Abstract
Within cellular structures, compartmentalization is the concept of spatial segregation of macromolecules, metabolites and biochemical pathways. This concept therefore bridges organellar structure and function. Mitochondria are morphologically complex, partitioned into several subcompartments by a topologically elaborate two-membrane system. They are also dynamically polymorphic, undergoing morphogenesis events with an extent and frequency that is just now being appreciated. Mitochondrial compartmentalization is therefore something that must be considered both spatially and temporally. Here we review new developments in how mitochondrial structure is established and regulated, the factors that underpin the distribution of lipids and proteins, and how they spatially demarcate locations of myriad mitochondrial processes. Consistent with its preeminence, disturbed mitochondrial compartmentalization contributes to the dysfunction associated with heritable and aging-related diseases.
Keywords: Mitochondria, Ultrastructure, Morphogenesis, Cristae, Bioenergetics, Macromolecular Trafficking
Mitochondrial Architecture Sets the Framework for Compartmentalization
The mitochondrion is an intricate organelle that is the main site of cellular energy production, and orchestrates many other cellular processes, including lipid metabolism, cellular ion homeostasis, and metal cofactor biosynthesis. This functional diversity is reflected in the complex mitochondrial ultrastructure (see Glossary) [1, 2] (Figure 1A). Unlike most organelles, mitochondria contain two membranes: an outer membrane (OM), which encompasses the organelle and interfaces directly with the cytosol, and a morphologically complex inner membrane (IM) that can be further divided into two parts: the inner boundary membrane (IBM) that is closely apposed to the OM, and the cristae membranes (CM) that produce lamellar or tubular protrusions into the interior of the organelle, depending on the morphogenic origin and metabolic state. This double membrane system delineates multiple aqueous compartments: the intermembrane space (IMS) between the OM and the IBM; the intracristal space (ICS) bound by the CM; and the matrix, which is the innermost compartment. Cristae themselves can be further parsed into functionally relevant regions, including the crista junction (CJ), which is an elliptical or circular structure that connects crista to the IBM, and the crista tip (CT) of the distal end of cristae.
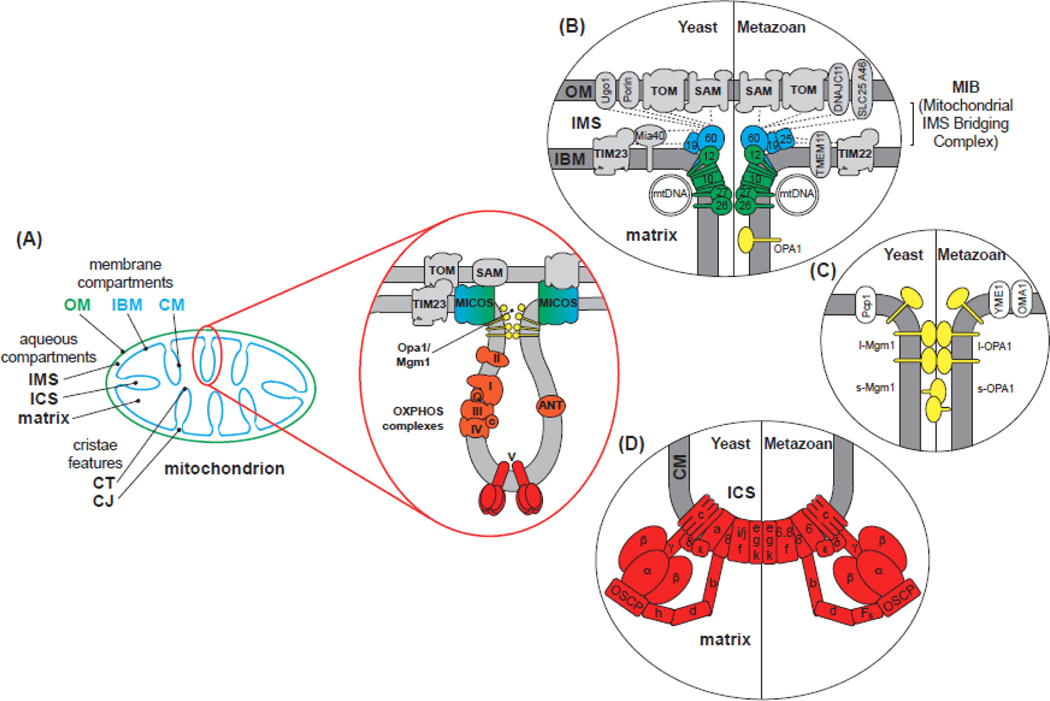
Figure 1. Mitochondrial Ultrastructure and Morphogenic Complexes.
A) Major mitochondrial compartments. Subcompartments are established by the outer membrane (OM; green) and inner membrane (IM; cyan), parsed into the inner boundary membrane (IBM) and cristae membrane (CM). Aqueous compartments include the intermembrane space (IMS) (between OM and IBM), the intracristal space (ICS) (enclosed by cristae), and matrix. Major cristae features include the crista junction (CJ) and the crista tip (CT). B-D) Assemblies that establish IM morphology. The subunits of each complex are shown for yeast (left) and for metazoans (right). B) The MICOS Complex establishes and stabilizes CJs. MICOS is composed of six (yeast) or seven (metazoan) known subunits, each named as MicX (X numbering based on molecular weight). The MIC60 subcomplex (cyan) consists of the central Mic60 (mitofilin) subunit and peripheral regulatory proteins Mic19/25, mediating a number of interactions (dashed lines) with IBM and OM proteins. The MIC10 subcomplex (green) contains the central Mic10 subunit, apolipoprotein O-related Mic26/27, which antagonistically regulate Mic10 assembly, and Mic12, which mediates MIC60-MIC10 subcomplex association. C) Mgm1/Opa1 Assemblies are GTPases that exist as long forms (l-Mgm1/l-Opa1) with an N-terminal transmembrane anchor, or as short forms (s-Mgm1/s-Opa1) that are proteolytically processed by IM proteases Pcp1 (yeast) or OMA1/YME1 (metazoan). Oligomers of l-Mgm1/l-Opa1 assemble in a cardiolipin-dependent manner to stabilize connections of cristae walls. D) The F1FO-ATP synthase consists of the F1 sector [catalytic headpiece (α3β3) and central stalk (γ,δ,ε)] and FO sector [rotor (c ring, a, 8), peripheral stalk (b, d, h/F6, OSCP), and supernumerary subunits (f, i/j/6.8, e, g, k)]. Homodimer assembly is mediated by subunits e, g and k (DAPIT in metazoans). Higher-order assembly occurs as extended dimer rows along cristae ridges.
Mitochondrial compartmentalization refers to the way in which macromolecules, metabolites, and biochemical processes are spatially distributed. Such distribution can be considered on different scales: (i) among membranes (OM, IBM, CM) or aqueous subcompartments (matrix, IMS, ICS); (ii) laterally along contiguous membranes (IBM, CJ, CM, CT); and (iii) between monolayers of a given membrane. Mitochondria are also highly pleomorphic, undergoing structural changes in accordance with the physiological state of the cell. They undergo constant fission and fusion [3] and form dynamic homotypic networks, as well as contacts with other organelles such as the endoplasmic reticulum (ER) [4]. Further, the IM displays remarkable structural plasticity capable of extensive cristae remodeling [1]. Compartmentalization of the mitochondrion, therefore, is not simply a question of spatial distribution, but one of spatiotemporal dynamics.
Herein, we first review key technologies that are enabling insights into mitochondrial structure with ever-increasing resolution. We then focus on the protein complexes and lipids that shape mitochondrial membranes to create its defining subcompartments. Having discussed ultrastructure and its determinants, we turn to current views on the compartmentalization of proteins and lipids within the mitochondrion, as well as mechanisms that underpin macromolecular distribution. We conclude with the functional implications of compartmentalization by illustrating new concepts that relate mitochondrial architecture and two critical processes, energy transduction and apoptosis.
Imaging Mitochondrial Ultrastructure: Brief History and Technical Advances
Our understanding of mitochondrial structure has co-evolved with technical advances in electron and light microscopy, with current imaging techniques providing unprecedented insights into ultrastructural features. Mitochondria are commonly depicted as tubules or ovoids with micron-scale dimensions (length ~1–5 μm; width ~0.5–1 μm). However, their ultrastructural features are in the nanometer spatial scale: the thickness of a typical membrane is ~6–8 nm; the combined thickness of OM and IBM is ~20 nm; CJ width ranges from 12–40 nm in diameter; and cristae lumen widths are ~25–30 nm [2]. The imaging resolution limit is defined as the shortest distance between clearly resolved objects. For traditional light microscopy, the resolution limit is ~200 nm (half the wavelength of emitted light) due to light diffraction and the point-spread function. Elucidation of mitochondrial ultrastructure has therefore required imaging techniques that surpass the resolution barrier of light microscopy.
Electron microscopy (EM) provides high resolution in the x-y dimensions due to the short wavelength of electrons (~0.01 nm or shorter, depending on accelerating voltage) and has rendered critical insights into mitochondrial fine structure. The pioneering work of Palade and Sjöstrand used transmission EM (TEM) to elucidate the basic two-membrane architecture of mitochondria [5]. Electron tomography (ET) is an extension of TEM used to render three-dimensional structures by imaging specimens over a range of tilt angles and computationally reconstructing the micrographs. Advances in mitochondrial ET by Frey, Manellla, and colleagues elucidated substructures including CJs and CTs [6]. Recent advances in cryogenic sample preparation and improvements in electron detector technology and image reconstruction have enabled high-resolution structural analysis. Transmission electron cryomicroscopy (cryo-EM) allows for determination of macromolecular structures at near-atomic resolution [7, 8] and electron tomography (cryoET) allows for three-dimensional reconstruction of whole organelles [9].
