Abstract
The transcriptional program of herpes simplex virus is regulated by the concerted action of three immediate-early (α) proteins, ICP4, ICP27, and ICP0. The experiments described in this study examine the role of the acidic amino terminus (amino acids 1 to 103) of ICP0 in gene activation. When tethered to a DNA binding domain, this sequence activates transcription in the yeast Saccharomyces cerevisiae. Deletion of these amino acids affects the ability of ICP0 to activate α-gene promoter reporters in transient expression assays, while it has little or no effect on a β- and a γ-gene reporter in the same assay. Viruses that express the deleted form of ICP0 (ICP0-NX) have a small-plaque phenotype on both Vero cells and the complementing cell line L7. Transient expression and immunofluorescence analyses demonstrate that ICP0-NX is a dominant negative form of ICP0. Immunoprecipitation of ICP0 from cells coinfected with viruses expressing ICP0-NX and ICP0 revealed that ICP0 oligomerizes in infected cells. These data, in conjunction with the finding that ICP0-N/X is dominant negative, provide both biochemical and genetic evidence that ICP0 functions as a multimer in infected cells.
Herpes simplex virus type 1 and 2 (HSV-1 and -2) infections proceed from a primary lytic infection in the periphery to a lifelong state of latency in sensory neurons characterized by episodes of virus reactivation and lytic infection. The regulatory mechanisms that control HSV gene expression, the establishment and maintenance of latency, and the reactivation of latent virus are complex.
The genes of HSV-1 and -2 are classified into three kinetic classes, immediate-early (α), early (β), and late (γ), based on their temporal expression during productive infections (references 30, 31, and 53 and references therein). The immediate-early genes (α0, α4, α27, α22, α47, and αX) (references 2 and 53 and references therein) are the first to be transcribed following delivery of the virus genome to the host cell nucleus. Subsequent expression of early genes, whose products are largely involved in replication of the virus genome, and late genes, which encode many of the structural components of the herpes simplex virion, requires the prior synthesis of α-gene products (11, 12, 41, 48, 51). Evidence suggests that three of the α-gene products, ICP0, ICP4, and ICP27, cooperatively regulate the expression of all kinetic classes of virus genes (19, 22, 40, 44, 48, 51, 54, 64). ICP4 appears to activate β- and γ-gene transcription while repressing its own expression as well as that of the α0 gene (11, 24, 36, 37, 48, 50, 52). ICP0, a promiscuous transcriptional activator (references 18, 22, and 53 and references therein), has been shown to affect the expression and synthesis of the essential α protein ICP27 and the expression of ICP0 and ICP4 at early times postinfection (38). ICP27 is required for the expression of β and γ genes during the herpesvirus life cycle. Although this protein appears to act primarily at the posttranscriptional level (28, 47, 56, 57, 60), its interaction with ICP4 supports a more direct role for ICP27 in the regulation of gene expression (46).
Mutational and biochemical analyses of ICP0 in transient expression assays and/or in the context of the virus genome led to the identification of domains essential for the activation of gene expression and virus replication (4, 6, 7, 15–17, 61). The role(s) of these domains in ICP0 function remains largely undefined. ICP0 contains an acidic NH2-terminal domain, an essential C3HC4 zinc finger (6, 7, 14–17, 20, 38), a nuclear localization signal, two centrally located proline-rich regions, and a serine tract which may represent a site of phosphorylation (references 7 and 53 and references therein; 66). ICP0 oligomerizes in infected cells (5, 10, 42) and interacts with ICP4, the major virus regulatory protein (43, 46, 67). ICP27 has been shown to influence the intracellular distribution of ICP0 in transfected cells (69). However, because ICP4 interacts directly with ICP0 and ICP27, ICP27-dependent effects on the intracellular localization of ICP0 may be mediated through its interaction with ICP4. Recent studies indicate that ICP0 also interacts with the cellular proteins cyclin D3 (33) and translation elongation factor eEF1Bα, a GTP exchange factor (referred to as EF-1δ in reference 32). However, the relevance of these interactions in the context of a productive infection is unknown.
This report presents a series of experiments that begin to characterize the role of the acidic NH2 terminus of ICP0 in the growth and replication of HSV-1. We demonstrate that this domain functions as a potent transcriptional activation domain in Saccharomyces cerevisiae. Deletion analysis in transient expression assays further demonstrates that this region of ICP0 is required for ICP0-mediated transcriptional activation of HSV-1 α, but not β or γ, promoters. In the context of the virus genome, deletion of the ICP0 NH2 terminus results in defects in the growth of HSV-1, small plaques, and high fluorescent focus unit (FFU)/PFU ratios in Vero cells and the ICP0-expressing cell line L7 (55). The ICP0 NH2-terminal deletion mutant EL0-N/X, which lacks amino acids (aa) 3 to 104, acts as a dominant negative inhibitor of ICP0 function through the formation of oligomers with wild-type ICP0. This analysis provides the first clear evidence that ICP0 contains a promoter-specific transcriptional activation domain and suggests that ICP0 function is dependent on the formation of dimers and/or higher-order oligomers in infected cells.
MATERIALS AND METHODS
Cells and viruses.
