Abstract
Virus replication in a human immunodeficiency virus (HIV)-infected individual, as determined by the steady-state level of plasma viremia, reflects a complex balance of viral and host factors. We have previously demonstrated that immunization of HIV-infected individuals with the common recall antigen, tetanus toxoid, disrupts this steady state, resulting in transient bursts of plasma viremia after immunization. The present study defines the viral genetic basis for the transient bursts in viremia after immune activation. Tetanus immunization was associated with dramatic and generally reversible shifts in the composition of plasma viral quasispecies. The viral bursts in most cases reflected a nonspecific increase in viral replication secondary to an expanded pool of susceptible CD4+ T cells. An exception to this was in a patient who harbored viruses of differing tropisms (syncytium inducing and non-syncytium inducing [NSI]). In this situation, immunization appeared to select for the replication of NSI viruses. In one of three patients, the data suggested that immune activation resulted in the appearance in plasma of virus induced from latently infected cells. These findings illustrate certain mechanisms whereby antigenic stimulation may influence the dynamics of HIV replication, including the relative expression of different viral variants.
The pathogenic mechanisms involved in the regulation of human immunodeficiency virus (HIV) replication in vivo are multifactorial and complex (27). Following primary HIV infection, most patients achieve a steady-state level of plasma viremia within 4 months to 1 year, which is referred to as the viral set point (39, 40, 46). However, HIV replication is a dynamic process in which the half-life of virus in plasma has been reported to be approximately 5.7 h (44). It has been suggested that approximately 99% of virus in the plasma is derived from recently infected CD4+ T cells that have a life span of approximately 2.2 days (36, 44, 52). HIV type 1 (HIV-1) replication is influenced by a combination of viral and host factors (26). Viral characteristics that sustain replication within a patient include replicative fitness, predilection for mutation, and cell tropism (26). Important host factors that can induce or suppress viral replication include the HIV-specific immune response, cellular activation, and the effects of endogenous cytokines and chemokines (26). Thus, the net level of viral replication, as reflected by plasma viremia, is the result of a complex balance of multiple forces. However, the major influences that drive HIV-1 replication in vivo within individual patients have not been fully delineated.
HIV-1 replication is enhanced by cellular activation (3, 8, 54). We and others have reported that immune activation resulting from exogenous stimuli such as vaccinations or coinfections induce substantial increases in plasma viremia (7, 9, 32, 35, 42, 48, 49). In this regard, immunization of HIV-infected individuals with the recall antigen tetanus toxoid induced transient bursts in plasma viremia over a 6-week period (48). The mechanism for this burst of virus in plasma is uncertain but is presumably due to a transient increase in the susceptible pool of activated CD4+ T cells and/or the secretion of proinflammatory cytokines that are capable of inducing the expression of HIV from already infected cells (12).
The present study investigates the viral genetic basis for the transient bursts in viremia after immune activation. We asked whether perturbations of the steady-state plasma viremia resulting from an activation stimulus such as immunization with a common recall antigen are also reflected in changes in the relative appearance of viral quasispecies. We also considered two hypotheses: (i) the viral burst reflects an increase in a selectively favored subpopulation of viruses, or (ii) the viral burst reflects a nonspecific increase in viral replication due to an increase in the pool of susceptible CD4+ T cells. In addition, we explored the role of induction of latent viruses after immunization in the perturbation of plasma viral quasispecies.
MATERIALS AND METHODS
Patients and samples.
The study was carried out according to a clinical protocol approved by the institutional review board of the National Institute of Allergy and Infectious Diseases. All patients were asymptomatic HIV-1-infected individuals who were receiving no antiretroviral medications. Briefly, participants were either given a 0.5-ml tetanus booster intramuscularly (patients 2 to 4) (Wyeth-Ayerst Laboratories, Marietta, Pa.) or were mock immunized (patient 1). Blood was drawn on the day of the injection and sampled at least six times thereafter over a 6-week period. Patient 2 underwent serial lymph node biopsies before and after immunization. Of note, patients 2 and 3 in this report correspond to patients 3 and 4, respectively, in a previous report (48).
Virus isolation, quantitation of plasma viremia, and mononuclear cell provirus.
Blood was collected in EDTA anticoagulant tubes (Becton-Dickinson, Mountain View, Calif.) and was processed within 2 h of sampling. Peripheral blood mononuclear cells (PBMC) were obtained by standard Ficoll centrifugation (LSM; Organon Teknika, Durham, N.C.). The samples were CD8+ T-cell bead depleted (Dynal, Lake Success, N.Y.) and cultured in the presence of 10 U of interleukin-2 (Boehringer-Mannheim, Mannheim, Germany) per milliliter. Cultures were monitored for 4 weeks for evidence of reverse transcriptase (RT) activity, as previously described (48). The ability of the isolated viruses to induce syncytia was assessed by coculture of 100 50% tissue culture infective doses of the isolate with 106 MT-2 cells as previously described (4). Plasma HIV-1 RNA levels were measured by a quantitative competitive reverse transcription-PCR assay as previously described (48). Proviral burden was determined by a semiquantitative PCR assay, using SK38 and SK39 primers to amplify the gag region as previously described (48).
DNA preparation, cDNA synthesis, and PCR.
DNA was prepared from PBMC by using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.). RNA was extracted from 500 μl of plasma with the QIAamp HCV kit (Qiagen, Chatsworth, Calif.). RNA was incubated with RNase-free DNAse (Boehringer-Mannheim, Mannheim, Germany) at 37°C for 20 min (1 U of RNA sample/μl) in DNase reaction buffer (50 mmol Tris-Cl [pH 7.5]/liter, 10 mmol of MgCl2/liter, 40 U of RNasin [Promega, Madison, Wis.]). The sample was then incubated at 80°C for 10 min to terminate the reaction. cDNA synthesis was carried out in a total volume of 20 μl with 7 μl of the RNA sample, 40 U of avian myeloblastosis virus (AMV) RT (Lifesciences, St. Petersburg, Fla.), 40 U of RNasin, deoxynucleoside triphosphate at a final concentration of 1 mM, and outer antisense primer (ED12) at 1.5 μM final concentration in AMV RT buffer (at a final concentration of 25 mM Tris-HCl, pH 8.3, 50 mM KCl, 2.0 mM dithiothreitol, 5.0 mM MgCl2), with an incubation at 42°C for 45 min, followed by 10 min at 70°C.
DNA and cDNA were amplified by using nested PCR by limiting dilution so that single copies of DNA or cDNA could be amplified. First-round primers were ED5 (5′-ATGGGATCAAAGCCTAAAGCCATGTG-3′, nucleotides 6556 to 6581, HIV-1 HXB2) and ED12 (5′-AGTGCTTCCTGCTGCTCCCAAGAACCCAAG-3′, nucleotides 7822 to 7792, HIV-1 HXB2). Second-round reactions used 1 μl of first-round product as the template and primers DR7 (5′-TCAACTCAACTGCTGTTAAATGGCAGTCTAGC-3′, nucleotides 6989 to 7020, HIV-1 HXB2) and DR8 (5′-CACTTCTCCAATTGTCCCTCATATCTCCTCC-3′, nucleotides 7637 to 7667, HIV-1 HXB2), as previously described (18, 19). The final reaction amplifies ∼650 bp of product spanning the C2 to V5 region of the HIV-1 envelope surface protein. PCRs and cycling conditions were performed as previously described (19).
