Abstract
Bacterial DNA and synthetic oligomers containing CpG dinucleotides activate the immune system through Toll-like receptor (TLR) 9. Here, we compare the immunostimulatory activity of three immunomers with different nucleotide sequences containing a synthetic cytosine-phosphate-2′-deoxy-7-deazaguanosine dinucleotide (CpR), called immunomodulatory oligonucleotides (IMOs), in mouse, human, and monkey systems. IMOs induced IL-12 and IFN-γ secretion more than a control non-CpG IMO in mice. All three IMOs activated HEK293 cells expressing TLR9 but not TLR3, -7, or -8. IMOs induced human B-cell proliferation and enhanced expression of CD86 and CD69 surface markers on B cells. The three IMOs induced CD86 expression on human plasmacytoid dendritic cells, but only IMOs that contained a 5′-terminal TCR nucleotide sequence induced IFN-α secretion. A sequence that forms a duplex structure also was required for IFN-α induction in human peripheral blood mononuclear cell cultures. IMOs induced chemokine and cytokine gene expression in human peripheral blood mononuclear cells. In monkeys, all three IMOs induced transient changes in peripheral blood leukocytes and lymphocytes and activated B and T lymphocytes. All three IMOs induced IFN-α in vivo in monkeys; the IMO sequence that forms a stable secondary structure induced the highest levels of IFN-α. These studies are, to our knowledge, the first comprehensive studies to compare the activity of IMOs containing synthetic stimulatory CpR dinucleotides in mouse, monkey, and human systems. These results suggest that IMOs induce strong and rapid immunostimulation and that the CpR dinucleotide is recognized by TLR9, leading to immune-cell activation and cytokine secretion in vitro and in vivo.
Keywords: CpG DNA, cytokines, immune stimulation, innate immunity, immunotherapy
The vertebrate immune system recognizes highly conserved, repeating molecular patterns that are present in pathogens through a number of pattern-recognition receptors, including Toll-like receptors (TLRs). One of the 11 TLRs identified in mammals, TLR9, recognizes cytosine-phosphate-guanine dinucleotide (CpG) dinucleotides present in bacterial DNA and synthetic oligonucleotides (CpG oligomers) (1). In general, CpG oligomers induce a T helper 1-type immune profile (2, 3), which includes secretion of IFN-α, IFN-γ, IL-12, TNF-α, and IL-6, with an increase in the levels of costimulatory surface molecules (2–7). Many CpG oligomers are being tested in clinical trials as therapies for various diseases, including cancers, infectious diseases, allergy, and asthma, and as adjuvant with vaccines and antigens (6, 7).
The immunostimulatory activity of CpG oligomers depends on the sequences flanking the CpG dinucleotide and the secondary structures they form (8–13). CpG oligomers that contain poly(dG) or palindromic sequences that allow formation of secondary structures, referred to as class A/D, activate dendritic cells (DCs) to mature and to secrete IFN-α, but they only weakly activate B cells (11). CpG oligomers that do not contain secondary-structure-forming sequences, referred to as class B/K, strongly activate B cells (9). Class B/K CpG oligomers stimulate maturation of DCs but do not induce secretion of IFN-α. One can incorporate sequence and structural motifs in the CpG oligomers, referred as class C, that can induce activation of B cells and DCs and also induce DCs to secrete IFN-α (12, 13).
Our previous studies on structural and chemical characteristics of CpG oligomers required for TLR9 activation showed that the receptor is specific for deoxyribonucleotides in the CpG and that ribonucleotides or 2′-O-alkyl ribonucleotides abrogate immunostimulation (14). We have identified critical structural features in the pentose sugar (14–18), phosphate backbone (14, 19), nucleobases (20, 21), and nucleosides (22) required for immunostimulatory activity. By incorporating synthetic pyrimidine (Y) and purine (R) bases in YpG and CpR motifs, we have determined the functional groups on cytosine and guanine required for receptor recognition (23).
The receptor requires that the CpG oligomer have a free 5′ end to induce immunostimulation; modifications incorporated at the 5′ end decrease the extent of immunostimulation depending on the size of the modification (24–29). CpG oligomers attached through their 3′ ends and containing two accessible 5′ ends (immunomers) have enhanced immunostimulatory activity (24–29).
Our previous studies showed that immunomers consisting of CpG or cytosine-phosphate-2′-deoxy-7-deazaguanosine dinucleotide (CpR) motifs, referred to as immunomodulatory oligonucleotides (IMOs), induce distinct cytokine profiles (29–31), although both motifs mediate immune responses through TLR9. In general, IMOs containing CpR dinucleotides induced higher or similar levels of IL-12 and lower IL-6 compared with IMOs containing CpG dinucleotide in vitro and in vivo (29–31). In the present study, we examined the ability of IMOs consisting of 3′-3′-linked structures and CpR dinucleotides in different sequences to induce immunostimulatory activity in human cell-based assays and in vivo in mice and monkeys. These studies suggest that TLR9 recognizes a variety of structurally similar nucleotide motifs and that the TLR response may be determined by nucleotide motif recognition patterns and the nucleotide sequence surrounding the CpR dinucleotide.
Materials and Methods
Synthesis and Purification of IMOs. IMOs and their CpG oligomer controls were synthesized on a 1- to 2-μmol scale by using β-cyanoethylphosphoramidite chemistry on a PerSeptive Biosystems 8909 Expedite DNA synthesizer (Framingham, MA) as described in refs. 26 and 29. The purity of full-length IMOs ranged from 90% to 95% with the remainder being shorter by 1 or 2 nt (n-1 and n-2) as determined by capillary gel electrophoresis and/or denaturing PAGE. All IMOs contained <0.5 units/ml endotoxin as determined by the Limulus assay (BioWhittaker) (for detailed protocols, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Assessment of Mouse Serum Cytokine Levels. Female C57BL/6 mice, 5 to 6 weeks old, were obtained from Taconic and maintained in accordance with the animal protocols of Hybridon's Institutional Animal Care and Use Committee. Mice (n = 5) were injected s.c. with IMOs at concentrations of 1 or 10 mg/kg (single dose). Serum was collected by retroorbital bleeding 4 h after IMO administration, and IL-12, IL-6, and IFN-γ were determined by sandwich ELISA as described in ref. 29. All reagents, including cytokine antibodies and standards, were purchased from Pharmingen.