Vero and L7 (55) cells were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Grand Island, N.Y.) containing 5% bovine calf serum (HyClone Laboratories Inc., Logan, Utah). For growth of L7 cells, DMEM was supplemented with 400 μg of G418 (Gibco BRL) per ml. 293T cells (13) were maintained in DMEM containing 10% fetal bovine serum (HyClone). The media were supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml) (Gibco BRL) unless otherwise noted.
The strains of wild-type HSV-1 used in this study were Glasgow 17 and KOS 1.1. The α0 cDNA virus vCPc0 (38, 46) and the α0 deletion virus dl1403 have been described elsewhere (61).
Plasmids. (i) α0 cDNA constructs.
Plasmid pDS17, which contains an α0 cDNA and its associated regulatory sequences (68), was constructed by replacing the MluI-EcoRI fragment of pDS16 (68) with the corresponding fragment from p0XB (7). Plasmid pDSE-17B was created by destruction of the Asp718 site located 3′ of the HSV-1 sequences in pDS17.
(ii) GAL4-BD–α0, LexA-α0, and GAL4-AD–α0 gene fusions.
Plasmid pGBT9 (Clontech, Palo Alto, Calif.) encodes the GAL4 DNA binding domain (GAL4-BD; aa 1 to 147) under the control of the yeast ADH1 promoter. Plasmid pBTM116 encodes the entire LexA protein coding sequence (aa 1 to 202) under the control of the yeast ADH1 promoter. Plasmid pXW2 (GAL4-BD–ICP0 aa 1 to 775) was constructed by insertion of the NcoI-BglII fragment of the α0 cDNA from pDS17 into pGBT9 digested with EcoRI and BamHI. In this construction, the NcoI and EcoRI sites were filled in with Klenow DNA polymerase. Plasmid pJY3 (GAL4-BD–ICP0 aa 1 to 105) was constructed by deletion of the XhoI-SalI fragment of the α0 gene from pXW2. pJY4 (GAL4-BD–ICP0 aa 1 to 212) was constructed by deletion of the Asp718-SalI fragment of pXW2. pJY7 (GAL4-BD–ICP0 aa 105 to 212) was constructed by insertion of the XhoI-PstI fragment of pJY4 into pGBT9 digested with SalI and PstI. Plasmid pCPC-YGal03-F (GAL4-BD–ICP0 aa 394 to 775) was constructed by deletion of the EcoRI-NotI fragment of pXW2. Plasmid pPC62-86 was constructed by replacing the SalI-BamHI fragment of pPC62 (8) with the corresponding fragment of pPC86 (8). pBD62-0105/769 (GAL4-BD–ICP0 aa 105 to 769) was constructed by insertion of the BglII fragment of pGAD0105/769 into pPC62-86. pGAD0105-769 (GAL4-AD–ICP0 aa 105 to 769) was constructed by insertion of the XhoI-EcoRI fragment of pGAD0 into pGAD10 (Clontech). pGAD0 (GAL4-AD–ICP0 aa 1 to 769) was constructed by insertion of the MluI-NotI and NotI-SalI fragments of pAD86-01 into pGAD10 digested with MluI and XhoI. Plasmid pGBT-VP16T was constructed by subcloning the EcoRI-BclI fragment of pGAL4-VP16 (a gift from Kathryn Calame, Department of Microbiology, Columbia University, New York, N.Y.) into pGBT9 digested with EcoRI and BamHI. Plasmids pJY5 (LexA-ICP0 aa 1 to 105) and pJY6 (LexA-ICP0 aa 1 to 212) were constructed by insertion of the EcoRI-PstI fragments of pJY3 and pJY4, respectively, into pBTM116. Plasmid pGAD10 (Clontech) encodes the GAL4 transcriptional activation domain (GAL4-AD; aa 768 to 881). pGAD0105 (GAL4-AD–ICP0 aa 1 to 105) was constructed by insertion of the MluI-XhoI fragment of pAD86-01 into pGAD10. pAD86-01 (GAL4-AD–ICP0 aa 1 to 775) encodes a fusion protein between the GAL4-AD and aa 1 to 775 of ICP0 in pPC86. All junctions between the GAL4-BD, LexA, or the GAL4-AD and regions of the α0 gene were verified by DNA sequence analysis.
