Abstract
In this study, we identified a new strain of the genus Neocypholaelaps from the beehives of Apis mellifera colonies in the Republic of Korea (ROK). The Neocypholaelap sp. KOR23 mites were collected from the hives of honeybee apiaries in Wonju, Gangwon-do, in May 2023. Morphological and molecular analyses based on 18S and 28S rRNA gene regions conclusively identified that these mites belong to the genus Neocypholaelaps, closely resembling Neocypholaelaps sp. APGD-2010 that was first isolated from the United States. The presence of 9 of 25 honeybee pathogens in these mite samples suggests that Neocypholaelaps sp. KOR23 mite may act as an intermediate vector and carrier of honeybee diseases. The identification of various honeybee pathogens within this mite highlights their significance in disease transmission among honeybee colonies. This comprehensive study provides valuable insights into the taxonomy and implications of these mites for bee health management and pathogen dissemination.
Introduction
During the current honeybee disease outbreak, parasites played a significant role in contributing to the sudden collapse of the colony. Several species of bee mites can coexist with honeybee (Apis mellifera) colonies for extended periods. In A. mellifera, some mite species may be not exhibit parasitic behavior (e.g. Forcellinia faini, Melichares dentriticus, Afrocypholaelaps africana mites) [1–4] or transmit diseases to bees (e.g. Varroa and Tropilaelaps mites) [5–7], a considerable number of mite species can cause stress to bees (e.g. Neocypholaelaps indica and Carpoglyphus lactis mites) [8, 9], resulting in reduced flight capacity and foraging efficiency. Additionally, certain parasitic mite species infest bees, causing injury or mortality in their hosts and facilitating the spread of harmful diseases. Obligate honeybee parasites like Varroa (Varroidae, Mesostigmata), Euvarroa (Varroidae, Mesostigmata), Tropilaelaps (Laelapidae, Mesostigmata), and Acarapis (Tarsonemidae, Prostigmata inflict direct harm and facilitate disease spread [10–12]. These mites have co-evolved with their hosts, developing an intimate dependence on honeybees for survival. While Varroa mites primarily parasitize bees in their brood cells [6, 13], Euvarroa and Tropilaelaps demonstrate host specificity towards Dwarf honeybees (Apis florea) and giant honeybees (Apis dorsata), respectively [7, 14–17]. The most pathogenic species within the genus Acarapis primarily parasitize A. mellifera but have also been found in Asian honeybee species [18–22]. Interestingly, the genus Tyrophagus (Acaridae: Sarcoptiformes) adds to the list of potentially detrimental mites, displaying parasitic behavior detrimental to both honeybees and bumblebees [23, 24].
The genus Neocypholaelaps, encompassing 22 species primarily concentrated in tropical regions, continues to pique the interest of researchers due to its diverse interactions with honeybees [25–40]. These Neocypholaelaps mites, belonging to the family Ameroseiidae, exhibit a relationship with their bee hosts, often associating with them both externally on their bodies and internally within their nests and hives [31]. Meanwhile, several Neocypholaelaps species, including N. favus Ishkawa, 1968, N. apicola Delfinado-Baker & Baker, 1983, N. geonomae Moraes & Narita, 2010, and N. indica Evans, 1963 have been identified in honeybee colonies across various geographical locations, spanning Europe, India, China, and even regions beyond the tropics, highlighting their adaptability [27, 32–35]. N. apicola Delfinado-Baker & Baker, 1983 was reported on the bodies of bees and within honeybee colonies in multiple European countries such as Greece [35], Denmark [36], Belgium [33], Slovakia [37], and Hungary [38], demonstrating its wider distribution. Another species, N. indica Evans, 1963, has been reported in both A. mellifera and A. cerana in Fuzhou, China, and southern Karnataka, India [39]. N. stridulans Evans, 1955 interacts with mason bees and coconut flower clusters in India [40]. In the Republic of Korea (ROK), the detection of Neocypholaelaps mites in honeybee colonies has also been reported in Gangwon and Gyeonggi provinces; however, species classification remains unclear.
Based on morphological characteristics, it is possible to determine the family to which a species belongs; however, accurate species-level identification can be challenging. Molecular methods, particularly DNA sequence-based identification, have proven to be the most reliable approaches for precise species determination. The cytochrome oxidase subunit I (CO1) gene has been successfully used in phylogenetic studies on mites [41–44]. However, these genes offer limited information for classifying Neocypholaelaps species within the genus and often have limited data in GenBank (https://www.ncbi.nlm.nih.gov) and BOLD systems (https://boldsystems.org). The nuclear ribosomal DNA genes, specifically 18S and 28S, hold promise for resolving higher-level mite phylogenies due to their combination of conserved and variable regions [45–48]. These genes contain conserved regions for identifying deeper branches and variable regions that can be used to discern relationships between closely related taxonomic units [45]. Furthermore, the combination of information from the 18S and 28S genes aids in a more accurate phylogenetic classification than using individual genes alone [47].
In this study, we employed morphological identification methods in conjunction with sequences of the 18S and 28S genes to elucidate the phylogenetic relationships between members of Neocypholaelaps sp. KOR23 and their related groups. Additionally, we provided insights into the potential pathogen reservoirs and transmission capabilities of these mites in honeybee colonies. This information offers a comprehensive perspective on the escalating disease prevalence in South Korean honeybees as well as its implications on a global scale.