Isolation of Human B Cells and Plasmacytoid DC (pDC). Peripheral blood mononuclear cells (PBMCs) from freshly drawn healthy volunteer blood (CBR Laboratories, Boston) were isolated by Ficoll density gradient centrifugation (Histopaque-1077, Sigma). B cells and pDCs were isolated from PBMCs by positive selection using the CD19 and BDCA4 cell isolation kits, respectively (Miltenyi Biotec, Auburn, CA), according to the manufacturer's instructions.
B Cell Proliferation Assay. Approximately 1 × 105 B cells were stimulated with different concentrations of IMO for 66 h, then pulsed with 0.75 μCi (1 Ci = 37 GBq) of [3H]thymidine, and harvested 8 h later. The incorporation of [3H]thymidine was measured by scintillation counter, and data were shown as cpm.
Stimulation of HEK293 Cells Expressing TLRs. HEK293 cells transfected with different TLRs (human TLR3, -7, or -8 or mouse TLR9) were obtained from InvivoGen (San Diego). HEK293 cells expressing TLRs were transiently transfected with reporter gene (Seap, InvivoGen) for 6 h and stimulated with IMOs at different doses (dose studies) for 24 h. TLR-dependent reporter gene expression was determined according to the manufacturer's protocol (InvivoGen).
Cytokine ELISAs. Human PBMCs were plated in 48-well plates with 5 × 106 cells per ml. pDCs and B cells were plated in 96-well dishes with 1 × 106 cells per ml. The IMOs dissolved in Dulbecco's PBS (pH 7.4; Mediatech, Washington, DC) were added to a final concentration of 10.0 μg/ml to the cell cultures. The cells then were incubated at 37°C for 24 h, and the supernatants were collected for ELISAs. The experiments were performed two or three times for each IMO in triplicate for each concentration. The secretion of IFN-α, IL-10, and/or IL-6 was measured by sandwich ELISA as described in ref. 29. The required reagents, including cytokine antibodies and standards, were purchased from Pharmingen and PBL Biomedical Laboratories (New Brunswick, NJ).
Flow Cytometry on Human PBMCs and DCs. Cell-surface markers, CD19, CD123, CD69, and CD86 were detected with a Coulter Epics-XL Flow Cytometer by using anti-human CD123-phycoerythrin, CD19-phycoerythrin, CD69-FITC, and CD86-FITC, which were purchased from Pharmingen. The PBMCs and pDCs were activated with 1 μg/ml IMOs at 37°C for 24 h. The cells were harvested and blocked with 10% human AB serum (Sigma) in staining buffer (PBS with 1% BSA and 0.1% NaN3, pH 7.4) at 4°C for 1 h. PBMCs (4 × 105) were stained with CD19-phycoerythrin plus CD69-FITC and CD86-FITC, and pDCs (2 × 105) were stained with CD123-phycoerythrin plus CD86-FITC. The data were acquired and analyzed with system II software (Coulter).
Gene Expression Profiles. Human PBMCs cultured at 10 million cells per ml in RPMI 1640 complete medium were stimulated with different IMOs for 24 h. Total RNA was extracted by using the Qiagen RNeasy Mini Kit (Valencia, CA), according to the manufacturer's protocol. Aliquots of 50 ng of total RNA in 50 μl of reaction medium were used for each RT-PCR with the one-step protocol. Amplification was performed in an Applied Biosystems Prism 7700 Sequence Detection System. The details of PCR protocols and primers and probes used are given in Supporting Materials and Methods.
Monkey Study. Six healthy young cynomolgus monkeys (Macaca fascicularis) weighing ≈2–5 kg were used in the study. All studies were conducted at WIL Research Laboratories (Ashland, OH) or MPI Research (Mattawan, MI) and were approved by their respective Animal Care and Use Committees. Animals were monitored daily by veterinarians. Each IMO was administered s.c. to three animals on day 1 at a concentration of 1 mg/kg. Blood samples (1 ml) were collected for plasma cytokine measurement at 0, 1, 2, 4, 8, and 24 h and, in some cases, at 48 and 72 h after administration of IMO. In addition, whole blood samples were collected for hematology evaluation (0.5 ml) after 24 h and for flow cytometry (5 ml) at 0, 24, and 168 h after administration of IMO. After collection of the 168-h blood sample, the animals were held without treatment (test article washout period) for 12–15 days before use in the next IMO study. All animals remained in good health throughout the experiment.
Hematology, Flow Cytometry, and Cytokine Analysis of Monkey Blood Samples. Blood cell count and differential measurements were conducted by WIL Research Laboratories or MPI Research. Flow cytometry was performed by using a FACS at FAST Systems (Gaithersburg, MD) (see Supporting Materials and Methods for details). Surface antigens on PBMC were stained as described above. Plasma samples were thawed on ice and tested in duplicate for IFN-α, IL-6, and IL-12 by using commercial kits obtained from PBL Biomedical Laboratories (human IFN-α), Pharmingen (human IL-6), and Bender MedSystems (Vienna) (monkey IL-12 total) according to the manufacturers' instructions. Sample volumes per assay well varied from 25 to 33 μl for IFN-α and IL-6 and 50 μlfor IL-12 detection.
Statistical Analysis. All data shown are mean ± SD. Statistical analysis was performed by using Student's t test for comparison between groups, and P < 0.05 was considered to be significant.