The mammalian GAL4-BD–ICP0 expression constructs were prepared as follows. pCPC-Gal0F (GAL4-BD–ICP0 aa 1 to 775) was constructed by subcloning the HindIII-PstI (blunted) fragment from pXW2 into pSG424 (26) digested with BamHI, filled, and digested with HindIII. pCPC-Gal07.6 (GAL4-BD–ICP0 aa 1 to 769) and pCPC-Gal07.1 (GAL4-BD–ICP0 aa 1 to 711) were made by deleting the SalI fragment and the BstEII-SalI fragment, respectively, from pCPC-Gal0F. pCPC-Gal05 (GAL4-BD–ICP0 aa 1 to 551) and pCPC-Gal03 (GAL4-BD–ICP0 aa 1 to 385) were constructed by inserting HindIII-AatII (blunted) and HindIII-SmaI fragments, respectively, from pXW2 into pSG424 that had been digested with HindIII-BamHI (filled). pCPC-Gal02 (GAL4-BD–ICP0 aa 1 to 212) was generated by deleting a 600-bp Asp 718 fragment from pCPC-Gal03, and pCPC-Gal01 (GAL4-BD–ICP0 aa 1 to 105) was made by subcloning a HindIII-PstI (blunted) fragment from pJY3 into pSG424 that had been digested with HindIII-BamHI (filled). pCPC-Gal03-F (GAL4-BD–ICP0 aa 392 to 775) and pCPC-Gal03-7.6 (GAL4-BD–ICP0 aa 392 to 769) were constructed by cloning the HindIII-PstI (blunted) and HindIII-SalI fragments, respectively, from pCPC-YGal03-F into pSG424 that had been digested either with HindIII-BamHI (filled) or HindIII-SalI. pCPC-Gal01-2 (GAL4-BD–ICP0 aa 105 to 212) and pCPC-Gal01-7.6 (GAL4-BD–ICP0 aa 105 to 769) were prepared by cloning the 750- and 2,400-bp HindIII-BamHI fragments from plasmids pBD62-0105/212 and pBD62-0105/769, respectively, into HindIII-BamHI-digested pSG424. pBD62-0105/212 was constructed by inserting a HindIII-PstI fragment of pJY7 (described above) into pPC62-86/3. pPC62-86/3 was constructed by inserting a BglII-BamHI fragment of pPC86 into pPC62-2, which was constructed by digesting pPC62 with SalI, filling the DNA ends, and ligating. pCPC-Gal01-5 (GAL4-BD–ICP0 aa 105 to 553) was constructed by inserting the 1,750-bp HindIII-SacI fragment from pBD62-0105/553 (GAL4-BD–ICP0 aa 105 to 553) into pSG424 digested with HindIII and SacI. pBD62-0105/553 was constructed by inserting the α0 sequences encoding aa 105 to 553 of HSV-1 ICP0 into pPC62.
(iii) α0 mutant constructs.
pEL0N/X, which encodes a mutant ICP0 lacking aa 3 to 104, was constructed as follows. Primers 0-N/X-5′3′ (CATGGAATTCGC) and 0-N/X-3′5′ (TCGAGCGAATTC) were annealed and ligated to pDSE-17B digested with NcoI and XhoI. The resulting deletion was verified by DNA sequencing. Plasmid pEL0-TIF, which encodes a fusion protein between the transcriptional activation domain of HSV-1 αTIF (αTIF-AD; aa 411 to 490) and ICP0 aa 105 to 775, was constructed as follows. The DNA sequence encoding aa 411 to 490 of αTIF was amplified from pRAB14 (2) by PCR (as described below), using primers (Gibco BRL) that generate an NcoI or XhoI site and maintain the reading frame of the α0 gene (VP16AD 5′3′ [GCCCATGGTGTCGACGGCCCCCCCGACC] and VP16AD 3′5′ [GCTCGAGACCCACCGTACTCGTCAATTCC], respectively). The resulting PCR product was cloned into pGEM-T, thus generating pGEM-T-VP16. The predicted DNA sequence of the αTIF-AD in pGEM-T-VP16 was confirmed by DNA sequence analysis. The NcoI-XhoI fragment of pGEM-T-VP16 was inserted into pDSE-17B digested with NcoI and XhoI, thus replacing the α0 sequence encoding ICP0 aa 2 to 104. Plasmid pEL17ΔM was constructed by insertion of the HindIII-MluI fragment of pDSE-17B into pIBI31.
(iv) Firefly luciferase reporter plasmids.
Plasmids pCPC-4P-LUC, pTK-LUC, and pEL-PgC-LUC have been described elsewhere (38). Plasmid pCPC-27P-LUC, containing the firefly luciferase reporter gene under the control of the HSV-1 (strain KOS 1.1) α27 gene regulatory sequences, was constructed in two steps. First, the BamHI-BglII fragment from plasmid pBSΔ27 (60) was cloned into pIC20H (39) digested with BamHI and BglII. The resulting plasmid, pCPC-27P, contains α27 gene sequences from −266 to −2 relative to the transcription initiation site. pCPC-27P-LUC was generated by cloning a BamHI fragment from p19LUC (63), containing the luciferase gene coding sequences, into the BglII site of pCPC-27P. Plasmid pCPC-22/47P-LUC was also made in two steps. First, pCPC-4/22P was constructed by subcloning the BamHI-BglII fragment from plasmid pIGA101 (23), containing the α4 and α22/47 gene 5′ regulatory sequences from HSV-1 strain F, into BamHI-BglII-digested pIC20H (39). pCPC-22/47P-LUC, containing the luciferase gene under the control of the α22/47 gene regulatory sequences (−724 to +33), was constructed by cloning the BamHI luciferase gene fragment from p19LUC into the BglII site of pCPC-4/22P. Plasmid pCPC-0P-LUC was constructed as follows. The SacI (filled)-NcoI (filled) fragment from pXW8 (37), containing the −789 to +147 α0 gene sequences from HSV-1 strain 17, was cloned into EcoRV-digested vector pZero2.1 (Stratagene, La Jolla, Calif.) to yield plasmid pCPC-0PL. A StuI fragment was deleted from pCPC-0PL to yield plasmid pCPC-0P, which contains the −537 to +147 region of the α0 gene. The BamHI fragment from p19LUC was cloned into BamHI-digested pCPC-0P to yield pCPC-0P-LUC. pG5-TK-LUC was a gift from Kathryn Calame.