Materials and methods
Neocypholaelaps mite collection and morphological identification
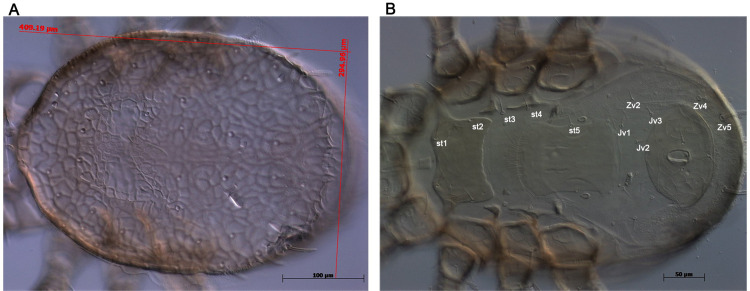
Neocypholaelaps sp. KOR23 mites were observed on beehive A. mellifera colonies in Wonju City, Gangwon Province, ROK, in 2023. Nine colonies from three apiaries were used to collect Neocypholaelaps sp. KOR23 mites. These mites were gathered from hive debris and stored in 50 mL tubes (Falcon) containing 95% ethanol that was labeled with hive number. The sample was immediately subjected to total nucleic acid extraction upon arrival the laboratory. The Neocypholaelaps mites were identified and imaged using a stereomicroscope (Discovery V8 Stereo, Germany) and a Leica DM1750M microscope (Figs 1 and S1). Identification at the genus level requires that genus classification characteristics are identified according to Delfinado and Baker (1983) [15] and Narita et al. (2013) [30].
Fig 1. Morphological characteristics of Neocypholaelaps sp. female mite.
Identity of acaroid mites was determined by analyzing morphological characteristics using a phase contrast microscope. (A) Dorsal view of the mite. (B) Ventral view of the mite.
Total nucleic acid extraction and amplification of CO1, 18S, and 28S regions
The total nucleic acid (TNA) of the mites were extracted using the Maxwell RSC Viral Total Nucleic Acid Purification Kit (Promega, USA) [14]. The hive debris samples were observed and pooled to collect Neocypholaelaps mites under a Discovery V8 Stereo microscope. Three Neocypholaelaps mite samples were collected from three apiaries. Ten adult mites from each apiary were transferred onto a Petri dish containing UltraPure™ distilled water (Invitrogen, USA) for washing. Mites were manually collected for extraction of TNA using a mounting needle under a dissecting microscope. Those mites were pooled and placed in 2 mL Eppendorf tubes containing 300 μL phosphate-buffered saline (1x) and 2.381 mm steel beads (Hanam, ROK) and used for TNA extraction [24]. The extracted TNA of Neocypholaelaps mite was stored at –20°C for PCR amplification of CO1, 18S, and 28S regions and checking honeybee pathogens.
Primers for the amplification of CO1, 18S, and 28S ribosomal DNA genes were designed based on the available sequences in GenBank (https://www.ncbi.nlm.nih.gov) and BOLD systems (https://boldsystems.org). Based on the morphological characteristics results observed under the microscope, the Neocypholaelaps mite was identified in this study. Then, the genetic analysis of CO1, 18S, and 28S genes was done for further identification of collected mites. Primers were designed based on the sequence information of Neocypholaelaps species available on GenBank and BOLD systems (Table 1). The available primers of CO1 gene were designed for N. indica Evans (1963), N. apicola Delfinado-Baker & Baker, 1983, and Neocypholaelaps sp. (NCBI accession nos.: LC522089.1, KP966315.1, and MF911280); The 18S and 28S gene were designed for Neocypholaelaps sp. (NCBI accession nos.: FJ911807.1 and FJ911742.1). These primers are listed in Table 1. The PCR reaction was performed in 50 μL of AccuPower® PCR preMix and Master Mix (Bioneer, ROK) containing 10 pmol of each primer and 20 ng of DNA as a template. The total volume was increased up to 50 μL with distilled water. The thermocycler protocol by Nguyen et al. was applied [24]. PCR amplification products were visualized by electrophoresis on a 1% agarose gel. The bands were excised under UV light and purified using a QIAquick Gel Extraction Kit (Qiagen, USA). Purified PCR products were sequenced by Cosmogentech (ROK).
Table 1. List of primers used for the amplification of CO1, 18S, and 28S genes in this study.
| Strains | Genes | Primers | Sequence (5’ → 3’) | Size (bp) |
|---|---|---|---|---|
| Neocypholaelaps sp. | 18S | Neo18-For | GGATGTGATTAGTTAATTGG | 1590 |
| Neo18-Rev | CTTCATTGCAAATAATACAGG | |||
| 28S | Neo28-For | TTAAATACAACAAGAGATGA | 1258 | |
| Neo28-Rev | CCTGCTGTCTTCAGCACTAAC | |||
| N. indica | CO1 | Neo1-For | GGTACTTTATATTTTATTT | 675 |
| Neo1-Rev | GGTGACCAAAAAATCAAAAT | |||
| N. apicola | CO1 | Neo2-For | GCTCATGCTTTTATTATAAT | 326 |
| Neo-Rev | AATAGTACAAATAAAATTAA | |||
| Neocypholaelaps sp. | CO1 | Neo3-For | TTATGGTGATGCCTGCTATA | 300 |
| Neo-Rev | AATAGTACAAATAAAATTAA |
Detection of honeybee pathogens
The total nucleic acid extracted from the mite samples was tested for honeybee pathogens [24, 49]. The honeybee pathogens that were detected in Korean honeybee colonies were employed to assess their presence in Neocypholaelaps mite. These pathogens include the following viral pathogens: AFB (American foulbrood), EFB (European foulbrood), ASCO (Ascosphaera apis), ASP (Aspergillus flavus), Nosema ceranae, Nosema apis, Spiroplasma (Spiroplasma sp. and S. apis), Trypanosoma spp., SBV (sacbrood virus), KSBV (Korea sacbrood virus), DWV (deformed wing virus), BQCV (black queen cell virus), KBV (Kashmir bee virus), ABPV (acute bee paralysis virus), IAPV (Israeli acute paralysis virus), AmFV (Apis mellifera filamentous virus), species of Lake Sinai virus (LSV1, LSV2, LSV3, and LSV4), VDV-1 (Varroa destructor virus 1), and VDV1-DWV (recombinant Varroa destructor virus 1 and deformed wing viruses). Honeybee pathogens were detected using RT-qPCR Kits [iNtRON Biotechnology, Inc., ROK], Pobgen bee pathogen detection Kit [Postbio, ROK], SsoAdvanced Universal SYBR Green Supermix [Bio-Rad, USA], and iTaq Universal SYBR Green One-Step Kit [Bio-Rad, USA]. The sequences of primers targeting honeybee pathogens for detection in Neocypholaelaps mite samples are provided in S1 Table. Melt-curve dissociation analysis was performed to verify the specificity of the PCR amplification. Negative and positive controls were included for each run. Samples with a Ct value of ≤ 35 and consistent melting curves were considered positive.
