Abstract
Expression of SHP-1 phosphatase, a key negative regulator of cell signaling, is lost in T cell lymphomas and other malignancies due to DNA methylation of the SHP-1 promoter by a currently undefined mechanism. We demonstrate that malignant T cells express DNA methyltransferase (DNMT) 1 and that constantly activated signal transducer and activator of transcription (STAT) 3 is capable of binding in vitro to DNA oligonucleotides corresponding to four STAT3 SIE/GAS binding sites identified in the SHP-1 promoter. STAT3, DNMT1, and histone deacetylase 1 form complexes and bind to the SHP-1 promoter in vivo. Treatment with pharmacologic grade DNMT1 anti-sense oligonucleotides and STAT3 small-interfering RNA induces in the malignant T cells DNA demethylation and expression of SHP-1 gene. These data indicate that STAT3 may, in part, transform cells by inducing epigenetic silencing of SHP-1 in cooperation with DNMT1 and, apparently, histone deacetylase 1. Reversal of such gene silencing represents an attractive aim for novel anticancer therapies.
Keywords: DNMT1, SHP-1, STAT3
Epigenetic gene silencing plays a key role in inhibiting tumor suppressor gene expression in cancer cells (1–3). The silencing affects gene promoter region and is executed by two principal mechanisms: methylation of DNA enriched in the CpG sequences and modification of histones. Whereas the CpG methylation is mediated by DNA methyltransferase 1 (DNMT1) and other members of the DNMT family, histone modifiers include histone deacetylases (HDACs).
T cell lymphomas are heterogeneous and include primary cutaneous T cell lymphomas (CTCL) and lymphomas that express ALK tyrosine kinase (ALK+ TCL). Persistent activation of cell signal transducer and activator of transcription (STAT) 3 seems to play a critical role in the pathogenesis of CTCL (4, 5), ALK+ TCL (6, 7) and many other malignancies (8).
SHP-1 tyrosine phosphatase is a key negative regulator of intracellular signaling (9–11). SHP-1 is ubiquitously expressed in hematopoietic and nonhematopoietic cells through transcription from two separate promoters (promoter 2 and 1, respectively). We found that malignant T cells derived from the large-cell type of CTCL fail to express SHP-1 due to CpG methylation of the promoter 2 (12). More recent studies by others (13, 14) demonstrated SHP-1 promoter methylation in a large spectrum of lymphoid and nonlymphoid malignancies. The mechanism of the SHP-1 gene silencing in the malignant cells is currently unknown.
Materials and Methods
Cell Lines. The cell lines used in this study have been described in detail before (4, 6, 12). In brief, PB-1, 2A, and 2B were established from a patient with a progressive CTCL. JB6 and SUDHL-1 were derived from two different ALK+ TCL and carry the t(2;5) chromosomal translocation involving ALK and NPM genes.
Quantitative RT-PCR. Total cellular RNA extracted with RNeasy kit (Qiagen, Valencia, CA) was converted to cDNA with SuperScript II reverse transcriptase (GIBCO/BRL) and purified as described in ref. 6. PCR was performed in duplicate for 30 cycles in the standard reaction and 40 cycles in the quantitative (real-time) PCR by using Applied Biosystems PRISM 7700 Sequence Detection System with the following sets of primers: SHP-1, 5′-AATGCGTCCCATACTGGCCCGA-3′ and 5′-CCCGCAGTTGGTCACAGAGT-3′; and DNMT1, 5′-CCAAAGCCCGAGAGAGTGCCTCAG-3′ and 5′-CCTTAGCAGCTTCCTCCTCCTT-3.
Western Blotting and Coimmunoprecipitation. These experiments were performed as described in refs. 4 and 6 by using enhanced chemiluminescence and antibodies against SHP-1, DNMT1, DNMT3A, STAT3, STAT5, SOCS3, BCL-XL, p300, CBP, and actin (all from Santa Cruz Biotechnology) and HDAC1 (Upstate Biotechnology, Lake Placid, NY).
Immunohistochemical Staining. The staining was performed as described in ref. 12 with formalin fixed tissue sections by using antigen retrieval and streptavidin–biotin complex techniques and the antibodies against SHP-1 and DNMT1 (Santa Cruz Biotechnology), HDAC1 (Upstate Biotechnology), and ALK (DAKO).
