellets were resuspended in 100 μl of PBS and freeze-thawed three times. A 20-μl volume was boiled in reducing SDS-PAGE sample buffer and then equally divided between two 12.5% polyacrylamide gels. Following electrophoresis, one gel was impregnated with diphenyloxazole (22.2% diphenyloxazole in dimethylsulfoxide [PPO-DMSO]), dried, and exposed to BioMax MR-2 film at −70°C (5). The other gel was transferred to nitrocellulose, and p37 was detected by antibodies and chemiluminescence as described above. The remaining 80 μl of infected cell lysates was diluted to 580 μl in 2× strength radioimmunoprecipitation assay buffer (RIPA; 1× solution is 1% [wt/vol] sodium deoxycholate, 1% [vol/vol] Triton X-100, 0.2% [wt/vol] SDS, 150 mM sodium chloride, 50 mM Tris-HCl [pH 7.4], and 1 U of Benzonase endonuclease per ml) and incubated on ice for 15 min. The samples were then heated to 70°C for 2 min, followed by centrifugation at 6,500 × g for 2 min. The supernatant was transferred to new microcentrifuge tubes, and the RIPA was adjusted to 1× concentration with water. One microliter of α-p37 was added to each sample, followed by incubation on ice for 2 h. Then, 40 μl of a 50% slurry of protein A-Sepharose beads in 1× RIPA was added, and incubation was continued for 18 h at 4°C with constant agitation. The immunoprecipitated proteins were washed three times with 1× RIPA, transferred to a new microcentrifuge tube, and washed again. The beads were pelleted a final time and resuspended in reducing sample buffer and boiled for 3 min. The proteins were resolved by SDS-PAGE utilizing 12.5% polyacrylamide gels as described above. Following electrophoresis, the gels were fluorographed by impregnation with PPO-DMSO, drying, and exposure to BioMax MR-2 film at −70°C.
Plaque formation and infectious titer assays.
Serial dilutions of purified virion preparations or infected cell lysates were made in MEM LG/GS and inoculated onto confluent monolayers of BSC40 cells in 35-mm wells of six-well tissue culture plates. Under duplicate infection conditions, one set of cells inoculated with vRB10 or vPA−p37 dilutions was simultaneously transfected with pRB21 to enhance plaque formation and facilitate determination of infectious titers for these samples. At 48 hpi, the cell monolayers were stained with 0.1% (wt/vol) crystal violet in 30% (vol/vol) ethanol and plaques were observed both macro- and microscopically.
Metabolic labeling and purification of virions.
RK13 cells (107) were seeded onto 150-mm tissue culture plates and cultured in MEM-10 LG/GS at 37°C in a 5% CO2 humidified atmosphere for 24 h. The cells were infected with either IHD-J, vRB10, vWTp37, or vPA−p37 at a MOI of 10 and cultured in MEM LG/GS until 4 hpi at 37°C in a 5% CO2 humidified atmosphere. The culture media were aspirated and replaced with methionine-deficient MEM containing 22.0 μCi/ml of [35S]methionine-cysteine and LG/GS. The infection continued until 24 hpi. The culture supernatants were transferred to 15-ml tubes and centrifuged at 700 × g and 4°C for 15 min to pellet free-floating cells. The supernatants were transferred to centrifuge tubes and subjected to ultracentrifugation at 100,000 × g and 4°C for 1 h. The pellets were resuspended in 1 ml of PBS and plaque assayed to determine infectious titers as described above.
Purification of virus from the infected RK13 cell lysates by sucrose gradients was as previously described (23). The cell monolayers were washed with PBS and then resuspended in 10 mM Tris-Cl (pH 8.0). The cells were allowed to swell on ice for 15 min and then Dounce homogenized to lyse them and release virus. The cellular debris was pelleted by centrifugation at 700 × g and 4°C for 15 min. The virus-containing supernatants were layered onto 6 ml of 36% sucrose cushions and subjected to ultracentrifugation at 55,000 × g and 4°C for 80 min. The pellets were resuspended in 1 ml of 10 mM Tris-Cl (pH 8.0) and Duall homogenized to disrupt aggregated material. They were then layered onto 25 to 40% sucrose gradients and subjected to ultracentrifugation at 33,000 × g and 4°C for 40 min. The virus bands were side-pulled by using a needle and syringe to withdraw the entire band in no more than 1 ml of solution. Each virus sample was diluted to 1 ml with water and plaque assayed to determine infectious titers as described above.
The remaining samples of extracellular and cell-associated virus were further purified by CsCl gradients (38). The gradients were prepared in ultracentrifuge tubes by first adding 5 ml of 1.2-g/ml CsCl solution followed by underlaying 4 ml of 1.25-g/ml and then 2.5 ml of 1.3-g/ml CsCl solutions through Pasteur pipettes. The virus samples were subjected to ultracentrifugation at 100,000 × g and 15°C for 2 h. Fractions of 500 μl each were collected from the bottom of the gradients, and the radioactivity per fraction was measured by scintillation counting. Peak fractions for IMV and EEV (as determined by relative positions in the gradient) were pooled, diluted in water to 12 ml, and subjected to ultracentrifugation at 100,000 × g and 4°C for 1 h. The virus pellets were resuspended in 100 μl of PBS and quantitated again by scintillation counting.
Proteinase K treatment of purified IMV and EEV.
Equivalent radioactive counts per minute of each EEV and IMV sample were treated with proteinase K at a final concentration of 2 μg/ml in PBS for 1 h at room temperature (44). Duplicate reactions were mock treated. All reactions were stopped by the addition of reducing SDS-PAGE sample buffer and boiling for 3 min. Samples were resolved by SDS-PAGE (12.5%) and blotted to nitrocellulose. The blots were probed with α-p37 and α-25K and then stripped and probed with α-L1R. Chemiluminescence detection of proteins was as described above.
Electron microscopy (EM).
BSC40 cells (107) in 100-mm tissue culture plates were infected with either IHD-J, vRB10, vWTp37, or vPA−p37 at a MOI of 10. At 16 hpi, the cells were scraped free from the plates and resuspended in the culture medium. The cells were pelleted by centrifugation at 700 × g and 4°C for 10 min. The cells were washed in PBS and pelleted again by centrifugation. The cells were fixed by resuspension in 2.5% paraformaldehyde (wt/vol)–200 mM cacodylate buffer (pH 7.4) and by incubation at 4°C for 8 h. The cells were pelleted by centrifugation and washed once with 200 mM cacodylate buffer (pH 7.4). The cells were left in 200 mM cacodylate buffer (pH 7.4) at 4°C for 16 h. The cells were pelleted by centrifugation, the cacodylate buffer aspirated and replaced with a 1% osmium (wt/vol) solution and then incubated at room temperature for 1 h. The samples were dehydrated by sequential 15-min incubations in 25, 50, 75, and 100% acetone. The 100% acetone incubation was repeated three times. The samples were then infiltrated with Spurr’s plastic. The samples were incubated at room temperature in solutions of one-third Spurr’s plastic–two-thirds acetone for 4 h followed by two-thirds Spurr’s plastic–one-third acetone for 16 h and, finally, 100% Spurr’s plastic for 6 h. The samples were placed under vacuum and heated to 70°C for 16 h. Sectioning was performed by personnel of the Oregon State University EM laboratory on a Sorvall Porter-Blum model MT2 ultramicrotome. Sections were poststained with Reynold’s lead citrate and uranyl acetate prior to examination by transmission EM.