Super-resolution microscopy (SRM), or nanoscopy, describes fluorescence-based techniques allowing resolution greater than that of conventional light microscopy. SRM reduces resolution limits to tens of nm or less, sufficient to image mitochondrial ultrastructure. SRM-based techniques have dual categorization. Deterministic SRM utilizes particular optical responses of fluorophores to excitation light to enhance resolution. This includes structured illumination microscopy (SIM), using non-uniform illumination patterns and Fourier reconstruction [10] and stimulated emission depletion (STED), which scans samples selectively de-exciting fluorophores outside the region of interest [11–13]. By comparison, stochastic SRM temporally resolves complex temporal behavior, and includes photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), whereby subsets of fluorophores are sequentially detected while limiting accumulation of active probes and imaging sequences are reconstructed. MINFLUX nanoscopy combines advantages of PALM and STED, using individually activated fluorophores with a structured excitation beam with a central zero-intensity point to achieve resolution of 5 nm or less [12, 14]. Advances in SRM technology have contributed greatly to the recent insights into mitochondrial compartmentalization described in this review.
Protein Complexes and Lipids that Shape Mitochondria
Before considering protein and lipid distribution within mitochondria, we will first review the factors that segregate the contiguous IM into subcompartments. Three protein complexes have emerged as key players in the establishment of cristae morphology. These are: (i) the mitochondrial contact site and cristae organizing system (MICOS) complex, (ii) assemblies of the mitochondrial genome maintenance (Mgm1, fungi) / optic atrophy (OPA1, metazoans), and (iii) the F1FO-ATP synthase (Figure 1B-D). The mechanisms by which cristae form, as well as their evolutionary origins, are matters of current investigation by several groups (Box 1).
Text Box 1. Cristae Diversity, Dynamics, and Origins.
Cristae are structurally diverse, broadly divided into two morphotypes including tubulo-vesicular and flat (lamellar or discoidal) [116]. Recent work has provided molecular-level insights into cristae morphogenesis. Neupert and colleagues proposed that in yeast, both tubular and lamellar cristae formation require MICOS and F1FO-ATP synthase; however, whereas lamellar cristae are formed from two different IBMs concomitant with Mgm1-depedent mitochondrial fusion and MICOS-delimited crista junctions (CJs), tubular cristae form by inward growth of the inner membrane (IM) independently of Mgm1 [117]. Tamura and coworkers expanded upon this model, showing that in yeast, the lipid transport protein Mdm35 functionally overlaps with Mgm1, demonstrating an interconnection between mitochondrial lipid biosynthesis and cristae morphogenesis [118]. By comparison, the Jakobs group demonstrated that in mammalian cells, crista formation is independent of mitochondrial fusion-fission dynamics and that MICOS assembly both initiated CJ formation and remodeled pre-existing cristae [12]. Further, work from the Battersby group has shown OPA1 processing is not linked to acute cristae structural alterations, supporting that this GTPase does not assume a direct role in cristae morphogenesis outside of apoptosis [119]. These studies highlight critical issues regarding common and divergent roles of different morphogenic complexes in the formation of cristae with different morphologies, as well as between different species and perhaps different tissues.
Innovations in live-scale nanoscopy [10, 13] have helped reveal that cristae undergo MICOS-dependent remodeling events on the time scale of seconds, providing evidence that individual cristae can not only retain physical separation from the inner boundary membrane (IBM) and other cristae, but can reversibly fuse to exchange contents [11, 120]. This supports a model in which the cristae of a given mitochondrion are not separate, fixed entities, but rather define a network of membrane-delimited structures capable of exchanging lipids, proteins, and metabolites.
Other avenues of research have shed light on the evolutionary provenance of cristae. Mitochondria are semi-autonomous, having originated as organelles some 1.5 billion years ago as once free-living prokaryotes, likely of the class Alphaproteobacteria, were retained in the host cell as endosymbionts [121]. Some groups of extant alphaproteobacteria contain cell membrane extensions that protrude into the cytoplasm, termed intracytoplasmic membranes. Although morphologically diverse, these membrane systems appear to form specialized compartments for energy transduction processes, providing expanded membrane surface area for enzyme complexes involved in photosynthesis, nitrification, or methanotrophy [122]. The recent discovery that members of alphaproteobacteria contain a homolog of the core MICOS subunit Mic60, termed alphaMic60, lends strong support to the pre-endosymbiotic origin of cristae [122].
The MICOS complex resides within the IM, predominantly at CJ sites, where it stabilizes contact sites with the OM, thereby scaffolding the organelle (Figure 1B) [15, 16]. MICOS is a multisubunit assembly that forms two functionally distinct subcomplexes. The MIC60 subcomplex establishes multiple contacts with the complexes of the OM and is required for the formation of CJs, whereas the MIC10 subcomplex maintains curvature at CJ sites and appears critical for lamellar crista formation [12]. Super-resolution microscopy has demonstrated that Mic60 is arranged in helical arrays along a mitochondrion [17], forming ringlike arrangements consistent with CJ openings [14].
Mgm1/OPA1 are GTPases of the dynamin family that exist as membrane-integral long forms (l-Mgm1/l-OPA1) or as soluble short forms (s-Mgm1/s-OPA1) in the IMS, determined by regulated proteolysis (Figure 1C). In addition to mediating the fusion of the IM, they also maintain cristae architecture through homotypic interactions that stabilize and tighten cristae necks [18–20]. Recent structural insights [7, 8, 21] have revealed how these dynamins assemble into dimers, trimers, and tetramers through different interactive interfaces, underscoring potential variation in how monomers assemble for different functions. These structures also present models by which Mgm1/Opa1 form helical lattices that utilize a GTPase power stroke to modulate membrane structure.
The F1FO-ATP synthase consists of a membrane-bound H+-driven rotary motor (the FO sector) and a matrix-localized catalytic domain (the F1 sector) connected by central and peripheral stalks (Figure 1D) [22, 23]. In addition to producing the majority of ATP in eukaryotic cells, mitochondrial F1FO-ATP synthases also shape crista morphology by assembling as arrays of dimers along the crest of lamellar cristae and along lengths of tubular cristae [24–27]. Dimers of ATP synthase place the monomers at angles of ~55º to 86º, sharply bending and stabilizing the strong curvature of the crista apex. Recent insights from live-cell microscopy show that the spatiotemporal distribution of F1FO-ATP synthases between CM/IBM depends on metabolic conditions [28].
Cristae compartments are delineated by regions of strong membrane curvature [29] (Box 2). In addition to key proteins, lipids themselves are determinants of mitochondrial ultrastructure. Mitochondrial membranes have a lipid composition that is predominantly (up to 95%) phospholipid, about half of which is the bilayer-forming lipid phosphatidylcholine (PC), and ~40% of which comprises nonbilayer (HII) lipids phosphatidylethanolamine (PE) and cardiolipin (CL). Given the inverted conical molecular geometry of PE and CL, these lipids stabilize regions of local negative curvature (ICS-facing leaflet of CTs, matrix-facing leaflet of CJs, and membrane contact sites) [30–33].
Text Box 2. Lipids and Membrane Proteins as Determinants of Membrane Features.
Specific lipids and proteins promote membrane curvature that is key to establishing compartments [29]. Lipids can promote ultrastructural features of membranes such as local curvature based on their inherent physicochemical properties, including charge distribution, acyl chain composition, and shape (Figure IA). Lipid molecular geometries can be classified as conical, cylindrical, or inverted conical depending on the relative effective volumes of the headgroups relative to the acyl chain region. The spontaneous curvature (c0) reflects this geometry, indicating the type of topological structure the lipid forms in aqueous solution. Cylindrical lipids, whose effective headgroup and nonpolar regions are roughly equal in volume (c0 ≈ 0), pack efficiently as flat bilayers, rendering flat lamellar (L) topology (gray). Conical lipids, whose headgroup volumes exceed their acyl regions (c0 > 0) form type I hexagonal (HI) tubes in isolation and promote positive curvature in biomembranes where headgroups face the convex aspect (cyan). Inverted conical lipids, whose nonpolar volumes exceed those of their headgroups (c0 < 0) form type II hexagonal (HII) tubes in isolation and promote negative curvature in biomembranes where headgroups face the concave aspect (red). Features that increase effective headgroup volume (e.g. ionic repulsion, increased hydration) promote conical (HI) geometry, whereas features that increase effective acyl volume (e.g. unsaturation, oxidative modifications) promote inverted conical (HII) geometry. Local enrichment of HI or HII lipids lowers the energetic cost of positive and negative membrane curvature, respectively, and can originate from lipid asymmetry between leaflets and/or localization of biosynthetic, remodeling, and trafficking enzymes.