DNA Methylation Analysis. The genomic DNA isolated with the DNeasy Tissue Kit (Qiagen) was modified by bisulfite treatment with the CpGenome DNA Modification Kit (Intergen, Purchase, NY) and amplified by PCR with two sets of SHP-1 promoter specific primer pairs that recognize either the methylated or unmethylated CpG sequences and analyzed by electrophoresis. For the DNA sequence analysis, PCR products obtained with the two sets of primers to cover the proximal SHP-1 promoter with 7 CpG sites, and the extended promoter region with 18 sites was separated on agarose gel, purified by using the QIAEX II gel purification kit (Qiagen), and cloned into pCR2.1 vector by using the TA Cloning Kit (Invitrogen). Products of the sequencing PCR performed with the T7 and M13 primers were analyzed on an automated DNA sequencer.
EMSA. The assays were performed as described in ref. 6. In brief, nuclear proteins were extracted and incubated with the 23-base-long, digoxigenin-labeled DNA oligonucleotides (ON) probes listed in Fig. 4B. In the supershift EMSA, anti-STAT3 and control ALK antibodies (Santa Cruz Biotechnology) were added to the cell lysates before their incubation with the ON probes.
Fig. 4.
Binding of malignant T cell-derived STAT3 to SHP-1 promoter 2 SIE/GAS sites. (A) EMSA with DNA ON that correspond to four STAT-binding sites designated SIE/GAS-1 to -4/wild type (WT) are listed in B. (B) Binding to the mutated STAT binding sites (the mutated bases are underlined). (C) Supershift EMSA with the depicted antibodies (Ab).
Chromatin Immunoprecipitation (ChIP) Assays. Chromatin-containing lysates were incubated with the antibodies listed above and in Fig. 5. DNA–protein immunocomplexes were precipitated with protein A-agarose beads and treated with RNase A and proteinase K; the DNA samples were extracted with phenol/chloroform, precipitated with ethanol and PCR amplified by using primers specific for SHP-1 promoter (5′-ATGATAAAGATAGCCCCTGTT-3′ and 5′-TCATCGAGTGAGTCCTGCTG-3′) and, as a control, exon 16 (5′-CTGACCCTGTGGAAGCATTT-3′ and 5′-AGTGATCCCAGGGCTTTATTT-3′). In the selected samples, the immunoprecipitation was followed by reimmunoprecipitation with a different antibody.
Fig. 5.
Binding of STAT3, DNMT1, and HDAC1 to SHP-1 promoter 2 in vivo. (A and B) Cell lysates from the listed malignant T cell and normal T cell-rich populations were examined in the ChIP assay by using the antibodies reactive with STAT3, DNMT1, HDAC1 (and, in A, p300 and CBP), and PCR primer pairs corresponding to SHP-1 promoter and exon 16 (Ex. 16). (C) STAT3 immunoprecipitate and supernatant from the 2A cell lysate were reimmunoprecipitated with anti-DNMT1 antibody or, as control, protein A-Sepharose beads.
Cell Transfections. DNMT1 AS-ON(1) (5′-AACCATGAGCACCGTTCTCC-3′) and DNMT1 AS-ON(2) (5′-TTCATGTCAGCCAAGGCCAC-3′), corresponding to exon 1 and 3′ untranslated region, respectively, and control, scrambled sequence oligonucleotides were HPLC-purified and FITC-labeled. STAT3 and control nonsense small interfering RNA (siRNA) mixtures were purchased from Upstate Biotechnology. Malignant T cells were transfected by using Lipofectamine (GIBCO/BRL).