Indirect immunofluorescence and fluorescence microscopy.
BSC40 cells were seeded onto microscope slide coverslips (microcover glasses [VWR]) at a density of 5 × 105 cells per 35-mm well in six-well tissue culture plates and cultured for 24 h at 37°C in a 5% CO2 humidified atmosphere. The cells were infected with either IHD-J, vRB10, vWTp37, or vPA−p37 at a MOI of 10. At 8 hpi, the cells were rinsed with ice-cold PBS followed by 100% methanol. The cells were prepared for immunofluorescence microscopy as described by Watkins (52). The cells were fixed by incubation in 3.7% formalin for 20 min at room temperature followed by permeabilization with 0.2% Triton X-100. The cells were rinsed twice for 5 min with ice-cold PBS. The cells were incubated with the primary antibodies α-p37 (used at a 1:2,000 dilution) or α-L1R (used at a 1:500 dilution) in PBS containing 2% bovine serum albumin (BSA) as a blocking agent for 1 h at 4°C. The cells were rinsed four times for 5 min each with ice-cold PBS. The secondary antibody was a goat-anti-rabbit IgG tetramethylrhodamine isothiocyanate conjugate (GαR-TRITC; Sigma) used at a 1:64 dilution in PBS containing 2% BSA. The secondary antibody incubations were for 1 h at 4°C in the dark. Forty minutes into the secondary antibody incubation, fluorescein isothiocyanate-conjugated phalloidin (Sigma) was added to a final concentration of 1 μg/ml. Twenty minutes later, the secondary antibody solution was aspirated and the cells were washed four times for 5 min with ice-cold PBS. The final wash was aspirated, and the cells were allowed to air dry for 10 min prior to mounting on slides and examination by confocal fluorescence microscopy.
Subcellular fractionation.
BSC40 cells were grown to be 95% confluent in 150-mm tissue culture plates (2 × 107 cells). The cells were infected with either IHD-J, vRB10, vWTp37, or vPA−p37 at a MOI of 10. The infected cells were cultured in MEM LG/GS at 37°C in a 5% CO2-humidified atmosphere for 16 h. The culture media were transferred to ultracentrifuge tubes and clarified by centrifugation at 100,000 × g and 4°C for 1 h. The cells were washed free of the plates with 10 ml of ice-cold PBS. The cells were pelleted by centrifugation at 700 × g and 4°C for 10 min followed by fractionation with differential centrifugation essentially as described by Child and Hruby (6). The PBS was aspirated, the cells were resuspended in 1.2 ml of hypotonic buffer (HB; 20 mM HEPES [pH 7.6], 5 mM potassium chloride, 1 mM magnesium chloride, 150 mM sodium chloride), and the suspension was incubated for 10 min on ice to swell the cells. All subsequent steps were at 4°C. The cells were then lysed by Dounce homogenization. A 200-μl volume of the cell lysates was set aside as the total cell extracts (TCE), while the remainder were subjected to centrifugation at 700 × g for 10 min. The supernatants (postnuclear supernatants [PNS]) were transferred to new microcentrifuge tubes for further fractionation, while the pellets from that centrifugation were resuspended in 1.0 ml of HB and set aside as the nuclear pellet (NP). The PNS were transferred to centrifuge tubes and diluted to 4.5 ml with HB and subjected to ultracentrifugation at 100,000 × g for 60 min. The pellets from this centrifugation (P100) were resuspended in 1 ml of HB and set aside as the particulate cytoplasmic fractions. The supernatants from this centrifugation and the clarified culture supernatants were adjusted to 10% trichloroacetic acid and subjected to centrifugation at 15,000 × g for 30 min. The pellets from this centrifugation were set aside as the soluble cytoplasmic fractions (S100) and the soluble culture supernatant fractions (SUP). All other fractions were adjusted to be 10% trichloroacetic acid and were subjected to centrifugation at 15,000 × g for 30 min. With the exception of the TCE fractions, which were resuspended in 20 μl, all precipitated pellets were resuspended in 100 μl of 1 M Tris (pH 10) to neutralize the acid and briefly sonicated to facilitate resuspension. Twenty microliters of each fraction was analyzed by SDS-PAGE (12.5% polyacrylamide gel) and immunoblotting with α-p37 as the primary antibody. For chemiluminescence detection, the primary antibody incubation was followed by incubation with GαR-HRP and then SuperSignal chemiluminescence substrate solutions, followed by exposure to BioMax MR-2 film. Relative quantitation of protein-antibody complexes was performed by film densitometry.
RESULTS
We have previously demonstrated that p37 is palmitylated on cysteine residues 185 and 186 of the 372-amino-acid protein (17). In addition, we, along with others (44), have confirmed that it is the fatty acyl modification and not the predicted transmembrane domain consisting of a hydrophobic alpha-helical region in the middle of the protein that mediates membrane interaction by p37. These studies have been performed either in vitro or by transient expression of p37. In order to understand the biological significance of the fatty acyl modification in an in vivo context, we sought to construct a recombinant VV expressing a nonpalmitylated p37 rather than the wild-type fatty acylated protein. Once the recombinant was constructed, we then performed a series of assays designed to compare its activity to that of the wild-type virus, IHD-J. p37 is known to be involved in the process of envelopment and release of virions (1); thus, we tested for plaque formation (a process dependent on enveloped virions), release of virions from infected cells, and retention of enveloped virions within infected cells. Also, the nucleation of actin to propel virions through and out of the cell is abrogated in the absence of p37 (7). It is not clear whether p37 is directly involved in actin nucleation or is merely a player in a prerequisite reaction. For that reason, we examined cells by fluorescence microscopy after fluorescently labeling VV antigens and F actin. Also, to confirm our previous studies, we analyzed infected cells and virions for p37 localization to determine if the palmitate moiety was important in targeting p37 to virions or intracellular membranes.
Construction of recombinant viruses.
To facilitate the construction of the recombinants used in this study, two plasmids were made (Fig. 1). One encodes the wild-type p37 (pDG5.0), and the other contains a mutated F13L sequence demonstrated to encode a nonpalmitylated p37 (pDG5.3) by transient expression (17). The plasmid pDG5.3 contains mutations that result in the replacement of the normally palmitylated cysteines (cysteines 185 and 186) with serines. The F13L ORF on both plasmids is transcriptionally regulated by the native F13L promoter and is flanked by sequences neighboring the F13L locus in the VV genome to allow for homologous recombination into its native site. The neo gene mediating G418 resistance, which is behind a synthetic VV early-late promoter, is also present on the plasmids to allow for selection of recombinants.