Integral and peripheral membrane proteins also promote local curvature (Figure IB). (i) Transmembrane proteins can bend membranes to conform to asymmetry in their shape, as proposed for the small, wedge-shaped Mic10 [34, 35, 37, 39]. (ii) Partitioning of a structural element such as an α-helix into the interface of one leaflet can disrupt headgroup packing to cause bending, as observed for an amphipathic helix of Mic60 as well the alpha-proteobacterial alphaMic60 [36, 38]. (iii) Proteins that oligomerize with a particular angle locally distort the membrane, as shown for F1FO ATP synthase dimer rows [24–27]. (iv) Membrane-interactive proteins can assemble as scaffolds whose curvature shapes the associated membrane, as proposed for assemblies of s-Mgm1 [7, 8, 21]. Additional proteins may play equally critical roles in modulating inner membrane (IM) morphology. For example, the BAR domain protein FAM92A1 was recently shown to potentiate cristae from the matrix-facing side of the IM [123].
Box 2, Figure I.
Membrane curvature determinants include (A) lipids and (B) membrane-interactive proteins that promote local positive (cyan) and negative (red) membrane curvature.
A key emerging concept is that the three cristae-shaping machineries interact specifically with lipids to cause localized membrane bending. The MICOS complex stabilizes negative (concave) curvature toward the matrix at CJs by the CL-binding and membrane-remodeling activity of the MIC10 and MIC60 subcomplexes [34–39]. By comparison, F1FO-ATP synthase assemblies stabilize positive (convex) curvature toward the matrix at cristae apices. The ensuing membrane deformation is a likely driving force for assembly of dimer rows [24, 40], a process that appears to be stabilized by CL [41], consistent with cardiolipin-interactive sites of the c-ring and interface wedge [42–44]. These dimer rows stabilize the highly curved region of the CT. Mgm1/OPA1 also assemble in a cardiolipin-dependent manner [45, 46]. Interestingly, s-Mgm1 can assemble into left- or right-handed helical lattices via distinct oligomerization interfaces on the positively- or negatively curved regions of membranes, respectively; the latter being consistent with cristae topology [7].
Finally, recent work highlights extensive crosstalk among morphogenic complexes. In keeping with the oppositely-oriented curvature that they promote, assembly of MICOS and F1FO-ATP synthase complexes are antagonistically related in yeast [47] and may be coordinated by the recently-discovered regulatory roles of Mic10/Mic27 [48, 49]. By comparison, in mammalian mitochondria MICOS and OPA1 complexes have a defined physical interaction in which OPA1 and MIC60 cooperatively define CJ abundance and physical dimensions of cristae [50, 51]. Interestingly, the OMA1 protease associates with MIC60 to regulate MICOS-mediated OM-IM contacts in a way that can be compensated by an artificial intermembrane tether [52].
Mitochondrial Compartmentalization and its Determinants
Mitochondrial Complexes have Regulated Spatial Distribution and Interaction
Like its bacterial equivalent, the OM of mitochondria contains both α-helical and β-barrel proteins and is porous to small molecules. By contrast, the IM of mitochondria, like the cell membrane of bacteria, contains only α-helical integral membrane proteins; importantly, the ion-impermeant IM contains respiratory and ATP synthase complexes, as well as the phospholipid CL, thus bearing the hallmarks of an energy-conserving membrane.
In the early 2000’s, immunolabeling and TEM demonstrated that respiratory Complex III and ATP synthase were highly enriched in CM of bovine heart mitochondria [53], supporting that cristae are the sites of oxidative phosphorylation (OXPHOS). Soon after, yeast mitochondria were shown to have subcompartment-specific enrichment of several complexes, including those in CM (mediating OXPHOS, Fe-S cluster biogenesis, and mitochondrial protein synthesis) and the IBM (mediating protein import and fusion/fission), that are sensitive to the physiological state of the mitochondrion [54, 55]. These studies showed that protein complexes and their associated biochemical processes are functionally compartmentalized, revealing a spatial division of labor.
At the next level of organization is the physical association of macromolecules that occurs within or between subcompartments, or the mitochondrial interactome. For example, respiratory complexes can exist as independent complexes or as respiratory supercomplexes (SCs) in yeast (III2IV1–2) and mammals (I1–2III2IV1–2) [56–58]. Advanced chemical crosslinking and mass spectrometry (MS)-based approaches [59] in isolated murine mitochondria, have established databases of >30% of the mitoproteome, revealing extensive interactions between OXPHOS and MICOS complexes [60] – an approach recently applied to intact tissues to evaluate OXPHOS supercomplex interactions [61]. Super-resolution, immunoelectron microscopy, and cryo-tomography have shown that OXPHOS complexes are primarily localized along the flat CM [62, 63]. Functionally relevant interactions can also span across subcompartments. For example, OXPHOS complexes of the CM form interactive networks with MICOS subunits of the CJ [60] and protein transport machinery of the OM (TOM complex) and IM (TIM complex) form supercomplexes in a substrate-dependent manner [64].
Microdomains within mitochondrial membranes define localized compartments of specific lipid and protein composition. Notably, CL plays a central role in stabilizing the quaternary structures of mitochondrial complexes, including respiratory SCs [65], protein import machinery [66], and the calcium uniporter [67]. Furthermore, recent advances in the polymer-based extraction of native mitochondrial lipoprotein complexes from mitochondria has shed light on the specialized lipid composition in the immediate vicinity of OXPHOS complexes [68]. The IM also harbors microdomains with specialized lipid and protein composition in regions bound by prohibitin proteins belonging to the SPFH superfamily. Prohibitins of two types, PHB1 and PHB2, assemble as heterodimeric ring-like structures on the IM (outer diameter up to 250 Å), scaffolding lipids and proteins with broad effects on mitochondrial metabolism [69].
General Determinants of Compartmentalization
Given the precise distribution patterns of mitochondrial complexes, the next question is: How is this compartmentalization established and regulated within this topologically complex organelle? The answer comes from multiple nonexclusive mechanisms (Figure 2). Compartmentalization primarily arises from barriers established within and among membranes across which there are temporal or physicochemical restrictions to movement (Figure 2A) but also includes routes allowing inter-compartmental passage (Figure 2B).
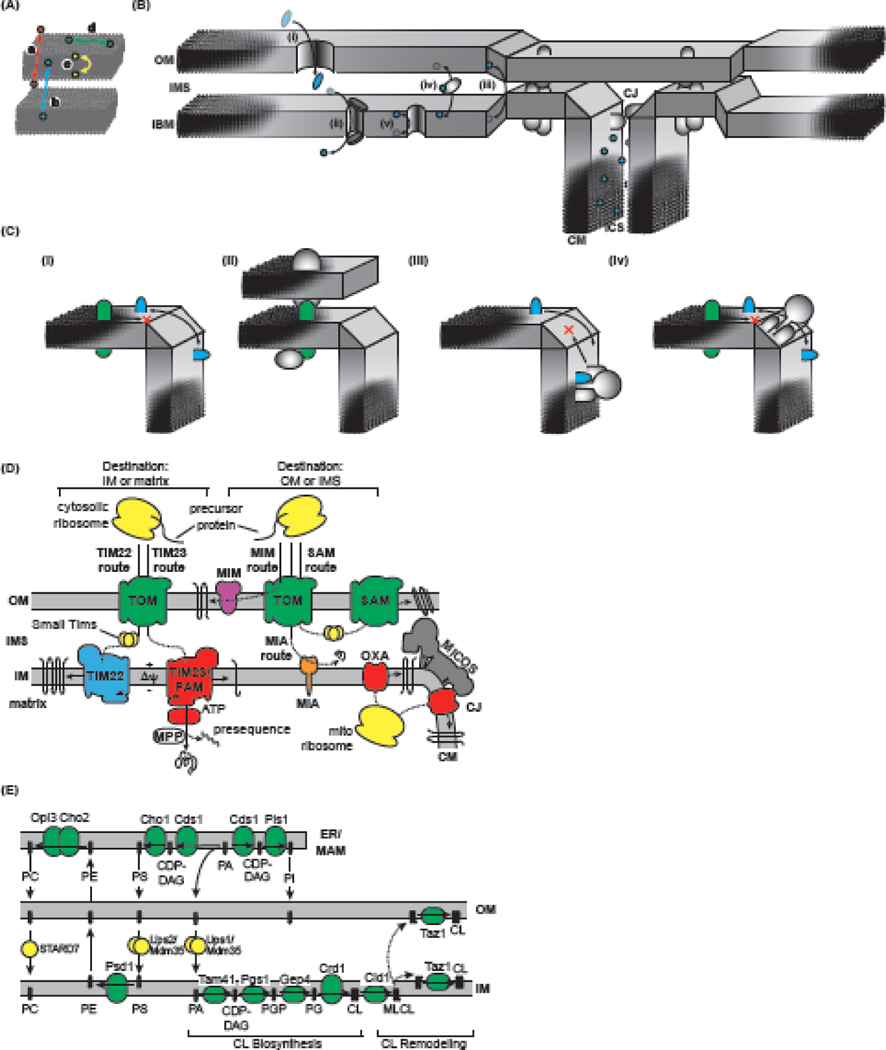
Figure 2. Determinants of Organellar Compartmentalization.
