Abstract
The membrane fusion reaction promoted by the paramyxovirus simian virus 5 (SV5) and human parainfluenza virus type 3 (HPIV-3) fusion (F) proteins and hemagglutinin-neuraminidase (HN) proteins was characterized when the surface densities of F and HN were varied. Using a quantitative content mixing assay, it was found that the extent of SV5 F-mediated fusion was dependent on the surface density of the SV5 F protein but independent of the density of SV5 HN protein, indicating that HN serves only a binding function in the reaction. However, the extent of HPIV-3 F protein promoted fusion reaction was found to be dependent on surface density of HPIV-3 HN protein, suggesting that the HPIV-3 HN protein is a direct participant in the fusion reaction. Analysis of the kinetics of lipid mixing demonstrated that both initial rates and final extents of fusion increased with rising SV5 F protein surface densities, suggesting that multiple fusion pores can be active during SV5 F protein-promoted membrane fusion. Initial rates and extent of lipid mixing were also found to increase with increasing influenza virus hemagglutinin protein surface density, suggesting parallels between the mechanism of fusion promoted by these two viral fusion proteins.
To date, much of our understanding of protein-mediated membrane fusion comes from studies of the viral fusion proteins, found in the membranes of enveloped viruses. Membrane fusion promoted by the influenza virus hemagglutinin (HA) proceeds through multiple steps, beginning when the HA molecule is triggered by low pH to undergo a dramatic set of structural changes (10, 61), an event that occurs within the endosome during viral entry. Considerable evidence suggests that multiple HA trimers then cluster to allow formation of a fusion pore (18, 20). Though the mechanism by which subsequent lipid mixing and complete fusion occur remains unclear, it is evident for HA that both the transmembrane domain of the protein and the structure of the surrounding lipids play an important role (13, 32).
The fusion (F) proteins of paramyxoviruses such as simian virus 5 (SV5) are known to promote fusion at neutral pH, allowing entry of the virus at the plasma membrane. However, despite the fact that low pH is not the trigger for fusion initiation, there appear to be many similarities between the paramyxovirus F proteins and the well-characterized HA protein. Both HA and F proteins are synthesized as biologically inactive precursor proteins which must be cleaved into disulfide-linked dimers to be biologically active (33, 52). Both form higher-order oligomers: X-ray diffraction studies show that HA is a trimer (61), and chemical cross-linking studies indicate that F is also a trimer (50). HA and F both contain a hydrophobic stretch, known as the fusion peptide, located at the N terminus of the transmembrane domain-containing segment that has been shown to insert into target membranes (2, 22, 39), and mutational analysis has indicated it is important for fusion (28, 48, 58). In addition, both HA and F have heptad repeats domains near the fusion peptide and the transmembrane domain; mutational analysis has indicated that these domains are also important for fusogenic activity (9, 48, 54). For HA, it has been shown that these heptad repeats are key domains in the transition from the metastable neutral-pH conformation to the low-pH conformation (10). Last, mutants which lack the cytoplasmic tail of the F protein (6) or the cytoplasmic tail and transmembrane domain of HA protein (32) have been shown to promote hemifusion, suggesting that the mechanisms of HA and F fusion promotion have common elements.
While HA, in addition to its fusion activity, has a role in the primary attachment of a virus to a cell, the F proteins of paramyxoviruses do not have a known role in the primary attachment of the virus to a cell surface. Instead, this function is performed by a second viral glycoprotein, the hemagglutinin-neuraminidase (HN) protein (26, 51). For many paramyxoviruses, the homotypic HN protein appears to be required for F protein-promoted membrane fusion (12, 19, 30, 38, 59, 60). However, the F protein of the paramyxovirus SV5 has been shown to promote fusion in the absence of its homotypic HN (5, 29). It has been suggested that for those paramyxoviruses requiring their homotypic HN protein for fusion, binding of the HN molecule to a sialic acid moiety on the target cell causes a conformational change in HN which in turn triggers the putative conformational change in the F protein necessary to initiate fusion (34, 53). For SV5, which does not require its homotypic HN for fusion activity, it has been hypothesized that either close contact with the target membrane or binding to an as yet unidentified receptor may induce the putative conformational change required to initiate fusion (34).
Whereas many regions of the paramyxovirus F protein important for fusion have been identified, the actual mechanism of F protein-mediated membrane fusion remains to be determined. Studies of the effects of varying the surface densities of HA on fusion have indicated that both the initial rate of fusion (18) and the lag phase prior to fusion initiation (14, 18) are dependent on the surface density of HA. The effect of HA surface density on extent of fusion varied depending on the assay used (18, 20, 36). These findings suggest that multiple fusion pores are opened during HA-mediated fusion and that several trimers of HA are likely involved in the formation of each pore. To further characterize the F protein-promoted fusion event, we have varied the surface densities of both F and HN proteins of the paramyxoviruses SV5 and human parainfluenza virus type 3 (HPIV-3). We show here that both the extent and initial rate of fusion are dependent on the surface density of the SV5 F protein but the extent of fusion is independent of the density of SV5 HN protein, indicating that HN serves only a binding function in the reaction. However, the extent of the HPIV-3 F protein-promoted fusion reaction was found to be dependent on the surface density of HPIV-3 HN protein, suggesting that the HPIV-3 HN protein is a direct participant in the fusion reaction. In our system, we find that the kinetics of fusion for influenza virus A/Udorn/72 (H3 subtype) HA are similar to those seen for the SV5 F protein, with both initial rate and extent of fusion varying with surface density, suggesting parallels in the fusion mechanism of these viral fusion proteins.
MATERIALS AND METHODS
Cells and viruses.
Monolayer cultures of the TC7 subclone of CV-1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS). The recombinant vaccinia virus vTF7-3, which expresses T7 RNA polymerase, was grown in CV-1 cells as described previously (21). SV5 and HPIV-3 stocks were grown and titers were determined as described previously (44).
Plasmid vectors.
The SV5 F cDNA (28, 41, 42) was subcloned into pGEM2X, a derivative of pGEM2 containing a XhoI site (4, 5). The SV5 HN cDNA (26, 41) and the HPIV-3 F and HPIV-3 HN cDNAs (23) were subcloned into pGEM3X. Plasmid pTF7.5 HA, containing the HA cDNA of influenza virus A/Udorn/72, was described previously (55). Plasmid pINT7 β-gal (40), containing the β-galactosidase cDNA, was kindly provided by Edward Berger and Bernard Moss (National Institutes of Health, Bethesda, Md.). All cDNAs were cloned in an orientation in pGEM such that mRNA-sense RNA transcripts could be synthesized by use of bacteriophage T7 RNA polymerase.
Expression of F and HN proteins.