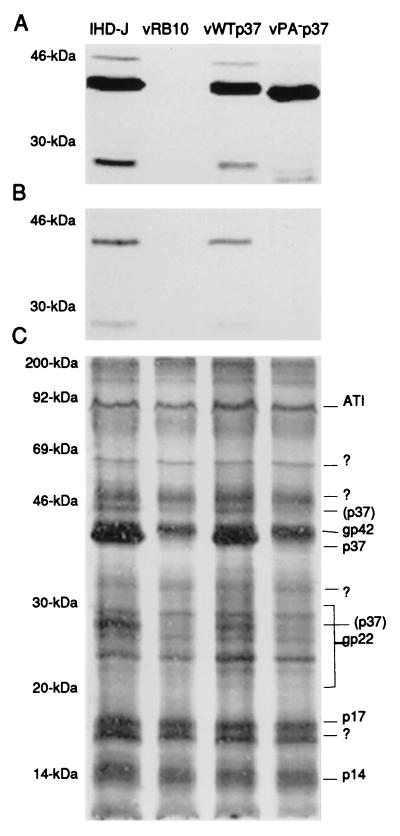
Cells were infected with vRB10, a virus that does not produce p37 due to an F13L deletion, and were transfected with either pDG5.0 or pDG5.3. The progeny were passed numerous times in the presence of G418 followed by a plaque purification of the wild-type p37-encoding virus, termed vWTp37, or a limiting dilution purification of the nonpalmitylated p37-encoding virus, termed vPA−p37. The viruses were tested for expression and palmitylation of p37 by labeling infected cells with [3H]PA, followed by immunoprecipitation of p37 from the cell extracts. The immunoprecipitates were resolved by SDS-PAGE and fluorographed to detect label. The tritium label was incorporated by p37 expressed from IHD-J or vWTp37 (Fig. 2B) but was not incorporated at detectable levels by vPA−p37-expressed p37, although the immunoblots of the same cell extracts demonstrate that it was expressed at comparable levels (Fig. 2A). We also noted that when total [3H]PA-labeled infected cell extracts were resolved by SDS-PAGE and fluorographed, there were three labeled protein bands in the extracts from IHD-J and vWTp37 that were absent from vRB10 and vPA−p37 (Fig. 2C). The same three bands were recognized by α-p37 in immunoblots, suggesting that they all represent some form of p37. The 37-kDa band was the predominant form of the protein, while the 46- and 28-kDa forms were present in reduced amounts. We have concluded that both vWTp37 and vPA−p37 express p37 at levels comparable to that of IHD-J and that vPA−p37 encodes a nonpalmitylated p37. Furthermore, p37 is present in at least two additional forms other than the well-characterized 37-kDa form, although the faster migrating species may represent a degradation product as we discuss below.
FIG. 2.
Expression and palmitylation of p37. The tritiated, palmitic acid-labeled extracts from cells that were infected with the indicated viruses were analyzed for p37 expression by immunoblotting (A) with anti-p37 antiserum and chemiluminescence development of the blot. p37 was analyzed for incorporation of the labeled palmitic acid by immunoprecipitation (B), resolution by SDS-PAGE, and fluorography. The efficiency of labeled palmitate incorporation by other VV proteins was demonstrated by SDS-PAGE and fluorography of the total infected cell extracts (C). Molecular weights are indicated beside each gel or blot. The previously described VV palmitylproteins are indicated to the right of panel C.?, unknown palmitylproteins; (p37), the 46- and 28-kDa forms of p37 described in the text.
Formation of enveloped virions by vPA−p37.
The formation of IEV and, subsequently of CEV and EEV is dependent on p37 (1). In the mutant virus vRB10, the F13L ORF was insertionally inactivated and 93% deleted by a gpt expression cassette mediating mycophenolic acid resistance. As a result, it is efficient at producing neither enveloped virions nor plaques on cell monolayers, for cell-to-cell spread is dependent on enveloped virions. The virus still produces normal amounts of infectious IMV, which are retained intracellularly unless released by cell lysis. Therefore, the ability to form plaques on cell monolayers stands as a simple test for enveloped virus production and release.
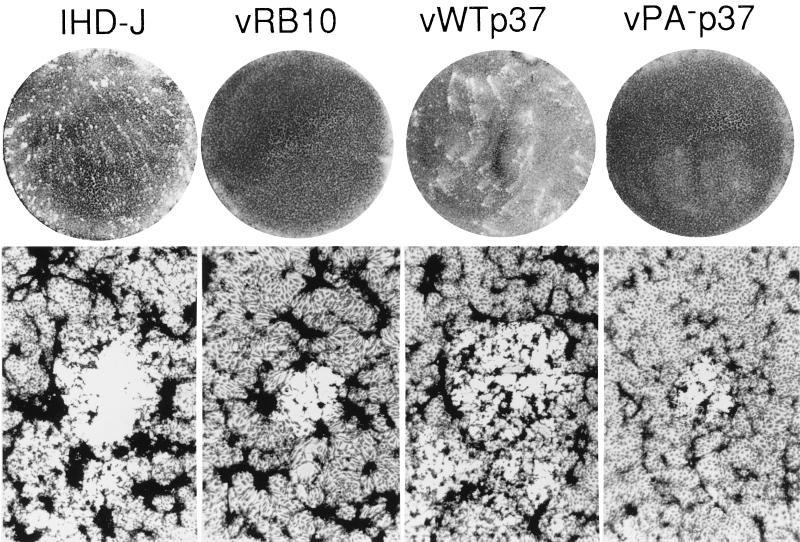
BSC40 cell monolayers were infected with serial dilutions of either IHD-J, vRB10, vWTp37, or vPA−p37 and cultured for 48 h. The cell monolayers were then stained with crystal violet to enhance visualization of the plaques. Both IHD-J and vWTp37 formed characteristic plaques with tails, making them comet-like in appearance (Fig. 3). This plaque morphology is the result of a single amino acid change in the A34R protein (4) and is typical of IHD-J and its derivative strains or mutants. Both vRB10 and vPA−p37 did not form visible plaques within 48 h, although very small plaques were visible to the naked eye between 72 and 96 hpi (data not shown). As confirmation that we had infected with vRB10 and vPA−p37, the cell monolayers were inspected microscopically. We were able to see microplaques forming at 48 hpi, but they were only approximately 1/10 the size of IHD-J or vWTp37 plaques (Fig. 3).
FIG. 3.
Plaque formation by wild-type and recombinant viruses. Monolayers of cells were infected at a low MOI to allow observation of individual plaques. At 48 hpi, the liquid overlay was aspirated and the cells were stained with crystal violet. The infecting viruses are indicated at the top. Magnifications of the macro- and microscopic views, ×1 and ×320, respectively.