Results
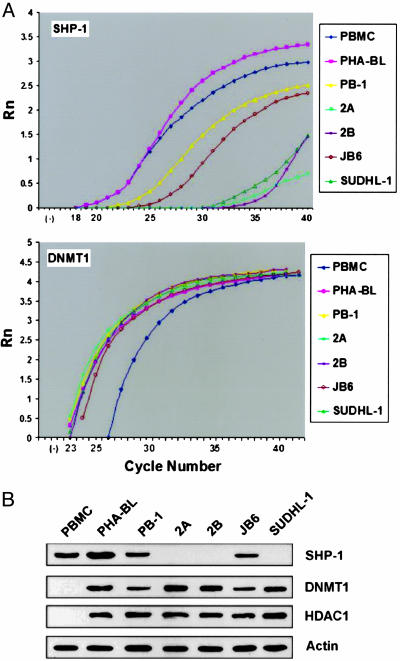
SHP-1, DNMT1, and HDAC1 Expression in T Cells. Real-time RT-PCR performed on the mRNA-derived cDNA showed that all T cell lymphoma cell lines examined displayed either a profound or moderate decrease in the amount of SHP-1 message as compared with normal peripheral blood mononuclear cells (PBMC) and phytohemagglutinin-activated T cell blasts (PHA-BL) that served as controls (Fig. 1A Upper). A similar pattern of SHP-1 loss was seen also on the protein level (Fig. 1B). In contrast, all T cell lymphoma cell lines expressed a robust DNMT1 message (Fig. 1A Lower) in the concentration comparable with PHA-BL and significantly higher than PBMC. The malignant T cells and PHA-BL also expressed DNMT1 protein (Fig. 1B). No DNMT1 protein could be detected in PBMC, suggesting either a very low concentration of the protein or the existence of posttranscriptional regulation of DNMT1 expression.
Fig. 1.
SHP-1, DNMT1, and HDAC1 expression in malignant T cells. (A) mRNA expression of SHP-1 (Upper) and DNMT1 (Lower) was determined by real-time RT-PCR. (B) Expression of SHP-1, DNMT1, and HDAC1 proteins.
Because HDAC1 can interact with DNMT1 (15), we examined its expression in the same set of malignant and normal T cells. Similar to DNMT1, HDAC1 protein was ubiquitously expressed by the malignant T cells (Fig. 1C). HDAC1 could be detected also in PHA-BL but was undetectable in PBMC.
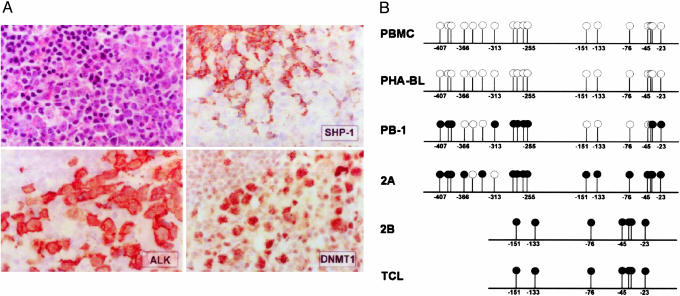
Expression of SHP-1, DNMT1, and HDAC1 in T Cell Lymphoma Tissues. To determine whether the loss of SHP-1 expression is associated with expression of DNMT1 and HDAC1 proteins also in the native T cell lymphomas, we performed immunohistochemical staining of the CTCL and ALK+ TCL tissue samples. Similar to the large-cell CTCL (12), ALK+ TCL demonstrated the lack of SHP-1 protein expression in essentially all malignant cells in all cases examined (Fig. 2A). In contrast, the ALK+ TCL and CTCL cells universally displayed strong expression of DNMT1 (Fig. 2A) and HDAC1 (data not shown).
Fig. 2.
SHP-1 and DNMT1 expression and SHP-1 promoter DNA methylation in T cell lymphoma tissues and cell lines. (A) Expression of ALK, SHP-1, and DNMT1 proteins in ALK+ TCL tissue. (B) DNA methylation of the SHP-1 promoter 2. ○, unmethylated CpG sites; •, methylated CpG sites.
DNA Methylation of SHP-1 Promoter 2 in Malignant T Cells. Methylation-sensitive PCR revealed methylation of the promoter in all five T-cell lymphoma cell lines and three CTCL tissues evaluated but not the PBMC and PHA-BL from three healthy individuals (data not shown). To examine in greater detail the extent of the methylation, we performed sequence analysis of the promoter (positions –200 to –20 and –400 to –20 that contained 7 and 18 CpG sites, respectively) by using three CTCL cell lines, one CTCL tissue, and normal PBMC and PHA-BL. As schematically depicted in Fig. 2B, neither PBMC nor PHA-BL displayed methylation of any of the 18 CpG sites. However, two malignant T cell lines, 2A and 2B, that express very little detectable SHP-1 mRNA (Fig. 1A) and no detectable SHP-1 protein (Fig. 1C) displayed essentially complete methylation of the CpG sites. PB-1 T cell line that shows the partial loss of SHP-1 mRNA and protein (Fig. 1 A and C, respectively), showed an intermediate degree of the CpG methylation with no appreciable common pattern among the evaluated clones. A tissue sample from the large-cell CTCL that also showed the lack of detectable SHP-1 protein (12) displayed complete methylation of the CpGs.