β-Galactosidase assays. (i) Filter assays.
S. cerevisiae Y190 (29) and CTY10-5d (Rolf Sternglanz, State University of New York at Stony Brook) were maintained on YPD medium at 30°C. Yeast cells were transformed by using 1 μg of plasmid DNAs and salmon sperm carrier DNA. When appropriate, transformants were grown at 30°C for 3 to 4 days on SD medium (Difco Laboratories, Detroit, Mich.) containing 25 mM 3-amino-1,2,4-triazole and lacking the appropriate amino acids (Trp or Leu and His) (27). Yeast colonies were lifted onto nitrocellulose membranes (Schleicher & Schuell, Keene, N.H.), and the cells were lysed by freezing them at −80°C for 15 min. After reaching room temperature, the nitrocellulose disks were placed into petri dishes containing 5 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.3% β-mercaptoethanol) containing 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml and incubated at 30°C for 1 to 5 h.
(ii) Liquid assays.
Clonal Y190 transformants were grown in SD medium without Trp or Leu and containing 0.05% glucose to an optical density at 600 nm (OD600) of approximately 0.6. The cells were collected by centrifugation at 1,500 × g for 5 min at 4°C and resuspended in ice-cold Z buffer. Then 200 μl of ice-cold acid-washed glass beads was added to each cell pellet, and the cells were lysed by vortexing them three times for 1 min each. The cell lysates were clarified by centrifugation at 2,500 × g for 5 min at 4°C, and the total protein concentrations were determined by the Bradford method. For β-Galactosidase assays, 150 μl of cell extract was diluted to 800 μl with Z buffer prior to the addition of 200 μl of 4 mg of o-nitrophenyl-β-d-galactopyranoside (ONPG) per ml and the mixture was incubated at 30°C until pale yellow (<30 min). Reactions were stopped by the addition of 500 μl of 1 M Na2CO3, and the OD420 was determined. Units of β-galactosidase per milligram of total protein were determined based on 1 U of β-galactosidase equaling the amount of enzyme required to hydrolyze 10−9 mol of ONPG per min.
Transfection and transient expression assays. (i) Transfections.
Vero cells were transfected with the indicated plasmids by the calcium phosphate precipitation method as previously described (38).
(ii) Luciferase assays.
Transfected Vero cell monolayers were harvested at 48 h posttransfection, and luciferase activities were quantified with a Berthold Lumat LB9501 luminometer (Wallac Inc., Gaithersburg, Md.) as described previously (3). Luciferase activities were determined from triplicate transfections in at least two independent experiments.
Construction of recombinant herpesviruses.
Recombinant herpesviruses were constructed as previously described (38). Briefly, linearized plasmid pEL0-N/X, pEL0-TIF, or pEL17ΔM and 100 PFU of herpesvirus nucleocapsids, purified from cells infected with the α0 null virus dl1403 (59), were transfected into Vero cells. Recombinant viruses vEL0-N/X, vEL0-TIF, and vEL0-NXR were isolated and plaque purified three times. vEL0-N/X encodes a mutant form of ICP0 lacking aa 3 to 104. vEL0-TIF encodes a mutant form of ICP0 in which aa 2 to 104 have been replaced with the HSV-1 αTIF-AD (aa 411 to 490); vEL0-NXR, which was constructed by using virus nucleocapsids isolated from cells infected with vEL0-N/X, encodes wild-type ICP0 from an α0 cDNA. The sequence arrangements of the α0 loci in these viruses were verified by Southern blot analysis. The DNA sequence of each mutation was verified.
Virus growth assays.
Vero cells were infected at the indicated multiplicities of infection (MOIs). Following virus adsorption, infected cell monolayers were washed twice with phosphate-buffered saline (PBS) and overlaid with fresh medium. At the indicated times, infections were halted by freezing the cells at −80°C. Virus yields were determined by titration on the ICP0-expressing cell line L7 (55). Growth curves represent the average of two independent infections, each titrated in duplicate.
Coimmunoprecipitations. (i) Transfections.
293T cells were transfected with either pDSE-17B (5 μg/10-cm-diameter plate), pEL0-N/X (5 μg/10-cm-diameter plate), or a mixture of both plasmids (5 μg of each plasmid/10-cm-diameter plate), using the calcium phosphate precipitation method (65). At 48 h posttransfection, the cells (four 10-cm-diameter plates per condition) were harvested and washed three times with ice-cold PBS containing 1 mM phenylmethylsulfonyl fluoride (PMSF). Each cell pellet was resuspended in 3 ml of ICP0 lysis buffer [20 mM HEPES-KOH (pH 7.9), 0.25% (vol/vol) Nonidet P-40, 400 mM NaCl, 10 mM MgCl2, 10% (vol/vol) glycerol, 1 mM PMSF, 0.1 mM l-1-chloro-3-(4-tosylamido)-4-phenyl-2-butanone (TPCK), 0.1 mM l-1-chloro-3-(4-tosylamido)-7-amino-2-heptanone (TLCK) (Boehringer Mannheim, Indianapolis, Ind.)] containing DNase I (50 μg/ml) and RNase I (50 μg/ml). Cell lysates were incubated for 20 min at 4°C, sonicated for 90 s in a Branson (Danbury, Conn.) Sonifier 450, and clarified by centrifugation at 5,000 × g for 15 min at 4°C followed by high-speed centrifugation at 16,000 × g for 15 min at 4°C.