Phylogenetic analysis
The amplified 18S and 28S ribosomal DNA gene sequences of Neocypholaelaps sp. KOR23 obtained in this study were deposited in the National Center for Biotechnology Information (NCBI) GenBank database under accession numbers OR576776.1 and OR576780.1, respectively. Those sequences were searched and compared using the nucleotide Basic Local Alignment Search Tool (BLASTn). The identified sequences of Neocypholaelaps mite were compared with those of the Mesostigmata mite family deposited in the NCBI database. Nucleotide sequences were aligned using ClustalW alignment in BioEdit version 7.0.0 software [50]. Sequence alignment was performed using the Molecular Evolutionary Genetics Analysis software (MEGA, version 11.0.13), and a phylogenetic tree was constructed based on the neighbor-joining method and a bootstrap probability score of 1000 [51].
Results
Morphological identification
The morphological identification of the collected mite showed similar characteristics to the genus of Neocypholaelaps adult mite as describe of N. favus Ishkawa, 1968, N. apicola Delfinado-Baker & Baker, 1983, N. geonomae n. sp. Moraes & Narita, 2010, and N. indica Evans, 1963 [26–28, 36]. Neocypholaelaps adult mite dorsal and ventral surfaces were observed under both microscope discovery V8 Stereo (S1A and S1B Fig) and electron microscopy Leica DM1750M (S1C and S1D Fig) for magnification. The genus Neocypholaelaps is characterized by its extremely small size, approximately 2060–2190 μm in length and 1322–1537 μm in width (S1A and S1B Fig). The dorsal shield is entirely reticulate, with reticula formed by simple lines, measuring 409–412 μm in length and 294–307 μm in width (Fig 1A). The sternal shield was wider than it was long, bearing setae st1 and st2, whereas st3 and st4 were situated on the unsclerotized cuticle (Fig 1B). The genital shield is smooth, 75–83 μm (average 79 μm) wide at its widest level, slightly convex posteriorly, and bearing st5. Moreover, it possesses a pair of pores and a pair of lyrifissures on the unsclerotized cuticle, located posterolateral to st5. The anal shield appeared smooth and oval, was equipped with three stouts and barbed setae, and featured a curved transverse line immediately anterior to the cribrum. Opisthogastric setae (Jv1–Jv5 and Zv2) were found on the unsclerotized cuticle (Fig 1B). In addition, a sclerotized transverse line was observed immediately posterior to the genital shield margin. Setae Jv4 and Jv5 are stout and barbed, whereas other ventral idiosomal setae are setiform and smooth.
Genetic identification
In this study show that using primers specific for the species Neocypholaelaps mites were unable to amplify the CO1 gene. This indicates that our isolated strains in the ROK do not belong to the known N. indica Evans, 1963, N. apicola Delfinado-Baker & Baker, 1983, or Neocypholaelaps sp. species found in honeybee colonies. Owing to the limited information on CO1 sequences in GenBank and BOLD systems, it is challenging to distinguish species-level differences based on CO1 sequences.
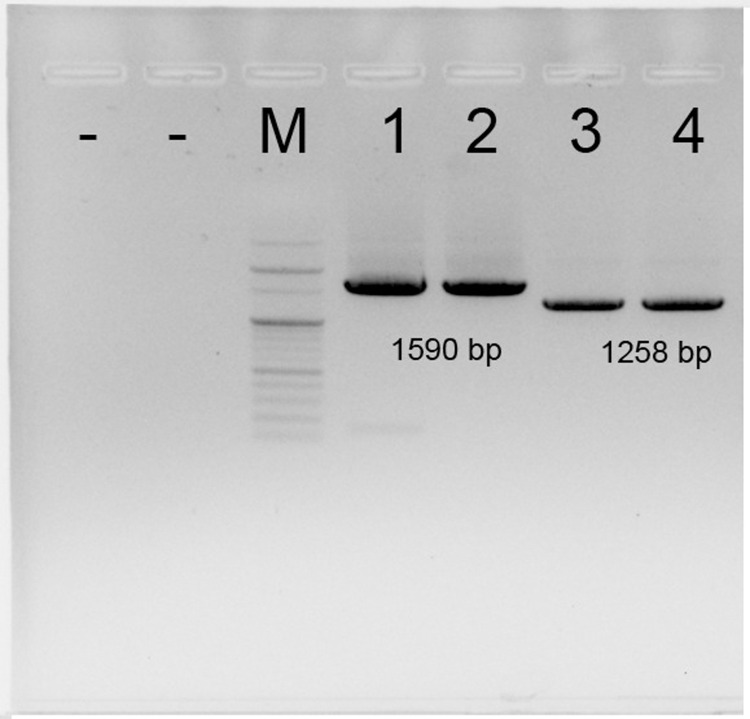
We successfully optimized the PCR products for 18S and 28S gene segments of 1590 and 1258 bp, respectively (Fig 2). After removing noisy sequences during read processing, the 18S and 28S sequences were subjected to BLASTn in GenBank, showing the highest similarity to Neocypholaelaps sp. APGD-2010 (NCBI accession nos.: FJ911807.1 and FJ911742.1) at 98.81 and 92.17%, respectively. These results demonstrate that the 18S gene region is much more conserved than the 28S region. The sequence similarity of 18S and 28S rRNA indicated a close relationship between this mite and the strain Neocypholaelaps sp. APGD-2010.