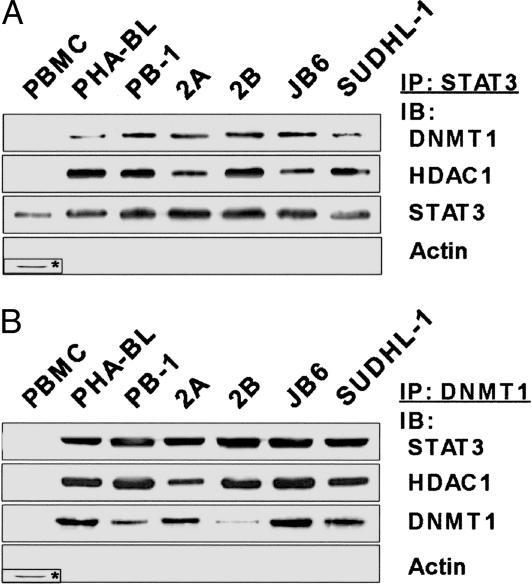
Binding of DNMT1 and HDAC1 to STAT3. Because neither DNMT1 nor HDAC1 binds DNA directly, we reasoned that the malignant T cells contain an activated transcription factor that anchors these enzymes to the SHP-1 promoter. We have focused on STAT3 because it has documented perpetual activation in both CTCL (4, 5) and ALK+ TCL (6, 7). Therefore, we tested next whether STAT3 formed complexes with DNMT1 and HDAC1 in the malignant T cells. As shown in Fig. 3, STAT3 did coprecipitate with both these proteins.
Fig. 3.
In vivo association of STAT3 with DNMT1 and HDAC1 in T cells. Cell lysates from malignant and normal T-cell populations were immunoprecipitated with an anti-STAT3 (A) and DNMT1 (B) antibodies and, in the case of PBMC, control anti-actin (*) antibody. Lysates were immunoblotted with all of the listed antibodies.
Binding of STAT3 to SHP-1 Promoter 2. We next analyzed the DNA sequence of the promoter (9) for potential STAT3 binding sites. All STATs bind to SIE/GAS DNA sequences that contain the TT and AA duplicates typically separated by five bases: 5′-TT(N5)AA-3′, where N identity may be quite diverse (16, 17). Whereas STAT5 displays a rather restricted binding specificity to the nanomers TTCC/TNG/AGAA (18), STAT3 binds not only to the TT(N5)AA nanomers, but also to TT(N4)AA octamers and TT(N6)AA decamers.
Our analysis identified four potential STAT3 binding sites depicted in Fig. 4. EMSA revealed that most malignant T cell lysates bound to all four ON probes (Fig. 4A). The binding capacity corresponded inversely with the expression of SHP-1 (Fig. 1C): protein isolates from all three cell lines (2A, 2B, and SUDHL1) that failed to express SHP-1 displayed the binding, whereas proteins isolated from the cells that show high to intermediate SHP-1 expression (PHA-BL, PB-1, and JB6) displayed nondetectable to weak binding.
To provide additional evidence of specificity of the binding, we performed mutational analysis of the identified STAT3 binding sites. Three types of mutants, designated M1, M2, and M3 and listed in Fig. 4B, were prepared for all four sites; M1 replaced the internal bases within the TT(N4–6)AA sequences, leaving intact the flanking TT and AA dimers and the remainder of the 23-mer ON. In the M2 mutants, both TT and AA dimers were replaced, whereas the rest of the oligonucleotide sequence remained unchanged. Finally, in M3, only one of the TT and AA dimers was substituted. All mutations changed binding properties of all four sequences. The degree of change varied substantially and was mutant type, rather then sequence specific. The M1 mutants displayed quite diverse, yet overall significantly weaker, binding. The M2 showed essentially no binding; M3 mutants were similarly inactive.
The selective expression of activated STAT3 in the malignant T cells (4, 6, 12) and the above described, STAT3-favoring features of the identified SIE/GAS sites, strongly suggested that STAT3 is the STAT that binds to the SHP-1 promoter. To provide the more direct evidence, we performed the EMSA supershift by using a STAT3-specific antibody. As shown in Fig. 4C, the antibody treatment led to diminished density of the basal band and to formation of the “supershifted” band; preincubation with the control, ALK-specific antibody had no effect.