Results
Design and Synthesis of IMOs. IMOs 2–4 were designed to contain two short oligomers linked through their 3′ ends (Table 1). All oligomers 1–6 used in the study were of the same length (22 nt). IMO 2 contained two CpR dinucleotides, one in each branch, in contrast to four CpG dinucleotides in conventional CpG oligomer 1. IMOs 3 and 4 contained a 5′-terminal TCR nucleotide sequence that was not present in IMO 2. IMO 4 had a self-complementary sequence that allowed duplex formation, in contrast to IMOs 2 and 3. IMOs 5 and 6 were control sequences with CpC and RpC dinucleotides, respectively. All of the IMOs were characterized by mass spectral analysis for their sequence integrity (Table 1) and by capillary and/or slab gel electrophoresis for purity.
Table 1. IMO sequences, chemical modifications, and mass spectral analysis data.
| Mol. wt.
|
|||
|---|---|---|---|
| Number | Sequence* | Found | Calcd |
| CpG oligomer 1 | 5′-TCGTCGTTTGTCGTTTGTCGTT-3′ | 7,054.8 | 7,056.4 |
| IMO 2 | 5′-TCTGTCRTTCT-X-TCTTRCTGTCT-5′ | 7,146.2 | 7,145.8 |
| IMO 3 | 5′-TCRTCRTTCTG-X-GTCTTRCTRCT-5′ | 7,194.8 | 7,193.9 |
| IMO 4 | 5′-TCRAACRTTCR-X-RCTT RCAARCT-5′ | 7,225.2 | 7,227.9 |
| Control IMO 5 | 5′-TCTCACCTTCT-X-TCTTCCACTCT-5′ | 6,996.3 | 7,005.7 |
| Control IMO 6 | 5′-TRCTRCTTCTG-X-GTCTTCRTCRT-5′ | 7,192.3 | 7,193.9 |
Mol. wt., molecular weight; Calcd, theoretically calculated values.
All oligonucleotides are phosphorthioate backbone modified. CG and CR dinucleotides are underlined; R and X stand for 2′-deoxy-7-deazaguanosine and glycerol linker, respectively.
In Vivo Cytokine Induction Profiles of IMOs in Mice. We measured the IL-12 and IFN-γ induction abilities of IMOs 2–4 delivered s.c. to mice at concentrations of 1 and 10 mg/kg. All three IMOs induced dose-dependent IL-12 secretion in vivo (Table 2). IFN-γ was not detected in mice that received IMOs at a concentration of 1 mg/kg, but all three IMOs induced high levels of IFN-γ at 10 mg/kg (Table 2). A control IMO 6 failed to induce secretion of IL-12 or IFN-γ, suggesting that the CpR dinucleotides present in IMOs 2–4 were responsible for the observed immune responses in mice. IMOs induced insignificant levels of IL-6 at either dose level (data not shown).
Table 2. Cytokine induction profiles of IMOs in vivo in C57BL/6 mice.
| IL-12
|
IFN-γ
|
||
|---|---|---|---|
| IMO | 1 mg/kg | 10 mg/kg | 10 mg/kg |
| PBS | 0.5 ± 0.4 | 0.8 ± 0.04 | 0 |
| IMO 2 | 11.8 ± 1.2 | 45.7 ± 13.7 | 3.3 ± 0.03 |
| IMO 3 | 19.3 ± 3.8 | 55.5 ± 16.5 | 2.8 ± 0 |
| IMO 4 | 18.3 ± 2.7 | 50.1 ± 13.6 | 4.2 ± 0 |
| IMO 6 | 0.8 ± 0.3 | NT | 0 |
NT, not tested. Values are in ng/ml and are ±SD.
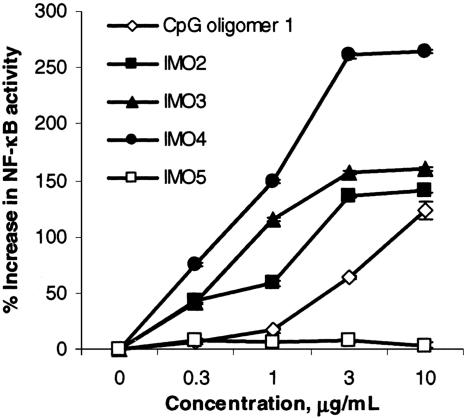
IMOs Mediated Immune Responses Through TLR9. To test whether IMOs are recognized by TLR9, we used HEK293 cells expressing mouse TLR9 transfected with a reporter gene. The results are presented as percentage increase in NF-κB activation (Fig. 1). The conventional CpG oligomer 1 produced a dose-dependent response, suggesting that TLR9 recognized oligomer 1 as expected (Fig. 1). IMOs 2–4, which contained CpR dinucleotides with two 5′ ends, activated NF-κB to a greater extent than did CpG oligomer 1 at all doses examined (Fig. 1). IMO 4 had greater activity in HEK293 cells than did IMOs 2 and 3, possibly because of the presence of the sequence AACRTT, which is optimally recognized by both mouse and human TLR9s. Control IMO 5 failed to activate TLR9, suggesting that TLR9 recognizes CpR dinucleotides in IMOs (Fig. 1). The HEK293 cells expressing an empty vector did not respond to any of the oligomers (see Fig. 8A, which is published as supporting information on the PNAS web site), suggesting that the observed response resulted from specific recognition of IMOs by TLR9. Additionally, both conventional CpG oligomer and IMOs failed to activate HEK293 cells expressing TLR3, -7, or -8 (Fig. 8B), suggesting that these were not receptors for them. TLR3- and TLR7/8-transfected HEK293 cells, however, showed NF-κB activation with poly(I)·poly(C) and R848, respectively, as expected (Fig. 8B).
Fig. 1.
Activation of HEK293 cells expressing mouse TLR9 with different concentrations of CpG DNA and IMOs. Data shown are representative of three independent experiments.