Fig 2. Amplification of 18S and 28S regions from DNA mite templates.
M is 100 bp DNA ladder (Enzynomics, ROK); 1 and 2 are PCR products of DNA templates using the 18S pair of primer; 3 and 4 are PCR products of DNA templates using the 28S pair of primer; “−” is negative control without a DNA template. Amplicon size from sample DNA is shown in base pair (bp).
Phylogenetic tree
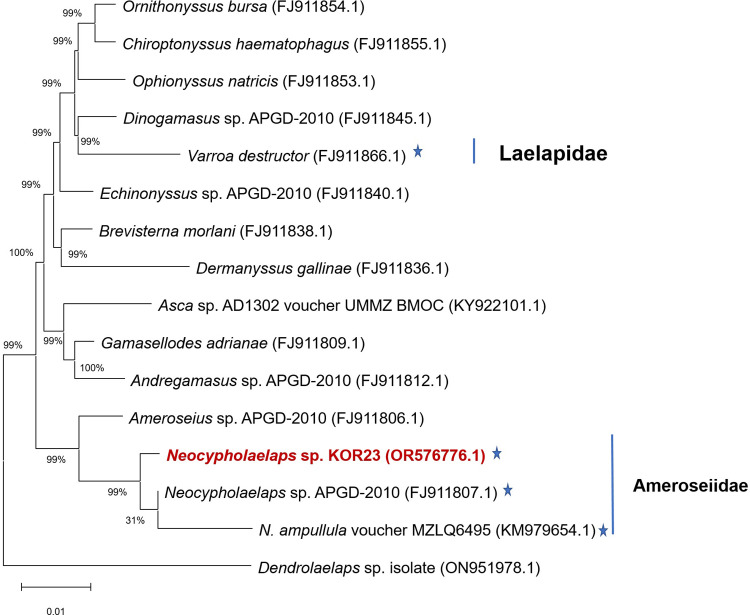
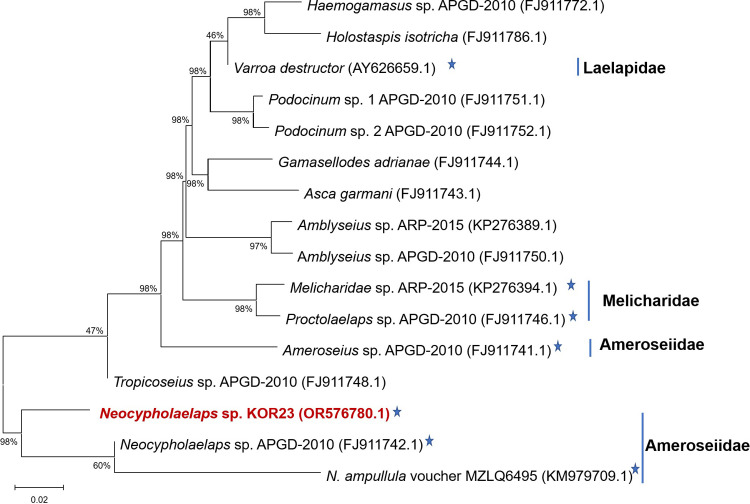
Based on 18S sequence-based phylogeny, two families (Laelapidae and Ameroseiidae) within the order Mesostigmata were found to be associated with honeybees. The 18S rRNA sequence of this mite species was similar to that of Neocypholaelaps sp. APGD-2010 and N. ampullula Berlese, 1910 voucher strain MZLQ6495 (Fig 3). In contrast, the 28S sequence-based phylogeny revealed three families (Laelapidae, Melicharidae, and Ameroseiidae) within the order Mesostigmata that are related to honeybees (Fig 4). Combining the 18S and 28S sequences of this mite species, it was identified as a member of the genus Neocypholaelaps but represents a distinct species. Our study newly provides the genetic information of a strain in the genus Neocypholaelaps isolated from beehives in the ROK in 2023 and was designed as Neocypholaelaps sp. KOR23.
Fig 3. The phylogenetic tree is based on the 18S sequences.
The used species were selected based on the level of nucleotide sequence similarity within the order Mesostigmata. Star (★) indicates species related to honeybees.
Fig 4. The phylogenetic tree is based on the 28S sequences.
The used species were selected based on the level of nucleotide sequence similarity within the order Mesostigmata. Star (★) indicates species related to honeybee colonies.
Testing of honeybee pathogens in Neocypholaelaps mites
As certain mite species present in beehives act as parasites on honeybees causing serious impact, this study aimed to investigate the presence of honeybee pathogens within newly isolated Neocypholaelaps species in ROK. The identities of the pathogens were confirmed by real-time RT-PCR (Table 2). Nine honeybee pathogens (AFB, ASCO, Spiroplasma sp., DWV, BQCV, CBPV, IAPV, KV, and LSV3) were detected in this mite species out of a total of 25 that were screened. Results indicated the presence of bacteria and viruses in Neocypholaelaps sp. KOR23 with high Ct values. Some honeybee pathogens findings suggest that Neocypholaelaps sp. KOR23 has the potential to harbor pathogens and may serve as a vector for honeybee infections, contributing to the rapid increase in disease outbreaks among honeybee colonies in the ROK. However, Nosema apis and N. ceranae and protozoa Trypanosoma spp. have not been identified in Neocypholaelaps sp. KOR23.