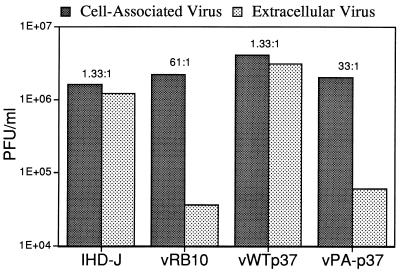
To assure ourselves that inefficient enveloped virus production was a bona fide vPA−p37 phenotype, we utilized another system to assay enveloped virus production. It has been demonstrated that the amino acid substitution on the A34R protein leading to the production of comet-like plaques is also responsible for the enhanced release of EEV from RK13 cells (36). RK13 cells were infected with either IHD-J, vRB10, vWTp37, or vPA−p37 at a MOI of 10 and cultured until 24 HPI. The virus purified from the culture supernatant, which at 24 hpi is almost 100% EEV, and the viruses purified from cell extracts were separately tested for infectious titer by a plaque assay. To enhance plaque formation by vRB10 and vPA−p37, cells were simultaneously transfected with pRB21, a plasmid that transiently expresses p37 in VV-infected cells. All viruses produced roughly equivalent amounts of cell-associated virus (predominantly IMV). While there was a ratio of cell-associated virus to extracellular virus of approximately 1:1 for IHD-J, vRB10 produced approximately 60- to 70-fold less extracellular virus (Fig. 4). The 1:1 ratio was restored for vWTp37 as expected, since the wild-type F13L gene was reinserted into the vRB10 genome. But vPA−p37 remained defective for release of virus, producing from 33 to more than 100 times more cell-associated virus than extracellular virus.
FIG. 4.
Comparison of virus production and release. RK13 cells were infected with the indicated viruses. At 24 hpi, viruses were purified from cell lysates (cell-associated virus) and the culture supernatants (extracellular virus) and were plaque titrated to determine infectious virus production. The ratios of cell-associated virus to extracellular virus are listed above the bars.
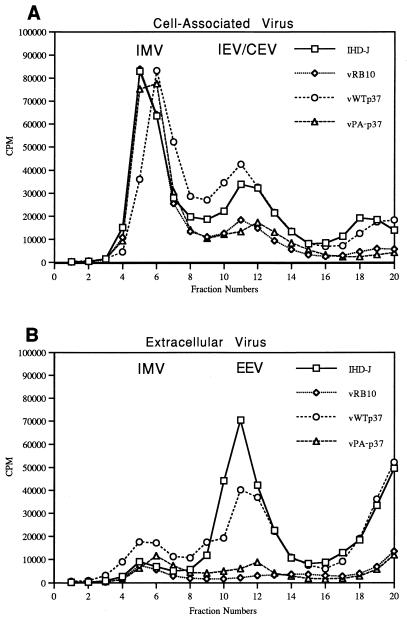
Both envelopment assays employed above are dependent on the production and release of infectious virus. Mutants of VV have been described that release normal amounts of EEV but with very low infectivity, resulting in a small plaque phenotype (30). We considered this a possibility for vPA−p37, necessitating the quantitation of virion particles produced. RK13 cells were infected as described above and labeled with [35S]methionine-cysteine after 4 hpi. At 24 hpi, virions were concentrated from the culture medium and purified from cell lysates as described in Materials and Methods. The labeled virus particles were purified by centrifugation in CsCl gradients, which were then fractionated, followed by determination of radioactivity per gradient fraction. IMV is separable from enveloped virions due to differing buoyant densities (38). While all viruses produced roughly equivalent amounts of IMV as determined by radioactive peaks in the gradient at 1.26 to 1.27 g/ml (Fig. 5A), IHD-J and vWTp37 had significantly higher peaks of radioactivity in fractions corresponding to densities at which enveloped virions sediment (1.22 to 1.24 g/ml). Detection of enveloped virions above background was possible in vRB10 or vPA−p37-infected cells, and both released a small amount of virus into the culture medium with buoyant densities characteristic of EEV (Fig. 5B), but in both cases they were drastically reduced from the normal levels. A small amount of IMV was detectable in the culture media of all viruses at 24 hpi and was probably due to cell lysis.
FIG. 5.
Quantitation of IMV and EEV by metabolic labeling and CsCl gradient purification. Virus from cells (A) or the culture media (B) of RK13 cells infected and labeled as outlined in Materials and Methods were separated according to density by CsCl gradient centrifugation. Fractions were collected from the bottoms of the gradients.
These experiments indicate that vPA−p37 is defective in the production and release of enveloped virions. vPA−p37 does not display an intermediate phenotype, suggesting that palmitylation is absolutely necessary for p37 function with regard to envelopment and release of virions.
EM examination of vPA−p37-infected cells.
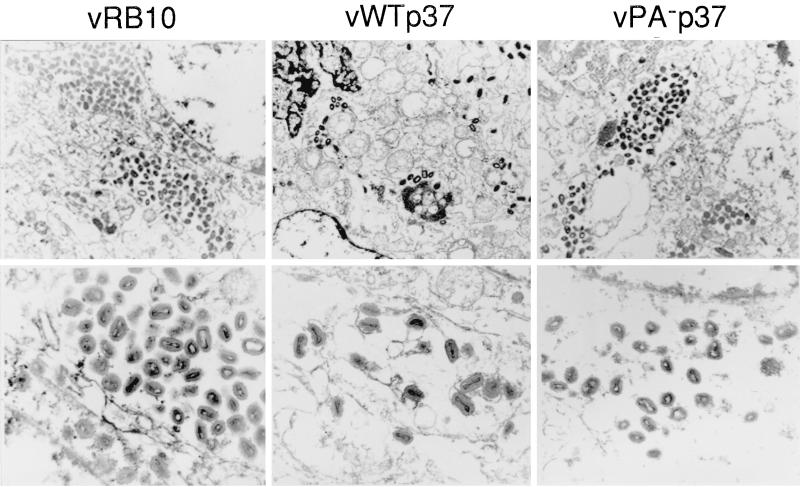
The morphogenesis of VV has been extensively characterized by transmission EM (9, 24, 41, 45, 46). The production of IMV occurs in discrete perinuclear foci, which are termed viroplasm, virosomes, or virus factories. At approximately 4 hpi, the first membrane crescents appear in the virus factories and are most likely derived from the membranes of the intermediate compartment or vesicles pinched off from it. The membrane crescents become closed circles as the core components are loosely packaged within. Then, the core condenses to the characteristic biconcave or dumbell shape, and the virion acquires a brick-like shape. At this time, it is possible to see by EM that the virion is in fact wrapped by two closely opposed membranes. The IMV particle is then targeted to the membranes of the trans-Golgi network, and, upon budding through the compartment, two additional membranes are acquired to form IEV. The formation of IEV is dependent on p37 as well as numerous other VV-encoded proteins. Since vPA−p37 appears defective for the production and release of enveloped virions, we examined infected cells by transmission EM to determine the stage at which envelopment was arrested.