Heteroduplex tracking assay (HTA).
Single-stranded probes were generated by subjecting DR7-DR8-derived PCR products to an additional five cycles of PCR in the presence of [α-32P]dTTP, 33 μmol of each deoxynucleoside triphosphate, primer DR8, and biotinylated primer DR7. After PCR, the labeled DNA was bound to streptavidin-coated magnetic beads (Dynal, Lake Success, N.Y.). Magnetic beads were washed, and the labeled strand was disassociated with 8 μl of 0.1 N NaOH for 10 min. The probe was neutralized by adding 40 μl of H2O, 4 μl of 0.2 N HCl, and 1 μl of 1 M Tris-HCl (pH 7.4). Driver sequences were obtained by nested PCR of cDNA or genomic DNA as described above. In PBMC samples with low proviral load, multiple, independent nested PCR products were pooled before analysis. Probes were mixed with the driver sequence at a ratio of 1:100 in annealing buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.8], 2 mM EDTA), denatured at 94°C for 3 min, and then placed on ice for 5 min, followed by heating to 55°C for 5 min to form heteroduplexes. The resulting reaction mixtures were electrophoresed in 5% polyacrylamide gels (acrylamide/bisacrylamide ratio, 37.5:1) at 250 V for 3 h (19), stained with ethidium bromide to assure even migration of bands, dried, and scanned in a Molecular Dynamics PhosphorImager (Sunnyvale, Calif.).
Sequencing.
PCR products were directly sequenced by using the ABI PRISM dye-terminator method and by using DR7 and DR8 primers for forward and reverse reactions, respectively, according to the manufacturer’s specifications (Perkin-Elmer, Foster City, Calif.). Automatic sequencing was performed on an Applied Biosystems Inc. (Foster City, Calif.) 373 sequencer. Sequence editing and assembly were performed by using Sequencher, version 3.0 (Gene Codes Corporation, Inc., Ann Arbor, Mich.).
Phylogenetic analysis.
To root our sequences, envelope (env) sequences from five patients from the United States were used as outgroups in the analysis of the envelope genes samples from each of the four immunized patients (three and one control). Sequences were aligned, using Clustal W (50) with default settings, and improved by hand. Phylogenetic reconstruction was performed using MOLPHY (1), which first produces an approximate phylogeny based on a neighbor-joining algorithm and then improves the topology using local rearrangements under a maximum likelihood model. The maximum likelihood algorithm uses the HKY85 matrix of nucleotide substitutions (34) and calculates bootstrap probabilities using the RELL procedure for local rearrangements. Genetic distances between plasma-derived sequences for each time point were calculated using DNADIST (28), based on Kimura’s two-parameter model. Mean diversity is calculated over all pairwise distances for each time point. Genetic diversity along the C2 to V5 region was calculated for each patient’s virus in plasma through time. A standard entropy formula for diversity was employed in which diversity = (ni/N) Σ ln (ni/N), where N is the total number of nucleotides at a given site across sequences of an alignment, and ni is the proportion of a nucleotide i (i = A, C, T, and G).
Nucleotide sequence accession numbers.
Nucleotide sequences obtained from patients 1 through 4 have been submitted to GenBank under accession no. AF080698 through AF081019.
RESULTS
Immunization of subjects.
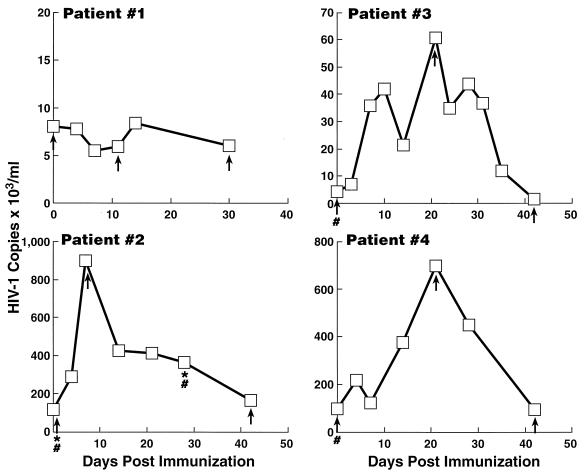
The clinical and laboratory profiles of the four patients are summarized in Table 1. Figure 1 shows the kinetics of plasma viremia in the mock-immunized (no. 1) and immunized (no. 2 through 4) patients. Patients 2 through 4 demonstrated significant increases in plasma viremia after immunization, ranging from 7- to 14-fold above baseline. Expression of activation markers on CD4+ T cells (i.e., HLA-DR and CD25) sampled from PBMC of patients 2 through 4 increased after immunization (data not shown). There were no significant changes in activation markers on CD4+ T cells of the unimmunized, HIV-infected control subject (data not shown).
TABLE 1.
Baseline characteristics of patients
| Patient | CD4+ T-cell count/μl | Duration (yr) HIV+ | Viremia (RNA copies/ml) | Provirus burden (DNA copies/ 106 PBMC) | Viral pheno- type |
|---|---|---|---|---|---|
| 1 | 406 | 7 | 8,000 | NDa | NSI |
| 2 | 350 | 8 | 120,000 | 1,000 | NSI |
| 3 | 531 | 8 | 4,200 | 10 | NSI |
| 4 | 389 | 9 | 100,000 | 360 | SI |
ND, not done.
FIG. 1.
Kinetics of plasma viremia postimmunization in patients 2 through 4. Patient 1 was mock immunized. Viral RNA copies (103/ml) of plasma are annotated on the ordinate, and days postimmunization are annotated on the abscissa. Plasma samples were obtained for sequencing of cDNA at baseline, during peak viremia, and at last follow-up and are indicated (↑). PBMC samples obtained for proviral sequencing at day 0 in patients 2 through 4 and at day 30 in patient 2 are indicated (#). LNMC samples obtained for proviral sequencing at days 0 and 30 postimmunization in patient 2 are indicated (∗). All patients were asymptomatic and were not receiving antiretroviral agents.
In order to study HIV sequence populations in vivo we amplified single molecules of virus (cDNA) or provirus after limiting dilution of the target sequence followed by a sensitive nested PCR method (18, 19, 47). The PCR product was then directly sequenced. This methodology has previously been validated by Simmonds et al. (47) to study the relative proportions of virus variants within patient samples. HIV cDNA sequences from plasma were obtained in all patients on the day of immunization, at the time of peak viremia, and at last follow-up when the level of plasma viremia had returned to baseline levels. As proviral DNA is largely composed of latent or nonreplicating viruses in a given patient (47), we were interested in examining baseline proviral sequences to determine if any of these variants would be induced from the activation of immunization. Thus proviral sequences from PBMC were obtained in patients 2, 3, and 4 on the day of immunization. Lymph node biopsies were performed before and after immunization in patient 2, and proviral sequences were determined on these samples on the day of immunization and 30 days later. At least 20 sequences were obtained per tissue compartment (e.g., plasma versus PBMC) at each sampled time point in all immunized patients. In some patients, nested PCR of undiluted cDNA or proviral DNA was performed and the products were analyzed by a heteroduplex tracking assay (see below) in order to confirm the nature of viral populations obtained by sequencing.