Table 2. Detection of honeybee pathogens in Neocypholaelaps sp. KOR23 mite.
| Pathogens | Name | Cycle threshold (*) | Average | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| Bacteria | AFB | 34.71 | 33.44 | 34.08 | 34.08 ± 0.64 |
| EFB | - | - | - | - | |
| ASP | - | - | - | - | |
| ASCO | 32.56 | 35.15 | 34.27 | 33.99 ± 1.31 | |
| Spiroplasma sp. | 32.25 | 31.12 | 30.95 | 31.44 ± 0.71 | |
| Spiroplasma apis | - | - | - | - | |
| Fungi | Nosema apis | - | - | - | - |
| Nosema ceranae | - | - | - | - | |
| Protozoa | Trypanosoma spp. | - | - | - | - |
| Viruses | AmFV | - | - | - | - |
| SBV | 40 | 35.39 | 34.53 | 36.64 ± 2.94 | |
| BQCV | 30.50 | 34.85 | 34.64 | 33.33 ± 2.45 | |
| CBPV | 36.13 | 30.90 | 30.62 | 32.55 ± 3.10 | |
| ABPV | - | - | - | - | |
| IAPV | 34.19 | 33.82 | 23.60 | 30.54 ± 6.01 | |
| KV | 35.05 | 35.27 | 34.02 | 34.78 ± 0.67 | |
| VDV1-DWV | - | - | - | - | |
| VDV-1 | - | - | - | - | |
| DWV | 33.14 | 35.20 | 35.06 | 34.46 ± 1.15 | |
| KSBV | - | - | - | - | |
| KBV | - | - | - | - | |
| LSV1 | - | - | - | - | |
| LSV2 | - | - | - | - | |
| LSV3 | 33.52 | 34.18 | 34.26 | 33.99 ± 0.41 | |
| LSV4 | - | - | - | - | |
(*) Data for cycle thresholds (Ct) of the three samples. The Ct value of ≤ 35 were considered positive.(-) No detection
Discussion
This study presents in detail the process of identification of Neocypholaelaps sp. KOR23 mite within honeybee colonies in Gangwon Province, in ROK. The species-level morphology of Neocypholaelaps mites were described and reported [28−30, 38]. According to the morphology of dorsal and ventral of adult mite, the new mite isolated from Korean honeybee colonies was observed belong to genus of Neocypholaelaps (Figs 1 and S1). The morphological information of Neocypholaelaps is limited in this study, making it challenging to compare with other species. Based on the general morphological and genetic characteristics of 18S and 28S genes, this new mite belongs to the genus Neocypholaelaps, with close similarity to Neocypholaelaps sp. APGD-2010 that was first isolated from the United States. The discovery of Neocypholaelaps in ROK, a temperate region, aligns with recent observations suggesting the genus’ ability to adapt to novel climatic conditions and potentially new host associations. The observed variation in the 18S and 28S ribosomal genes suggests they may play a role in adaptation to novel climatic conditions and potentially new host associations. Further research is needed to gain a deeper understanding of the distribution patterns and hosts of Neocypholaelaps species. To accurately classify the subspecies level of Neocypholaelaps sp. KOR23, detailed morphological characteristics need to be studied, comparing them with strains isolated from temperate regions such as N. favus Ishikawa, 1968, and N. apicola Delfinado-Baker & Baker, 1983, as reported [34, 38]. Additionally, decoding the entire gene sequence of this mite species would be essential.
In honeybee colonies, bees nesting in cavities play a crucial role as hosts for a diverse range of species, including both pathogenic and non-pathogenic species. The presence of bee mites can have serious consequences on honeybee populations. Infestation of honeybees with hemolymph-feeding mites, such as Varroa destructor, Tropilaelaps, and Tyrophagus mites, has been linked to increased colony loss during the winter season and the transmission of honeybee diseases [6, 14, 23, 24, 52]. The coexistence of these parasitic characteristics within honeybee colonies facilitates the exchange of parasites. Certain mite species in this genus, such as N. phooni Baker & Delfinado-Baker, 1985 and N. malayensis Delfinado-Baker, Baker & Phoon, 1989 are associated with stingless bees (Heterotrigona, Geniotrigona, Tetragonula, and Meliponula). Their life cycle occurs entirely inside the bee colonies, but there is no evidence of parasitism in adult or brood bees [31]. In contrast, in Japan, an astonishing number of 3000 N. favus Ishikawa, 1968 mites were found in a single colony, resulting in considerable harm to honeybees [34]. Similarly, as many as 400 N. indica Evans, 1972 mites were observed in a single A. cerana individual [8]. High infestation rates of N. indica Evans, 1972 and its invasion on the body surfaces of three honeybee species were also recorded in India in 2021 [27]. This study identified the presence of Neocypholaelaps sp. KOR23 in honeybee colonies within Gangwon province at high densities. Further research is crucial to understand its potential role in honeybee health and develop appropriate management strategies.
Analysis of the presence of honeybee pathogens within these mites showed that nine of twenty-five pathogens were found, indicating the prevalence of honeybee diseases in this mite. The presence of several honeybee pathogens in Neocypholaelaps species suggests that these mites pose a risk as potential carriers and transmitters of honeybee diseases. Possibly, this could be one of the factors contributing to severe damage to honeybee colonies. This study is the first to demonstrate the presence of honeybee pathogens in Neocypholaelaps sp. mite. Recent research has also reported the appearance of honeybee pathogens in parasites from honeybee colonies. Almost honeybee pathogens have been detected in Varroa mite [5, 6, 53, 54]; DWV and BQCV were found on the small hive beetle (Aethina tunida) [55]; DWV and Trypanosoma spp. were detected on Tyrophagus curvipenis [24]. Some of viruses and bacteria pathogen of honeybee were detected in Neocypholaelaps sp. KOR23. However, microsporidian (Nosema apis and N. cernaena) and protozoa (Trypanosoma spp.) were not seen in Neocypholaelaps sp. KOR23. Further study on the detection of the same pathogens in the honeybee colonies where the mites were detected might be needed to confirm the role of Neocypholaelaps sp. KOR23 mite in pathogen transmission.