F and HN proteins from SV5 and HPIV-3 and influenza virus (A/Udorn/72 [H3 subtype]) HA were expressed transiently by use of the recombinant vaccinia virus-T7 RNA polymerase expression system (21). Subconfluent monolayers of CV-1 cells were infected with recombinant vaccinia virus vTF7-3 at approximately 10 PFU per cell and incubated at 37°C for 30 min. The virus inoculum was then removed, and cells were transfected with plasmid DNA by using cationic liposomes prepared as described previously (49). A total of 7.5 μg of plasmid DNA and 15 μl of liposomes in 2.0 ml of OPTI-MEM (GIBCO-BRL, Gaithersburg, Md.) was used for a 6-cm-diameter tissue culture dish. At 5 h posttransfection, cells were washed once in phosphate-buffered saline (PBS) and incubated overnight at 33°C in DMEM with 10% FCS.
Quantification of surface density by flow cytometry.
Following overnight incubation, transfected CV-1 cells were chilled on ice for 10 min and treated for flow cytometric analysis as described previously (28). Monoclonal antibody (MAb) F1a specific for SV5 F, MAb HN4b specific for SV5 HN (47), MAb 145/50 specific for HPIV-3 F protein, MAb 66/4 specific for the HPIV-3 HN protein (16, 17), and MAb D6/1 specific for influenza virus A/Udorn/72 HA (gift from Kathleen Coelingh) were used as primary antibodies, and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G was used as the secondary antibody. All antibodies were used in conditions of antibody excess (data not shown). Fluorescence intensity of 10,000 cells was measured by flow cytometry (FACSCaliber; Becton Dickinson, Mountain View, Calif.). As a control for transfection variability, CV-1 cells were infected the day prior to flow cytometric analysis with 10 PFU of SV5 or HPIV-3 per cell. At 18 h postinfection (p.i.), infected cell cultures were processed for flow cytometry together with DNA-transfected samples. Mean fluorescent intensities (MFI) were compared to those observed in SV5- or HPIV-3-infected cells.
Quantification of molar ratios of F and HN molecules on the surface of HPIV-3-infected cells.
CV-1 cells were incubated with 10 PFU of HPIV-3 per cell for 1 h at 37°C, and the medium was replaced with 90% Cys- and Met-deficient DMEM–10% DMEM containing a final amount of 2% FCS and 40 μCi of [35S]methionine (Amersham International, Arlington Heights, Ill.) per ml. Following a 17-h incubation, cells were either lysed and immunoprecipitated as previously described (35) or placed at 4°C for antibody capture. In both procedures, 4 μl of MAb C191/9 to HPIV-3 F or 40 μl of tissue culture supernatant to HPIV-3 HN (each amount determined previously to provide antibody excess conditions) was used as the primary antibody, and 1 μl of rabbit anti-mouse immunoglobulin G was used as a secondary antibody. Plates for antibody capture were washed twice with cold PBS and incubated for 1 h with rocking in antibody solution in PBS–1% bovine serum albumin. Cells were then washed five times with cold PBS, lysed, and subjected to immunoprecipitation as described above. Samples were analyzed on a 15% acrylamide gel, and radioactivity was quantified on a FujiBAS 1000 bioimager (Fuji Medical Systems, Stamford, Conn.).
β-Galactosidase assay for content mixing.
Following overnight incubation, transfected cells were assayed for content mixing activity by an assay using activation of the reporter gene β-galactosidase (5, 40). Cell fusion was measured by a colorimetric lysate assay for β-galactosidase as described previously (40), and the results were read by a plate reader (ELX800; Bio-tek Instruments, Inc., Winooski, Vt.). For experiments in which the effect of trypsin treatment was tested, after neuraminidase treatment, cells were washed and either mock treated or treated for 1 h with TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (5 μg/ml) in OPTI-MEM containing 2.5 mM CaCl2.
Spectrofluorometric analysis of lipid mixing.
Fresh human erythrocytes (RBCs) were labeled with the lipid probe octadecyl rhodamine B (R18) as described previously (5). Following overnight incubation, transfected cells were washed with PBS and incubated for 1 h at 37°C with 50 mU of neuraminidase (Clostridium perfringens type V; Sigma Chemical Co., St. Louis, Mo.) per ml in DMEM. Cells were then washed twice with PBS and incubated with 3 ml of R18-labeled RBCs (0.02 or 0.05% hematocrit) at 4°C for 30 min with occasional gentle rocking to ensure even spread. Excess unbound RBCs were removed with multiple washes with ice-cold PBS, and the RBC-cell complexes were removed from the dish by incubation for 30 min at 4°C in a solution of PBS containing 50 mM EDTA. The RBC-cell complexes were washed with cold PBS and placed on ice until further use. Fifty microliters of RBC-cell complexes was added to 3 ml of prewarmed PBS, and the fluorescence was measured continuously in a spectrofluorometer (Aminco Bowman series 2) with 1-s time resolution at 560- and 590-nm excitation and emission, respectively. To reduce scattering, a 570-nm-cutoff filter was placed in the emission optical pathway. The percent fluorescence was calculated as 100(F − F0/Ft − F0), where F0 and F are the fluorescence intensities at time zero and at a given time point, respectively, and Ft is the fluorescence intensity in the presence of 0.1% Triton X-100, taken as fluorescence when no self-quenching is observed (7).
Confocal microscopy of fusion of R18-labeled RBCs with transfected cells.
Transfections were performed on CV-1 cells in 6-cm-diameter dishes containing a coverslip, the cells were incubated overnight, and washing and neuraminidase treatment were carried out as described above. Cells were then incubated with 5 μM SYTO 12 nucleic acid dye (Molecular Probes, Eugene, Oreg.) in phosphate-free DMEM (GIBCO-BRL) for 30 min at 37°C and washed with ice-cold PBS, and R18-labeled RBCs were bound to the CV-1 cells as described above. Excess unbound RBCs were removed by washing with PBS, and plates were stored at 4°C. Fusion was initiated by replacement of cold PBS with PBS prewarmed to 37°C, and the cells were incubated at 37°C for various times. Fusion was stopped by replacement with ice-cold PBS. Fusion was visualized in a confocal microscope system (Zeiss LSM 410; Carl Zeiss, Inc., Thornwood, N.Y.), with dual images recorded on both fluorescein and rhodamine channels.
RESULTS
Expression of varying surface densities of the viral glycoproteins.