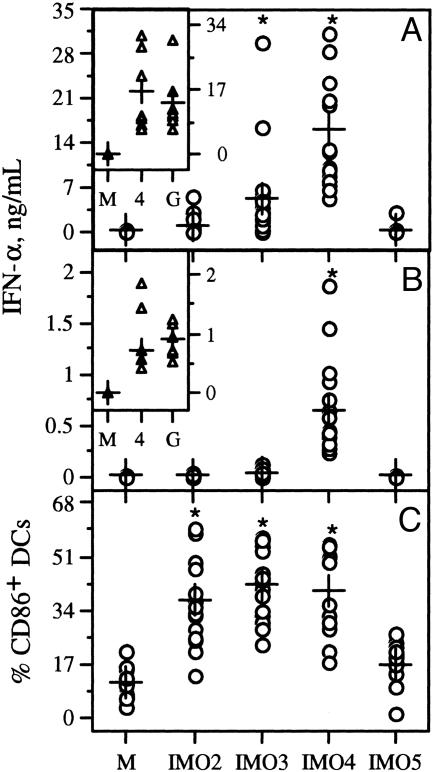
IMOs Activated Human pDCs and PBMCs and Induced IFN-α Secretion. IMO 4 induced the highest levels of IFN-α production by both human pDCs and PBMCs among the three IMOs (Fig. 2 A and B). The levels of IFN-α induced by IMO 4 in human pDC and PBMC cultures matched the levels of IFN-α induced by a poly(dG) CpG oligomer (class A) (Fig. 2 A Inset and B Inset, respectively). IMO 3 induced strong IFN-α secretion compared with medium and control non-CpG IMO 5 in pDC cultures (Fig. 2A) and low levels of IFN-α in PBMC cultures (Fig. 2B). IMO 2 induced insignificant levels of IFN-α, as has been reported for conventional class-B CpG oligomers such as 1 (12, 13). All three IMOs increased expression of the early-activation marker CD86 on pDCs compared with control IMO 5 (Fig. 2C). These results suggest that all three IMOs activated pDCs, but only IMOs 3 and 4 induced IFN-α secretion in pDCs.
Fig. 2.
IMOs induce IFN-α secretion and activate pDCs. IFN-α induction profiles of IMOs in pDCs isolated from human PBMCs (A) and human PBMCs obtained from the blood of 15–18 healthy human volunteers (B). Cells were stimulated with IMOs at 10 μg/ml concentration for 24 h. Insets show comparison of IFN-α secretion levels by IMO 4 and a class-A CpG oligomer represented as G. Results shown are for 8–12 donors. (C) Stimulation of isolated pDCs with IMOs up-regulated expression of costimulatory molecule CD86. M indicates medium control without IMO stimulation. Each data point represents one donor. ±, mean value; *, significance compared with control IMO 5.
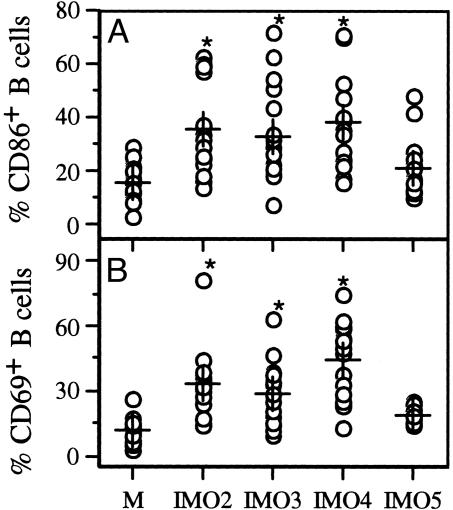
Activation of Human B Cells by IMOs. IMOs 2–4 enhanced B-cell proliferation in a dose-dependent fashion at lower doses and inhibited proliferation at higher doses (see Fig. 9, which is published as supporting information on the PNAS web site), resulting in a bell-shaped dose-response curve. Control IMOs 5 and 6 induced low levels of nonspecific B-cell proliferation, as has been reported with phosphorothioate oligomers. Consistent with the ability of IMOs to induce B-cell proliferation, expression of CD86 and CD69 increased after IMO stimulation compared with medium or control IMO 5 (Fig. 3). In contrast to IFN-α induction in pDC cultures, all three IMOs induced similar levels of B-cell proliferation at a concentration of 1 μg/ml. Further, IMOs 2–4 induced IL-6 secretion in isolated B-cell cultures (data not shown), whereas control non-CpG IMO 5 did not, suggesting that all three IMOs activated B cells to proliferate and secrete cytokines and enhanced the expression of surface markers.
Fig. 3.
Stimulation of PBMCs with IMOs up-regulated expression of costimulatory molecules CD86 (A) and CD69 (B) on B cells. M indicates medium control without IMO stimulation. Each data point represents one donor; + indicates mean value; * indicates statistical significance compared with control IMO 5.
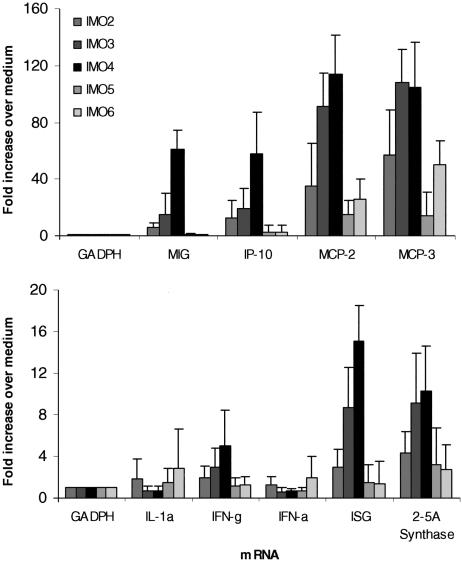
IMOs Enhanced Chemokine and Cytokine Gene Expression in PBMCs. The stimulation of PBMCs with IMOs 2–4 potently up-regulated the expression of chemokines, IP-10, MCP-2, MCP-3, and MIG compared with control IMOs 5 and 6 (Fig. 4A); the levels of gene expression induced by the three IMOs varied. None of the IMOs significantly induced IL-1α gene expression compared with control IMOs (Fig. 4B), but expression of IFN-inducible genes, such as 2′,5′-A-synthetase and ISG, was increased (Fig. 4B).