Quasispecies analysis.
In the analysis of our phylogenetic trees, we defined a phyletic group as a topological cluster of at least three sequences that generally but not always had bootstrap values exceeding 90%. In all instances, differing phyletic groups within a patient corresponded to distinct differences in the amino acid length and/or composition of V3, V4, and/or V5 regions, suggesting that viruses belonging to a particular group have distinct phenotypes in vivo.
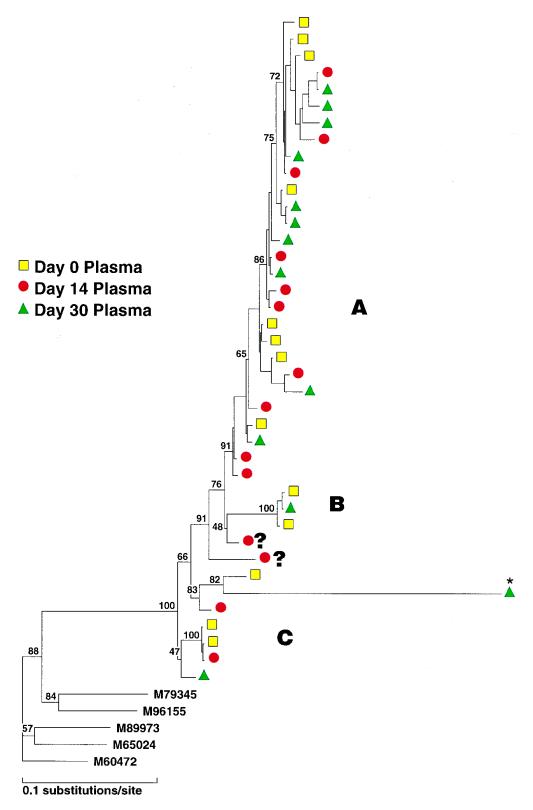
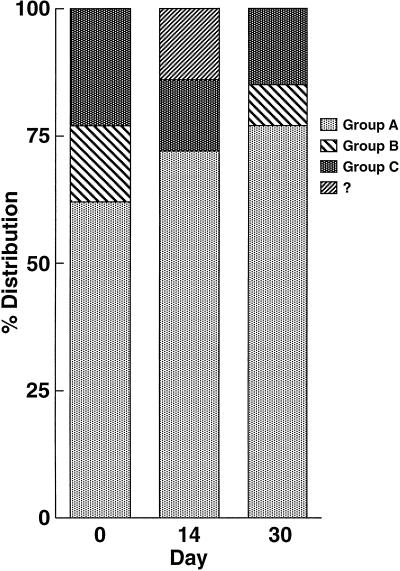
The phylogenetic reconstruction of plasma cDNA sequences obtained from patient 1, who was mock immunized, is shown in Fig. 2, left. The majority of viruses in plasma samples at all time points belonged to a large group (A), and a minority of viruses formed two smaller clusters (B and C). A small number of sequences did not clearly fit into any defined subgroup (identified as ?). There was overall stability in the distribution of the predominating variants in this unimmunized patient over a 30-day period (Fig. 2, right), with group A viruses making up 62, 72, and 77% of plasma sequences sampled at 0, 14, and 30 days post-mock immunization, respectively. The differences in group A frequency were not statistically significant between time points as determined by Fisher’s exact test. Group B sequences at the 14-day time point were not detected by sequence analysis. This was in all likelihood due to the fact that fewer sequences (<15 per time point) were sampled in this patient. Of note, HTAs performed on PCR products obtained from plasma cDNA samples using probes derived from variant B and C sequences revealed the presence of these minor groups at all time points (data not shown).
FIG. 2.
(Left) Phylogenetic analysis of the C2 to V5 region of all sequences (cDNA) sampled from patient 1. Viral sequences are shown by colored symbols according to their sampling time and tissue compartment. Three groups are indicated as A, B, and C; unclassified sequences are indicated by ?. Bootstrap probabilities are shown by percentages. ∗, This sequence has an artifactually long branch length due to a deletion of the V3 region. The following prototype HIV env subtype B strains were used as outgroups: M79345 (derived from a primary isolate), M96155 (from isolate 89.6), M89973 (YU-2), M65024 (HIVSFAAA), and M60472 (ADA). (Right) Frequency of detection of sequence variants in patient 1 (mock immunized) in sequential plasma samples. All sequences were derived from RNA.
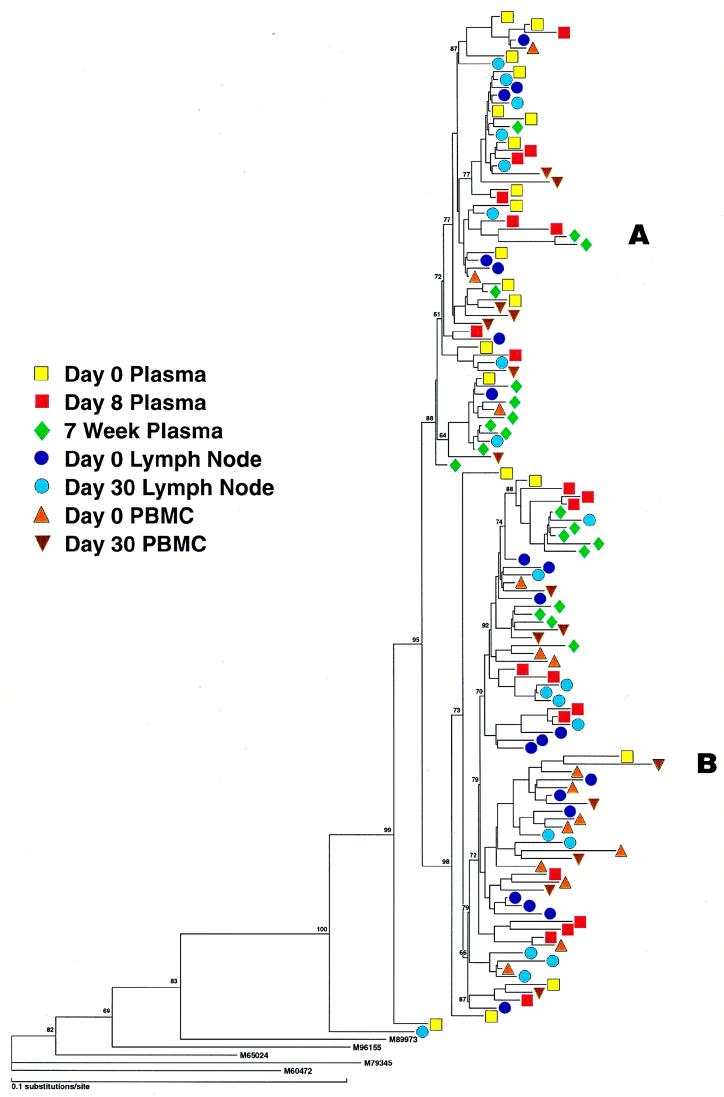
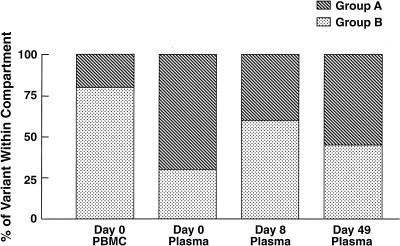
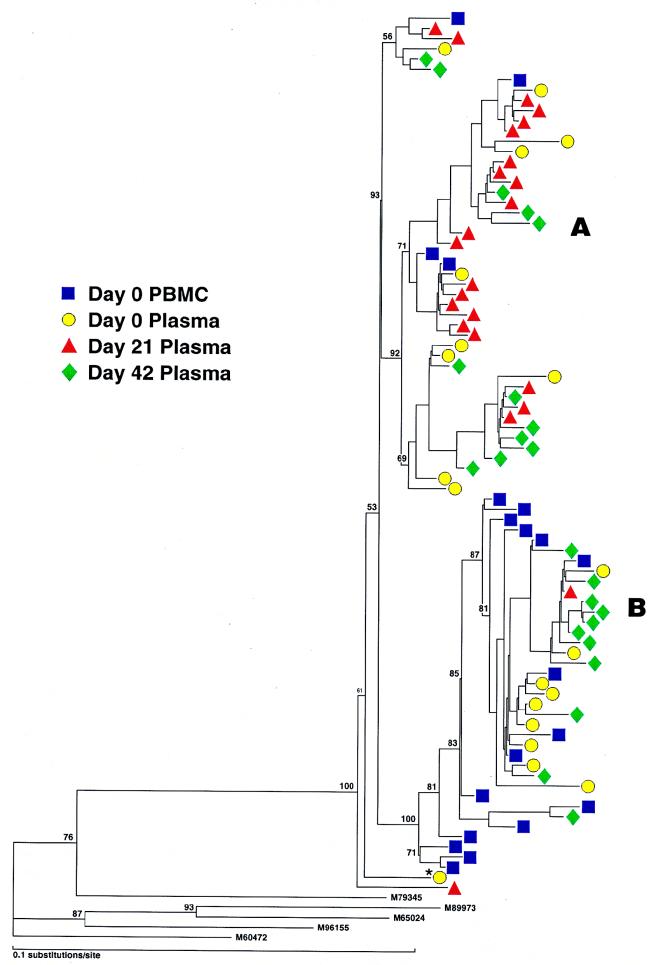
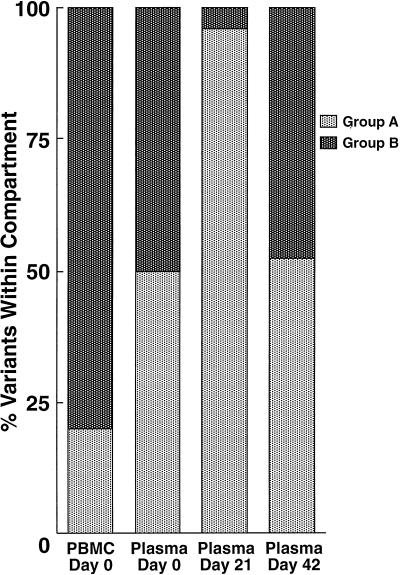