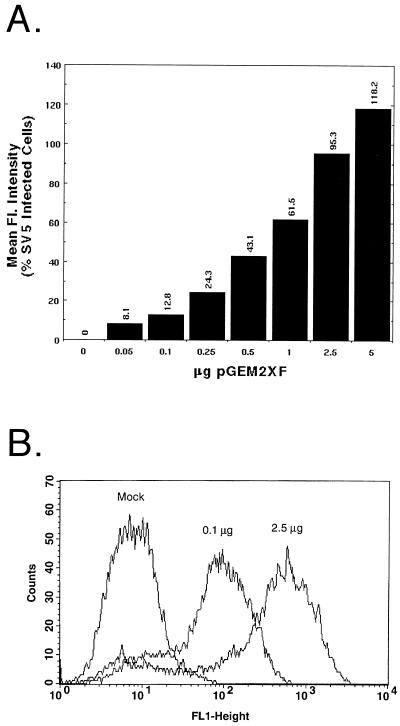
As expression of both the SV5 F protein and an attachment protein such as SV5 HN or influenza virus HA is required to detect fusion by the quantitative fusion assays used in this study, a method of transient expression that would readily permit variation in the surface densities of two proteins was sought. Therefore, the relative surface expression level of SV5 F protein was determined when varying amounts of plasmid encoding F were transfected into cells and expressed using the vaccinia virus-T7 RNA polymerase system. The total DNA amount was kept constant at 7.5 μg per 6-cm-diameter dish, and the amount of plasmid pGEM2XSV5F (SV5 F DNA) was varied from 0.05 to 5.0 μg. Transfected cells were treated with the F-specific MAb F1a, and cell surface fluorescence was analyzed by flow cytometry. To control for variation in transfection efficiencies among experiments, MFI was routinely compared to that of SV5-infected cells at 18 h p.i. As shown in Fig. 1A, addition of increasing quantities of SV5 F DNA resulted in a rise in surface density of the SV5 F protein, with the rate of increase slowing significantly above 2.5 μg of plasmid DNA per 6-cm-diameter dish. The flow cytometry data also indicated that the percentage of cells expressing F (peak height [Fig. 1B]) was largely unaffected by increasing the quantity of SV5 F DNA and that the distribution of expression levels within an overall population of cells expressing different levels of F (peak shape [Fig. 1B]) was not skewed in different populations.
FIG. 1.
Expression of varying surface densities of the SV5 F protein. (A) Duplicate plates of vTF7-3-infected CV-1 cells were transfected (as described in Materials and Methods) with varying quantities of SV5 F DNA and pGEM3X DNA to give a final DNA amount of 7.5 μg per 6-cm-diameter plate. Samples were processed for flow cytometric analysis using MAb F1a as described in Materials and Methods. The MFI was compared to that observed in SV5-infected cells at 18 h p.i., with the MFI of mock-transfected samples subtracted. (B) Example of raw data from flow cytometric analysis, showing data for mock-infected cells or cells transfected with 0.1 μg of SV5 F DNA and 2.5 μg of SV5 F DNA.
Effect of increasing amounts of SV5 F protein on extent of membrane fusion.
To determine the effect of increasing F protein surface density on the extent of F protein-promoted membrane fusion, duplicate plates of vTF7-3-infected CV-1 cells were transfected with 2.5 μg of pGEM3XHN (SV5 HN DNA) and varying amounts of SV5 F DNA. MFI, determined by flow cytometry using F-specific MAb F1a, was determined as a percentage of that observed in SV5-infected cells at 18 h p.i. The extent of fusion was measured by the β-galactosidase reporter gene activation fusion assay. At low F surface densities, the extent of fusion was found to increase in parallel with increasing surface densities of F, suggesting the additional F trimers allow an increased number of fusion events (Fig. 2). The increase in extent of fusion with F expression levels began to plateau at surface densities representing approximately 50% of those found in an SV5-infected cells. Fusion was found to decrease reproducibly at levels of SV5 F-protein expression above 100% of that found in SV5-infected cells at 18 h p.i. Surface densities of F protein at the inhibitory level did not result in significant decreases in HN protein expression (data not shown), suggesting that loss of HN was not the cause of the decrease in extent of fusion. The inhibition at high surface densities of F protein was also not due to the presence of uncleaved F protein, as treatment with exogenous trypsin did not relieve the inhibition (data not shown).
FIG. 2.
Effect of increasing amounts of SV5 F protein on extent of membrane fusion. vTF7-3-infected CV-1 cells were transfected with 2.5 μg of SV5 HN DNA and various amounts of SV5 F DNA and pGEM3X DNA to give a final amount of 7.5 μg of DNA per 6-cm-diameter plate. Flow cytometry using MAb F1a was performed in duplicate, and MFI was compared to that observed in SV5-infected cell at 18 h p.i. The β-galactosidase fusion assay was performed in triplicate as described in Materials and Methods.
Effect of SV5 HN protein surface density on fusion promoted by the SV5 F protein.
To determine whether the surface density of the SV5 HN protein used to provide binding function affected the extent of fusion promoted by the SV5 F protein, triplicate sets of CV-1 cells were transfected to give expression of a constant amount of the SV5 F protein and varying levels of SV5 HN surface density. Flow cytometry was performed with either the SV5 F-specific MAb F1a or the SV5 HN-specific MAb HN4b. The extent of fusion was determined by use of the β-galactosidase fusion assay. While no fusion was observed in the absence of the SV5 HN protein (needed to provide a binding function), maximal fusion extents were obtained with the lowest amount of SV5 HN protein tested, corresponding to 30% of that seen in an SV5-infected cell (Fig. 3). Increases of HN protein surface density of 15-fold, to >200% of that seen on an SV5-infected cell surface, did not increase the extent of fusion promoted by the SV5 F protein. Under these conditions, the surface density of the F protein was not significantly affected by expression of increasing amounts of SV5 HN DNA (Fig. 3). In addition, variation in HN expression did not affect F-promoted fusion when F surface densities of 80 to 150% of that found in an SV5-infected cell were examined (data not shown). Thus, these data indicate that the ratio of F protein to HN protein, which by biotinylation analysis appears to be approximately one F trimer per HN tetramer on the surface of infected cells (18a), is unimportant for SV5 mediated fusion, providing further evidence that the HN protein allows binding but does not directly participate in the fusion reaction.
FIG. 3.
Effect of SV5 HN protein surface density on fusion promoted by the SV5 F protein. vTF7-3 infected CV-1 cells were transfected with 2.5 μg of SV5 F DNA and varying amounts of SV5 HN DNA and pGEM3X DNA to give a final amount of 7.5 μg of DNA per 6-cm-diameter plate. Flow cytometry was performed in duplicate with either MAb F1a or MAb HN4b. The β-galactosidase fusion assay was performed in triplicate.
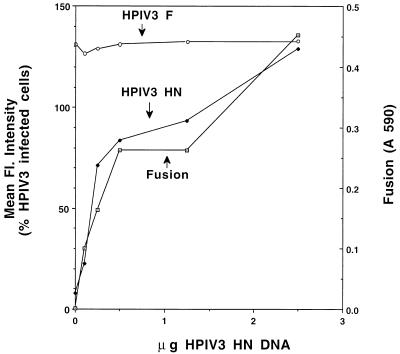
Effect of HPIV-3 HN protein surface density on fusion promoted by the HPIV-3 F protein.