Binding of STAT3, DNMT1, and HDAC1 to SHP-1 Promoter in Vitro. We performed ChIP assays by using STAT3, DNMT1, and HDAC1 antibodies. Binding of two closely related proteins p300 and CBP that display histone acetylase activity (19), interact with STATs (20), and are expressed by malignant T cells and associate with STAT3 (data not shown) was also examined. Immunoprecipitation was followed by PCR with primers SHP-1 promoter 2-specific primers; SHP-1 gene exon 16-detecting primers served as a control. As shown in Fig. 5A, STAT3, DNMT1, and HDAC1 were detected at the SHP-1 promoter of the two SHP-1-negative T cell lines 2A and 2B. In contrast, binding of p300 and CBP could not be detected, suggesting that they interact with STAT3 at promoters of other genes, likely the ones that are activated, rather than silenced, by STAT3.
To determine whether the binding of STAT3, DNMT1 and HDAC1 proteins is confined to the SHP-1-negative cells, we also examined in the ChIP assay the SHP-1-high (PBMC and PHA-BL) and -intermediate (PB-1 and JB6) cells. As shown in Fig. 5B, no binding to the SHP-1 promoter of any of these proteins can be detected in PBMC; PHA-BL demonstrated only traces of the STAT3 binding to the promoter and no detectable association of either DNMT1 or HDAC1. Both PB-1 and JB6 showed weak binding of STAT3, DNMT1, and HDAC1. These findings indicate that the degree of association of STAT3 and the other two proteins with the SHP-1 promoter correlates well with the epigenetic silencing of the SHP-1 gene. To provide even more direct evidence that SHP-1-negative T cells STAT3 forms complexes with DNMT1 and HDAC1 at the SHP-1 promoter, we performed two-step precipitation “re-ChIP” experiments in which cell homogenates were consecutively precipitated with the anti-STAT3 antibody and either the DNMT1 or HDAC1 antibody. STAT3 could be coprecipitated with DNMT1 (Fig. 5C) and HDAC1 (data not shown) at the SHP-1 promoter.
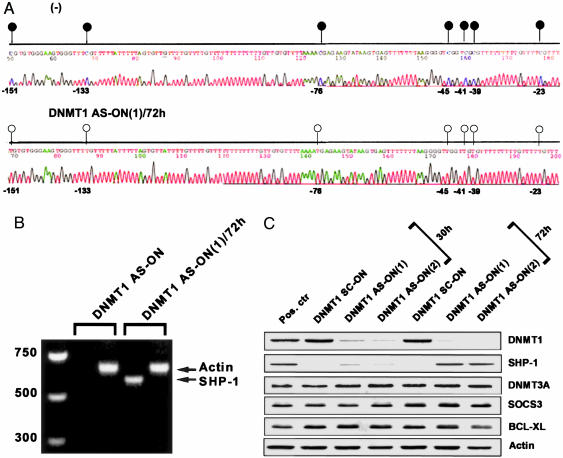
Effect of DNMT1 Depletion on SHP-1 Gene Expression. To demonstrate that DNMT1 is functionally involved in methylation of the SHP-1 gene promoter and inhibition of the gene expression, we treated malignant T cells with two different antisense ON designated DNMT1 AS-ON(1) and DNMT1 AS-ON(2), described in Materials and Methods and in refs. 21 and 22, and two control, scrambled ON, DNMT1 SC-ON(1) and (2). The DNMT1 AS-ON incorporation led at 72 h to the demethylation of the SHP-1 promoter (Fig. 6A) and expression of the SHP-1 mRNA (Fig. 6B). It also resulted in expression of SHP-1 protein at the 72-h, but not the 30-h, time point (Fig. 6C). This expression was preceded by the loss of expression of DNMT1 protein that seemed essentially complete by 30 h. The expression of several proteins used as controls of the DNMT1 AS-ON target specificity: DNMT3A, actin, and SOCS3 and BCL-XL, which are transcriptionally activated rather than inhibited by STAT3 (8), were unaffected by the treatment.
Fig. 6.
The effect of down-regulation of DNMT1 expression on SHP-1 gene silencing. (A) Promoter methylation status in 2B cells before (Upper) and after (Lower) treatment with DNMT1 AS-ON. (B) RNA expression. (C) Expression of DNMT1, SHP-1, and the control proteins.