A) Defined physical compartments include those that exist (a) between membrane-separated aqueous compartments, (b) between membranes, (c) between leaflets of a given membrane, and (d) between lateral zones of a membrane. B) Inter-compartment transit routes include (i) passive or (ii) active transporters that allow movement of molecules between aqueous compartments; (iii) transient and/or regulated contact sites between membranes, (iv) carrier proteins that ferry lipids between distinct membranes; and (v) transporters that mediate transbilayer lipid diffusion. C) Lateral compartmentalization between the inner boundary membrane (IBM) and cristae membrane (CM) originates from several nonexclusive mechanisms, including (i) restricted diffusion related to membrane curvature, (ii) compartment-specific localization by protein interactions, (iii) active sorting by crista junction (CJ) complexes (e.g., MICOS), and (iv) assembly of otherwise mobile subunits into large quaternary assemblies. IMS, intermembrane space; ICS, intracristal space. D) Protein trafficking routes include the Translocon of the Outer Mitochondrial Membrane (TOM) and Sorting and Assembly Machinery (SAM) complexes of the outer membrane (OM), and the Translocons of the Inner Mitochondrial Membrane 23 and 22 (TIM23 and TIM22), the Presequence Translocase-Associated Motor (PAM) complex, the Mitochondrial Intermembrane Space Assembly (MIA) and Oxidase Assembly (OXA) translocase. Targeted polypeptides (precursors) contain targeting information as cleavable presequences or in the protein itself that is recognized by cognate transport machineries to sort proteins to their correct destinations. E) Mitochondrial lipid homeostasis requires the uptake of lipids that are mostly produced in the mitochondrialassociated membrane (MAM) subcompartment of the ER. CDP-DAG, cytidine diphosphate glycerol; Cho, PS synthase; Cds1, CDP-DAG synthase; Opi3/Cho2, PE methyltransferases; PI, phosphatidylinositol; Pis1, PI synthase; Psd1, PS decarboxylase 1.
Membranes provide low-dielectric barriers between aqueous compartments; however, the mitochondrial OM contains passive (β-barrel) pores (Figure 2B-i) and the IM contains primary and secondary active transporters (Figure 2B-ii) that selectively compartmentalize aqueous constituents. Membrane-specific composition is a fundamental type of compartmentalization, as membrane-bound lipids, proteins, and metabolites do not freely exchange among non-contiguous membranes. However, mitochondria contain multiple routes that link otherwise disparate bilayers. First, mitochondria contain membrane contact sites (MCSs) that facilitate inter-membrane traffic (Figure 2B-iii). Several MCSs have been identified that mediate close OM-IM apposition, including those formed by TOM-TIM23 complexes [64], but recent evidence supports MICOS as a key MCS required for protein and lipid trafficking (see below). Furthermore, lipid transfer proteins ferry specific lipids between membranes (Figure 2B-iv). Finally, transmembrane compartmentalization also arises from the distinct composition on either leaflet of a membrane, originating from a combination of protein sorting mechanisms and the energetic barriers to intermonolayer diffusion. Although energy-requiring flippases and floppases have not been identified in mitochondria, the Ca2+-dependent scramblase PLSCR3 is anticipated to mediate the equilibration of CL, and potentially other phospholipids, between IM monolayers (Figure 2B-v); however, this activity has yet to be experimentally established.
One of the most critical types of mitochondrial compartmentalization occurs laterally among regions in contiguous and topologically equivalent membranes – most notably between the IBM and CM. Several mechanistic models could account for the maintenance of distinct lipid and protein composition between these otherwise connected compartments (Figure 2C). The first arises from the fact that the IM has regions of unusually strong curvature (Box 2), particularly at CJs. Recent modeling studies indicate that local regions of strong membrane curvature pose sizeable kinetic barriers to diffusion [70]; therefore, the nonplanar geometry of the CJ itself could serve as a sorting barrier (Figure 2C-i). Specific protein interactions could stabilize particular proteins in a given subcompartment (Figure 2C-ii). Steric bulk could also limit passage across the CJ, which could regulate the free trafficking of unassembled protein complex subunits but prevent their back-diffusion toward the IBM after assembly into complexes at the CM (Figure 2C-iii), a mechanism consistent with the slow diffusion kinetics measured for OXPHOS complexes in CM [62]. Finally, MICOS could assume a more active role in permitting the passage of some lipids or proteins across the CJ but not others (Figure 2C-iv). Which of these compartmentalization mechanisms are responsible for the distribution of lipids and proteins of the IM awaits experimental determination.
Factors Controlling Mitochondrial Protein Trafficking
The mitoproteome consists of ~1500 proteins in human and ~1000 proteins in yeast. The vast majority are encoded in nuclear DNA (nDNA), synthesized on cytosolic ribosomes, and targeted to mitochondria by specific targeting signals [71] (Figure 2D). The TOM complex of the OM is the general entry gate for nuclear-encoded proteins. Polypeptides destined for the IM or matrix follow the TIM23 or TIM22 pathways, both of which use the membrane potential (ΔΨm) Yas an energy source. The TIM23 pathway the major route for precursor proteins synthesized with cleavable, amino-terminal presequences, mediating the lateral integration of some proteins into the IM and the import of soluble proteins into the matrix in association with the PAM complex. By comparison, substrates of the TIM22 pathway are polytopic membrane proteins with internal targeting signals that are maintained in a soluble state in the IMS by small TIM chaperones prior to membrane integration. Proteins destined for the OM include β-barrels, precursors of which are transported into the IMS where they are escorted by small TIMs and inserted by the SAM complex, and α-helical proteins recognized by the TOM complex receptors and subsequently integrated by the MIM pathway. Many soluble IMS-destined precursors follow the MIA pathway, which catalyzes the oxidative folding of small, thiol-rich proteins. Finally, a small subset of mitochondrial proteins are encoded by the mitochondrial genome (mtDNA) and translated on mitochondrial ribosomes. Among them, the majority are membrane-bound OXPHOS complex subunits that are cotranslationally integrated via the OXA machinery.
Recent work has illuminated key aspects of the compartment-specific sorting of mitochondrial proteins. The first involves the role of the MICOS complex. MICOS is proximal to all import complexes and deletion of its subunits inhibits SAM, MIA, TIM22, and TIM23 pathways [72–74], consistent with active TOM-TIM23 translocation located near CJs [75]. The MICOS-dependence of import may be species-specific, facilitating TIM23-mediated import in yeast and TIM22-mediated import in human [73]. By stabilizing an interactive network at CJs, MICOS likely renders protein trafficking more efficient by maintaining proximity among import complexes. Whether MICOS plays a more active role in protein sorting awaits determination.
Another key insight involves the coordination of nuclear and mitochondrial genomes. OXPHOS complexes (except CII) contain subunits encoded by both nDNA and mtDNA, involving an assembly process that includes chaperone assembly factors and CL remodeling [76, 77]. Given the enrichment of IM import complexes at the IBM, a critical question is where OXPHOS complexes assemble to reach their destination in CMs. In yeast, newly-translated subunits of CIII and CIV can assemble in the IBM prior to moving to CM, whereas those of CV assemble directly in CM [78]. However, in human mitochondria, the majority of mitochondrial protein synthesis is confined to CM [79]. Future work on the spatial regulation of protein trafficking sites will be critical toward understanding how protein compartmentalization is established and maintained.
Factors Controlling Mitochondrial Lipid Compartmentalization
Mitochondrial membranes are built through a combination of their own lipid biosynthetic capacity and their uptake of lipids made in other organelles [80], most notably the ER (Figure 2E). CL and PE are made on opposite sides of the IM by cardiolipin synthase (Crd1) [81] and phosphatidylserine decarboxylase 1 (Psd1) [82], respectively. CL biosynthesis requires movement of phosphatidic acid (PA) to the matrix-facing IM leaflet following its production in the ER, the OM, or the IMS-side of the IM [80]. For Psd1 to make PE, phosphatidylserine (PS) generated in the ER must gain access to the IM or be exposed on the IMS-leaflet of the OM [83, 84].
Mitochondria acquire lipids made elsewhere through the assistance of inter-organelle MCSs that may have direct roles in lipid transport [85–88]. Once in the cytosol-facing leaflet of the OM, lipids destined for other mitochondrial compartments need to traverse to the IMS-facing OM leaflet, move across the aqueous IMS and into the IMS-side of the IM, and possibly flip to its matrix-side. Very little is known about how lipids are distributed between leaflets of either mitochondrial membrane. In contrast, proteins that mediate the movement of specific phospholipids across the IMS and into the IM – STARD7 for PC [89], Ups2/Mdm35 for PS [90], and Ups1/Mdm35 for PA [91] – or that facilitate PE synthesis in the OM (MICOS) [83]) have recently been identified. In the net, the combinatorial actions of biosynthetic and trafficking processes ultimately shape the distinct lipid compositions of the mitochondrial OM and IM [92]. Acquiring the correct balance of phospholipids is necessary for each mitochondrial membrane to function optimally. For instance, in either the absence of Ups1-mediated PA trafficking or when key CL biosynthetic enzymes are missing, TIM23 activity and assembly is reduced [66, 93–95]. IM proteins such as the ADP/ATP carrier have evolved to be functionally dependent on the presence of CL in the IM [96]. Moreover, PE made in the IM by Psd1 is of special importance for complex III function [68]. Membrane proteins also impact phospholipids. An exciting example of this is that CL acyl chain composition and stability is positively impacted by the extreme protein density of the IM [76]. On the flip side, CL has a major role in facilitating numerous macromolecular interactions that occur in this compartment, including SCs [65, 96]. This fascinating symbiotic relationship between lipid and membrane proteins is the crux of what makes compartmentalization so important for the structure of mitochondria.