BSC40 cells were infected with virus at a MOI of 10. At 16 hpi, the cells were treated as outlined in Materials and Methods to prepare them for transmission EM. Upon comparing IHD-J and vRB10, two differences were noted. (i) While IHD-J virions had disseminated throughout the cell to the periphery (data not shown), vRB10 virions remained concentrated in perinuclear clusters distinct from but near the virus factories (Fig. 6) (1). (ii) At higher magnification, it was not difficult to detect IHD-J virions in association with intracellular membranes, in the process of being enveloped by them, and fully enveloped as IEV, but all vRB10 virions had a distinct IMV morphology (Fig. 6). As expected, vWTp37 was indistinguishable from IHD-J at high and low magnifications due to restoration of the wild-type F13L to the vRB10 genome. Within vPA−p37-infected cells, only IMV could be observed, and, like vRB10, most virions were concentrated in perinuclear clusters. It seems, therefore, that palmitylated p37 is necessary not only for envelopment of IMV to form IEV but also for movement away from the perinuclear location of virion production.
FIG. 6.
EM of infected cells. Thin sections of BSC40 cells infected with virus as indicated above the electron micrographs were examined by transmission EM. Note virion clusters, in vRB10- and vPA−p37-infected cells (magnification, ×16,400), while vWTp37 virions are well disseminated throughout the cell. Note also envelopment of vWTp37 virions by intracellular membranes (magnification, ×73,000).
Fluorescence microscopic analysis of the actin stress network and localization of VV antigens.
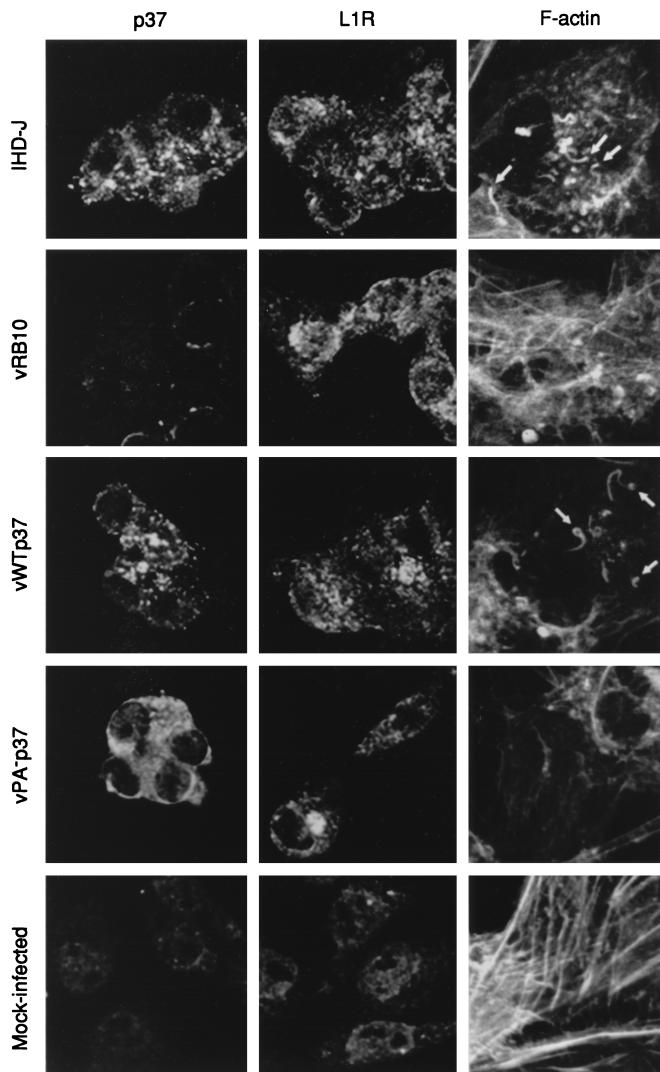
VV uses actin polymerization for intracellular motility and projection outside the cell, presumably into neighboring uninfected cells. This is observed by fluorescence microscopy as a reorganization of the cellular actin stress network to form thick actin tails up to 0.74 μm in length that are tipped with a single IEV particle (8). Drugs or mutations that block the production of IEV also prevent the nucleation of actin tails (7); thus, we would expect vPA−p37 to be defective for this process. Additionally, by transient expression of a nonpalmitylated p37 within infected cells, we have demonstrated by immunofluorescence microscopy and subcellular fractionation that the mutated protein is at least partially soluble (17). Transient expression results in the overexpression of the protein in some cells, while in others it is not expressed. Altered expression levels may have resulted in discrepancies between the wild-type protein and the mutant with regard to localization. As an extension of our previous work, we repeated our assays for p37 localization using the recombinant viruses vWTp37 and vPA−p37, which express p37 in a manner indistinguishable from that of the wild-type IHD-J.
BSC40 cells were infected with virus at a MOI of 10 for 8 h. The cells were fixed and stained with polyclonal sera to p37 and L1R, an IMV membrane-associated protein, and fluorescence secondary antibodies. F actin was stained with fluorescein-labeled phalloidin. Confocal fluorescence microscopy was utilized to observe localization of VV antigens and polymerized actin. Mock-infected cells had very little background fluorescence specific for the VV antigens while maintaining an intact actin cytoskeleton (Fig. 7). At 8 hpi, both IHD-J and vWTp37 had completely reorganized the actin cytoskeleton. Numerous thick actin tails were visible and appeared to be tipped with virions based on p37 and L1R localization. Since it is known that when p37 is virion associated it is specific for the enveloped forms of the virus (IEV, CEV, and EEV), it is likely that these represent IEV. The IMV outer membrane protein, L1R, is somewhat concentrated in the perinuclear region of IHD-J- and vWTp37-infected cells, but is also distributed throughout the cytoplasm in a slightly punctate pattern. On the other hand, p37 is distinctly punctate throughout the cytoplasm of IHD-J- and vWTp37-infected cells, as would be expected for a TGN-associated protein. Within vRB10-infected cells, the actin cytoskeleton has been broken down but is not reorganized into the thick filaments typical of the wild-type virus. Based on the localization of L1R, which is concentrated near the nucleus with little staining in the cytoplasm, virions appear to have remained near the nucleus, in agreement with our EM observations. As expected, p37-specific fluorescence was not above background due to the deletion of F13L from vRB10. In agreement with our previous findings, p37 was distributed diffusely throughout the cytoplasm of vPA−p37-infected cells, as would be expected for a soluble protein, while L1R-specific staining was concentrated near the nucleus as was observed for vRB10. Like vRB10, the actin cytoskeleton was disassembled but not reorganized in vPA−p37-infected cells.
FIG. 7.
Immunofluorescence microscopy of infected cells. The localizations of VV antigens, p37 and L1R, and F actin were determined by indirect fluorescence labeling and confocal microscopy. The fluorescently labeled antigens are indicated at the top of each column, and the infecting viruses are indicated to the left of each row. All images were captured through a magnification of ×100 with an oil immersion objective. Images of F-actin staining were zoomed at ×1.5. Thick actin-containing microfilaments are indicated by arrows.
Subcellular fractionation of VV-infected cells.