Fig. 4.
Profiles of chemokine and cytokine gene expression modulation by IMOs in human PBMCs. PBMCs from three to six healthy human donors were cultured for 24 h with 10 μg/ml IMOs, and mRNA was isolated and analyzed by TaqMan RT-PCR. Each value is mean ± SD.
High levels of IFN-α were detected by ELISA at 24 h in PBMC cultures with IMO 4, but IFN-α mRNA was not detected (Fig. 4B). We therefore studied the kinetics of expression of IFN-α mRNA after IMO stimulation of human PBMCs. The kinetics data suggest that IFN-α mRNA appeared 3 h after stimulation of PBMC with IMOs, and levels peaked by 10 h and returned to basal levels in 24 h (see Table 3, which is published as supporting information on the PNAS web site). IFN-γ mRNA levels also increased by 10 h after stimulation and returned to low levels at 24 h (Table 3).
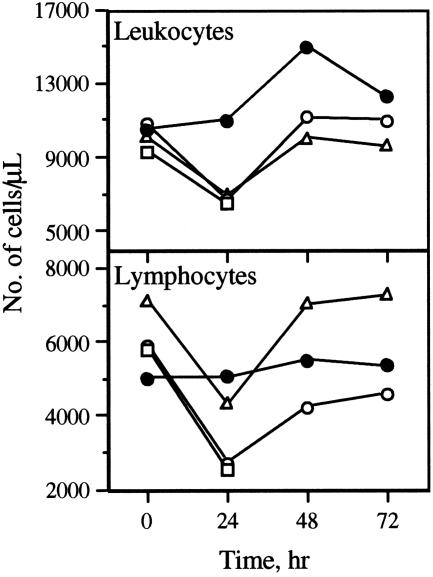
Effect of IMOs on Peripheral Blood Immune Cells of Monkeys. Within 24 h after s.c. administration of IMOs at a dosage of 1 mg/kg, total numbers of circulating leukocytes and lymphocytes decreased in monkeys that received IMOs 2–4 compared with those that received control IMO 6 (Fig. 5). The migration of immune cells from circulation was transient, however, because the numbers of both leukocytes and lymphocytes returned to preadministration levels in the next 24–48 h (Fig. 5). Administration of control IMO 6 failed to produce such dynamic changes in either leukocyte or lymphocyte numbers in the peripheral blood of monkeys. Less significant changes in neutrophil and monocyte numbers were observed in monkeys that received IMOs 2–4 (data not shown).
Fig. 5.
Monkey in vivo immunostimulatory profiles of IMOs. Changes in leukocyte and lymphocyte numbers were as determined by hematological assessment at different time intervals after s.c. administration of 1 mg/kg IMO 2 (□), IMO 3 (○), IMO 4 (▵), or IMO 6 (•). Each value is an average of three monkeys.
The administration of IMOs 2–4 resulted in a decrease in all populations of lymphocytes in whole blood 24 h after IMO administration as determined by FACS analysis (see Fig. 10, which is published as supporting information on the PNAS web site). All three IMOs caused a transient decrease in CD3+ T lymphocytes, CD3+/CD4+ helper/inducer T lymphocytes, CD3+/CD8+ suppressor/cytotoxic T lymphocytes, CD3–/CD16+ natural killer (NK) cells, and CD20+ B lymphocytes (Fig. 9). In contrast, control IMO 6 failed to produce any changes in total lymphocytes, except that the number of CD20+ B lymphocytes decreased 24 h after IMO 6 administration.
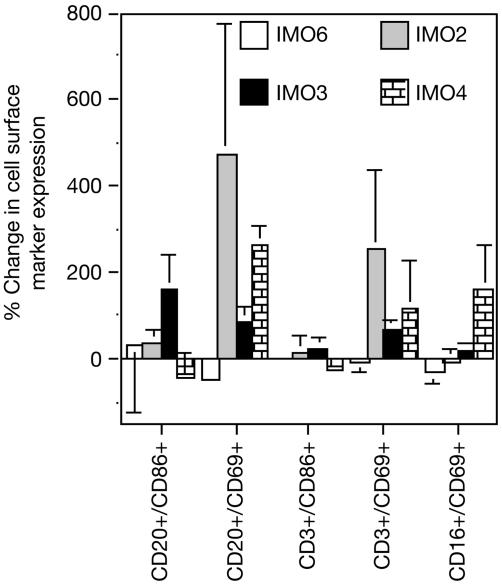
Immune Cell Activation by IMOs in Monkeys. All three IMOs induced significant activation of CD86 and/or CD69 surface markers on CD20+ B lymphocytes, CD3+ T lymphocytes, and CD16+ NK cells as determined by FACS analysis (Fig. 6). IMO 3 produced a greater induction of CD86 on CD20+ B lymphocytes than did IMOs 2 and 4. All three IMOs induced CD69, but not CD86, expression on CD3+ T lymphocytes. IMO 4 induced significant expression of CD69 on CD16+ NK cells. Control IMO 6 induced minimal or no activation of surface markers on lymphocytes (Fig. 6).
Fig. 6.
Activation of cell surface markers on B, T, and NK cells 24 h after s.c. administration of 1 mg/kg IMOs in monkeys.