The Functional Implications of Compartmentalization
The evolutionary conservation of mitochondrial compartmentalization underscores its functional importance. For instance, the establishment of specialized compartments is key to mitochondrial metabolic regulation as a means of concentrating or separating enzymes and metabolites, creating specialized redox compartments such as the IMS, whose highly oxidizing environment is critical for the function of resident proteins [97], and as a protective mechanism for the confinement of oxidative damage [98]. Here we review two key functional outcomes of compartmentalization in bioenergetics and apoptosis.
Keeping the Powerhouse Charged
Mitochondria are the central regulators of eukaryotic energy metabolism. CMs are the chemiosmotic membranes that mediate the two coupled processes of OXPHOS: the generation of an electrochemical proton potential () by respiratory complexes, and the -dependent synthesis of ATP by the F1FO-ATP synthase (Figure 3A). Among metazoan tissues, there is a positive correlation between IM surface area and energy demand, where respiration is more a function of cristae density than of mitochondrial volume [99]. To a first approximation, then, cristae are subcompartments that serve to maximize CM surface area for OXPHOS enzymes and therefore energy output within the limits of cell volume.
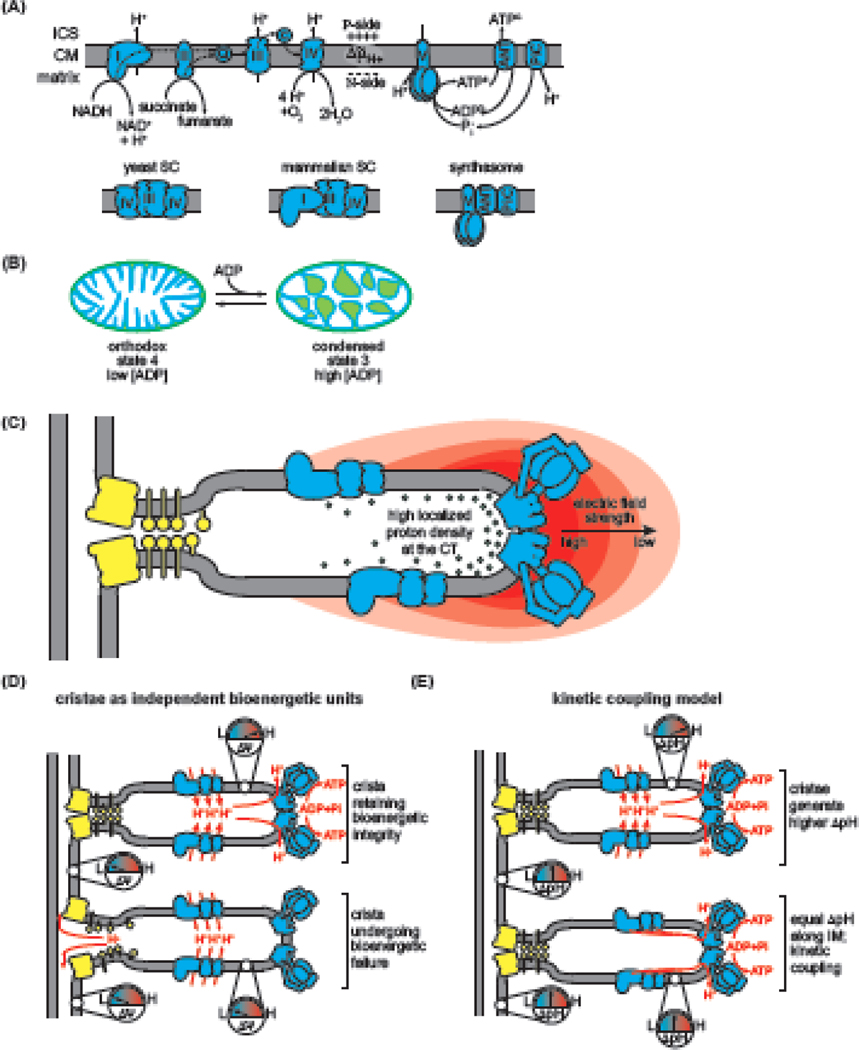
Figure 3. Mitochondrial Compartmentalization and Energy Transduction.
A) The OXPHOS system comprises the electron transport chain of respiratory complexes CI-CIV, which accept reducing equivalents and pass them through a series of redox centers, vectorially pumping H+ toward the intracristal space (ICS) at CI, CIII, and CIV to establish the across the cristae membrane (CM). ATP synthesis involves the F1FO-ATP synthase, transporters that mediate ADP/ATP exchange (ANT in mammals, Aac in yeast), and H+/Pi symport (PiC), together defining the synthasome supercomplex (SC). The provides a reservoir of H+ that flows toward the matrix across the membrane-bound rotary motor of the FO sector of ATP synthase, driving ATP synthesis on the catalytic F1 sector. Below, structural organization of the yeast and metazoan respiratory SCs and the metazoan synthasome. B) Inner membrane (IM) plasticity coupled to the metabolic state shown by the transition from orthodox topology (matrix expanded, ICS contracted) to condensed topology (matrix contracted, ICS expanded) when respiration rate increases upon ADP addition. C) Cristae geometry enhances the electrochemical proton potential, illustrated by a greater electric field (gradient of red arcs) greater localized proton density (green spheres) at the highly curved regions of the crista tip (CT) (adapted from [27]). D) Hetero-potential model [106]. Upper, the Δψ of individual cristae are electrochemically insulated from the inner boundary membrane (IBM) and from each other, which enhances the energetic reserves of cristae. Lower, bioenergetic crisis in individual cristae are isolated from other crista. E) Models of ΔpH distribution. Upper, cristae are modeled as domains of enhanced ΔpH relative to IBM. Lower, ΔpH does not differ significantly between CM and IBM, but OXPHOS efficiency is enhanced by lateral membrane diffusion of protons between proton gradient sources and sinks (kinetic coupling model of [107]).
But beyond maximizing membrane surface area, ample evidence points to ways in which cristae compartmentalization regulates bioenergetic efficiency. First, cristae morphology optimizes OXPHOS enzyme efficiency. Hackenbrock first observed that cristae morphology was coupled to energetic state [100, 101] (Figure 3B). Work since has sought the functional connection between cristae structure and bioenergetics. Mitochondrial shape mediated by OPA1 regulates the assembly of respiratory SCs and respiratory capacity [102] and as regulated diffusion barriers, CJs are bottlenecks for the diffusion of the soluble electron carrier cyt c out of the ICS [103]. Further, cristae geometry modulates the . The curvature of the crista apex strengthens the local electrostatic field and therefore the proton gradient at the site where the is consumed by F1FO-ATP synthase [27], and several modeling studies support that cristae morphology enhances bioenergetic efficiency [104, 105] (Figure 3C).
One bioenergetic consequence of compartmentalization may be that the magnitude of membrane energization differs among compartments. In support of this, it was recently shown that: (i) IM architecture stabilized by MICOS and OPA1 maintains electrochemical discontinuity between the CM (where ΔΨm is higher) and IBM (where ΔΨm is lower), and (ii) individual cristae act as discrete bioenergetic units electrically insulated from the IBM and other cristae, potentially by a physical barrier to proton movement through CJs [106] (Figure 3D). In this hetero-potential model, the energetic autonomy of each crista could keep local bioenergetic crisis in one crista from immediately collapsing the potential of all interconnected cristae, as it would if the IM were an equipotential continuum. By contrast, another recent study found no difference in pH gradients across the IM of cristae relative to IBM, suggesting that cristae morphology does not enhance transmembrane energetics, and concluding instead that the driving force for ATP synthesis is promoted by a kinetic coupling mechanism [107] (Figure 3E). In this model, protons pumped across the CM by respiratory complexes (ΔpH sources) quickly diffuse along lateral membrane paths to the proximal F1FOATP synthase (ΔpH sink) before they can equilibrate with the bulk [108]. Reconciling these two models will be critical for understanding the role of cristae morphology in energy transduction and will require clarification of the different roles of the ΔpH and ΔΨm in ATP synthesis.
Executing the Cellular Death Sentence
At the onset of intrinsic apoptosis, cyt c is released from the ICS to the cytosol in a process that involves OM rupture and massive cristae remodeling [1]. Attendant with this remodeling is the destabilization of an extensive protein network composed mostly of OPA1 and MICOS subunits [50]. In the context of elevated levels of reactive oxygen species, cyt c gains CL peroxidase activity [109] with two important outcomes. First, CL peroxidation decreases its interaction with cyt c thus favoring the release of this apoptogenic factor [110]. Second, upon its peroxidation, a proportion of CL moves to the OM with the help of PLSCR3 [111] and nucleoside diphosphate kinase D [112], where it recruits and activates the NLRP3 inflammasome to initiate pyroptosis [113], a pro-inflammatory form of apoptosis [114]. These examples highlight an extra benefit of mitochondrial compartmentalization that has evolved in metazoans: its regulated collapse can be exploited to trigger functional outcomes beyond the mitochondrion that are essential for organismal physiology. Indeed, recent work has shown that apoptosis is associated with massive IM restructuring and fragmentation tied to dissociation of F1FO-ATP synthase dimers [115].
Concluding Remarks
Contrasting the historical textbook view of the mitochondrion as a rather static organelle, recent technology-driven insights have shown unexpected complexity and dynamism of its ultrastructure. Continued refinement of current models describing morphogenesis – particularly how morphogenic complexes interact together and with lipids – will be necessary to understand how compartmentalization is established and regulated. Consistent with the concept that ‘form follows function’, it will be critical to gain further insights into how the spatiotemporal division of labor within mitochondria directly bears on its functionality and how defective compartmentalization causes mitochondrial dysfunction (see Outstanding Questions).