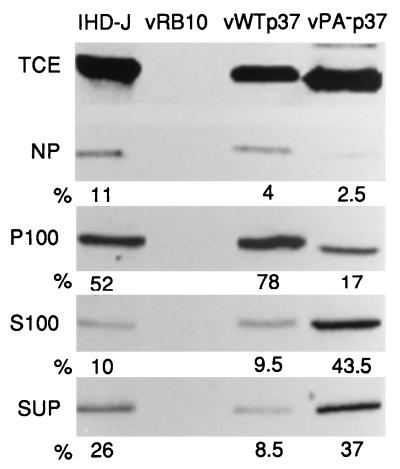
As we have discussed previously, the membrane interaction of p37 is mediated by the palmitate moiety. This was first demonstrated by Schmutz et al. (44) and was confirmed by site-directed mutagenesis and transient expression analysis of a nonpalmitylated p37 (17). Within infected cells, p37 is targeted to the membranes of the TGN and colocalizes with other EEV-specific antigens (43). If cellular membrane fractions containing p37 are treated with neutral hydroxylamine, cleaving palmitate from p37, then p37 becomes soluble. By indirect immunofluorescence and differential centrifugation subcellular fractionation, transiently expressed nonpalmitylated p37 behaves like a soluble protein and does not colocalize with other EEV-specific proteins as efficiently as the wild-type protein. We sought to confirm and extend these findings.
BSC40 cells were infected with virus at a MOI of 10 for 16 h (Fig. 8). The infected cells were then fractionated by differential centrifugation to yield a nuclear fraction (NP) and a cytoplasmic fraction (PNS) that was further fractionated to yield a particulate cytoplasmic fraction (P100) and a soluble cytoplasmic fraction (S100). In addition, the culture media (SUP) were clarified by a 100,000 × g centrifugation, concentrated by addition of trichloroacetic acid, and assayed along with the other fractions for p37 by SDS-PAGE and immunoblotting. Following chemiluminescence development of the blots, the relative protein concentrations were obtained by film densitometry. While vPA−p37 expressed p37 at levels similar to those of IHD-J and vWTp37 as determined by comparison of TCE, it was depleted from both the NP and P100. The majority of vPA−p37-expressed p37 was found in the S100 and SUP fractions, while very little IHD-J- or vWTp37-expressed p37 was found in the soluble fractions. This confirms our previous finding of increased solubility for p37 when it is not palmitylated.
FIG. 8.
Subcellular fractionation of infected cells. Infected cells (as indicated above the topmost gel) were fractionated by differential centrifugation to yield a TCE, a nuclear fraction (NP), a particulate cytoplasmic fraction (P100), and a soluble cytoplasmic fraction (S100). The culture supernatants (SUP) along with the subcellular fractions were assayed for p37 by SDS-PAGE, and immunoblots were developed by chemiluminescence. Relative amounts of p37 were determined by film densitometry, and the percentages of the total (NP plus P100 plus S100 plus SUP) for each virus are shown below the gel of each fraction. Note the increased percentages of p37 in the S100 and SUP fractions of vPA−p37.
Analysis of virion-associated p37.
Schmutz et al. (44) have demonstrated that p37 decorates the cytoplasmic face of the TGN and, after virion envelopment, is situated on the core-proximal face of the outermost envelope of EEV. It may be that p37 is EEV associated only by default due to TGN association, but this is unlikely considering that p37 is necessary for envelopment of virus. A TGN-associated receptor for the IMV particle has not been identified but seems likely to exist, since the IMV particle is specifically wrapped by TGN membranes. We considered the possibility that p37 is the TGN receptor for IMV and that it is not found to be IMV associated due to strong TGN interaction distal from the site of IMV production.
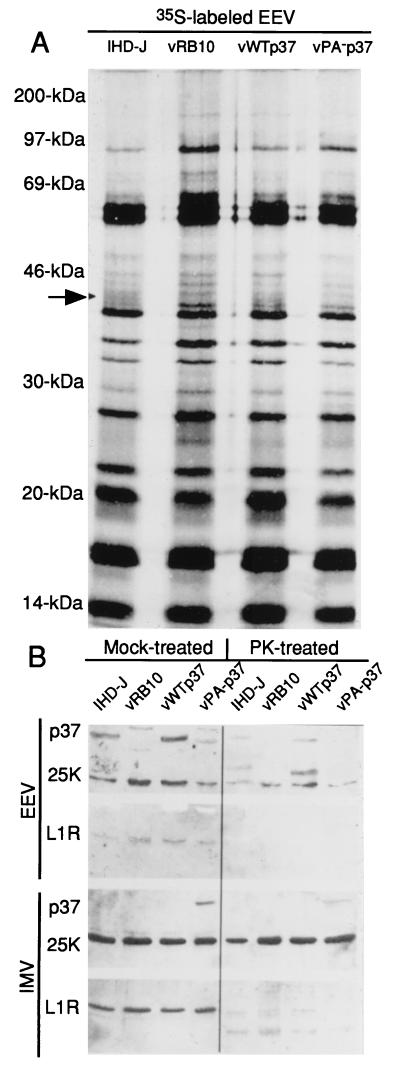
EEV and IMV were purified from cultures of metabolically labeled RK13 cells infected with either IHD-J, vRB10, vWTp37, or vPA−p37. Equivalent amounts of EEV and equivalent amounts of IMV, as determined by radioactive counts per sample, were resolved by SDS-PAGE, blotted to nitrocellulose, and probed with antibodies to p37, L1R, and the 25-kDa core protein, followed by secondary antibody incubations and chemiluminescence development (Fig. 9B). As expected, p37 was found to be specific for EEV produced by IHD-J and vWTp37. We also found p37 to be vPA−p37 EEV associated, but in relatively smaller amounts than that for wild-type p37. Surprisingly, vPA−p37-produced p37 was also IMV associated, leading us to believe that it was the IMV receptor on the TGN. We also observed the L1R and the 25-kDa core protein to be both IMV and EEV associated, as has been previously shown (39, 50). If p37 is the TGN-associated ligand for IMV, then it should decorate the outside of IMV when it is soluble. To test this, equivalent amounts of EEV and equivalent amounts of IMV were treated with 2 μg of proteinase K (PK) per ml for 1 h at room temperature to degrade any exposed proteins. Following PK treatment, the reactions were stopped by the addition of SDS-PAGE sample buffer and boiling for 3 min. The reactions were treated as described above to detect p37, L1R, and the 25-kDa core protein. The 25-kDa core protein was PK resistant on both IMV and EEV, while L1R was PK sensitive on both IMV and EEV. The vPA−p37 EEV-associated p37 was PK sensitive, while a portion of the IMV-associated p37 was PK resistant. This suggests that p37 most likely is not exposed on the outer face of vPA−p37 IMV but that it is protected within the core of the particle. Two major p37-derived degradation products were recognized by the p37 polyclonal antiserum. The larger fragment, of approximately 29 kDa, comigrates with one of the three bands detectable by α-p37 in total cell extracts, suggesting that some p37 is degraded by cellular or viral proteases in a manner similar to that for PK proteolysis. The smaller proteolytic product is detectable by α-p37 as a diffuse band running between 11 and 14 kDa.