A total of 130 sequences were studied in patient 2; these consisted of cDNA from plasma (days 0 and 8 and 7 weeks postimmunization), provirus from PBMC (days 0 and 30 postimmunization), and provirus from lymph node mononuclear cells (LNMC) (days 0 and 30 postimmunization). Alignment of all sequences revealed the presence of two distinct populations of variants (A and B) in each tissue compartment at all time points studied (Fig. 3, left). Both groups could be easily distinguished by comparison of amino acid translations of the V4 regions (data not shown). Prior to immunization, the relative frequencies of groups A and B in plasma were significantly different from those in proviral LNMC (see the legend to Fig. 3, right) and PBMC (P < 0.01 for each, Fisher’s exact test). Group A made up 75% of sequences in plasma but was the minor population in sequences derived from proviral PBMC and LNMC (Fig. 3, right). Conversely, group B comprised the major proviral population within PBMC and LNMC and thus was more likely a reflection of the archival population of viruses (47, 52). In this patient, group A presumably represented a more recently evolved variant circulating in the plasma, although viruses from group B were also actively replicating and circulating in the plasma at the time of immunization. At the time of peak viremia (day 8), there was a reversal in the relative proportions of virus populations in the plasma, with group B predominating (P < 0.025, Fisher’s exact test). In contrast to group A which increased 4-fold in the plasma following immunization, group B had increased 18-fold. By 7 weeks after immunization, at a time when viremia approached baseline values, there was a trend toward a return to the original distribution of plasma viruses. There was considerable similarity of the V3 loop in all sequences, which had a macrophage-tropic (M-tropic) pattern consistent with the isolation of non-syncytium-inducing (NSI) viruses in culture (data not shown).
FIG. 3.
(Left) Phylogenetic analysis of sequences (cDNA and proviral) from patient 2. Samples from plasma represent RNA sequences, and those from PBMC and LNMC are proviral. Note two major groups, A and B, which are found in all compartments and time points sampled. Variant A was predominant in plasma, and variant B was predominant in PBMC at baseline. (Right) Frequency distribution of RNA variants in plasma and proviral variants in PBMC before and after immunization. For comparative purposes the proviral distributions of LNMC and PBMC at day 30 are not illustrated but are as follows: day 30 PBMC, group A, 35%, and group B, 65%; day 0 LNMC, group A, 35% and group B, 65%; day 30 LNMC group A, 45% and group B, 55%.