Effect of STAT3 Depletion on SHP-1 Expression. To demonstrate directly that STAT3 is involved in the silencing of the SHP-1 gene, we treated the SHP-1-negative 2A cells with a STAT3 siRNA. As shown in Fig. 7A, STAT3 siRNA-treated cells demonstrated at 72 h demethylation of most (>70%) of the examined 18 CpG sites. As presented in Fig. 7B, by 48 h, there was a profound inhibition of STAT3 protein expression. This decrease correlated with a marked reexpression of SHP-1 protein at 72 h. STAT3 depletion led also to the gradual abrogation of SOCS3 and BCL-XL expression, whereas expression of the control STAT5 and actin remained unaffected.
Fig. 7.
Effect of down-regulation of STAT3 expression on SHP-1 gene silencing. (A) Promoter methylation status in the 2A cells treated with nonsense (NS) siRNA (Upper) and STAT3-specific siRNA (Lower). (B) Protein expression of STAT3, SHP-1, and the control proteins. (C) Association of the STAT3/DNMT1/HDAC1 complex with the SHP-1 promoter after 72 h of cell treatment medium alone (–), STAT3 siRNA, or DNMT1 AS-ON(1) detected in the ChIP assay.
Discussion
This study focused on the mechanism of epigenetic silencing of the SHP-1 tyrosine phosphatase gene that occurs in a large spectrum of malignancies (12–14). We demonstrate that cell lines and tissues from two distinct types of T cell lymphoma, CTCL and ALK+ TCL, express two members of the epigenetic gene silencing machinery: DNMT1 and HDAC1. Furthermore, STAT3, which is persistently activated in malignant T cells in CTCL (4, 5) and ALK+ TCL (6, 7), binds to the SHP-1 promoter and brings these two proteins to the promoter to silence the SHP-1 gene. Whereas we documented functional involvement of DNMT1 in the SHP-1 gene silencing, the exact role of HDAC1 in the process remains to be elucidated.
Although STAT3 seems to act mainly as transcription activator (8), transcriptional repression by STAT3 has also been described in refs. 23 and 24 with the mechanism(s) of the repression remaining largely undefined. This report provides the evidence that oncogenic STAT3 promotes epigenetic gene silencing. Importantly, we show that STAT3 used this inhibitory mechanism to target SHP-1 tyrosine phosphatase, a well recognized tumor suppressor (9). Because in normal cells SHP-1 down-regulates signaling mediated by a spectrum of cytokines, growth factors, chemokines, antigens and other molecules (9–11), loss of SHP-1 renders the malignant cells hypersensitive to a whole array of extra- and intracellular stimuli. Noteworthy, activation of STAT3 by tyrosine 705 phosphorylation, and the simultaneous expression of DNMT1 and HDAC1 is insufficient to mediate the fully effective SHP-1 gene silencing. Both normal, mitogen-activated T cells (PHA-BL) and certain populations of malignant T cells (PB-1 and JB6) express the phospho-STAT3, DNMT1, HDAC1 (Fig. 1C), yet do not at all (for the former) or only partially (for the latter) methylate the SHP-1 promoter (Fig. 2) and suppress the gene expression (Fig. 1 A and C). This feature seems to stem from the inability of STAT3 in these cells to bind effectively to SHP-1 promoter (Figs. 4A and 5B). Whether, in turn, the capacity of STAT3 seen in the other CTCL and ALK+ TCL populations not only to effectively bind (Figs. 4A and 5B) to but also induce marked DNA methylation of the SHP-1 promoter (Figs. 2 and 7) is due to additional posttranscriptional modification(s) of the STAT protein that might change its conformation (25), expression of a corepressor that enhances STAT3 binding to the promoter, mutation(s) of the STAT3 gene, or other reasons remains to be determined.
The exact mechanisms of the epigenetic gene silencing in both normal immature and malignant cells are the subject of intense investigations (26). Binding of transcription factors to a gene promoter, as demonstrated here for STAT3, could be the step that provides an anchor for the proteins that directly mediate DNA methylation and chromatin remodeling (15, 27). A study (27) has implicated a chimeric PML/RARα transcription factor in an induction of the epigenetic gene silencing of the target RARε gene. However, the exact role, if any, of RARε silencing in the PML/RARα-mediated cell transformation is unclear. Here we demonstrate that STAT3, which is perpetually activated by continuous phosphorylation by upstream tyrosine kinases (4, 6, 7), rather than chromosomal translocation, induces epigenetic gene silencing and targets a key tumor suppressor gene. Considering that translocations are relatively rare, whereas persistent activation of transcription factors seems universal in cancer, our study suggests that similar, transcription factor-mediated epigenetic silencing of tumor suppressor genes might play a role in pathogenesis of a whole spectrum of malignancies.