For the majority of paramyxoviruses other than SV5, the HN protein has been implicated as being involved in the fusion reaction. Therefore, the effect of varying the surface density of the HN protein from the paramyxovirus HPIV-3 on fusion promoted by the HPIV-3 F protein was determined. CV-1 cells were transfected with HPIV-3 F and HN DNAs to yield expression of a constant amount of HPIV-3 F protein and varying amounts of the HPIV-3 HN protein. Flow cytometric analysis, using HPIV-3 F-specific MAb 145/50 or HPIV-3 HN-specific MAb 66-4, and the β-galactosidase fusion assay were performed. The extent of fusion was found to increase with increasing surface densities of HPIV-3 HN (Fig. 4), until F and HN were present on the surface in ratios similar to that found on an HPIV-3 infected cell. Maximal fusion at similar ratios of the HPIV-3 F and HN proteins was also observed when different amounts of the HPIV-3 F protein were expressed (data not shown). Analysis by antibody capture of the molar ratios of F and HN molecules on the surface of HPIV-3-infected cells indicated that the ratio of HN tetramers to F trimers is approximately 1:1 (data not shown). These data suggest that this HN protein likely plays a direct role in the fusion reaction promoted by the HPIV-3 F protein, with fusion most efficiently promoted when the F and HN oligomers are present at the surface in equimolar ratios.
FIG. 4.
Effect of HPIV-3 HN protein surface density on fusion promoted by the HPIV-3 F protein. vTF7-3-infected CV-1 cells were transfected with 0.5 μg of HPIV-3 F DNA (the amount required to yield approximately 100% of F protein expression observed in an HPIV-3-infected cell) and varying amounts of HPIV-3 HN DNA and pGEM3X DNA to give a final amount of 7.5 μg of DNA per 6-cm-diameter plate. Flow cytometric analysis was performed in duplicate with either MAb 145/50 specific for HPIV-3 F or MAb 66/4 specific for HPIV-3 HN. The β-galactosidase fusion assay was performed in triplicate.
Effect of increasing surface densities of SV5 F protein on rates of fusion.
To determine whether changes in surface density of the SV5 F protein affected the initial rate of fusion as well as the extent of fusion, duplicate sets of CV-1 cells were transfected to yield expression of various surface densities of the SV5 F protein and a constant amount of a sialic acid binding protein, in this case influenza virus HA. One set of plates was processed for flow cytometry using F-specific MAb F1a so that surface expression levels could be determined. The other set of plates was used to determine the kinetics of fusion by monitoring the fluorescence dequenching of R18 upon fusion. R18-labeled human RBCs were bound to CV-1 cells at 4°C to give an average of one to two RBCs per cell, fusion was initiated by injection of 40 μl of RBC-acceptor cell complexes into a cuvette containing 3 ml of PBS prewarmed to 37°C, and the kinetics of fluorescence dequenching were determined in a fluorometer. Both the extent and initial rate of fusion were found to increase with higher surface densities of the SV5 F protein (Fig. 5A). However, at very high F surface densities (120% of that found in an SV5-infected cell), a decrease in both extent and initial rate was observed (data not shown), consistent with results found when the β-galactosidase fusion assay was used.
FIG. 5.
Effect of increasing surface densities of SV5 F protein on rates of fusion. (A) vTF7-3-infected CV-1 cells were transfected with 2.5 μg of pTF7.5 HA DNA and various amounts of SV5 F DNA and pGEM3X DNA to yield final amounts of 7.5 μg of DNA per 6-cm-diameter plate. Flow cytometric analysis with MAb F1a was performed with duplicate samples. Spectrofluorometry assays were performed as described in Materials and Methods. Individual curves shown are representative of three independent experiments. (B) Spectrofluorimetry and flow cytometric analysis were performed as for panel A except that SV5 HN DNA was substituted for pTF7.5 HA DNA. (C) vTF7-3-infected CV-1 cells were transfected with varying amounts of HA (A/Udorn/72 [H3 subtype]) DNA and pGEM3X DNA to yield a final amount of 7.5 μg per 6-cm-diameter plate. Flow cytometric analysis with MAb D6/1 was performed on duplicate samples; results are shown as a percentage of the value for the highest-expressing sample.
The kinetics of fusion with varying amounts of the SV5 F protein were also examined by using SV5 HN as the binding protein. Under conditions found to yield optimal fusion with influenza virus HA as the binding protein (one to two RBCs per cell), we found that F protein-promoted fusion with SV5 HN as the binding protein was too rapid to measure with confidence. Therefore, the RBC binding level was increased to an average of five to eight RBCs per cell, as it has been shown previously that an increased number of RBCs per cell resulted in lower initial rates of fusion (18). Under these conditions, using SV5 F and HN proteins, an increase in initial rate and extent of fusion was observed with surface densities of the SV5 F protein of up to 75% of that found in an SV5-infected cell (Fig. 5B); initial rates for surface densities above 75% of that of an SV5-infected cell were found to be too fast to measure accurately.
To compare directly the kinetics of SV5 F protein-promoted membrane fusion to those seen with influenza virus HA, duplicate sets of plates were transfected to yield various surface densities of influenza virus A/Udorn/72 (H3 subtype) HA. One set was processed for flow cytometry, using MAb D6/1, to determine relative surface densities of the influenza virus HA. The other set was used for determination of fluorescence dequenching of R18-labeled RBCs, with an average of six to eight RBCs bound per cell. Fusion was initiated by addition of 0.25 M citric acid at 60 s to lower the sample pH to 5.0. As has been demonstrated previously (18), the initial rate of fusion was found to increase with rising surface density of (Fig. 5C). No lag phase was seen, even when fusion was examined at 29°C (data not shown), consistent with the high levels of protein expression achieved with the vaccinia virus-T7 polymerase system. In addition, the extent of fusion was found to vary, similar to that seen for the SV5 F protein but different from that reported for the H2 subtype HA (A/Japan/305/57) (18). Variation from 0.02 to 0.1% added RBCs, giving rise to different average numbers of RBCs per cell, did not affect the variation in extent of fusion observed (data not shown).
Examination of fusion by confocal microscopy.