In conclusion, the novel Neocypholaelaps sp. KOR23 mite was detected in honeybee colonies in Gangwon Province, ROK. Our phylogenetic analysis, conducted using the 18S and 28S regions (NCBI accession nos.: OR576776.1 and OR576780.1), enabled the precise identification of the genus of Neocypholaelaps mite and its close relationship with Neocypholaelaps sp. APGD-2010 (NCBI accession nos.: FJ911807.1 and FJ911742.1). The 18S and 28S genes could be applied for appearance Neocypholaelaps sp. from beehive honeybee colonies in different provinces in ROK. The presence of some honeybee pathogens in this mite species highlights the significance of disease transmission within honeybee colonies. However, a thorough analysis and comparison of the presence of the pathogen in this mite and corresponding honeybee colonies is required. Such an analysis will provide a better assessment of infection levels, disease reservoir potential, and the role of the mite as an intermediate vector in the transmission of diseases to honeybee colonies.
Supporting information
(A, B) Optical microscope Discovery V8 Stereo (Germany) with a magnification of 5.0× (Dorsal and ventral of Neocypholaelaps sp. adult mite). (C, D) Dorsal and ventral view of Neocypholaelaps sp. adult mite under a Leica DM1750M microscope at 10× magnification.
(TIF)
(DOCX)
(TIF)
Acknowledgments
We would like to thank Animal and Plant Quarantine Agency for approval to conduct this research. Our heartfelt thanks go to the beekeepers who facilitated the sampling. We extend our appreciation to all members of our laboratories for their unwavering dedication and diligent efforts.
Data Availability
The amplified 18S and 28S ribosomal DNA gene sequences of Neocypholaelaps sp. KOR23 obtained in this study were deposited in the National Center for Biotechnology Information (NCBI) GenBank database under accession numbers OR576776.1 and OR576780.1, respectively.
Funding Statement
This work was supported by Animal and Plant Quarantine Agency (Grant No. N-1543081-2021-25-03).
References
- 1.Baily L, Ball BV. Parasite mite. In: Baily L, Ball BV, editors. Honey bee pathology. 2nd ed. Academic Press. London, United Kingdom. 1991. pp. 78–105. [Google Scholar]
- 2.Fain A, Gerson U. Notes on two astigmatic mites (Acari) living in beehives in Thailand. Acarologia. 1990;31(4):381–4. [Google Scholar]
- 3.Sammataro D, Gerson U, Needham G. Parasitic mites of honey bees: life history, implications, and impact. Annu Rev Entomol. 2000;45:519–48. doi: 10.1146/annurev.ento.45.1.519 [DOI] [PubMed] [Google Scholar]
- 4.Seeman OD, Walter DE. Life history of Afrocypholaelaps africana (Evans) (Acari: Ameroseiidae), a mite inhabiting mangrove flowers and phoretic on honeybees. J Austral Entomol Soc. 1995;34:45–50. 10.1111/j.1440-6055.1995.tb01277.x [DOI] [Google Scholar]
- 5.Forfert N, Natsopoulou ME, Frey E, Rosenkranz P, Paxton RJ, Moritz RF. Parasites and Pathogens of the Honeybee (Apis mellifera) and Their Influence on Inter-Colonial Transmission. PLoS One. 2015;10(10):e0140337. doi: 10.1371/journal.pone.0140337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Posada-Florez F, Ryabov EV, Heerman MC, Chen Y, Evans JD, Sonenshine DE, et al. Varroa destructor mites vector and transmit pathogenic honey bee viruses acquired from an artificial diet. PLoS One. 2020;15(11):e0242688. doi: 10.1371/journal.pone.0242688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chantawannakul P, Ramsey S, vanEngelsdorp D, Khongphinitbunjong K, Phokasem P. Tropilaelaps mite: an emerging threat to European honey bee. Curr Opin Insect Sci. 2018;26:69–75. doi: 10.1016/j.cois.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 8.Ramanan RV, Ghai S. Observations on the mite Neocypholaelaps indica Evans and its relationship with the honey bee Apis cerana indica Fabricius and the flowering of Eucalyptus trees. Entomon. 1984;9:291–2. [Google Scholar]
- 9.Baker E, Delfinado M. Notes on the driedfruit mite Carpoglyphus lactis (Acarina: Carpoglyphidae) infesting honeybee combs. J Apicul Res. 1978;17(1):52–4. 10.1080/00218839.1978.11099902 [DOI] [Google Scholar]
- 10.Chantawannakul P, de Guzman LI, Li J, Williams GR. Parasites, pathogens, and pests of honeybees in Asia. Apidologie. 2016;47:301–24. 10.1007/s13592-015-0407-5 [DOI] [Google Scholar]
- 11.Warrit N, Lekprayoon C. Asian honeybee mites. In: Hepburn H, Radloff S, editors. Honeybees of Asia: Springer; 2010. pp. 347–68. 10.1007/978-3-642-16422-4_16 [DOI] [Google Scholar]
- 12.Zheng H, Cao L, Huang S, Neumann P, Hu F. Current status of the beekeeping industry in China. In: Chantawannakul P, Williams G, Neumann P, editors. Asian beekeeping in the 21st century: Spring; 2018. pp. 129–58. [Google Scholar]