The involvement of STAT3, DNMT1 and, apparently, HDAC1 in the epigenetic silencing of the SHP-1 gene may have therapeutic implications. Inhibitors of DNMT (28) and HDAC (29) are evaluated in various malignancies with promising results. Whereas most of the studies used small molecule inhibitors, such as 5-aza-2-deoxycytidine, that targets not only DNMT1 but also other DNMTs (30), the DNMT1-specific antisense ON used in this study have undergone clinical evaluation (22). Direct targeting of STAT3 in malignant tumors may represent another important therapeutic aim (8). Inhibitors that interfere with STAT3 function (31) or induce its ubiquitination (32) have been identified, paving the road for development of clinically suitable drugs.
Acknowledgments
This work was supported, in part, by the National Cancer Institute Grants R01-CA89194, R01-CA96856, and P50-CA93371.
Author contributions: Q.Z. and M.A.W. designed research; Q.Z., H.Y.W., M.M., P.N.R., and T.N. performed research; and M.A.W. wrote the paper.
Abbreviations: ALK+ TCL, T cell lymphoma expressing anaplastic lymphoma kinase; ChIP, chromatin immunoprecipitation; CTCL, cutaneous T-cell lymphoma; DNMT, DNA methyltransferase; HDAC, histone deacetylase; ON, oligonucleotide; PBMC; peripheral blood mononuclear cells; PHA-BL, phytohemagglutinin-activated T cell blasts; siRNA, small interfering RNA; STAT, signal transducer and activator of transcription.
References
- 1.Jones, P. A. & Baylin, S. B. (2002) Nat. Rev. Genet. 3, 415–428. [DOI] [PubMed] [Google Scholar]
- 2.Robertson, K. D. (2002) Oncogene 21, 5361–5379. [DOI] [PubMed] [Google Scholar]
- 3.Herman, J. G. & Baylin, S. B. (2003) N. Engl. J. Med. 349, 2042–2054. [DOI] [PubMed] [Google Scholar]
- 4.Zhang, Q. Nowak, I., Vonderheid, E. C., Rook, A. H., Kadin, M. E., Nowell, P. C., Shaw, L. M. & Wasik, M. A. (1996) Proc. Natl. Acad. Sci. USA 93, 9148–9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen, M., Kaltoft, K., Nordahl, M., Ropke, C., Geisler, C., Mustelin, T., Dobson, P., Svejgaard, A. & Odum, N. (1997) Proc. Natl. Acad. Sci. USA 94, 6764–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, Q., Raghunath, P. N., Xue, L., Majewski, M., Carpentieri, D. F., Odum, N., Morris, S., Skorski, T. & Wasik, M. A. (2002) J. Immunol. 168, 466–474. [DOI] [PubMed] [Google Scholar]
- 7.Zamo, A., Chiarle, R., Piva, R., Howes, J., Fan, Y., Chilosi, M., Levy, D. E. & Inghirami, G. (2002) Oncogene 21, 1038–1047. [DOI] [PubMed] [Google Scholar]
- 8.Yu, H. & Jove, R. (2004) Nat. Rev. Cancer 4, 97–105. [DOI] [PubMed] [Google Scholar]
- 9.Wu, C., Sun, M., Liu, L. & Zhou, G. W. (2002) Gene. 306, 1–12. [DOI] [PubMed] [Google Scholar]
- 10.Stebbins, C. C., Watzl, C., Billadeau, D. D., Leibson, P. J., Burshtyn, D. N. & Long, E. O. (2003) Mol. Cell. Biol. 23, 6291–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, C., Burkhardt, C., Imhof, D., Ringel, J., Zschornig, O., Wieligmann, K., Zacharias, M. & Bohmer, F.-D. (2004) J. Biol. Chem. 279, 11375–11383. [DOI] [PubMed] [Google Scholar]