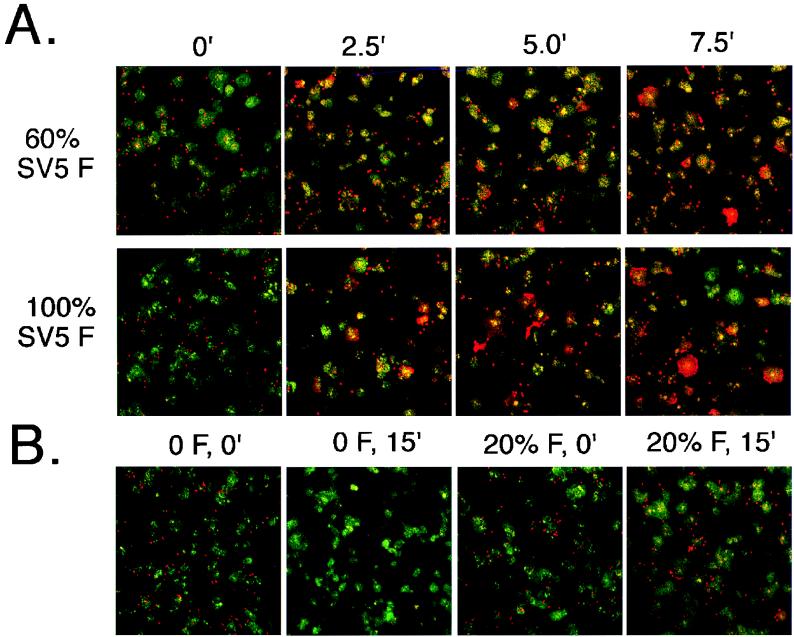
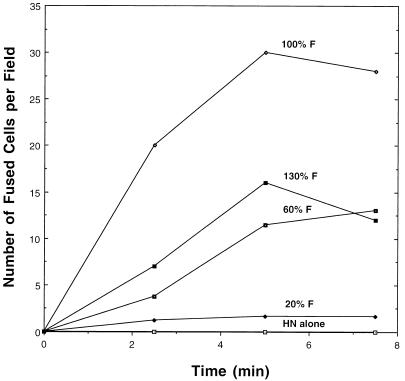
To confirm the data obtained from the fluorometric assay, fusion of R18-labeled RBCs with cells expressing the SV5 F protein was examined by confocal microscopy. Duplicate sets of CV-1 cells were transfected such that a constant amount of the SV5 HN protein and varying amounts of the SV5 F protein (0 to 130% of that found in SV5-infected cells) were expressed. One set of cultures was processed for flow cytometric analysis to determine the F protein surface density. For the microscopy assay, the second set of cultures on coverslips was treated with neuraminidase for 60 min at 37°C and then stained for 30 min at 37°C with SYTO 12 (5 μg/ml; Molecular Probes), a nucleic acid-specific probe which permits staining of all cells. R18-labeled RBCs were then bound to the cultures, fusion was initiated by addition of prewarmed PBS and incubation at 37°C for various periods, and the reaction was terminated by transfer to 0°C. Coverslips were examined on a confocal microscope, using the fluorescein channel for SYTO 12 staining and the rhodamine channel for R18 staining. Prior to initiation of fusion at 37°C, cells expressing HN alone or cells expressing HN and increasing surface densities of F protein (20, 60, or 100% of the amount found in SV5-infected cells at 18 h p.i.) showed similar levels of RBC binding (Fig. 6 and data not shown). The one exception was for the highest surface density of F (130%), which routinely gave somewhat less RBC binding. After incubation at 37°C for 2.5 min, fusion was detected at all surface densities of F examined (Fig. 6A and 7). The number of fusion events (Fig. 7) and the rate of spread to the acceptor cells (Fig. 6A) were found to increase with surface densities of up to 100% of that seen in SV5-infected cells. An increase in extent of R18 dye transfer and its spread was seen after 5 min at 37°C, with little additional fusion detected after this time. One interesting and unanticipated observation was that for cells expressing HN alone, RBCs eluted from the cells, with almost complete loss after 15 min at 37°C, whereas for cells expressing amounts of F protein that caused only low extents fusion (20% of F of SV5-infected cells at 18 h p.i.), the unfused RBCs remained bound for up to 15 min at 37°C (the longest time examined) (Fig. 6B). This observation suggests that the presence of F protein prevented the release of the RBCs from the cell, even when detectable fusion had not occurred.
FIG. 6.
Examination of fusion by confocal microscopy. Six-centimeter-diameter dishes of vTF7-3-infected CV-1 cells, either with or without coverslips, were transfected with 2.5 μg of SV5 HN DNA and increasing amounts of SV5 F DNA and pGEM3X DNA to yield a final amount of 7.5 μg of DNA. Flow cytometric analysis using MAb F1a was performed in duplicate on plates lacking coverslips. Confocal microscopy analysis was performed as described in Materials and Methods. (A) Representative portions (one-quarter of complete field) of confocal images at various time points (minutes) for cells expressing either 60 or 100% of the SV5 F-protein surface density observed in an SV5-infected cell at 18 h p.i. (B) Representative confocal images from cells expressing HN alone (no SV5 F) or cells expressing 20% of the SV5 F-protein surface density observed in SV5-infected cells at 18 h p.i.
FIG. 7.
Quantitation of fusion by confocal microscopy. The number of fusion events was determined by analysis of images made with a confocal microscope as shown in Fig. 6. The data from the rhodamine channel were used during the counting of fusion events so that the cases where even small amounts of dye spread could be properly quantified. At least three complete fields were counted per time point.
DISCUSSION
While much has been learned concerning the mechanism by which the influenza virus HA protein promotes membrane fusion at low pH (reviewed in reference 25), the mechanism by which viruses fuse with the plasma membrane at neutral pH remains much less understood. One of the better-characterized model systems for fusion promoted at neutral pH is that of paramyxovirus-mediated cell fusion. The data presented here, however, make it clear that there is a major difference among the paramyxoviruses in the relative importance of a proper ratio between the F and HN proteins needed for efficient fusion promotion. For SV5, differences in the HN protein surface density of over 15-fold had no effect on the extent of fusion promoted by the SV5 F protein. These data indicate that for SV5, the HN protein does not serve a direct role in the fusion process, a conclusion consistent with previous observations indicating that the SV5 F protein can promote fusion in the absence of its homotypic HN protein (3, 5, 6, 29, 43, 45). However, examination of the kinetics of fusion presented here and previously (3, 5) suggests that the SV5 HN protein can enhance the initial rate of fusion promoted by the SV5 F protein. As the SV5 F to HN protein ratio is unimportant for fusion, and because other binding proteins can substitute for HN (reference 5 and Fig. 5), it seems reasonable to suggest that the enhancement in initial fusion rates observed on coexpression of the SV5 HN protein is due to the ability of HN to optimize conditions for fusion promotion by the SV5 F protein, perhaps by providing a optimal distance between target and acceptor cells.