Outstanding Questions.
What is the functional relevance of the recently observed rapid exchange of lipids, proteins, and metabolites among cristae compartments? Could this have implications for macromolecular trafficking or strategies for handling oxidatively damaged proteins or lipids?
Are there distinct distributions of lipids between distinct inner membrane (IM) compartments, and if so, how are these gradients generated and what functions to they serve?
Given that alphaMic60 is an evolutionary progenitor of cristae-forming complexes, why have eukaryotic innovations (Mgm1/OPA1 and F1FO-ATP synthase dimer ribbons) played such a critical role in the evolution of mitochondrial cristae morphogenesis?
What is the cause of the structural divergence in mitochondrial morphogenic complexes between yeast and metazoans and how might they differently regulate mitochondrial ultrastructure?
What are the mechanisms that regulate the distribution of membrane-bound macromolecules between the inner boundary and cristae membranes (IBM versus CM) and soluble molecules between the intermembrane and intracristal spaces (IMS versus ICS)? Does MICOS play more of an active or passive role?
What is the molecular basis for the newly discovered differences in cristae morphogenesis and protein trafficking/assembly between yeast and metazoans?
What is the cause-and-effect relationship between alterations in mitochondrial ultrastructure and mitochondrial dysfunction?
What is the role of cristae architecture in mitochondrial bioenergetics?
What molecular mechanisms regulate the distribution of lipids in each subcompartment of the mitochondrion?
Text Box 3. The Pathophysiology of Disrupted Compartmentalization.
Mitochondrial dysfunction is commonly associated with aberrant ultrastructure and associated changes in compartmentalization. Such morphological changes in mitochondria are associated with aging [124] and with complex diseases including myopathy, cancers, metabolic syndrome, and a range of neurodegenerative diseases [125, 126]. Although mitochondrial dysfunction can affect any organ, its effect is greatest in tissues with high energetic demand, including cardiac and skeletal muscle, the brain and visual system. The correlation between reduced mitochondrial function and abnormal ultrastructure raises a critical question of causality. Namely, do defects in mitochondrial ultrastructure cause deficits in functional properties of the organelle? Or, conversely, does compromised functionality lead to abnormal mitochondrial morphology?
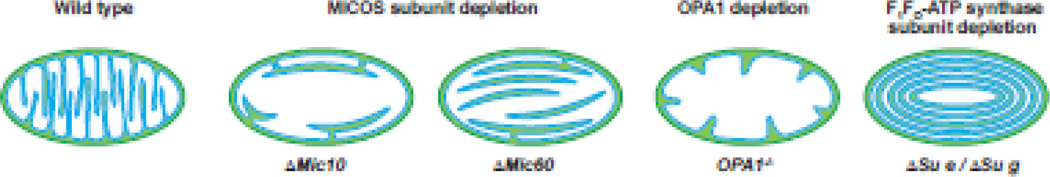
Disruption of complexes that define subcompartment boundaries strongly affects mitochondrial ultrastructure (Figure I). First, disruption of MICOS by deletion of key subunits results in loss of crista junctions (CJs) with cristae membranes appearing as arcs or stacks largely disconnected from the inner boundary membrane (IBM) [12]. Furthermore, mutations in MICOS subunits are correlated with known pathologies; for example, aberrations in or loss of MIC60 are linked to Parkinson’s disease [127], reduced expression of MIC19/CHCHD3 is linked to type II diabetes [128], and QIL1/MIC13 mutations are linked to early-onset mitochondrial encephalomyopathy with liver disease [129, 130]. Second, downregulation, deletion or improper processing of OPA1 causes widening of cristae and CJs, reduction of cristae number, and, as would be expected with compromised IM fusion activity, fragmentation [1]. Indeed, multiple missense and frameshift mutations in the OPA1 gene are known to cause type 1 optic atrophy (from which the gene was named), inherited in an autosomal dominant manner [131]. OPA1 defects result in defective mitochondrial structure that particularly affects retinal ganglion cells, resulting in atrophy of optic nerves and vision loss, but may also include ataxia, hearing loss and myopathy. Finally, deletion of the F1FO-ATPsynthase dimer-stabilizing subunits Su e, Su g, and Su k (ATP5I, ATP5L, and DAPIT, respectively, in mammals) cause aberrant cristae architecture with loss of cristae rims and cristae membranes as concentric onion-like rings [26, 40]. Fungal aging models have revealed that age-related degradation of cristae morphology is associated with the disassembly of ATP synthase dimer ribbons into monomers [132, 133] and mutations in Su k/DAPIT result in altered cristae morphology and a form of Leigh syndrome [134]. Thus, regardless of whether morphological abnormalities are the primary causes or downstream outcomes of defective mitochondrial function, loss of compartmentalization is likely an underpinning factor in many mitochondrial diseases.
Box 3, Figure I.
Schematic illustration of wild type mitochondrial ultrastructure with lamellar cristae in comparison with those deficieint in MICOS subunits, OPA1, and dimer-stabilizing subunits of F1FO-ATP synthase as indicated.
Highlights.
Mitochondria contain two membranes that partition the organelles into compositionally and functionally distinct subcompartments that are defined by a topologically complex ultrastructure.
In addition to their morphological complexity, mitochondria are pleomorphic, undergoing morphogenesis events with an extent and frequency that is just now becoming appreciated.
The protein complexes that define inner membrane morphology form an interactive network with lipid interactions, and new insights are illuminating how they establish and regulate compartmentalization.
The general determinants of compartmentalization as well as the factors that govern protein and lipid distribution have recently been identified.
Novel research on the functional relevance of compartmentalization has highlighted a key role of regulated cristae subcompartment structure in bioenergetics and in human diseases.
Acknowledgements
This work was supported by the National Institutes of Health (R01GM136975 to S.M.C and N.N.A).
Glossary
- BAR domain
crescent-shaped domain that interacts with curved membrane surfaces to both promote and detect local membrane curvatures, named after BIN/Amphiphysin/Rvs proteins in which they are found
- Chemiosmotic
attribute of a semi-permeable barrier that allows the selective flux of ions down their electrochemical gradients, typically energetically coupled to another process
- Electrochemical proton potential
the difference in proton electrochemical potential (). The potential across the CM of actively respiring mitochondria has a major contribution from the electric potential (ΔΨm ~150 mV, matrix negative) and a minor contribution from the proton concentration difference (ΔpH ~ 1 unit, matrix alkaline)
- Flippase/floppase
ABC transporters that utilize the energy provided by ATP hydrolysis to move specific phospholipids against their gradient from the outer to the inner membrane leaflet (flippase) or from the inner to the outer leaflet (floppase). Together, they help generate lipid asymmetry in membranes
- Interactome
a network of physically interacting molecules defining a specific biochemical function or process
- Intrinsic apoptosis
a controlled process of cell death initiated by pro-apoptotic effectors (e.g. Bax/Bak) that interact with mitochondria to release factors (e.g. cyt c) that propagate a proteolytic cascade
- Membrane contact site
regions of close apposition between two membranes, generally composed of interacting protein complexes, that facilitate signaling and the passage of small molecules. Such sites can be inter-organellar, mediating connections that are homotypic (between the same organelles) or heterotypic (between different organelles). They can also exist between membranes of a single organelle
- Membrane curvature
the physical bending of a biomembrane to produce positively (convex) and negatively (concave) curved surfaces
- Pleomorphic
displaying plasticity in structure and size
- Scramblase
Ca2+-dependent transporters that equilibrate phospholipids between membrane leaflets. Unlike filppases and floppases, scramblases do not need an external energy source to transport lipids
- SPFH superfamily
protein family named after primary members (stomatin, prohibitin, flotillin, HflK/C), which commonly associate on membranes to form lipid raft microdomians that recruit specific protein complexes
- Supercomplex (SCs)
assembly of the respiratory complexes (CI, CIII, and CIV) into supramolecular structures. This solid-state arrangement likely enhances metabolic efficiency compared with a fluid-state model in which individual complexes are connected by freely-diffusing electron carriers
- Ultrastructure
the structure of cellular or subcellular objects that requires higher magnification than standard optical microscopy, typically observable by electron microscopy or super-resolution microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cogliati S. et al. (2016) Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem Sci 41 (3), 261–273. [DOI] [PubMed] [Google Scholar]
- 2.Quintana-Cabrera R. et al. (2018) Who and how in the regulation of mitochondrial cristae shape and function. Biochem Biophys Res Commun 500 (1), 94–101. [DOI] [PubMed] [Google Scholar]
- 3.Giacomello M. et al. (2020) The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol 21 (4), 204–224. [DOI] [PubMed] [Google Scholar]
- 4.Li W. et al. (2020) Dynamic organization of intracellular organelle networks. Wiley Interdiscip Rev Syst Biol Med, e1505. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen N. (1995) Mitochondrial structure and the practice of cell biology in the 1950s. J Hist Biol 28 (3), 381–429. [DOI] [PubMed] [Google Scholar]
- 6.Frey TG et al. (2002) Insight into mitochondrial structure and function from electron tomography. Biochim Biophys Acta 1555 (1–3), 196–203. [DOI] [PubMed] [Google Scholar]
- 7.Faelber K. et al. (2019) Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Nature 571 (7765), 429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang D. et al. (2020) Cryo-EM structures of S-OPA1 reveal its interactions with membrane and changes upon nucleotide binding. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhlbrandt W. (2015) Structure and function of mitochondrial membrane protein complexes. BMC Biol 13, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu C. et al. (2020) OPA1 and MICOS Regulate mitochondrial crista dynamics and formation. Cell Death Dis 11 (10), 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondadi AK et al. (2020) Cristae undergo continuous cycles of membrane remodelling in a MICOS-dependent manner. EMBO Rep 21 (3), e49776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephan T. et al. (2020) MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J 39 (14), e104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephan T. et al. (2019) Live-cell STED nanoscopy of mitochondrial cristae. Sci Rep 9 (1), 12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pape JK et al. (2020) Multicolor 3D MINFLUX nanoscopy of mitochondrial MICOS proteins. Proc Natl Acad Sci U S A 117 (34), 20607–20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rampelt H. et al. (2017) Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim Biophys Acta Mol Cell Res 1864 (4), 737–746. [DOI] [PubMed] [Google Scholar]