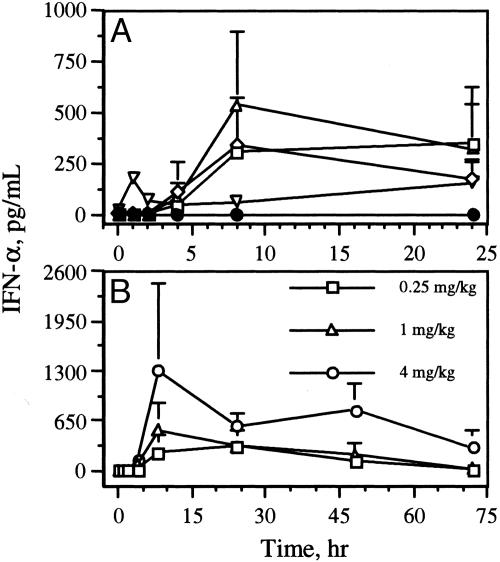
Cytokine Profiles of IMOs in Monkeys. IMOs 2–4 induced higher levels of IFN-α secretion in vivo than did control IMO 6 (Fig. 7A). These in vivo results of IMO2 are in contrast with the results of human cell-culture assays. With all three IMOs, IFN-α levels peaked by 8 h; at 24 h, significantly higher levels of IFN-α were still detected than before administration. At 72 h after administration of IMO 4, IFN-α levels had returned nearly to basal values (Fig. 7B). The induction of IFN-α by IMOs 3 (data not shown) and 4 (Fig. 7B) was dose-dependent. IMOs 2–4 induced minimal levels of IL-6 compared with IMO 6 (see Fig. 11, which is published as supporting information on the PNAS web site). No significant changes in IL-12 levels were observed with any of the IMOs (Fig. 11), but in general IL-12 levels dropped slightly after IMO administration and returned to normal levels within 4 h (Fig. 11).
Fig. 7.
IFN-α induction profiles of IMOs in monkeys. (A) Plasma IFN-α levels induced by CpG DNA 1 (▿), IMO 2 (□), IMO 3 (⋄), IMO 4 (▵), and control IMO 6(•) in monkeys. (B) Dose-dependent IFN-α induction by IMO 4 in monkeys.
Discussion
CpG oligomers promote B-cell proliferation, Ig production, and the secretion of a number of T helper 1-type cytokines, including IL-12, IFN-γ, IL-6, and TNF-α, from B-cells, monocytes/macrophages, DCs, and NK cells. TLR9 is shown to be engaged in the recognition of CpG oligomer and the subsequent initiation of a signaling cascade in a MyD88-dependent fashion (1) leading to the activation of the transcription factors NF-κB (32) and AP-1 (33). Although a CpG dinucleotide is essential for TLR9 activation, many other factors, such as the nucleotides flanking the CpG dinucleotide, the presence of an accessible 5′ end, the position of the CpG dinucleotide in the oligomer, and the secondary structure of the oligomer, play a critical role in the activation of immune cells.
Our earlier studies showed that TLR9 recognizes a CpR dinucleotide motif in oligomers and induces immune responses (23, 29–31). Our studies also demonstrated that TLR9 required an accessible 5′ end and that oligomers containing two 5′ ends, referred to as immunomers, had greater activity than did those with a single 5′ end (24–31, 34), which was later confirmed by other research groups (35, 36). In a systematic study, we showed that immunomers containing 11 nt in each branch were optimal for TLR9 recognition and immune stimulation (26). In the present study, we examined the immunostimulatory properties of IMOs containing CpR dinucleotide in different sequence contexts in vitro and in vivo. The results obtained in experiments with HEK293 cells expressing TLR9 suggested that both CpR dinucleotide in IMOs and CpG dinucleotide in oligomer 1 signal through TLR9. Although both motifs signal through TLR9, our earlier studies showed that IMOs containing CpR dinucleotide induce higher or similar IL-12 and lower IL-6 secretion compared with CpG-containing IMOs (29–31). Consistent with earlier observations, in the present study IMOs 2–4 also induced IL-12 and IFN-γ, but not IL-6, secretion in mice.
In human cell-culture assays, IMOs induced IFN-α secretion and surface marker (CD86) expression by pDCs in a sequence-dependent manner. Although CD86 expression was stimulated to a similar extent by all three IMOs that contained CpR dinucleotides, IMO 4, which contained both a terminal TCR nucleotide sequence and a palindromic sequence, induced higher levels of IFN-α than did IMO 3, which contained a terminal TCR nucleotide sequence but no palindromic sequence. IMO 2, which contained neither a terminal TCR nor a palindromic sequence, induced no IFN-α production in human cell-culture assays. Only IMO 4 induced IFN-α secretion in PBMC cultures.
We compared the IFN-α levels induced by the CpR-dinucleotide-containing IMOs 3 and 4 with those of a class-A CpG oligomer (G in Fig. 2 Insets) that was known to induce higher levels of IFN-α. These results demonstrate that IMOs containing CpR dinucleotides induced IFN-α secretion as strongly as the class A CpG oligomer and did so in a sequence- and secondary structure-dependent fashion. Although a 5′-TCR nucleotide sequence provides a stimulus for IFN-α secretion by pDCs, it is not sufficient to induce secretion of IFN-α in PBMC cultures. The results obtained with IMO 4 indicate that a secondary structure is necessary for IFN-α induction in PBMC cultures.
All three IMOs induced human B-cell proliferation to a similar extent, and the proliferation activity depended on the concentration of the IMO and the presence of a CpR dinucleotide. Comparison of the B-cell proliferation induced by control IMOs 5 and 6 suggests that the R moiety by itself did not contribute to immune stimulation, and it was immunostimulatory only when present in the G position of a CpG dinucleotide (CpR) through TLR9 activation.