Fusion promoted by the HPIV-3 F protein, at least in its initial stages, appears to be mechanistically different from that observed with SV5. It has been shown previously that HPIV-3 F protein requires its homotypic HN protein for fusion promotion (5, 29, 30). Data presented here demonstrate that the ratio of the two proteins directly affects the extent of fusion promoted by the HPIV-3 F protein, with fusion most efficiently promoted when the proteins are present on the surface in equimolar ratios, as is found in HPIV-3-infected cells. It has been suggested that the binding of HN to its receptor sialic acid may in turn trigger the conformational change in F necessary to release the fusion peptide (reviewed in reference 34), and it seems reasonable that this HN-F interaction occurs most efficiently when the proteins are present at the cell-cell junction in equimolar ratios.
Several aspects of the SV5 F protein-promoted fusion reaction appear to be similar to those found for the influenza virus HA-promoted fusion reaction. The initial rate of cell-cell fusion was found to increase as the surface density of the SV5 F protein was increased. Increases in the initial rate of fusion with higher surface densities of the influenza virus HA protein have also been observed (reference 18 and Fig. 5C). This finding suggests that more fusion pores are open at the higher surface densities of the F protein. As the extent of fusion also increases, it is not possible to determine from the spectrofluorometric data whether the multiple pores forming during fusion promoted by the SV5 F protein are between single or multiple RBCs and the acceptor cell. However, both the number of RBC-cell events and the rate of spread of dye from individual RBCs were seen to increase with increasing surface densities of SV5 F protein when fusion was examined by confocal microscopy, suggesting that the increase in initial rates of fusion is due to both multiple pores between individual RBCs and the acceptor cell and also multiple RBC-acceptor cell fusion events.
The observation that expression of F protein on the cell surface can prevent release of RBCs from cells expressing both F and HN (Fig. 6B) suggests that the SV5 F protein interacts with the target cell prior to fusion initiation. This interaction could represent interaction of the F protein with a specific receptor or could result from insertion of the F-protein fusion peptide into the target membrane, an interaction that has been shown with influenza virus HA to lead to stable cell-cell interactions in the absence of sialic acid binding (13). At low surface densities, there is sufficient F protein interacting with the target cell to facilitate this initial interaction but not enough to efficiently promote fusion, suggesting that, as is proposed for influenza virus HA-mediated fusion, the SV5 F protein requires multiple oligomers to promote fusion.
In analyzing fusion kinetics, we did not observe a lag prior to initiation of fusion at any of the surface densities examined even when spectrofluorometry was carried out at 30°C, the lowest temperature at which SV5 F protein-promoted cell-cell fusion was measurable (data not shown). A lag prior to initiation of fusion has been shown for fusion mediated by influenza virus HA (14, 18, 20, 37, 57), Semliki Forest virus E2E1 (8), and vesicular stomatitis virus G protein (15) and during fusion of Sendai virus with cells (27). The lag phase of influenza virus HA-promoted membrane fusion has been shown to be dependent on the surface density of HA (14, 18) and has been hypothesized to represent the time necessary for trimers to accumulate at the fusion pore after the low-pH-induced conformational change. It is likely that the surface densities of the SV5 F protein tested here are high enough to prevent detection of a lag phase, as no lag phase was detected when examining influenza virus HA expressed with this system. This finding is consistent with previous observations demonstrating that the lag phase for fusion between influenza virus particles, containing a high surface density of HA, and target cells is extremely brief and, like the lag phase of vesicular stomatitis virus G-protein-mediated fusion, can be detected only after examination with stop-flow techniques (14, 15). Finally, it is possible that any required association of SV5 F protein trimers occurs during the incubation at 4°C, leading to immediate fusion upon transfer to 37°C. In this regard, it has been shown that influenza virus HA can promote fusion at a very low rate at 0°C (57), indicating that association of some viral fusion proteins can occur at low temperatures.
The extent of fusion promoted by the SV5 F protein is clearly dependent on the surface density of the protein, as judged by three different methods of quantitation of fusion (Fig. 2, 6, and 7). It has been demonstrated previously that lateral mobility of the F protein of Sendai virus, another paramyxovirus, is essential for cell-cell fusion (24) and that the Sendai F protein accumulates transiently in areas of cell-cell contact (1), with maximum accumulation seen 5 min after cell-cell contact, followed by dispersal of the molecules. Our results suggest that a threshold local density of the SV5 F protein must be present to have sufficient accumulation of trimers in the region of contact, as there are many trimers of the F protein present on the cell surface at even the lowest surface density examined, yet a fusion event is rarely seen. In addition, as differences in extent were seen in fusion assays of both long duration (β-galactosidase) and short duration (spectrofluorometer and confocal examination), the results indicate that additional time does not overcome the effects of lowered F surface density on extent of fusion. In addition, the results of multiple assays indicated that expression of higher levels of the SV5 F protein is inhibitory to both extent and initial rate of fusion. This inhibition is not a result of the presence of uncleaved F protein at the surface, as treatment with exogenous trypsin did not relieve the inhibitory effect of high levels of F protein, nor was a decrease in the level of HN protein detected at high levels of F protein. Slightly lower amounts of RBC binding in confocal assays were detected at levels of F protein above 100% of levels in an SV5-infected cell, possibly suggesting that high levels of F protein may interfere with the function of the HN protein. Alternatively, it is possible that at high concentrations, the F protein either has slower mobility or self-associates, leading to fewer fusion events. Finally, it is possible that at high surface densities of the F protein, an as yet unidentified cellular receptor needed for trimeric F protein activation becomes saturated.
As was seen for the SV5 F protein, the extent of fusion was found to vary with increasing surface densities of influenza virus A/Udorn/72 HA (Fig. 5C). Variations in extent of fusion have also been seen for influenza virus X-117 HA, where increasing temperature, pH, or urea concentrations were used to give rise to different amounts of activated HA (11). Fluorescence dequenching experiments with the H2 subtype HA (A/Japan/305/72) suggested that a difference in surface density had no effect on the extent of fusion (18), though in experiments using liposome fusion assays with cells lines expressing the H2 subtype HA a difference in fusion extent was observed (20), suggesting assay-to-assay variation. Furthermore, it is possible that differences between the results presented here and those of Danieli and coworkers (18) are a result either of the higher surface densities examined here or of lipid mobility differences between the two systems. However, it also seems probable that the subtype of HA used is an important factor. Influenza virus H2 subtype HA is not inactivated by incubation at low pH (20), and protease sensitivity and MAb reactivity studies suggest that this HA (H2 subtype) can exist as a stable intermediate (46) in the absence of target membranes. Influenza virus H3 subtype HA, which includes HA of strains Udorn, X-31, and X-117, is inactivated by incubation at low pH (31, 56), and in the case of X-31 HA, this inactivation has been demonstrated to correlate with a conformational change which occurs subsequent to the initial low-pH-induced conformational change (57). We suggest that for the SV5 F protein, as for the H3 subtypes of influenza virus HA, a long period of time between the putative initial conformational change and subsequent trimer association is not tolerated, and that either formation of the fusion pore proceeds within a short period or the F protein proceeds to an inactive conformation which is unable to associate with other F-protein trimers. This “do-or-die” scenario would prevent a single inappropriate conformational change from leading to a fusion pore complex. Further work is needed to confirm this hypothesis and to determine whether the mechanism by which other neutral-pH fusion proteins promote membrane fusion conforms to this paradigm.