- 16.Wollweber F. et al. (2017) Mitochondrial contact site and cristae organizing system: A central player in membrane shaping and crosstalk. Biochim Biophys Acta Mol Cell Res 1864 (9), 1481–1489. [DOI] [PubMed] [Google Scholar]
- 17.Stoldt S. et al. (2019) Mic60 exhibits a coordinated clustered distribution along and across yeast and mammalian mitochondria. Proc Natl Acad Sci U S A 116 (20), 9853–9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ban T. et al. (2017) Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol 19 (7), 856–863. [DOI] [PubMed] [Google Scholar]
- 19.Frezza C. et al. (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126 (1), 177–89. [DOI] [PubMed] [Google Scholar]
- 20.Meeusen S. et al. (2006) Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 127 (2), 383–95. [DOI] [PubMed] [Google Scholar]
- 21.Yan L. et al. (2020) Structural analysis of a trimeric assembly of the mitochondrial dynamin-like GTPase Mgm1. Proc Natl Acad Sci U S A 117 (8), 4061–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhlbrandt W. (2019) Structure and Mechanisms of F-Type ATP Synthases. Annu Rev Biochem 88, 515–549. [DOI] [PubMed] [Google Scholar]
- 23.Nirody JA et al. (2020) ATP synthase: Evolution, energetics, and membrane interactions. J Gen Physiol 152 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blum TB et al. (2019) Dimers of mitochondrial ATP synthase induce membrane curvature and selfassemble into rows. Proc Natl Acad Sci U S A 116 (10), 4250–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudkina NV et al. (2006) Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett 580 (14), 3427–32. [DOI] [PubMed] [Google Scholar]
- 26.Paumard P. et al. (2002) The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J 21 (3), 221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strauss M. et al. (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J 27 (7), 1154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salewskij K. et al. (2020) The spatio-temporal organization of mitochondrial F1FO ATP synthase in cristae depends on its activity mode. Biochim Biophys Acta Bioenerg 1861 (1), 148091. [DOI] [PubMed] [Google Scholar]
- 29.Bozelli JC Jr. and Epand RM (2020) Membrane Shape and the Regulation of Biological Processes. J Mol Biol 432 (18), 5124–5136. [DOI] [PubMed] [Google Scholar]
- 30.Basu Ball W. et al. (2018) The role of nonbilayer phospholipids in mitochondrial structure and function. FEBS Lett 592 (8), 1273–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltran-Heredia E. et al. (2019) Membrane curvature induces cardiolipin sorting. Commun Biol 2, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyd KJ et al. (2017) Buckling Under Pressure: Curvature-Based Lipid Segregation and Stability Modulation in Cardiolipin-Containing Bilayers. Langmuir 33 (27), 6937–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elias-Wolff F. et al. (2019) Curvature sensing by cardiolipin in simulated buckled membranes. Soft Matter 15 (4), 792–802. [DOI] [PubMed] [Google Scholar]
- 34.Barbot M. et al. (2015) Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab 21 (5), 756–63. [DOI] [PubMed] [Google Scholar]
- 35.Bohnert M. et al. (2015) Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab 21 (5), 747–55. [DOI] [PubMed] [Google Scholar]
- 36.Hessenberger M. et al. (2017) Regulated membrane remodeling by Mic60 controls formation of mitochondrial crista junctions. Nat Commun 8, 15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rampelt H. et al. (2018) Assembly of the Mitochondrial Cristae Organizer Mic10 Is Regulated by Mic26-Mic27 Antagonism and Cardiolipin. J Mol Biol 430 (13), 1883–1890. [DOI] [PubMed] [Google Scholar]
- 38.Tarasenko D. et al. (2017) The MICOS component Mic60 displays a conserved membrane-bending activity that is necessary for normal cristae morphology. J Cell Biol 216 (4), 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber TA et al. (2013) APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS One 8 (5), e63683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies KM et al. (2012) Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci U S A 109 (34), 13602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acehan D. et al. (2011) Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys J 100 (9), 2184–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duncan AL et al. (2016) Cardiolipin binds selectively but transiently to conserved lysine residues in the rotor of metazoan ATP synthases. Proc Natl Acad Sci U S A 113 (31), 8687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muhleip A. et al. (2019) Structure of a mitochondrial ATP synthase with bound native cardiolipin. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spikes TE et al. (2020) Structure of the dimeric ATP synthase from bovine mitochondria. Proc Natl Acad Sci U S A 117 (38), 23519–23526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeVay RM et al. (2009) Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol 186 (6), 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ban T. et al. (2010) OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum Mol Genet 19 (11), 2113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabl R. et al. (2009) Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol 185 (6), 1047–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eydt K. et al. (2017) Cristae architecture is determined by an interplay of the MICOS complex and the F1FO ATP synthase via Mic27 and Mic10. Microb Cell 4 (8), 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rampelt H. et al. (2017) Mic10, a Core Subunit of the Mitochondrial Contact Site and Cristae Organizing System, Interacts with the Dimeric F1Fo-ATP Synthase. J Mol Biol 429 (8), 1162–1170. [DOI] [PubMed] [Google Scholar]
- 50.Glytsou C. et al. (2016) Optic Atrophy 1 Is Epistatic to the Core MICOS Component MIC60 in Mitochondrial Cristae Shape Control. Cell Rep 17 (11), 3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrera M. et al. (2016) OPA1 functionally interacts with MIC60 but is dispensable for crista junction formation. FEBS Lett 590 (19), 3309–3322. [DOI] [PubMed] [Google Scholar]
- 52.Viana MP et al. (2021) Protease OMA1 modulates mitochondrial bioenergetics and ultrastructure through dynamic association with MICOS complex. iScience 24 (2), 102119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilkerson RW et al. (2003) The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett 546 (2–3), 355–8. [DOI] [PubMed] [Google Scholar]
- 54.Vogel F. et al. (2006) Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol 175 (2), 237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wurm CA and Jakobs S. (2006) Differential protein distributions define two sub-compartments of the mitochondrial inner membrane in yeast. FEBS Lett 580 (24), 5628–34. [DOI] [PubMed] [Google Scholar]
- 56.Gu J. et al. (2016) The architecture of the mammalian respirasome. Nature 537 (7622), 639–43. [DOI] [PubMed] [Google Scholar]
- 57.Rathore S. et al. (2019) Cryo-EM structure of the yeast respiratory supercomplex. Nat Struct Mol Biol 26 (1), 50–57. [DOI] [PubMed] [Google Scholar]
- 58.Wu M. et al. (2020) Research journey of respirasome. Protein Cell 11 (5), 318–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koshiba T. and Kosako H. (2020) Mass spectrometry-based methods for analysing the mitochondrial interactome in mammalian cells. J Biochem 167 (3), 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schweppe DK et al. (2017) Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc Natl Acad Sci U S A 114 (7), 1732–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chavez JD et al. (2018) Chemical Crosslinking Mass Spectrometry Analysis of Protein Conformations and Supercomplexes in Heart Tissue. Cell Syst 6 (1), 136–141 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkens V. et al. (2013) Restricted diffusion of OXPHOS complexes in dynamic mitochondria delays their exchange between cristae and engenders a transitory mosaic distribution. J Cell Sci 126 (Pt 1), 103–16. [DOI] [PubMed] [Google Scholar]
- 63.Davies KM et al. (2011) Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci U S A 108 (34), 14121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chacinska A. et al. (2010) Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol Cell Biol 30 (1), 307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfeiffer K. et al. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem 278 (52), 52873–80. [DOI] [PubMed] [Google Scholar]
- 66.Malhotra K. et al. (2017) Cardiolipin mediates membrane and channel interactions of the mitochondrial TIM23 protein import complex receptor Tim50. Sci Adv 3 (9), e1700532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghosh S. et al. (2020) An essential role for cardiolipin in the stability and function of the mitochondrial calcium uniporter. Proc Natl Acad Sci U S A 117 (28), 16383–16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calzada E. et al. (2019) Phosphatidylethanolamine made in the inner mitochondrial membrane is essential for yeast cytochrome bc1 complex function. Nat Commun 10 (1), 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tatsuta T. and Langer T. (2017) Prohibitins. Curr Biol 27 (13), R629–R631. [DOI] [PubMed] [Google Scholar]
- 70.Rojas Molina R. et al. (2021) Diffusion on Membrane Domes, Tubes, and Pearling Structures. Biophys J 120 (3), 424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song J. et al. (2021) Quality control of the mitochondrial proteome. Nat Rev Mol Cell Biol 22 (1), 54–70. [DOI] [PubMed] [Google Scholar]
- 72.Bohnert M. et al. (2012) Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol Biol Cell 23 (20), 3948–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Callegari S. et al. (2019) A MICOS-TIM22 Association Promotes Carrier Import into Human Mitochondria. J Mol Biol 431 (15), 2835–2851. [DOI] [PubMed] [Google Scholar]
- 74.von der Malsburg K. et al. (2011) Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell 21 (4), 694–707. [DOI] [PubMed] [Google Scholar]