- 12.Zhang, Q., Raghunath, P. N., Vonderheid, E., Odum, N. & Wasik, M. A. (2000) Am. J. Path. 157, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka, T., Ouchida, M., Koyama, M., Ogama, Y., Takada, S., Nakatani, Y., Tanaka, T., Yoshino, T., Hayashi, K. & Ohara, N. (2002). Cancer Res. 62, 6390–6394. [PubMed] [Google Scholar]
- 14.Chim, C. S., Fung, T. K., Cheung, W. C., Liang, R. & Kwong, Y. L. (2004) Blood 103, 4630–4635. [DOI] [PubMed] [Google Scholar]
- 15.Robertson, K. D. (2000) Nat Genet. 25, 338–342. [DOI] [PubMed] [Google Scholar]
- 16.Ehret, G. B., Reichenbach, P., Schindler, U., Horvath, C. M., Fritz, S., Nabholz, M. & Bucher, P. (2001). (2001) J. Biol. Chem. 276, 6675–6688. [DOI] [PubMed] [Google Scholar]
- 17.Aaronson, D. S. & Horvath, C. M. (2002) Science 296, 1653–1655. [DOI] [PubMed] [Google Scholar]
- 18.Soldaini, E., John, S., Moro, S., Bollenbacher, J., Schindler, U. & Leonard, W. J. (2000) Mol. Cell. Biol. 20, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blobel, G. A. (2000) Blood 95, 745–753. [PubMed] [Google Scholar]
- 20.Paulson, M., Pisharody, S., Pan, L., Guadagno, S., Mui, A. L. & Levy, D. E. (1999) J. Biol. Chem. 274, 25343–25349. [DOI] [PubMed] [Google Scholar]
- 21.Robert, M. F., Morin, S., Beaulieu, N., Gauthier, F., Chute, I. C., Barsalou, A. & MacLeod, A. R. (2003) Nat. Genet. 33, 61–65. [DOI] [PubMed] [Google Scholar]
- 22.Stewart, D. J., Donehower, R. C., Eisenhauer, E. A., Wainman, N., Shah, A. K., Bonfils, C., MacLeod, A. R., Besterman, J. M. & Reid, G. K. (2003) Ann. Oncol. 14, 766–774. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov, V. N., Bhoumik, A., Krasilnikov, M., Raz, R., Owen-Schaub, L. B., Levy, D., Horvath, C. M. & Ronai, Z. (2001) Mol. Cell 7, 517–528. [DOI] [PubMed] [Google Scholar]
- 24.Kim, H., Suh, J. M., Hwang, E. S., Kim, D. W., Chung, H. K., Song, J. H., Hwang, J. H., Park, K. C., Ro, H. K. & Jo, E. K. (2003) J. Immunol. 171, 616–627. [DOI] [PubMed] [Google Scholar]
- 25.Kabotyanski, E. B. & Rosen, J. M. (2003) J. Biol. Chem. 278, 17218–17227. [DOI] [PubMed] [Google Scholar]
- 26.Fahrner, J. A. & Baylin, S. B. (2003) Genes Dev. 17, 1805–1812. [DOI] [PubMed] [Google Scholar]
- 27.Di Croce, L., Raker, V. A., Corsaro, M., Fazi, F., Fanelli, M., Faretta, M., Fuks, F., Lo Coco, F., Kouzarides, T. & Nervi, C. (2002) Science 295, 1079–1082. [DOI] [PubMed] [Google Scholar]
- 28.Claus, R. & Lubbert, M. (2003) Oncogene 22, 6489–6496. [DOI] [PubMed] [Google Scholar]
- 29.Johnstone, R. W. (2002) Nat. Rev. Drug Discov. 1, 287–299. [DOI] [PubMed] [Google Scholar]
- 30.Weisenberger, D. J., Velicescu, M., Cheng, J. C., Gonzales, F. A., Liang, G. & Jones, P. A. (2004) Mol. Cancer Res. 2, 62–72. [PubMed] [Google Scholar]
- 31.Turkson, J., Kim, J. S., Zhang, S., Yuan, J., Huang, M., Glenn, M., Haura, E., Sebti, S., Hamilton, A. D. & Jove, R. (2004) Mol. Cancer Ther. 3, 261–269. [PubMed] [Google Scholar]
- 32.Ulane, C. M., Rodriguez, J. J., Parisien, J. P. & Horvath, C. M. (2003) J. Virol. 77, 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]