- 13.Ravoet J, De Smet L, Wenseleers T, de Graaf DC. Vertical transmission of honey bee viruses in a Belgian queen breeding program. BMC Vet Res. 2015;11(1):1–6. doi: 10.1186/s12917-015-0386-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truong A-T, Yoo M-S, Yun B-R, Kang JE, Noh J, Hwang TJ, et al. Prevalence and pathogen detection of Varroa and Tropilaelaps mites in Apis mellifera (Hymenoptera, Apidae) apiaries in South Korea. J Apic Res. 2023;62(4):804–12. 10.1080/00218839.2021.2013425 [DOI] [Google Scholar]
- 15.Delfinado-Baker M, Baker EW. A new species of Neocypholaelaps (Acari: Amereseiidae) from brood combs of the Indian Honey bee. Apidologie. 1983;14(1):1–7. [Google Scholar]
- 16.Forsgren E, De Miranda JR, Isaksson M, Wei S, Fries I. Deformed wing virus associated with Tropilaelaps mercedesae infesting European honey bees (Apis mellifera). Exp Appl Acarol. 2009;47(2):87–97. 10.1007/s10493-008-9204-4 [DOI] [PubMed] [Google Scholar]
- 17.Gao J, Ma S, Wang X, Yang Y, Luo Q, Wang X, et al. Tropilaelaps mercedesae parasitism changes behavior and gene expression in honey bee workers. PLoS Pathog. 2021;17(7):e1009684. doi: 10.1371/journal.ppat.1009684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denmark H, Cromroy H, Sanford MT. Honey bee tracheal mite, Acarapis woodi (Rennie) (Arachnida: Acari: Tarsonemidae). DPI Entomology Circular, University Florida, IFAS Extension. 2000. [Google Scholar]
- 19.Gary NE, Page RE, Lorenzen K. Effect of age of worker honey bees (Apis mellifera) on tracheal mite (Acarapis woodi) infestation. Exp Appl Acarol. 1989;7:153–60. 10.1007/BF01270435 [DOI] [Google Scholar]
- 20.Kojima Y, Toki T, Morimoto T, Yoshiyama M, Kimura K, Kadowaki T. Infestation of Japanese native honey bees by tracheal mite and virus from non-native European honey bees in Japan. Microb Ecol. 2011;62:895–06. doi: 10.1007/s00248-011-9947-z [DOI] [PubMed] [Google Scholar]
- 21.Maeda T. Infestation of honey bees by tracheal mites, Acarapis woodi, in Japan. J Acarol Soc Japan. 2015;24(1):9–17. [Google Scholar]
- 22.Otis GW, Scott-dupree CD. Effects of Acarapis woodi on overwintered colonies of honey bees (Hymenoptera: Apidae) in New York. J Econ Entomol. 1992;85(1):40–6. 10.1093/jee/85.1.40 [DOI] [Google Scholar]
- 23.Texeira ÉW, dos Santos LG, Matioli AL, Alves MLTMF. First report in Brazil of Tyrophagus putrescentiae (Schrank)(Acari: Acaridae) in colonies of Africanized honey bees (Apis mellifera L.). Interciencia. 2014;39(10):742–4. [Google Scholar]
- 24.Nguyen T-T, Yoo M-S, Truong A-T, Lee JH, Youn SY, Lee S-J, et al. First identification of Tyrophagus curvipenis (Acari: Acaridae) and pathogen detection in Apis mellifera colonies in the Republic of Korea. Sci Rep. 2023;13(1):9469. doi: 10.1038/s41598-023-36695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mašán P. A revision of the family Ameroseiidae (Acari, Mesostigmata), with some data on Slovak fauna. Zookeys. 2017;(704):1–228. doi: 10.3897/zookeys.704.13304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evas GO. The genus Neocypholaelaps Vitzthum (Acari: Mesostigmata). Ann Mag Nat Hist. 1963;6:209–230. 10.1080/00222936308651345 [DOI] [Google Scholar]
- 27.Poorna Chandra S, Srinivas Reddy K, Chinnamade Gowda C, Eswarappa G. Mites associated with honey bee colonies with special reference to phoretic mite, Neocypholaelaps indica Evans in selected locations of Southern Karnataka. J Entomol Zool Stud. 2021;9(1):629–35. [Google Scholar]
- 28.De Moraes GJ, Narita JP. Description of a new species of Neocypholaelaps (Acari: Ameroseiidae) from Brazil, with a key to the world species. Zootaxa. 2010;2554(1):37–44. 10.11646/ZOOTAXA.2554.1.3 [DOI] [Google Scholar]
- 29.Narita JP, De Moraes GJ, Fernando LC. Two species of Neocypholaelaps from Sri Lanka (Acari: Ameroseiidae), with description of a new species. Zootaxa. 2011;2741(1):59–65. 10.11646/zootaxa.2741.1.3 [DOI] [Google Scholar]
- 30.Narita JPZ, Pedelabat M, Moraes Gd. A new species of Neocypholaelaps vitzthum (Acari: Ameroseiidae), with notes on the cheliceral lobes and ventral pore-like structures of mites of this family. Zootaxa. 2013;3666(1):1–15. doi: 10.11646/zootaxa.3666.1.1 [DOI] [PubMed] [Google Scholar]
- 31.Baker EW, Delfinado-Baker M. An unusual new species of Neocypholaelaps (Acari: Ameroseiidae) from the nests of stingless bees (Apidae: Meliponinae). Int J Acarol. 1985;11(4):227–32. 10.1080/01647958508683421 [DOI] [Google Scholar]
- 32.Haragsim O, Samšiňák K, Vobrázková E. The mites inhabiting the bee‐hives in ČSR. Z Angew Entomol. 1978;87(1‐4):52–67. 10.1111/j.1439-0418.1978.tb02425.x [DOI] [Google Scholar]