(ii) Infections.
Vero cells were infected with wild-type HSV-1, vEL0-N/X, or both viruses at an MOI of 5. At 7 h postinfection, the cells were washed three times with ice-cold PBS containing 1 mM PMSF and cell lysates were prepared as described above.
(iii) Immunoprecipitation.
Immunoprecipitations were performed with a rabbit polyclonal antibody (CLU7) that recognizes epitopes within ICP0 between aa 312 and 400 (37) or an affinity-purified rabbit polyclonal antibody (α0-N18) that recognizes the NH2-terminal 18 aa of HSV-1 ICP0. A 500-μl aliquot of each supernatant was mixed with 5 μl of the indicated anti-ICP0 antibody and incubated at 4°C for 1 h with constant mixing prior to the addition of 30 μl of a GammaBind Plus Sepharose (Pharmacia, Piscataway, N.J.) suspension (50% [vol/vol] in ICP0 lysis buffer), and incubation was continued for another 1 h at 4°C. The beads were washed as previously described (38), resuspended in 30 μl of 1.5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and boiled for 5 min. The immunoprecipitated proteins were separated by SDS-PAGE and detected by Western blot analysis, as described below, using antibody CLU7 (37).
Western blot analysis. (i) Transfected cells.
293T cells were transfected, in 10-cm-diameter plates, with 15 μg of plasmid DNAs as previously described (38, 46). Forty-eight hours posttransfection, the cells were scraped into ice-cold PBS and collected by centrifugation at 2,000 × g for 5 min at 4°C. Cell pellets were solubilized in 1.5× SDS-PAGE sample buffer as described previously (38). The proteins were separated in 7.5% denaturing polyacrylamide gels (34) and electrophoretically transferred to nitrocellulose membranes (62). ICP0 was detected by using the rabbit polyclonal antibody CLU7 as previously described (38).
(ii) HSV-1-infected cells.
Vero cells were infected with 1 FFU per cell. At 8 h postinfection, the cells were scraped into ice-cold PBS and collected by centrifugation. Infected cell pellets were solubilized in 1.5× SDS-PAGE sample buffer, and the proteins were separated by SDS-PAGE as previously described (38). Immunodetection of virus proteins was performed as described elsewhere (45). The following antibodies were used to detect the indicated HSV-1 proteins: ICP0, CLU7 (37); ICP4, mouse monoclonal antibody H1114 (Goodwin Institute for Cancer Research, Plantation, Fla.); ICP27, rabbit polyclonal antibody CLU38 (37); glycoprotein B(gB), rabbit polyclonal antibody R69 (provided by G. Cohen, University of Pennsylvania); and VP5, rabbit polyclonal antibody NC-1 (also provided by G. Cohen). The secondary antibodies used in this study were horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G and goat anti-mouse immunoglobulin G (Sigma Chemical, St. Louis, Mo.). Immunoblots were developed as previously described (38).
Immunofluorescence.
Vero cells were either transfected with 5 μg of plasmid DNAs encoding wild-type or mutant forms of ICP0 or infected with the indicated herpesviruses. At 48 h posttransfection or 8 h postinfection, immunodetection of ICP0 was performed as previously described (38), using either monoclonal antibody H1083, polyclonal antibody CLU7, or affinity-purified anti-ICP0 antibody α0-N18.
Fluorescent-focus assays.
Vero cells (106 per 35-mm-diameter plate) were infected at an MOI of 0.01 or 0.001 with wild-type HSV-1 strain Glasgow 17, vCPc0, vEL0-N/X, vEL0-NXR, vEL0-TIF, or dl1403. At 8 and 24 h postinfection, cell monolayers were washed with PBS and fixed in 90% methanol for 10 min. ICP4 and ICP0 were detected with mouse monoclonal antibody 58S (58) and rabbit polyclonal antibody CLU7 (37), respectively, diluted 1:200 in PBS. The average number of FFU or PFU per cell was determined at 8 or 24 h postinfection, respectively. Thus, the FFU/PFU ratio represents the number of positively staining cells at 8 h postinfection relative to the number of infected cell foci at 24 h postinfection per field of cells. The FFU/PFU ratio for each virus was determined in three independent experiments.
RESULTS
The acidic NH2 terminus of ICP0 can activate transcription in S. cerevisiae when fused to a heterologous DNA binding domain or protein.