- 75.Gold VA et al. (2014) Visualizing active membrane protein complexes by electron cryotomography. Nat Commun 5, 4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Y. et al. (2019) Assembly of the complexes of oxidative phosphorylation triggers the remodeling of cardiolipin. Proc Natl Acad Sci U S A 116 (23), 11235–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang JX et al. (2020) Mitochondrial OXPHOS Biogenesis: Co-Regulation of Protein Synthesis, Import, and Assembly Pathways. Int J Mol Sci 21 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stoldt S. et al. (2018) Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat Cell Biol 20 (5), 528–534. [DOI] [PubMed] [Google Scholar]
- 79.Zorkau M. et al. (2021) High-resolution imaging reveals compartmentalization of mitochondrial protein synthesis in cultured human cells. Proc Natl Acad Sci U S A 118 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Acoba MG et al. (2020) Phospholipid ebb and flow makes mitochondria go. J Cell Biol 219 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schlame M. and Haldar D. (1993) Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J Biol Chem 268 (1), 74–9. [PubMed] [Google Scholar]
- 82.Tamura Y. et al. (2012) Phosphatidylethanolamine biosynthesis in mitochondria: phosphatidylserine (PS) trafficking is independent of a PS decarboxylase and intermembrane space proteins UPS1P and UPS2P. J Biol Chem 287 (52), 43961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aaltonen MJ et al. (2016) MICOS and phospholipid transfer by Ups2-Mdm35 organize membrane lipid synthesis in mitochondria. J Cell Biol 213 (5), 525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sam PN et al. (2021) Impaired phosphatidylethanolamine metabolism activates a reversible stress response that detects and resolves mutant mitochondrial precursors. iScience 24 (3), 102196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kornmann B. et al. (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325 (5939), 477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elbaz-Alon Y. et al. (2014) A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell 30 (1), 95–102. [DOI] [PubMed] [Google Scholar]
- 87.Kawano S. et al. (2018) Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J Cell Biol 217 (3), 959–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li P. et al. (2020) Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J Cell Biol 219 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saita S. et al. (2018) PARL partitions the lipid transfer protein STARD7 between the cytosol and mitochondria. EMBO J 37 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miyata N. et al. (2016) Phosphatidylserine transport by Ups2-Mdm35 in respiration-active mitochondria. J Cell Biol 214 (1), 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Connerth M. et al. (2012) Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338 (6108), 815–8. [DOI] [PubMed] [Google Scholar]
- 92.Zinser E. et al. (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol 173 (6), 2026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tamura Y. et al. (2009) Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol 185 (6), 1029–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tamura Y. et al. (2006) Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J Cell Biol 174 (5), 631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tamura Y. et al. (2013) Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab 17 (5), 709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Senoo N. et al. (2020) Cardiolipin, conformation, and respiratory complex-dependent oligomerization of the major mitochondrial ADP/ATP carrier in yeast. Sci Adv 6 (35), eabb0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu J. et al. (2008) The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J Biol Chem 283 (43), 29126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scorrano L. et al. (2002) A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell 2 (1), 55–67. [DOI] [PubMed] [Google Scholar]
- 99.Nielsen J. et al. (2017) Plasticity in mitochondrial cristae density allows metabolic capacity modulation in human skeletal muscle. J Physiol 595 (9), 2839–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hackenbrock CR (1966) Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J Cell Biol 30 (2), 269–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hackenbrock CR (1968) Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. J Cell Biol 37 (2), 345–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cogliati S. et al. (2013) Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155 (1), 160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mannella CA et al. (1994) The internal compartmentation of rat-liver mitochondria: tomographic study using the high-voltage transmission electron microscope. Microsc Res Tech 27 (4), 278–83. [DOI] [PubMed] [Google Scholar]
- 104.Lee JW (2020) Protonic Capacitor: Elucidating the biological significance of mitochondrial cristae formation. Sci Rep 10 (1), 10304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garcia GC et al. (2019) Mitochondrial morphology provides a mechanism for energy buffering at synapses. Sci Rep 9 (1), 18306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wolf DM et al. (2019) Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J 38 (22), e101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Toth A. et al. (2020) Kinetic coupling of the respiratory chain with ATP synthase, but not proton gradients, drives ATP production in cristae membranes. Proc Natl Acad Sci U S A 117 (5), 2412–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rieger B. et al. (2014) Lateral pH gradient between OXPHOS complex IV and F(0)F(1) ATP-synthase in folded mitochondrial membranes. Nat Commun 5, 3103. [DOI] [PubMed] [Google Scholar]
- 109.Aluri HS et al. (2014) Electron flow into cytochrome c coupled with reactive oxygen species from the electron transport chain converts cytochrome c to a cardiolipin peroxidase: role during ischemia-reperfusion. Biochim Biophys Acta 1840 (11), 3199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nomura K. et al. (2000) Mitochondrial phospholipid hydroperoxide glutathione peroxidase inhibits the release of cytochrome c from mitochondria by suppressing the peroxidation of cardiolipin in hypoglycaemia-induced apoptosis. Biochem J 351 (Pt 1), 183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chu CT et al. (2013) Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol 15 (10), 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schlattner U. et al. (2015) Mitochondrial NM23-H4/NDPK-D: a bifunctional nanoswitch for bioenergetics and lipid signaling. Naunyn Schmiedebergs Arch Pharmacol 388 (2), 271–8. [DOI] [PubMed] [Google Scholar]
- 113.Elliott EI et al. (2018) Cutting Edge: Mitochondrial Assembly of the NLRP3 Inflammasome Complex Is Initiated at Priming. J Immunol 200 (9), 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu Q. et al. (2018) The role of mitochondria in NLRP3 inflammasome activation. Mol Immunol 103, 115–124. [DOI] [PubMed] [Google Scholar]
- 115.Ader NR et al. (2019) Molecular and topological reorganizations in mitochondrial architecture interplay during Bax-mediated steps of apoptosis. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Panek T. et al. (2020) Returning to the Fold for Lessons in Mitochondrial Crista Diversity and Evolution. Curr Biol 30 (10), R575–R588. [DOI] [PubMed] [Google Scholar]
- 117.Harner ME et al. (2016) An evidence based hypothesis on the existence of two pathways of mitochondrial crista formation. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kojima R. et al. (2019) Maintenance of Cardiolipin and Crista Structure Requires Cooperative Functions of Mitochondrial Dynamics and Phospholipid Transport. Cell Rep 26 (3), 518–528 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Richter U. et al. (2019) Mitochondrial stress response triggered by defects in protein synthesis quality control. Life Sci Alliance 2 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang C. et al. (2019) A photostable fluorescent marker for the superresolution live imaging of the dynamic structure of the mitochondrial cristae. Proc Natl Acad Sci U S A 116 (32), 15817–15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roger AJ et al. (2017) The Origin and Diversification of Mitochondria. Curr Biol 27 (21), R1177–R1192. [DOI] [PubMed] [Google Scholar]
- 122.Munoz-Gomez SA et al. (2017) The Origin of Mitochondrial Cristae from Alphaproteobacteria. Mol Biol Evol 34 (4), 943–956. [DOI] [PubMed] [Google Scholar]
- 123.Wang L. et al. (2019) FAM92A1 is a BAR domain protein required for mitochondrial ultrastructure and function. J Cell Biol 218 (1), 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brandt T. et al. (2017) Changes of mitochondrial ultrastructure and function during ageing in mice and Drosophila. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vincent AE et al. (2016) The Spectrum of Mitochondrial Ultrastructural Defects in Mitochondrial Myopathy. Sci Rep 6, 30610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Colina-Tenorio L. et al. (2020) Shaping the mitochondrial inner membrane in health and disease. J Intern Med 287 (6), 645–664. [DOI] [PubMed] [Google Scholar]
- 127.Van Laar VS et al. (2018) Potential Role of Mic60/Mitofilin in Parkinson’s Disease. Front Neurosci 12, 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hwang H. et al. (2010) Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes 59 (1), 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guarani V. et al. (2016) QIL1 mutation causes MICOS disassembly and early onset fatal mitochondrial encephalopathy with liver disease. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kishita Y. et al. (2020) A novel homozygous variant in MICOS13/QIL1 causes hepato-encephalopathy with mitochondrial DNA depletion syndrome. Mol Genet Genomic Med 8 (10), e1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Delettre C. et al. (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet 26 (2), 207–10. [DOI] [PubMed] [Google Scholar]
- 132.Daum B. et al. (2013) Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci U S A 110 (38), 15301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rampello NG et al. (2018) Impact of F1Fo-ATP-synthase dimer assembly factors on mitochondrial function and organismic aging. Microb Cell 5 (4), 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Siegmund SE et al. (2018) Three-Dimensional Analysis of Mitochondrial Crista Ultrastructure in a Patient with Leigh Syndrome by In Situ Cryoelectron Tomography. iScience 6, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]