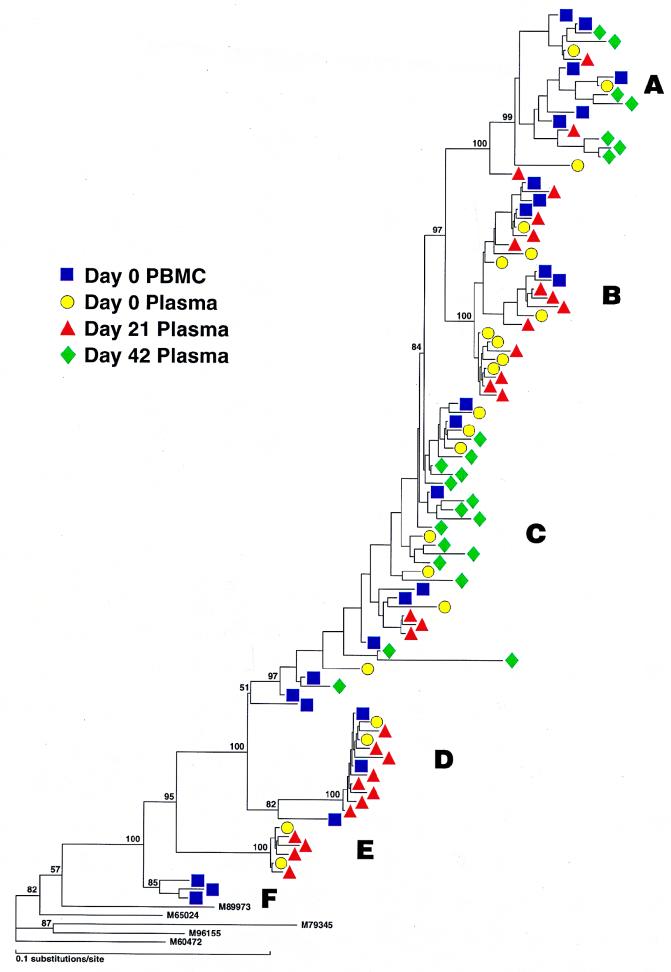
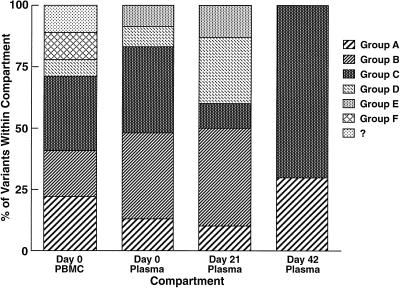
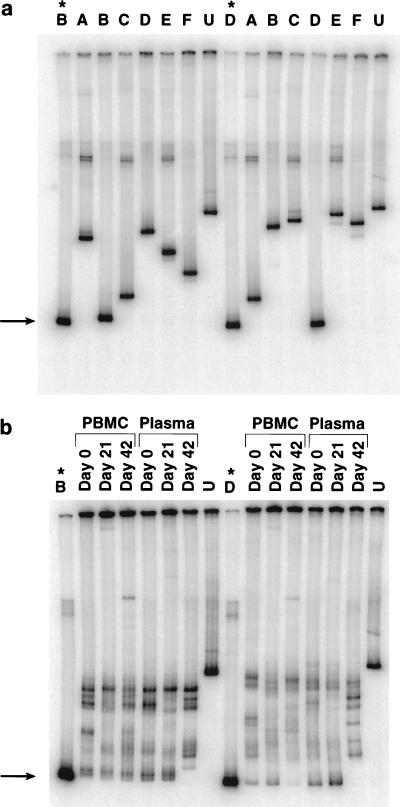
Patient 3, who was a slow progressor, showed a much more complex viral phylogenetic tree (Fig. 4, left) that comprised six distinguishable groups (arbitrarily termed A through F). At least five groups were identified in the PBMC proviral compartment (A, B, C, D, and F) as well as sequences which did not clearly fit into any group (Fig. 4, right). Twenty-two percent of proviral PBMC sequences were not found in plasma and likely represented archival virus within latently infected cells (groups F and ?). Since groups A, B, C, and D were also detected in the plasma, these proviral sequences were also likely contributing to active viral replication. At the time of immunization, five groups could be identified in plasma, with groups B and C predominating. The composition of variants in plasma before immunization and at peak viremia was somewhat similar, although group C variants decreased and group D variants expanded at peak viremia. By day 42, however, a dramatic change in quasispecies makeup was seen, in which 80% of virus variants that were circulating at day 21 (groups B, D, and E) were replaced by groups A and C. In order to confirm these quasispecies shifts in patient 3, we performed HTAs on PCR products from undiluted samples obtained from plasma and PBMC at multiple time points from immunization. We elected to perform HTAs by using probes made from group B and D viruses, since these variants comprised 70% of viruses at day 21 and were not seen at day 42. Probes made from representative group B and D sequences could distinguish sequences obtained from other groups (Fig. 5a). These probes were then employed against PCR products from undiluted samples taken from PBMC (proviral DNA) and plasma (cDNA) at days 0, 21, and 42 (Fig. 5b). By HTA, group B and D viruses were not identified in plasma from day 42, as illustrated by slower migration of heteroduplexes at these time points (Fig. 5), thus confirming our sequence analysis. Interestingly, a reduced signal intensity for variant D was observed in PBMC sampled at day 42, suggesting early clearance of this variant from the PBMC compartment.
FIG. 4.
(Left) Phylogenetic analysis of sequences (cDNA and proviral) from patient 3. Major groups are indicated as A through F. (Right) Frequency distribution of viral variants in plasma and PBMC preimmunization and in plasma postimmunization in patient 3.
FIG. 5.
(a) HTA of viral sequences (C2 through V5) obtained from patient 3. After referring to the phylogenetic tree, a representative sequence from group B or D was selected as a probe against other variants. Probes derived from group B or D sequences were able to easily distinguish sequences of the same group (B or D) from a panel of sequences obtained from different groups in the same patient or from an unrelated sample (lane U). The experiment was repeated three times, using different representative sequences for probe and panel, with similar results. ∗, probe lane; arrow, position of homoduplex migration. (b) The effect of tetanus immunization on viral quasispecies turnover in patient 3, as displayed by HTA. In order to determine the turnover of groups B and D, an HTA was performed with the probes described above. For plasma samples, RT PCR products amplified from approximately 100 copies of HIV-1 RNA templates were used as the driver. For PBMC samples, PCR products amplified from about 40 copies fo HIV-1 genomic DNA were used as the driver. Arrow, position of homoduplex migration.
The phylogenetic reconstruction of sequences obtained from patient 4 revealed the presence of two major variants, A and B (Fig. 6, left). Groups A and B were clearly distinguishable in the V3 region: the group A consensus V3 loop was CTRPNNNTRRGIHIGPGSAFYATGDIIGDIRQAHC; the group B consensus V3 loop was CTRPSINKRRHIHIGPGRAFYATDITGDIRQAHC. The V3 loop of B variants differed from that of A variants (at positions 5, 6, 8, 11, 18, 24, and 27 from the cysteine on the 5′ side of the V3 region) by having a greater number of positively charged amino acids and the loss of a potential N-linked glycosylation site (position 6) in group B. These changes have previously been shown to be consistent with a switch from an envelope of an NSI to that of a syncytium-inducing (SI) virus (11, 21, 23, 31). The existence of SI viruses in patient 4 was confirmed by PBMC culture (Table 1). One sequence obtained from plasma at day 0 was a recombinant of both variants, comprising an A V3 region and a B V4/V5 region (data not shown and Fig. 6, left). Prior to immunization, patient 4 had an approximately equal distribution of variants A and B in the plasma (Fig. 6, right). At the time of peak viremia (day 21), variant A (NSI) made up 95% of viruses (P < 0.005, Fisher’s exact test). By day 41, as plasma viremia returned to baseline levels, the original distribution of viruses was observed. By extrapolating from absolute plasma viral levels (Fig. 1), the NSI variants had increased 13-fold following immunization, whereas the SI variants had essentially remained unchanged. Of note, viruses of variant B (SI) made up the greatest proportion in the proviral PBMC compartment.
FIG. 6.
(Left) Phylogenetic analysis of sequences from patient 4. Two major groups, A and B, are identified. ∗, a recombinant of groups A and B. (Right) Frequency distribution of groups A and B in plasma and PBMC preimmunization and in plasma postimmunization.
We did not observe the emergence of new groups of variants during the course of immunization in any of the patients studied. We also did not observe the emergence of SI-type envelopes in those patients who had NSI viruses in culture at baseline (patients 1, 2, and 3).
Diversity analysis.