FIG. 9.
Analysis of virion-associated p37. Viruses (as indicated above each gel lane) were purified by CsCl gradients from the culture supernatants of infected RK13 cells. (A) Resolution of EEV polypeptides by SDS-PAGE and autoradiography. Arrow, position of p37 in IHD-J and vWTp37 lanes. (B) PK treatment of purified EEV and IMV. Mock-treated or PK-treated (as outlined in Materials and Methods) EEV and IMV were resolved by SDS-PAGE and blotted to nitrocellulose. The blots were sequentially probed for p37 and 25K and then L1R, each of which is indicated to the left of the gels. Note the presence of undigested p37 in the PK-treated IMV sample.
DISCUSSION
Palmitylation of p37 was first observed by Hiller and Weber (20). Since then, the protein has been characterized extensively both biochemically and biologically. It is palmitylated and/or oleated (17, 35) on cysteines 185 and 186 of the 372-amino-acid polypeptide. The fatty acyl modification serves as a membrane anchor, for without it the protein is soluble (17, 44). Within infected cells, p37 is targeted to the TGN (43), where it plays an important role in the envelopment and release of virions (1). The mutant virus, vRB10, does not express p37 and as a consequence produces very little IEV, CEV, or EEV, while producing normal amounts of IMV. Normally, VV reorganizes the actin cytoskeleton using actin polymerization for intercellular and perhaps intracellular movement (7). Possibly because IEVs are not formed, virus-tipped thick actin filaments are not found in vRB10-infected cells.
Recent findings suggest that p37 is a member of the phospholipase D superfamily based on the presence of a conserved motif (48). The HXKXXXXD motif (with the single-letter amino acid code) and an additional conserved serine are only partially conserved in p37, but mutagenesis of any of the conserved residues results in a loss of function for the protein. These findings all indicate an important role for p37 in the late stages of the VV life cycle but do not entirely disclose the significance of the palmitate modification.
In this report, we have described the construction and characterization of a VV mutant that expresses a nonpalmitylated p37 rather than the wild-type fatty acylated form. Because the known role of p37 is to mediate (along with other VV proteins) the envelopment of IMV, our assays were designed to examine that aspect of VV biology. In most assays, the nonpalmitylated p37 mutant virus is indistinguishable from the F13L deletion mutant vRB10, suggesting that the fatty acyl moiety is indispensable for protein function.
Most often the purpose of palmitylation is membrane targeting and anchoring of proteins, but occasionally it serves another purpose (26, 28, 31). We considered the possibilities that palmitylation of p37 regulated protein-protein interactions or that the protein cycled between an inactive nonpalmitylated pool and an active palmitylated pool as a means of regulating protein function. It has not been conclusively demonstrated that p37 has any protein partners, although it is known to colocalize with numerous VV proteins in infected cells (43). With what is known about p37, we can only conclude that palmitylation does not serve as a mediator of protein-protein interactions. Likewise, palmitylation does not appear to serve as a regulatory mechanism for p37 function. As we have demonstrated by both subcellular fractionation and immunofluorescence microscopy, nearly 100% of the wild-type p37 is a component of the particulate fraction of cells, suggesting efficient membrane interaction. We have also demonstrated the protein-membrane interaction to be dependent on palmitylation as well as being necessary for p37 function. If palmitylation served as a regulatory function, we would expect a significant fraction of the protein to be nonpalmitylated. Instead, we find that less than 1% of the protein is not palmitylated (17). It seems that the sole purpose of p37 palmitylation is to target and anchor the protein in the membranes of the TGN. By our analyses presented here, it is apparent that p37 interaction with the TGN is absolutely necessary for function.
In their analysis of p37, Baek et al. purified bacterially expressed p37 and tested it for lipase activity, which they found. We have not observed palmitylation of p37 in bacterial cells (unpublished data). This suggests that palmitylation is not necessary for enzymatic activity and that the defect in the nonpalmitylated p37 is solely targeting. Considering our findings along with the findings of Baek et al. and Sung et al., it appears that the lipase activity of p37 is required for the formation of IEV and that it must be specifically localized to the TGN in order to be biologically relevant. The in vivo substrates for this enzyme are unknown.
In summary, we have demonstrated palmitylation of p37 to be necessary for targeting p37 to the membranes of the TGN. The association of p37 with the TGN membranes is an absolute requirement for the function of p37. The primary biological activity of p37 is best demonstrated by the F13L deletion mutant vRB10, which is deficient for all aspects of envelopment and release of virus while producing normal amount of IMV. When the wild-type F13L sequence is restored to the vRB10 genome, the mutant is rescued. But when the mutated F13L sequence, encoding a nonpalmitylated p37, is inserted into vRB10, the resulting recombinant virus is indistinguishable from vRB10 in the assays described here. The lack of an intermediate phenotype suggests a complete loss of function for the protein.
VV continues to stand out as a uniquely capable model system for the analysis of mammalian protein processing. In addition to acylation, VV polypeptides are subject to proteolytic processing, glycosylation, phosphorylation, ADP ribosylation, disulfide cross-linking (51), and sulfation (35). We have yet to decipher all of the intricacies of these modifications in VV or eukaryotic systems, and palmitylation of proteins is one of the least understood of these processes. Perhaps exploitation of the VV system will allow us not only to examine various functions of protein palmitylation but to identify factors mediating the process, including those responsible for the modification reaction and molecular properties of the modified protein.
ACKNOWLEDGMENTS
This work has been supported by the National Institutes of Health grant no. AI-21335.
We are also thankful to Bernard Moss for providing vRB10, Riccardo Wittek for providing anti-p37 antibody, and Anne-Marie Gerrard (Central Services Laboratory of the Center for Gene Research and Biotechnology at Oregon State University) for assistance with confocal microscopy.