The most interesting and surprising results arose in the studies of IMOs administered in vivo to nonhuman primates. IMO 2 did not induce IFN-α production in human cell-culture assays, but it induced significantly greater IFN-α production in monkeys than did control IMO 6. IMO 4 induced the highest levels of IFN-α of the three IMOs. These results confirm that (i) synthetic CpR dinucleotide containing IMOs induce IFN-α secretion in humans and nonhuman primates, and (ii) whereas in vitro studies provide a guidance, they do not always predict in vivo effects.
As has been shown with recombinant cytokines in humans (37, 38), IMO administration to monkeys resulted in a transient decrease in lymphocyte and leukocyte numbers in peripheral blood. Such changes in immune-cell numbers in blood were not observed with the administration of control IMO 6, however, suggesting that these changes were related to the immunostimulatory activity of IMOs 2–4. Consistent with human cell-culture results, the administration of IMOs 2–4 to monkeys resulted in an increase in the expression of CD86 and CD69 surface markers on B, NK, and T cells. These results demonstrate the immunostimulatory effects of IMOs that contain previously undescribed DNA structures and synthetic CpR dinucleotides.
In summary, our current results demonstrate that IMOs containing two 5′ ends induce rapid and higher levels of cytokine secretion in vivo in mice. Activation of the immune system by IMOs containing CpR dinucleotides occurs through TLR9, as is the case of bacterial DNA and synthetic oligomers containing natural unmethylated CpG dinucleotides. Although all three IMOs with CpR dinucleotides activated human B cells to proliferate, express surface markers, and secrete cytokines, and activated human pDC to express higher levels of CD69 and CD86 markers, only IMOs containing a 5′-TCR nucleotide sequence induced IFN-α secretion in pDC cultures. In PBMC cultures, all three IMOs showed up-regulation of cytokine and chemokine gene expression that depend on the sequence- and secondary-structure-forming nature of the IMOs.
In monkeys, IMOs containing CpR dinucleotides induced a transient decrease in lymphocytes in the circulating blood and increased CD69 and CD86 surface-marker expression on B, NK, and T cells. In contrast to cell-culture studies, all three IMOs induced IFN-α production in vivo in monkeys. However, the IMO that has a nucleotide sequence that allows formation of a secondary structure induced higher and more sustained secretion of IFN-α (up to 72 h) than did IMOs that did not have secondary structure-forming sequence. These previously undescribed IMOs that induce distinct cytokine profiles may be useful in the treatment of cancer, infectious diseases, asthma, and allergies either alone or in combination with antigens, vaccines, mAbs, and chemotherapeutic agents.
Supplementary Material
Acknowledgments
We thank the members of WIL Research Laboratories, MPI Research, and FAST Systems for help with monkey experiments and hematological and flow cytometry analysis of the samples.
Author contributions: E.R.K. and S.A. designed research; T.S. was involved with nonhuman primate studies; L.B., Y.L., D.Y., D.W., Y.-P.C., S.S.S., and J.X.T. performed research; E.R.K., L.B., Y.L., D.Y., and D.W. analyzed data; and E.R.K. wrote the paper.
Abbreviations: CpG, cytosine-phosphate-guanine dinucleotide; CpR, cytosine-phosphate-2′-deoxy-7-deazaguanosine dinucleotide; DC, dendritic cell; pDC, plasmacytoid DC; IMO, immunomodulatory oligonucleotide; NK, natural killer; PBMC, peripheral blood mononuclear cell; TLR, Toll-like receptor.
References
- 1.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K. & Akira, S. (2000) Nature 408, 740–745. [DOI] [PubMed] [Google Scholar]
- 2.Tokunaga, T., Yamamoto, H., Shimada, S., Abe, H., Fukuda, T., Fujisawa, Y., Furutani, Y., Yano, O., Kataoka, T., Sudo, T., et al. (1984) J. Natl. Cancer Inst. 72, 955–962. [PubMed] [Google Scholar]
- 3.Messina, J. P., Gilkeson, G. S. & Pisetsky, D. S. (1991) J. Immunol. 147, 1759–1764. [PubMed] [Google Scholar]
- 4.Klinman, D. M., Yi, A. K., Beaucage, S. L., Conover, J. & Krieg, A. M. (1996) Proc. Natl. Acad. Sci. USA 93, 2879–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao, Q., Temsamani, J., Zhou, R. Z. & Agrawal, S. (1997) Antisense Nucleic Acid Drug Dev. 7, 495–502. [DOI] [PubMed] [Google Scholar]
- 6.Kandimalla, E. R. & Agrawal, S. (2005) in Toll and Toll-Like Receptors: An Immunologic Perspective, ed. Rich, T. (Kluwer Academic/Plenum, New York) pp. 181–212.