ACKNOWLEDGMENTS
We thank Bernard Moss and Edward Berger (NIH) for providing vTF7-3 and pINTT7 β-gal, Brian Murphy and Kathleen Coelingh for supplying MAbs, and Reay Paterson, Grace Lin, Andrew Pekosz, and George Leser for helpful discussions.
This work was supported by research grant AI-23173 from the National Institute of Allergy and Infectious Diseases. R.E.D. was supported by Public Health Service NRSA F32 AI-09607. R.E.D. and S.B.J. were Associates and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aroeti B, Henis Y I. Accumulation of Sendai virus glycoproteins in cell-cell contact regions and its role in cell fusion. J Biol Chem. 1991;266:15845–15849. [PubMed] [Google Scholar]
- 2.Asano K, Asano A. Why is a specific amino acid sequence of F glycoprotein required for the membrane fusion reaction between envelope of HVJ (Sendai virus) and target cell membranes? Biochem Int. 1985;10:115–122. [PubMed] [Google Scholar]
- 3.Bagai S, Lamb R A. A glycine to alanine substitution in the paramyxovirus SV5 fusion peptide increases the initial rate of fusion. Virology. 1997;238:283–290. doi: 10.1006/viro.1997.8858. [DOI] [PubMed] [Google Scholar]
- 4.Bagai S, Lamb R A. Individual roles of N-linked oligosaccharide chains in intracellular transport of the paramyxovirus SV5 fusion protein. Virology. 1995;209:250–256. doi: 10.1006/viro.1995.1251. [DOI] [PubMed] [Google Scholar]
- 5.Bagai S, Lamb R A. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol. 1995;69:6712–6719. doi: 10.1128/jvi.69.11.6712-6719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagai S, Lamb R A. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J Cell Biol. 1996;135:73–84. doi: 10.1083/jcb.135.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bagai S, Puri A, Blumenthal R, Sarkar D P. Hemagglutinin-neuraminidase enhances F protein-mediated membrane fusion of reconstituted Sendai virus envelopes with cells. J Virol. 1993;67:3312–3318. doi: 10.1128/jvi.67.6.3312-3318.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bron R, Wahlberg J M, Garoff H, Wilschut J. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 1993;12:693–701. doi: 10.1002/j.1460-2075.1993.tb05703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckland R, Malvoisin E, Beauverger P, Wild F. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J Gen Virol. 1992;73:1703–1707. doi: 10.1099/0022-1317-73-7-1703. [DOI] [PubMed] [Google Scholar]
- 10.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 11.Carr C M, Chaudhry C, Kim P S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cattaneo R, Rose J K. Cell fusion by the envelope glycoproteins of persistent measles viruses which cause lethal human brain disease. J Virol. 1993;67:1493–1502. doi: 10.1128/jvi.67.3.1493-1502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chernomordik L V, Leikina E, Frolov V, Bronk P, Zimmerberg J. An early stage of membrane fusion mediated by the low pH conformation of influenza hemagglutinin depends upon membrane lipids. J Cell Biol. 1997;136:81–93. doi: 10.1083/jcb.136.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clague M J, Schoch C, Blumenthal R. Delay time for influenza virus hemagglutinin-induced membrane fusion depends on hemagglutinin surface density. J Virol. 1991;65:2402–2407. doi: 10.1128/jvi.65.5.2402-2407.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clague M J, Schoch C, Zech L, Blumenthal R. Gating kinetics of pH-activated membrane fusion of vesicular stomatitis virus with cells: stopped-flow measurements by dequenching of octadecylrhodamine fluorescence. Biochemistry. 1990;29:1303–1308. doi: 10.1021/bi00457a028. [DOI] [PubMed] [Google Scholar]
- 16.Coelingh K L V, Winter C C, Jorgensen E D, Murphy B R. Antigenic and structural properties of the hemagglutinin-neuraminidase glycoprotein of human parainfluenza virus type 3: sequence analysis of variants selected with monoclonal antibodies which inhibit infectivity, hemagglutination, and neuraminidase activities. J Virol. 1987;61:1473–1477. doi: 10.1128/jvi.61.5.1473-1477.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coelingh K V, Tierney E L. Antigenic and functional organization of human parainfluenza virus type 3 fusion glycoprotein. J Virol. 1989;63:375–382. doi: 10.1128/jvi.63.1.375-382.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danieli T, Pelletier S L, Henis Y I, White J M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Dutch, R. E., and R. A. Lamb. Unpublished results.