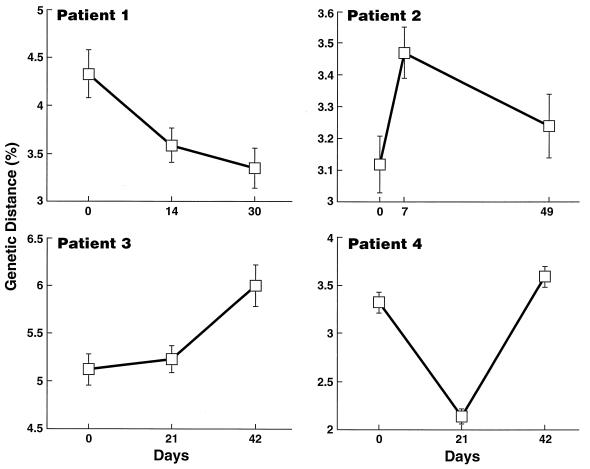
No consistent trends in the mean virus population diversity within plasma samples was observed with time (Fig. 7); however, reversible changes were noted in two of three immunized patients. For example, in patient 2, virus diversity had significantly increased at peak viremia (3.1% at day 0 to 3.5% at day 8), whereas in patient 4, virus diversity had decreased at the time of peak viremia (3.3% at day 0 to 2.1% at day 21). These patterns of diversity were also reflected in the topology of the phylogenetic trees. In patient 2, viruses from group B, which was predominant at peak viremia, had longer branch lengths than group A viruses, thus reflecting the mean diversity of the plasma virus populations at this time point. In patient 4, a single group made up the majority of the plasma viral population at peak viremia, which accounted for the decrease in diversity at day 21.
FIG. 7.
Mean diversity of plasma viral sequences over time in patients 1 through 4. Error bars represent standard error of the mean.
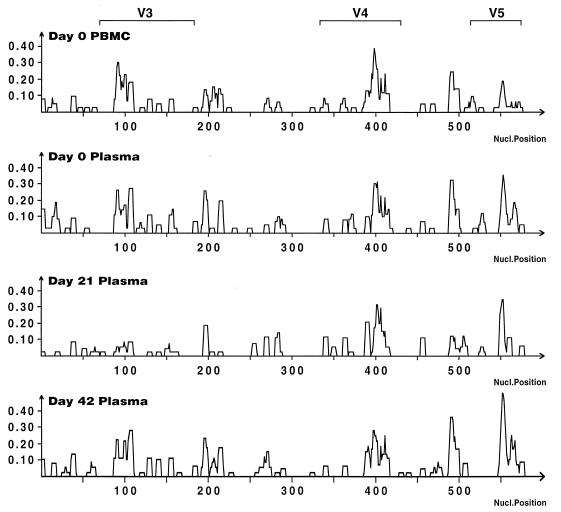
In order to identify regions of the envelope that were responsible for overall changes in mean diversity within samples, entropy was plotted against nucleotide position for all within sample sequences. No specific region along the envelope in patients 1 and 3 clearly accounted for the majority of changes in mean diversity. In patient 2, the increased diversity at peak viremia was due to changes in the V4 region (data not shown). Interestingly, in patient 4, the decrease in diversity seen at peak viremia was localized to the V3 region (Fig. 8) and reflected the decrease of SI variants.
FIG. 8.
Diversity along env over time in patient 4. Entropy (ordinate) is plotted against nucleotide position.
Accumulation rates of synonymous and nonsynonymous substitutions.
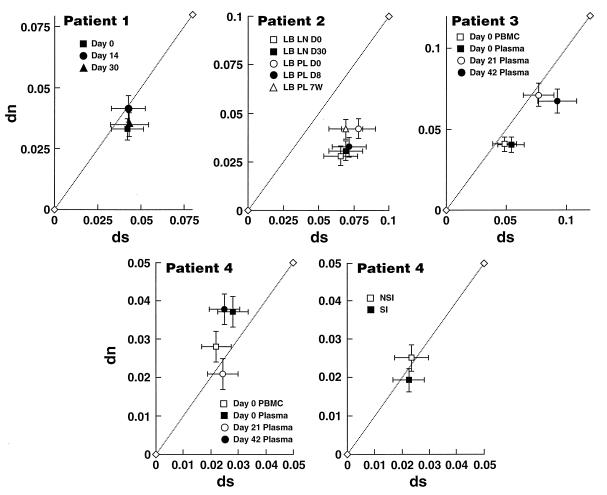
Selection pressures on HIV-1 quasispecies were determined during the course of immunization by analyzing the synonymous (amino acid preserving) and nonsynonymous (amino acid changing) nucleotide substitution patterns (41). The mean numbers of nucleotide substitutions per synonymous site, ds, and per nonsynonymous site, dn, for all pairwise comparisons within sequences sampled at each time point were determined. Thus, all comparisons were of intrasample sequences against a consensus sequence from the same sample. Generally, a dn/ds ratio of <1 implies that purifying selection for replication fitness predominates, whereas a dn/ds ratio of >1 implies strong positive selection for amino acid diversification, as might be imposed by the immune system or by the availability of specific target cells (37).
The unimmunized patient, no. 1, revealed dn/ds ratios that indicated no selection (neutrality) at variable sites (Fig. 9). In patient 2, dn/ds ratios at all time points in plasma and lymph nodes were consistent with a pattern for purifying selection throughout the course of immunization. Patient 3 revealed a pattern similar to that of mock-immunized patient 1. The most interesting findings were in patient 4, who had both NSI and SI viruses. Prior to immunization and at last follow-up, when the viremia was at steady state, dn/ds ratios were >1, indicating strong positive selection for amino acid change. During peak viremia, the dn/ds ratio was close to unity, implying neutral drift at variable sites and suggesting a loss of the selective pressure that was driving the fixation of nonsilent mutations prior to immunization. We considered the possibility that the high dn/ds ratios observed at days 0 and 42 were a consequence of the presence of T-tropic (SI) viruses within the sample. It has been previously postulated that SI viruses may be under greater immunologic pressure than NSI viruses within a given patient, thus driving a higher nonsynonymous substitution rate in SI viruses (6). However, upon comparison of dn and ds values between all M-tropic (group A) and T-tropic variants (group B) obtained from all time points in this patient, both groups showed dn/ds ratios consistent with neutral evolution (Fig. 9). Thus, the increased dn/ds ratios observed in plasma sequences obtained from baseline and at last follow-up are not the sole consequence of the presence of SI viruses in the samples but reflect selection in some subset of the sequences analyzed.
FIG. 9.
Effect of tetanus immunization on synonymous (ds) and nonsynonymous (dn) substitution rates. Intrasample ds and dn values for patients 1 through 4. Significant dn/ds ratios away from neutrality, i.e., dn/ds either <1 or >1, are defined as those plots with error bars which have significantly deviated either below or above the x = y line, respectively.
DISCUSSION
The present study has characterized viral sequence variation and evolutionary relationships in three HIV-infected patients after immunization with a common recall antigen. Antigen-specific immune activation induces changes in the host characterized by proliferation of antigen-responsive cells, elaboration of cytokines, and subsequent proliferation of bystander cells (10, 33, 51). Such a state of immune activation results in an increase in the number of available activated CD4+ T cells that serve as susceptible targets for HIV. We tested this hypothesis in some detail by searching for any systematic trends in the growing virus population. We asked whether immunization had temporally influenced the source of plasma virus (phylogenetic analysis), virus diversity (diversity plots), selection pressures (ds/dn), and precise positions of new substitutions along the env sequence (entropy).