- 33.Fain A, Hosseinian SH. Observations on the mites (Acari) infesting hives of Apis mellifera race carnica (Insecta Apidae) in Belgium. Bull Ann Soc r belge Ent. 2000;136(1/6):32–33. (in French). [Google Scholar]
- 34.Ishikawa K, editor Studies on the mesostigmatid mites associated with the insects in Japan (I). Rep Res Matsuyama Shinonome Junior Coll. 1968;3:197–218. [Google Scholar]
- 35.Emmanouel N, Pelekassis C, Santas L. Harmful mesostigmatic mites ectoparasitic to honey bees. Entomol Hellenica. 1983;1:17–23. 10.12681/eh.13889 [DOI] [Google Scholar]
- 36.Schousboe C. Rare mite in Danish hives (Neocypholaelaps favus) Tidsskr Biavl. 1986;120:322–5. (In Dutch). [Google Scholar]
- 37.Fenďa P, Lukáš J. First records of mites (Acari: Mesostigmata) from Slovakia. Folia Faun Slovaca. 2014;19(2):171–5. [Google Scholar]
- 38.Kontschán J, TÓbiás I, Bozsik G, SzŐcs G. First record of Neocypholaelaps apicola from beehives in Hungary (Acari: Mesostigmata: Ameroseiidae): re-description and DNA barcoding. Acta Zool Acad Sci Hung. 2015;61(3):237–45 [Google Scholar]
- 39.Fan Q-H, Jiang F. The tiny flower fairies Neocypholaelaps indica Evans, 1963 (Acari: Ameroseiidae). Syst Appl Acarol. 2014;19(2):248–9. [Google Scholar]
- 40.Haq M, editor. Life cycle and behaviour of the coconut mite Neocypholaelaps stridulans (Evans)(Acari: Ameroseiidae) in India. In: Proceedings of the 10th international congress of Acarology. CSIRO Publishing; 2001. pp. 361–4. [Google Scholar]
- 41.Roy L, Dowling AP, Chauve CM, Buronfosse T. Diversity of phylogenetic information according to the locus and the taxonomic level: an example from a parasitic mesostigmatid mite genus. Int J Mol Sci. 2010;11(4):1704–34. doi: 10.3390/ijms11041704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H, Chen T, Dong W. Divergence time of mites of the family Laelapidae based on mitochondrial barcoding region. PLoS One. 2023;18(2):e0279598. doi: 10.1371/journal.pone.0279598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng J, Guo H, Huang S, Lin Y, Huang W, Duan X. Molecular identification of Neocypholaelaps indica Evans based on mitochondrial COI gene. J Environ Entomol. 2019;41(2):380–6. [Google Scholar]
- 44.Antonovskaia A. Using DNA markers in studies of chigger mites (Acariformes, Trombiculidae). Entomol Rev. 2018;98:1351–68. 10.1134/S0013873818090130 [DOI] [Google Scholar]
- 45.Cruickshank RH. Molecular markers for the phylogenetics of mites and ticks. Syst Appl Acarol. 2002;7(1):3–14. 10.11158/saa.7.1.1 [DOI] [Google Scholar]
- 46.Klompen H, Lekveishvili M, Black IV WC. Phylogeny of parasitiform mites (Acari) based on rRNA. Mol Phylogenet Evol. 2007;43(3):936–51. doi: 10.1016/j.ympev.2006.10.024 [DOI] [PubMed] [Google Scholar]
- 47.Sourassou NF, De Moraes GJ, Júnior ID, Corrêa AS. Phylogenetic analysis of Ascidae sensu lato and related groups (Acari: Mesostigmata: Gamasina) based on nuclear ribosomal DNA partial sequences. Syst Appl Acarol. 2015;20(3):225–40. 10.11158/saa.20.3.1 [DOI] [Google Scholar]
- 48.Dowling AP, O’Connor BM. Phylogenetic relationships within the suborder Dermanyssina (Acari: Parasitiformes) and a test of dermanyssoid monophyly. Int J Acarol. 2010;36(4):299–312. 10.1080/01647951003604569 [DOI] [Google Scholar]
- 49.Truong A-T, Yoo M-S, Seo SK, Hwang TJ, Yoon S-S, Cho YS. Prevalence of honey bee pathogens and parasites in South Korea: A five-year surveillance study from 2017 to 2021. Heliyon. 2023;9(2). doi: 10.1016/j.heliyon.2023.e13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall T. BioEdit version 7.0.0; 2004. Distributed by the Author, website. Available from: www.mbio.ncsu.edu/BioEdit/bioedit.html
- 51.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 52.Parmentier L, Smagghe G, de Graaf DC, Meeus I. Varroa destructor Macula-like virus, Lake Sinai virus and other new RNA viruses in wild bumblebee hosts (Bombus pascuorum, Bombus lapidaries, and Bombus pratorum). J Invertebr Pathol. 2016;134:6–11. doi: 10.1016/j.jip.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 53.Lester PJ, Felden A, Baty JW, Bulgarella M, Haywood J, Mortensen AN, et al. Viral communities in the parasite Varroa destructor and in colonies of their honey bee host (Apis mellifera) in New Zealand. Sci Rep. 2022;12(1):8809. doi: 10.1038/s41598-022-12888-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daughenbaugh KF, Martin M, Brutscher LM, Cavigli I, Garcia E, Lavin M, et al. Honey Bee Infecting Lake Sinai Viruses. Viruses. 2015;7(6):3285–309. doi: 10.3390/v7062772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoo M-S, Truong A-T, Choi Y-S, Hong K-J, Hwang TJ, Seo SK, et al. Pathogen detection and phylogenetic analysis of Aethina tumida Murray in South Korea. J Apicul Sci. 2022;66(1):45–55. 10.2478/jas-2022-0004 [DOI] [Google Scholar]