- 19.Ebata S N, Cote M J, Kang C Y, Dimock K. The fusion and hemagglutinin-neuraminidase glycoproteins of human parainfluenza virus 3 are both required for fusion. Virology. 1991;183:437–441. doi: 10.1016/0042-6822(91)90162-5. [DOI] [PubMed] [Google Scholar]
- 20.Ellens H, Bentz J, Mason D, Zhang F, White J M. Fusion of influenza hemagglutinin-expressing fibroblasts with glcophorin-bearing liposomes: role of hemagglutinin surface density. Biochemistry. 1990;29:9697–9707. doi: 10.1021/bi00493a027. [DOI] [PubMed] [Google Scholar]
- 21.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harter C, Bachi T, Semenza B, Brunner J. Hydrophobic photolabeling identifies BHA2 as the subunit mediating the interaction of bromelain-solubilized influenza virus hemagglutinin with liposomes at low pH. Biochemistry. 1988;27:1856–1864. doi: 10.1021/bi00406a010. [DOI] [PubMed] [Google Scholar]
- 23.Heminway B R, Yu Y, Galinski M S. Paramyxovirus mediated cell fusion requires co-expression of both the fusion and hemagglutinin-neuraminidase glycoproteins. Virus Res. 1994;31:1–16. doi: 10.1016/0168-1702(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 24.Henis Y I, Herman-Barhom B, Aroeti B, Gutman O. Lateral mobility of both envelope protein (F and HN) of Sendai virus in the cell membrane is essential for cell-cell fusion. J Biol Chem. 1989;264:17119–17125. [PubMed] [Google Scholar]
- 25.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 26.Hiebert S W, Paterson R G, Lamb R A. Hemagglutinin-neuraminidase protein of the paramyxovirus simian virus 5: nucleotide sequence of the mRNA predicts an N-terminal membrane anchor. J Virol. 1985;54:1–6. doi: 10.1128/jvi.54.1.1-6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoekstra D, Klappe K, de Boer T, Wilschut J. Characterization of the fusogenic properties of Sendai virus: kinetics of fusion with erythrocyte membranes. Biochemistry. 1985;24:4739–4745. doi: 10.1021/bi00339a005. [DOI] [PubMed] [Google Scholar]
- 28.Horvath C M, Lamb R A. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J Virol. 1992;66:2443–2455. doi: 10.1128/jvi.66.4.2443-2455.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath C M, Paterson R G, Shaughnessy M A, Wood R, Lamb R A. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol. 1992;66:4564–4569. doi: 10.1128/jvi.66.7.4564-4569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu X, Ray R, Compans R W. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Junankar P R, Cherry R J. Temperature and pH dependence of the haemolytic activity of influenza virus and of the rotational mobility of the spike glycoproteins. Biochim Biophys Acta. 1986;854:198–206. doi: 10.1016/0005-2736(86)90111-2. [DOI] [PubMed] [Google Scholar]
- 32.Kemble G W, Danieli T, White J M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 33.Klenk H-D, Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 34.Lamb R A. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 35.Lamb R A, Choppin P W. Determination by peptide mapping of the unique polypeptides in Sendai virions and infected cells. Virology. 1978;84:469–478. doi: 10.1016/0042-6822(78)90263-5. [DOI] [PubMed] [Google Scholar]
- 36.Melikyan G B, Niles W D, Cohen F S. The fusion kinetics of influenza hemagglutinin expressing cells to planar bilayer membranes is affected by HA density and host cell surface. J Gen Physiol. 1995;106:783–802. doi: 10.1085/jgp.106.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris S J, Sarkar D P, White J M, Blumenthal R. Kinetics of pH-dependent fusion between 3T3 fibroblasts expressing influenza hemagglutinin and red blood cells. J Biol Chem. 1989;264:3972–3978. [PubMed] [Google Scholar]
- 38.Morrison T, McQuain C, McGinnes L. Complementation between avirulent Newcastle disease virus and a fusion protein gene expressed from a retrovirus vector: requirements for membrane fusion. J Virol. 1991;65:813–822. doi: 10.1128/jvi.65.2.813-822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick S L, Hoekstra D. Membrane penetration of Sendai virus glycoproteins during the early stage of fusion with liposomes as determined by hydrophobic affinity labeling. Proc Natl Acad Sci USA. 1988;85:7433–7437. doi: 10.1073/pnas.85.20.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paterson R G, Harris T J R, Lamb R A. Analysis and gene assignment of mRNAs of a paramyxovirus, simian virus 5. Virology. 1984;138:310–323. doi: 10.1016/0042-6822(84)90354-4. [DOI] [PubMed] [Google Scholar]
- 42.Paterson R G, Harris T J R, Lamb R A. Fusion protein of the paramyxovirus simian virus 5: nucleotide sequence of mRNA predicts a highly hydrophobic glycoprotein. Proc Natl Acad Sci USA. 1984;81:6706–6710. doi: 10.1073/pnas.81.21.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paterson R G, Hiebert S W, Lamb R A. Expression at the cell surface of biologically active fusion and hemagglutinin-neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc Natl Acad Sci USA. 1985;82:7520–7524. doi: 10.1073/pnas.82.22.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paterson R G, Lamb R A. The molecular biology of influenza viruses and paramyxoviruses. In: Davidson A, Elliott R M, editors. Molecular virology: a practical approach. Oxford, England: IRL Oxford University Press; 1993. pp. 35–73. [Google Scholar]
- 45.Paterson R G, Shaughnessy M A, Lamb R A. Analysis of the relationship between cleavability of a paramyxovirus fusion protein and length of the connecting peptide. J Virol. 1989;63:1293–1301. doi: 10.1128/jvi.63.3.1293-1301.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puri A, Booy F P, Doms R W, White J M, Blumenthal R. Conformation changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J Virol. 1990;64:3824–3832. doi: 10.1128/jvi.64.8.3824-3832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Randall R E, Young D F, Goswami K K A, Russell W C. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J Gen Virol. 1987;68:2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- 48.Reitter J N, Sergel T, Morrison T G. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J Virol. 1995;69:5995–6004. doi: 10.1128/jvi.69.10.5995-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. Bio/Technology. 1991;10:520–525. [PubMed] [Google Scholar]
- 50.Russell R, Paterson R G, Lamb R A. Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology. 1994;199:160–168. doi: 10.1006/viro.1994.1108. [DOI] [PubMed] [Google Scholar]
- 51.Scheid A, Caliguiri L A, Compans R W, Choppin P W. Isolation of paramyxovirus glycoproteins. Association of both hemagglutinating and neuraminidase activities with the larger SV5 glycoprotein. Virology. 1972;50:640–652. doi: 10.1016/0042-6822(72)90418-7. [DOI] [PubMed] [Google Scholar]
- 52.Scheid A, Choppin P W. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology. 1974;57:475–490. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- 53.Sergel T, McGinnes L W, Peeples M E, Morrison T G. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology. 1993;193:717–726. doi: 10.1006/viro.1993.1180. [DOI] [PubMed] [Google Scholar]
- 54.Sergel-German T, McQuain C, Morrison T. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J Virol. 1994;68:7654–7658. doi: 10.1128/jvi.68.11.7654-7658.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson D A, Lamb R A. Alterations to influenza virus hemagglutinin cytoplasmic tail modulate virus infectivity. J Virol. 1992;66:790–803. doi: 10.1128/jvi.66.2.790-803.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stegmann T, Booy F P, Wilschut J. Effects of low pH on influenza virus: activation and inactivation of the membrane fusion capacity of the hemagglutinin. J Biol Chem. 1987;262:17744–17749. [PubMed] [Google Scholar]
- 57.Stegmann T, White J M, Helenius A. Intermediates in influenza induced membrane fusion. EMBO J. 1990;9:4231–4241. doi: 10.1002/j.1460-2075.1990.tb07871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steinhauer D A, Wharton S A, Skehel J J, Wiley D C. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanabayashi K, Takeuchi K, Okazaki K, Hishiyama M, Yamada A. Expression of mumps virus glycoproteins in mammalian cells from cloned cDNAs: both F and HN proteins are required for cell fusion. Virology. 1992;187:801–804. doi: 10.1016/0042-6822(92)90482-5. [DOI] [PubMed] [Google Scholar]
- 60.Wild T F, Malvoisin E, Buckland R. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J Gen Virol. 1991;72:439–442. doi: 10.1099/0022-1317-72-2-439. [DOI] [PubMed] [Google Scholar]
- 61.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–375. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]