A common feature was that the perturbation in plasma viremia induced by the activating stimulus of the immunization was consistently associated with significant perturbations in the relative predominance of viral quasispecies detectable in the plasma of immunized subjects. Although the unimmunized patient (no. 1) demonstrated a shift in a minor plasma virus variant over time, this change was not as dramatic as that seen in the immunized patients. The combined observations of the relative stability of the major quasispecies variant in the mock-immunized subject (patient 1) and the transient and apparently reversible nature of the plasma quasispecies shifts in patients 2 and 4, strongly suggest that the immunization had actually influenced the quasispecies distributions in our patients. The quasispecies changes observed in patient 3 are noteworthy. We were able to confirm, using the HTA, that 80% of virus variants that were present during the time of peak viremia (day 21) were cleared from the plasma by day 42. This rate of clearance of variants over a 3-week period is not altogether surprising given our current understanding of viral turnover (36, 44, 52).
We asked whether immune activation had provided a replicative advantage to certain virus variants. In most cases we were unable to demonstrate a consistent pattern of selection. We could not demonstrate consistent changes in diversity, nor were we able to demonstrate an increase in the dn/ds ratio as might be expected with an increase following the emergence of an escape mutant. No common set of amino acid substitutions along env could explain the increase in viremia. A possible explanation for this is that during immunization a subset of CD4+ cells becomes activated and that virus release from these cells is amplified, irrespective of the virus genotypes they contain. This leads to a change in the distribution of variants but not to any evidence of selection and raises the notion that the shifting waves of diversity observed during infection are attributable to various selection pressures on the CD4+ T cells themselves and not on the specific virus that they harbor. For example, immunization will activate tetanus-specific CD4+ T cells and resident virions will incidentally become amplified. This idea has recently been supported by Cheynier et. al. (13) who demonstrated in the macaque model that the dynamics of simian immunodeficiency virus quasispecies after BCG immunization was a reflection of those viruses found in circulating BCG-specific CD4+ T cells.
Specific selection of viruses, may, however, be operating in those patients who harbor variants of differing tropisms, as seen in patient 4. Prior to immunization, equal proportions of NSI and SI viruses were circulating in the plasma, whereas after immunization the induced burst in viremia was due to an expansion of NSI viruses. This was an unexpected finding since SI viruses generally replicate to a greater degree than NSI viruses in tissue culture (4). In this patient, diversity of the V3 region transiently decreased after immunization. The higher rates of nonsynonymous substitutions observed (dn/ds of >1) in plasma prior to immunization and at last follow-up (day 42) suggest that the selective pressures that might have been driving the fixation of new mutants at baseline were relaxed during the burst of viremia. These findings suggest that prior to immunization, strong selective pressures were acting on both the NSI and SI viruses and that the sudden availability of a new population of activated CD4+ T cells following immunization allowed the NSI population to expand without constraint. The explanation for why NSI viruses would have an apparent replicative advantage over SI viruses in the setting of immune activation is not clear. A similar situation has been described in patients who, during acute HIV infection, were initially infected with mixtures of SI and NSI viruses and then were subsequently shown to harbor only NSI strains (17). The NSI virus phenotype has been correlated with macrophage tropism and usage of the CCR5 HIV coreceptor, whereas the SI phenotype has been correlated with T-cell-line tropism and usage of the CXCR4 HIV coreceptor (2, 14, 16, 22, 24, 25, 29, 45). It has been demonstrated that CCR5 expression is markedly upregulated and CXCR4 expression is downregulated on activated CD4+ T cells (5, 43). This observation lends credence to the possibility that tetanus immunization transiently increased the pool of activated CD4+ T cells that express CCR5 and little or no CXCR4, thus favoring the replication of viruses that solely use CCR5 as a coreceptor, i.e., M-tropic (NSI) viruses. Although material was not available to study coreceptor expression in patient 4, we have subsequently observed modest transient increases in CCR5 expression and decreases in CXCR4 expression in both HIV-infected individuals and healthy volunteers after tetanus immunization (data not shown). In addition, we are currently determining the exact coreceptor usage of the envelope variants obtained in patient 4 in order to address this issue further. It is also unclear what role the recent discovery of other coreceptors that appear to be used by NSI and SI strains play in the setting of immune activation (2, 14, 20, 24, 29, 38).
We asked what contribution latent viruses play in the induced viremia postimmunization. Studies by Simmonds et al. (47) and Wei et al. (52) have demonstrated that turnover of proviral DNA in PBMC lags behind turnover of plasma cDNA and that the majority of proviral DNA in PBMC reflects archival or latent viruses that had previously appeared in the plasma. It is also important to recognize that sources of provirus within lymph nodes include latently infected CD4+ T cells, long-lived macrophages, and dendritic cells, which may all potentially be induced to produce virus (15, 36). Of the three immunized patients described in this report, patients 2 and 4 manifested a quasispecies distribution in PBMC (and LNMC in patient 2) proviral samples that could be easily distinguished from that in plasma, thus confirming previous findings (47). In patient 3, a small group of proviral variants (∼20%) could be distinguishable from those in plasma. If immune activation did indeed reactivate latently infected cells to a significant extent, one might then have expected to detect the predominating proviral PBMC variants in the plasma after immunization. This was in fact the case with patient 2, in whom the predominant variant (B) within the proviral PBMC (and LNMC) compartment emerged as the predominant variant in the plasma during peak viremia. The higher baseline proviral load seen in patient 2 (Table 1) might have allowed a greater opportunity for proviruses to become reactivated and to significantly contribute to the actively replicating pool of viruses in the plasma. This pattern of quasispecies shift was not observed in either patient 3 or patient 4; thus, in these two patients, the preexisting major variants in the plasma were expanded after immunization, suggesting that the transient increase in plasma viremia that resulted from immune activation was merely acceleration of a process that was already accounting for the bulk of plasma virus as opposed to a shift from one source of virus replication to another.
These observations may have considerable relevance to the pathogenesis of HIV disease. Our observations confirm the dynamic nature of HIV replication in vivo (36, 44, 52). The fact that immune activation appears to, in most cases, nonspecifically amplify viruses of differing genotypes likely contributes to the great diversity observed in vivo. The exception to this may be in patients who harbor viruses of differing tropisms; the observation that immune activation favors viruses of an NSI phenotype over SI strains may explain in part the paucity of SI strains observed during the early stages of HIV. Finally, the fact that latently infected cells may contribute to plasma viremia under certain circumstances such as immune activation is of particular interest in light of the recent observations that a stable reservoir of resting, latently infected CD4+ T cells that can be induced to express replication-competent virus persists over extended periods of time in patients whose plasma viremia has been driven to below detectable levels by highly active antiretroviral therapy (15, 30, 53).
ACKNOWLEDGMENTS
We thank Tae-Wook Chun, Susan Moir, Lin Qi Zhang, Raj Shankarappa, and Jim Arthos for helpful discussions. We thank MaryBeth Daucher and Colombe Chappey for help with DNA sequencing. We are also grateful to the four patients who committed their time and effort to this study.
This work was supported by NIH grants to G.L. (A132885 and A127757).