- 7.Klinman, D. M. (2004) Nat. Rev. Immunol. 4, 249–258. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto, S., Kuramoto, E., Shimada, S. & Tokunaga, T. (1988) Jpn. J. Cancer Res. 79, 866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krieg, A. M., Yi, A. K., Matson, S., Waldschmidt, T. J., Bishop, G. A., Teasdale, R., Koretzky, G. A. & Klinman, D. M. (1995) Nature 374, 546–549. [DOI] [PubMed] [Google Scholar]
- 10.Sato, Y., Roman, M., Tighe, H., Lee, D., Corr, M., Nguyen, M. D., Silverman, G. J., Lotz, M., Carson, D. A. & Raz, E. (1996) Science 273, 352–354. [DOI] [PubMed] [Google Scholar]
- 11.Verthelyi, D., Ishii, K. J., Gursel, M., Takeshita, F. & Klinman, D. M. (2001) J. Immunol. 166, 2372–2377. [DOI] [PubMed] [Google Scholar]
- 12.Marshall, J. D., Fearon, K., Abbate, C., Subramanian, S., Yee, P., Gregorio, J., Coffman, R. L. & vanNest, G. (2003) J. Leukocyte Biol. 73, 781–792. [DOI] [PubMed] [Google Scholar]
- 13.Hartman, G., Battiany, J., Poeck, H., Wagner, M., Kerkmann, M., Lubenow, N., Rothenfusser, S. & Endres, S. (2003) Eur. J. Immunol. 33, 1633–1641. [DOI] [PubMed] [Google Scholar]
- 14.Zhao, Q., Temsamani, J., Iadarola, P. L., Jiang, Z. & Agrawal, S. (1996) Biochem. Pharmacol. 51, 173–182. [DOI] [PubMed] [Google Scholar]
- 15.Zhao, Q., Yu, D. & Agrawal, S. (1999) Bioorg. Med. Chem. Lett. 9, 3453–3458. [DOI] [PubMed] [Google Scholar]
- 16.Zhao, Q., Yu, D. & Agrawal, S. (2000) Bioorg. Med. Chem. Lett. 10, 1051–1054. [DOI] [PubMed] [Google Scholar]
- 17.Yu, D., Kandimalla, E. R., Zhao, Q., Cong, Y. & Agrawal, S. (2002) Nucleic Acids Res. 30, 1613–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal, S. & Kandimalla, E. R. (2001) Curr. Cancer Drug Targets 1, 197–209. [DOI] [PubMed] [Google Scholar]
- 19.Yu, D., Kandimalla, E. R., Zhao, Q., Cong, Y. & Agrawal, S. (2001) Bioorg. Med. Chem. 9, 2803–2808. [DOI] [PubMed] [Google Scholar]
- 20.Yu, D., Kandimalla, E. R., Zhao, Q., Cong, Y. & Agrawal, S. (2001) Bioorg. Med. Chem. Lett. 11, 2263–2267. [DOI] [PubMed] [Google Scholar]
- 21.Yu, D., Kandimalla, E. R., Zhao, Q., Bhagat, L., Cong, Y. & Agrawal, S. (2003) Bioorg. Med. Chem. 11, 459–464. [DOI] [PubMed] [Google Scholar]
- 22.Yu, D., Kandimalla, E. R., Cong, Y., Tang, J., Tang, J. Y., Zhao, Q. & Agrawal, S. (2002) J. Med. Chem. 45, 4540–4548. [DOI] [PubMed] [Google Scholar]
- 23.Kandimalla, E. R., Yu, D., Zhao, Q. & Agrawal, S. (2001) Bioorg. Med. Chem. 9, 807–813. [DOI] [PubMed] [Google Scholar]
- 24.Yu, D., Zhao, Q., Kandimalla, E. R. & Agrawal, S. (2000) Bioorg. Med. Chem. Lett. 10, 2585–2588. [DOI] [PubMed] [Google Scholar]
- 25.Kandimalla, E. R., Bhagat, L., Yu, D., Cong, Y., Tang, J. & Agrawal, S. (2002) Bioconjug. Chem. 13, 966–974. [DOI] [PubMed] [Google Scholar]
- 26.Yu, D., Kandimalla, E. R., Bhagat, L., Tang, J. Y., Cong, Y., Tang, J. & Agrawal, S. (2002) Nucleic Acids Res. 30, 4460–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, D., Zhu, F. G., Bhagat, L., Wang, H., Kandimalla, E. R., Zhang, R. & Agrawal, S. (2002) Biochem. Biophys. Res. Commun. 297, 83–90. [DOI] [PubMed] [Google Scholar]
- 28.Bhagat, L., Zhu, F. G., Yu, D., Tang, J., Wang, H., Kandimalla, E. R., Zhang, R. & Agrawal, S. (2003) Biochem. Biophys. Res. Commun. 300, 853–861. [DOI] [PubMed] [Google Scholar]
- 29.Kandimalla, E. R., Bhagat, L., Wang, D., Yu, D., Tang, J., Wang, H., Huang, P., Zhang, R. & Agrawal, S. (2003) Nucleic Acids Res. 31, 2393–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agrawal, D. K., Edwan, J., Kandimalla, E. R., Yu, D., Bhagat, L., Wang, D. & Agrawal, S. (2004) Int. Immunopharmacol. 4, 127–138. [DOI] [PubMed] [Google Scholar]
- 31.Wang, D., Li, Y., Yu, D., Song, S. S., Kandimalla, E. R. & Agrawal, S. (2004) Int. J. Oncol. 24, 901–908. [PubMed] [Google Scholar]
- 32.Stacey, K. J., Sweet, M. J. & Hume, D. A. (1996) J. Immunol. 157, 2116–2122. [PubMed] [Google Scholar]
- 33.Yi, A.-K. & Krieg, A. M. (1998) J. Immunol. 161, 4493–4497. [PubMed] [Google Scholar]
- 34.Kandimalla, E. R., Bhagat, L., Cong, Y.-P., Yu, D., Zhu, F.-G., Wang, D., Tang, J. X., Tang, J.-Y., Knetter, C. F., Lien, E. & Agrawal, S. (2003) Proc. Natl. Acad. Sci. USA 100, 14303–14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall, J. D., Hessel E. M., Gregorio, J., Abbate, C., Yee, P., Chu, M., vanNest, G., Coffman, R. L. & Fearon, K. L. (2003) Nucleic Acids Res. 31, 5122–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayanan, S., Dalpke, A. H., Siegmund, K., Heeg, K. & Richert, C. (2003) J. Med. Chem. 46, 5031–5044. [DOI] [PubMed] [Google Scholar]
- 37.Aulitzky, W. E., Tilg, H., Vogel, W., Aulitzky, W., Berger, M., Gastl, G., Herold, M. & Huber, C. (1991) Ann. Hematol. 62, 25–31. [DOI] [PubMed] [Google Scholar]
- 38.Robertson, M. J., Pelloso, D., Abonour, R., Hromas, R. A., Nelson, R. P., Jr., Wood, L. & Cornetta, K. (2002) Clin. Cancer Res. 8, 3383–3393. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.