Proteins that mediate the activation of gene expression at the level of transcription often contain modular DNA binding and transcriptional activation domains, i.e., discrete domains that retain function outside of their native context. For example, the well-characterized transcriptional activation domains of the GAL4 and HSV-1 αTIF proteins activate gene expression when fused to heterologous DNA binding domains (references 9 and 49 and references therein). Accordingly, in an effort to identify transcriptional activation domains within the HSV-1 immediate-early regulatory protein ICP0, regions of the α0 gene were fused to sequences encoding the GAL4-BD or the Escherichia coli LexA protein. The ability of each of the resulting fusion proteins to activate transcription was examined in two strains of S. cerevisiae, Y190 and CTY10-5d. Y190 contains lacZ and HIS3 reporter genes under the control of a GAL4 upstream activation sequence, and CTY10-5d contains lacZ under the control of a GAL1 promoter containing four LexA binding sites. Thus, the ability of various regions of ICP0 to function as transcriptional activation domains when fused to the GAL4-BD or LexA protein was determined by growth of Y190 in the absence of histidine and the expression of β-galactosidase in Y190 or CTY10-5d (Fig. 1).
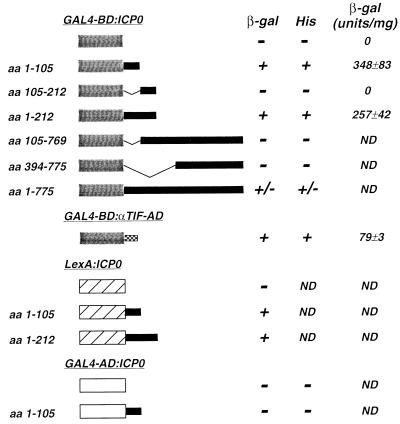
FIG. 1.
Analysis of transcriptional activation by ICP0 fusion proteins in S. cerevisiae. Plasmids directing the expression of fusion proteins containing the yeast GAL4-BD or the bacterial LexA protein fused to regions of ICP0 or the minimal HSV-1 αTIF-AD were constructed. The activation of reporter genes by these fusion proteins was determined in the yeast S. cerevisiae as described in Materials and Methods. A schematic representation of these fusion proteins is shown. The activation of reporter gene expression was determined by growth of strain Y190 in the absence of histidine (His) and by the expression of β-galactosidase (β-gal) as determined by filter lift and liquid assays. ND, not determined.
Fusion proteins containing the GAL4-BD and ICP0 aa 1 to 105 or 1 to 212 activated expression of both lacZ and HIS3 in Y190, as did the minimal HSV-1 αTIF-AD when fused to the GAL4-BD (Fig. 1). The GAL4-BD alone, or fusion proteins containing the NH2-terminal 105 or 212 aa of ICP0 fused to the GAL4-AD, failed to activate lacZ expression or allow growth of Y190 in the absence of histidine (Fig. 1). Fusion proteins lacking the NH2-terminal 105 aa of ICP0 also failed to mediate reporter gene expression (Fig. 1); however, as several of these proteins could not be detected in transformed yeast cell extracts by Western blot analysis (data not shown), their inability to activate reporter gene expression may result from their failure to accumulate. These findings suggest that the acidic NH2-terminal 105 aa of ICP0 constitute a transcriptional activation domain in S. cerevisiae when fused to the GAL4-BD.
To eliminate the possibility that the NH2 terminus of ICP0 was capable of activating reporter gene expression exclusively in the context of a fusion protein with the GAL4-BD, plasmids encoding fusion proteins containing aa 1 to 105 or 1 to 212 of ICP0 fused to the bacterial LexA protein were constructed (Fig. 1). These fusion proteins were tested for activation of a GAL1 promoter containing four LexA binding sites in S. cerevisiae CTY10-5D (Fig. 1). The bacterial LexA protein alone did not activate lacZ expression. However, LexA fusion proteins containing ICP0 aa 1 to 105 or 1 to 212 activated lacZ expression in CTY10-5D (Fig. 1). Thus, the NH2-terminal 105 aa of ICP0, when fused to either the GAL4-BD or the LexA protein, function as a transcriptional activator.
Quantitative β-galactosidase assays revealed that the NH2 terminus of ICP0 (aa 1 to 105) activated reporter gene expression in S. cerevisiae to levels threefold greater than that of the minimal HSV-1 αTIF-AD when fused to the GAL4-BD (Fig. 1). We further note that the activation of reporter gene expression was dependent on fusion of this domain of ICP0 to a DNA binding domain (data not shown). Collectively, these results are consistent with the conclusion that the acidic NH2-terminal 105 aa of HSV-1 ICP0 constitute a transcriptional activation domain in yeast. They do not, however, exclude the possibility that ICP0 contains other transcriptional activation domains.