REFERENCES
- 1.Adachi J, Hasegawa M. MOLPHY, version 2.3. Programs for molecular phylogenetics based on maximum likelihood. Tokyo, Japan: A publication of the Institute of Statistical Mathematics; 1996. [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Ascher M S, Sheppard H W. AIDS as immune system activation. II. The panergic imnesia hypothesis. J Acquired Immune Defic Syndr. 1990;3:177–191. [PubMed] [Google Scholar]
- 4.Asjo B, Albert J, Karlsson A, Morfeld-Manson L, Biberfeld G, Lidman K, Fenyo E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 5.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonhoeffer S, Holmes E C, Nowak M A. Causes of HIV diversity. Nature. 1995;376:125. doi: 10.1038/376125a0. [DOI] [PubMed] [Google Scholar]
- 7.Brichacek B, Swindells S, Janoff E N, Pirruccello S, Stevenson M. Increased plasma human immunodeficiency virus type 1 burden following antigenic challenge with pneumococcal vaccine. J Infect Dis. 1996;174:1191–1199. doi: 10.1093/infdis/174.6.1191. [DOI] [PubMed] [Google Scholar]
- 8.Bukrinsky M I, Stanwick T L, Dempsey M P, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush C E, Donovan R M, Markowitz N P, Kvale P, Saravolatz L D. A study of HIV RNA viral load in AIDS patients with bacterial pneumonia. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:23–26. doi: 10.1097/00042560-199609000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Carding S, Allan W, McMickle A, Doherty P. Activation of cytokine genes in T cells during primary and secondary murine influenza pneumonia. J Exp Med. 1993;177:475–482. doi: 10.1084/jem.177.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheynier R, Henrichwark S, Hadida F, Pelletier E, Oksenhendler E, Autran B, Wain-Hobson S. HIV and T cell expansion in splenic white pulps is accompanied by infiltration of HIV-specific cytotoxic T lymphocytes. Cell. 1994;78:373–387. doi: 10.1016/0092-8674(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 13.Cheynier R, Gratton S, Halloran M, Stahmer I, Letvin N L, Wain-Hobson S. Antigenic stimulation by BCG vaccine as an in vivo driving force for SIV replication and dissemination. Nat Med. 1998;4:421–427. doi: 10.1038/nm0498-421. [DOI] [PubMed] [Google Scholar]
- 14.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 15.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornelissen M, Mulder K G, Veenstra J, Zorgdrager F, Kuiken C, Hartman S, Dekker J, van der Hoek L, Sol C, Coutinho R, Goudsmit J. Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population. J Virol. 1995;69:1810–1818. doi: 10.1128/jvi.69.3.1810-1818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delwart E C, Herring B, Rodrigo A G, Mullins J I. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. In: Laboratory C S H, editor. PCR methods and applications. supplement. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1995. pp. S202–S216. [DOI] [PubMed] [Google Scholar]
- 19.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 20.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di M P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 21.De Wolf F, Hogervorst E, Goudsmit J, Fenyo E, Rubsamen-Waigmann H, Holmes H, Galvao-Castro B, Karita E, Wasi C, Sempala S D K, Baan E, Zorgdrager F, Lukashov V, Osmanov S, Kuiken C, Cornelissen M the WHO Network for HIV Isolation and Characterization. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: phenotypic and genotypic characteristics. AIDS Res Hum Retroviruses. 1994;10:1398–1400. doi: 10.1089/aid.1994.10.1387. [DOI] [PubMed] [Google Scholar]
- 22.Doms R, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 23.Donaldson Y K, Bell J E, Holmes E C, Hughes E S, Brown H K, Simmonds P. In vivo distribution and cytopathology of variants of human immunodeficiency virus type 1 showing restricted sequence variability in the V3 loop. J Virol. 1994;68:5991–6005. doi: 10.1128/jvi.68.9.5991-6005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 25.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 26.Fauci A S. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 27.Fauci A S. Multifactorial nature of human immunodeficiency virus disease: implications for therapy. Science. 1993;262:1011–1018. doi: 10.1126/science.8235617. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. PHYLIP (phylogeny inference package), 3.5c ed. Seattle, Wash: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 29.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 30.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 31.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goletti D, Weissman D, Jackson R W, Graham N M H, Vlahov D, Klein R S, Munsiff S S, Ortona L, Cauda R, Fauci A S. Effect of Mycobacterium tuberculosis on HIV replication, role of immune activation. J Immunol. 1996;157:1271–1278. [PubMed] [Google Scholar]
- 33.Grossman Z, Paul W. Adaptive cellular interactions in the immune system: the tunable activation threshold and the significance of subthreshold responses. Proc Natl Acad Sci USA. 1992;89:10365–10369. doi: 10.1073/pnas.89.21.10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasegawa M, Iida Y, Yano T, Takaiwa F, Iwabuchi M. Phylogenetic relationships among eukaryotic kingdoms inferred from ribosomal RNA sequences. J Mol Evol. 1985;22:32–38. doi: 10.1007/BF02105802. [DOI] [PubMed] [Google Scholar]
- 35.Ho D D. HIV-1 viraemia and influenza. Lancet. 1992;33:1549. doi: 10.1016/0140-6736(92)91321-x. [DOI] [PubMed] [Google Scholar]
- 36.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 37.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. p. 394. [Google Scholar]
- 38.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C J. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 40.Mellors J W, Rinaldo C J, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 41.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 42.O’Brien W A, Grovit-Ferbas K, Namazi A, Ovcak-Derzic S, Wang H J, Park J, Yeramian C, Mao S-H, Zack J A. Human immunodeficiency virus-type 1 replication can be increased in peripheral blood of seropositive patients after influenza vaccination. Blood. 1995;86:1082–1089. [PubMed] [Google Scholar]
- 43.Ostrowski, M. A., S. J. Justement, T. Chun, A. Catanzaro, C. A. Hallahan, L. A. Ehler, S. B. Mizell, P. N. Kumar, J. Mican, and A. S. Fauci. Expression of chemokine receptors CXCR4 and CCR5 in HIV-1 infected and uninfected individuals. J. Immunol., in press. [PubMed]
- 44.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 45.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 46.Schacker T W, Hughes J P, Shea T, Coombs R W, Corey L. Biological and virologic characteristics of primary HIV infection. Ann Intern Med. 1998;128:613–620. doi: 10.7326/0003-4819-128-8-199804150-00001. [DOI] [PubMed] [Google Scholar]
- 47.Simmonds P, Zhang L Q, McOmish F, Balfe P, Ludlam C A, Leigh Brown A J. Discontinuous sequence change of human immunodeficiency virus (HIV) type 1 env sequences in plasma viral and lymphocyte-associated proviral populations in vivo: implications for models of HIV pathogenesis. J Virol. 1991;65:6266–6676. doi: 10.1128/jvi.65.11.6266-6276.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanley S K, Ostrowski M A, Justement J S, Gantt K, Hedayati S, Mannix M, Roche K, Schwartzentruber D J, Fox C H, Fauci A S. Effect of immunization with a common recall antigen on viral expression in patients infected with human immunodeficiency virus type 1. N Engl J Med. 1996;334:1222–1230. doi: 10.1056/NEJM199605093341903. [DOI] [PubMed] [Google Scholar]
- 49.Staprans S I, Hamilton B L, Follansbee S E, Elbeik T, Barbosa P, Grant R M, Feinberg M B. Activation of virus replication after vaccination of HIV-1-infected individuals. J Exp Med. 1995;182:1727–1737. doi: 10.1084/jem.182.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unutmaz D, Pileri P, Abrignani S. Antigen-independent activation of naive and memory resting T cells by a cytokine combination. J Exp Med. 1994;180:1159–1164. doi: 10.1084/jem.180.3.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini I A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 53.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 54.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]