REFERENCES
- 1.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco R, Moss B. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene. 1995;158:157–62. doi: 10.1016/0378-1119(95)00149-z. [DOI] [PubMed] [Google Scholar]
- 4.Blasco R, Sisler J R, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonner W M, Laskey R A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- 6.Child S J, Hruby D E. Evidence for multiple species of vaccinia virus-encoded palmitylated proteins. Virology. 1992;191:262–271. doi: 10.1016/0042-6822(92)90188-u. [DOI] [PubMed] [Google Scholar]
- 7.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 8.Cudmore S, Reckmann I, Griffiths G, Way M. Vaccinia virus: a model system of actin-membrane interactions. J Cell Sci. 1996;109:1739–1747. doi: 10.1242/jcs.109.7.1739. [DOI] [PubMed] [Google Scholar]
- 9.Dubochet J, Adrian M, Richter K, Garces J, Wittek R. Structure of intracellular mature vaccinia virus observed by cryoelectron microscopy. J Virol. 1994;68:1935–1941. doi: 10.1128/jvi.68.3.1935-1941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earl P L, Cooper N, Moss B. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates & Wiley Interscience; 1992. pp. 16.16.1–16.16.7. [Google Scholar]
- 12.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 13.Franke C A, Reynolds P L, Hruby D E. Fatty acid acylation of vaccinia virus proteins. J Virol. 1989;63:4285–4291. doi: 10.1128/jvi.63.10.4285-4291.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke C A, Rice C M, Strauss J H, Hruby D E. Neomycin resistance as a dominant selectable marker for selection and isolation of vaccinia virus recombinants. Mol Cell Biol. 1985;5:1918–1924. doi: 10.1128/mcb.5.8.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke C A, Wilson E M, Hruby D E. Use of a cell-free system to identify the vaccinia virus L1R gene product as the major late myristylated virion protein M25. J Virol. 1990;64:5988–5996. doi: 10.1128/jvi.64.12.5988-5996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 17.Grosenbach D W, Ulaeto D O, Hruby D E. Palmitylation of the vaccinia virus 37-kDa major envelope antigen. Identification of a conserved acceptor motif and biological relevance. J Biol Chem. 1997;272:1956–1964. doi: 10.1074/jbc.272.3.1956. [DOI] [PubMed] [Google Scholar]
- 18.Hiller G, Eibl H, Weber K. Characterization of intracellular and extracellular vaccinia virus variants: N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine interferes with cytoplasmic virus dissemination and release. J Virol. 1981;39:903–913. doi: 10.1128/jvi.39.3.903-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiller G, Jungwirth C, Weber K. Fluorescence microscopical analysis of the life cycle of vaccinia virus in chick embryo fibroblasts. Virus-cytoskeleton interactions. Exp Cell Res. 1981;132:81–87. doi: 10.1016/0014-4827(81)90085-9. [DOI] [PubMed] [Google Scholar]
- 20.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 22.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hruby D E, Guarino L A, Kates J R. Vaccinia virus replication. I. Requirement for the host-cell nucleus. J Virol. 1979;29:705–715. doi: 10.1128/jvi.29.2.705-715.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hung T, Chou C, Fang C, Chang Z. Morphogenesis of vaccinia virus in the process of envelopment as observed by freeze-etching electron microscopy. Intervirology. 1980;14:91–100. doi: 10.1159/000149168. [DOI] [PubMed] [Google Scholar]
- 25.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson C S, Zlatine P, Bano C, Kabouridis P, Mehul B, Parenti M, Milligan G, Ley S C, Magee A I. Dynamic protein acylation and the regulation of localization and function of signal-transducing proteins. Biochem Soc Trans. 1995;23:568–571. doi: 10.1042/bst0230568. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Magee A I, Newman C M, Giannakouros T, Hancock J F, Fawell E, Armstrong J. Lipid modifications and function of the ras superfamily of proteins. Biochem Soc Trans. 1992;20:497–499. doi: 10.1042/bst0200497. [DOI] [PubMed] [Google Scholar]
- 29.Martin K H. Ph.D. dissertation. Corvallis: Oregon State University; 1997. [Google Scholar]
- 30.McIntosh A A, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milligan G, Parenti M, Magee A I. The dynamic role of palmitoylation in signal transduction. Trends Biochem Sci. 1995;20:181–187. doi: 10.1016/s0968-0004(00)89004-0. [DOI] [PubMed] [Google Scholar]
- 32.Moss B. Poxviruses. In: Fields B N, Chanock R M, Hirsch M S, Melnick J, Monath T P, Roizman B, editors. Virology. 2nd ed. Vol. 2. New York, N.Y: Raven Press; 1990. pp. 2079–2111. [Google Scholar]
- 33.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a M(r) 43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 34.Payne L. Polypeptide composition of extracellular enveloped vaccinia virus. J Virol. 1978;27:28–37. doi: 10.1128/jvi.27.1.28-37.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne L G. Characterization of vaccinia virus glycoproteins by monoclonal antibody precipitation. Virology. 1992;187:251–260. doi: 10.1016/0042-6822(92)90313-e. [DOI] [PubMed] [Google Scholar]
- 36.Payne L G. Identification of the vaccinia hemagglutinin polypeptide from a cell system yielding large amounts of extracellular enveloped virus. J Virol. 1979;31:147–155. doi: 10.1128/jvi.31.1.147-155.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payne L G. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J Gen Virol. 1980;50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 38.Payne L G, Norrby E. Presence of haemagglutinin in the envelope of extracellular vaccinia virus particles. J Gen Virol. 1976;32:63–72. doi: 10.1099/0022-1317-32-1-63. [DOI] [PubMed] [Google Scholar]
- 39.Ravanello M P, Hruby D E. Characterization of the vaccinia virus L1R myristylprotein as a component of the intracellular virion envelope. J Gen Virol. 1994;75:1479–1483. doi: 10.1099/0022-1317-75-6-1479. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez J F, Smith G L. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 1990;18:5347–5351. doi: 10.1093/nar/18.18.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roos N, Cyrklaff M, Cudmore S, Balsco R, Krijnse-Locker J, Griffiths G. A novel immunogold cryelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996;15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 42.Roper R L, Moss B M. Role of vaccinia A33R outer envelope glycoprotein in virus release and spread. Oral Presentation W41-2. Bozeman: Montana State University; 1997. [Google Scholar]
- 43.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmutz C, Rindisbacher L, Galmiche M C, Wittek R. Biochemical analysis of the major vaccinia virus envelope antigen. Virology. 1995;213:19–27. doi: 10.1006/viro.1995.1542. [DOI] [PubMed] [Google Scholar]
- 45.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van ’t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stern W, Pogo B G, Dales S. Biogenesis of poxviruses: analysis of the morphogenetic sequence using a conditional lethal mutant defective in envelope self-assembly. Proc Natl Acad Sci USA. 1977;74:2162–2166. doi: 10.1073/pnas.74.5.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stokes G V. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J Virol. 1976;18:636–643. doi: 10.1128/jvi.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung T C, Roper R L, Zhang Y, Rudge S A, Ternel R, Hammond S M, Morris A J, Moss B, Engebrecht J, Frohman M A. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 1997;16:4519–4530. doi: 10.1093/emboj/16.15.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.VanSlyke J K, Hruby D E. Immunolocalization of vaccinia virus structural proteins during virion formation. Virology. 1994;198:624–635. doi: 10.1006/viro.1994.1074. [DOI] [PubMed] [Google Scholar]
- 51.VanSlyke J K, Hruby D E. Posttranslational modification of vaccinia virus proteins. Curr Top Microbiol Immunol. 1990;163:185–206. doi: 10.1007/978-3-642-75605-4_7. [DOI] [PubMed] [Google Scholar]
- 52.Watkins S. Immunohistochemistry. In: Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 14.22–14.29. [Google Scholar]
- 53.Whitehead S S, Hruby D E. Differential utilization of a conserved motif for the proteolytic maturation of vaccinia virus proteins. Virology. 1994;200:154–161. doi: 10.1006/viro.1994.1174. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolffe E J, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]