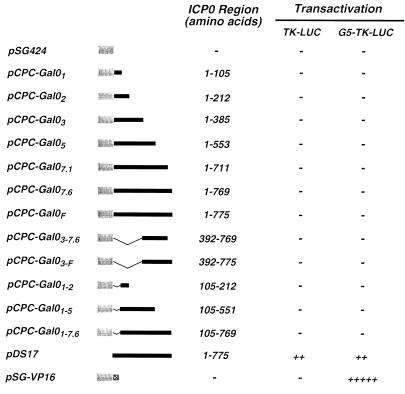
Having shown that the NH2-terminal region of ICP0 can function as a transcriptional activation domain in yeast, we next asked whether this region could also activate gene expression in mammalian cells when fused to the GAL4-BD. Vero cells were cotransfected with plasmids encoding the indicated fusion proteins under the control of the simian virus 40 early promoter (Fig. 2) and reporter plasmids encoding the firefly luciferase gene under the control of the HSV-1 thymidine kinase (TK) promoter or the TK promoter containing five concatemerized GAL4 binding sites (G5-TK) (Fig. 2). Plasmids encoding wild-type ICP0 or a fusion protein containing the GAL4-BD and the minimal HSV-1 αTIF-AD were included as controls (Fig. 2). Consistent with its ability to activate gene expression from a wide range of viral and cellular gene promoters, wild-type ICP0 activated gene expression from both the TK and G5-TK promoters, whereas the GAL4-BD–αTIF-AD fusion protein activated only the G5-TK promoter (Fig. 2). Fusion proteins containing the GAL4-BD and regions of ICP0 universally failed to activate reporter gene expression (Fig. 2). This result was not due to transcriptional “squelching” (25), as varying the concentration of effector plasmids over a wide range did not alter this result. The lack of luciferase expression also could not be explained by low abundance of these GAL4-ICP0 fusion proteins (Fig. 2), as all of these proteins were readily detected in transfected Vero cells by indirect immunofluorescence (data not shown). These analyses, however, revealed that GAL4-BD–ICP0 fusion proteins containing the NH2-terminal 105 or 212 aa of ICP0 were excluded from the nuclei of transfected cells despite the presence of a nuclear localization signal in the GAL4-BD (data not shown). This observation may explain the failure of these fusion proteins to activate reporter gene expression. However, it does not address why full-length ICP0 was unable to activate reporter gene expression when fused to the GAL4-BD as this protein accumulated in the nuclei of transfected cells (data not shown).
FIG. 2.
Analysis of transcriptional activation by GAL4-ICP0 fusion proteins in mammalian cells. The yeast GAL4-BD was fused to regions of ICP0, and the ability of each of these proteins to activate gene expression in Vero cells was determined as described in Materials and Methods. Plasmids directing the expression of the indicated fusion proteins were constructed and cotransfected into cells with reporter plasmids encoding the firefly luciferase gene under the control of either the HSV-1 TK promoter (pTK-LUC) or a TK promoter containing five concatemerized GAL4 binding sites (pG5-TK-LUC). The absence (−) or presence (+) of activation of reporter gene expression was determined by luciferase assays as described in Materials and Methods. Plasmids pDS17, which directs the expression of wild-type HSV-1 ICP0, and pSG-VP16, which directs the expression of a GAL4–αTIF-AD fusion protein, were used as controls.
Deletion of the NH2 terminus of ICP0 affects the level of transactivation of immediate-early virus gene promoters.
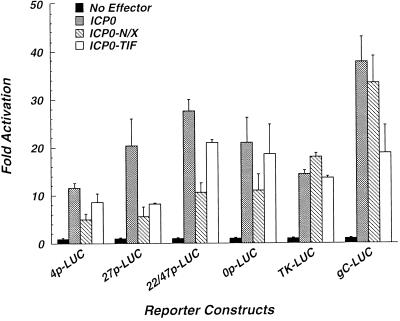
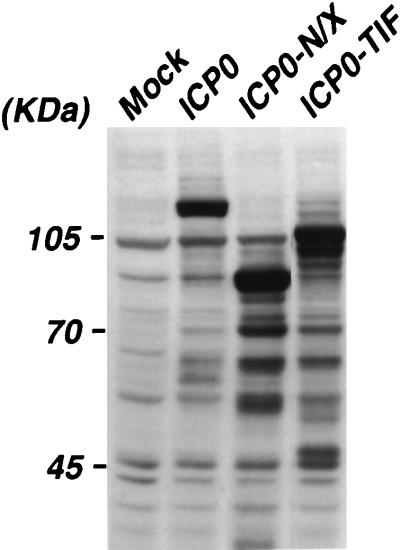
The data presented above raised the possibility that the NH2-terminal 105 aa of ICP0 constitute a transcriptional activation domain. However, they also underscored the complications that can arise from the use of heterologous fusion proteins in the analysis of protein function. Therefore, the α0 cDNA plasmid pEL0-N/X, which encodes a truncated form of ICP0 lacking aa 3 to 104 (ICP0-N/X) under the control of the endogenous α0 promoter/regulatory sequences, was constructed. The effect(s) of this deletion on the ability of ICP0 to activate transcription from each of the three major temporal classes of herpesvirus promoters was assessed in transient expression assays. Briefly, Vero cells were cotransfected with plasmids encoding wild-type ICP0 or ICP0-N/X and reporter constructs directing expression of the firefly luciferase gene under the control of the α4, α27, α22/47, or α0 promoter, the β-TK promoter, or the γ-gC promoter (Fig. 3). This analysis demonstrates that deletion of aa 3 to 104 results in a significant reduction in activation from all of the herpesvirus α-gene promoters examined (Fig. 3). This result cannot be attributed to gross structural alterations in ICP0-N/X, as it activated reporter gene expression from the TK and gC promoters to wild-type levels (Fig. 3). Western blot analysis demonstrated that ICP0-N/X accumulated to wild-type levels in transfected cells (Fig. 4). These data, in combination with our finding that the NH2 terminus of ICP0 activates gene expression in yeast when fused to a heterologous DNA binding domain, support a role for this domain in the activation of transcription from HSV-1 α-gene promoters.
FIG. 3.
Activation of HSV-1 promoters by wild-type and mutant ICP0. Vero cells were cotransfected with plasmids (1 μg) directing the expression of wild-type HSV-1 ICP0, or the indicated mutant ICP0 proteins, and the appropriate reporter plasmids (1 μg). ICP0-N/X lacks aa 3 to 104 of ICP0. ICP0-TIF contains the HSV-1 αTIF-AD in place of aa 2 to 104 of ICP0. Reporter plasmids direct expression of the firefly luciferase gene under the control of the α4 (4p-LUC), α27 (27p-LUC), α22/47 (22/47p-LUC), α0 (0p-LUC), TK (TK-LUC), or gC (gC-LUC) promoter from HSV-1. Total cell extracts were prepared at 48 h posttransfection, and luciferase activities were quantified as described in Materials and Methods. Cells transfected with reporter plasmids only are indicated (No Effector). Levels of transcriptional activation are expressed relative to the basal expression levels derived for each reporter plasmid.
FIG. 4.
Accumulation of wild-type and mutant forms of ICP0 in transfected cells. Vero cells were either mock transfected (Mock) or transfected with plasmids directing the expression of wild-type ICP0 or the indicated mutant proteins (ICP0-N/X or ICP0-TIF). Cell extracts were prepared at 48 h posttransfection, and the relative abundance of these proteins was determined by Western blot analysis using rabbit polyclonal antibody CLU7 as described in Materials and Methods.
To further address the role of the ICP0 NH2 terminus in the activation of gene expression, we next asked whether a heterologous transcriptional activation domain could functionally substitute for this region of ICP0. Accordingly, a construct directing the expression of a fusion protein containing only the αTIF-AD (aa 411 to 490) fused to aa 105 to 775 of ICP0 (ICP0-TIF) was generated. This protein accumulated in transfected Vero cells to wild-type levels (Fig. 4) and activated gene expression from α-gene promoters in transient expression assays to higher levels than ICP0-N/X (Fig. 3). ICP0-TIF activated the TK promoter to wild-type levels but exhibited defects in the activation of the gC promoter (Fig. 3). To address the possibility that the αTIF-AD functioned merely as a stuffer peptide, a construct encoding a fusion protein containing aa 761 to 861 of E. coli β-galactosidase fused to aa 104 to 775 of ICP0 (ICP0-βgal) was generated. In transient expression assays, ICP0-βgal failed to activate reporter gene expression (data not shown). These data suggest that the well-characterized transcriptional activation domain of HSV-1 αTIF can functionally replace the NH2 terminus of ICP0 and further support the conclusion that ICP0 contains an NH2-terminal, promoter-specific transcriptional activation domain.
Construction and analysis of a herpesvirus that directs the expression of ICP0-N/X.
While the results described above suggest a role for the NH2 terminus of ICP0 in the activation of gene expression, they do not establish whether this region is important for the growth and replication of HSV-1. To address this issue, the mutant α0 cDNA pEL0-N/X was introduced into the HSV-1 genome by homologous recombination to generate a recombinant herpesvirus (vEL0-N/X) that directs the synthesis of ICP0-N/X protein during the course of an infection. The recombinant virus vEL0-TIF, which directs the synthesis of ICP0-TIF, was constructed in a similar fashion. The sequence arrangements of the α0 loci of these viruses were verified by Southern blot analysis and DNA sequencing (data not shown).
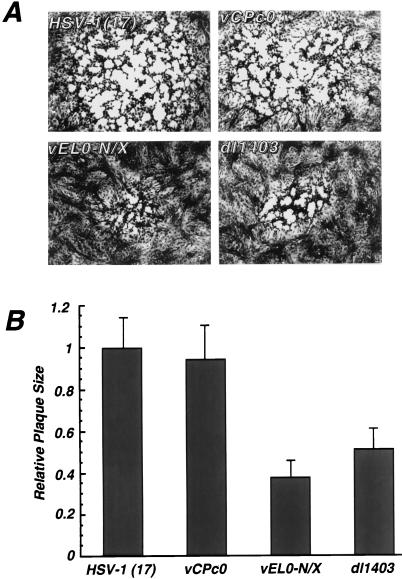
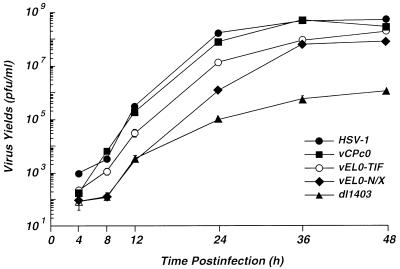
vEL0-N/X exhibits a small-plaque phenotype on Vero cells relative to wild-type HSV-1 or the α0 cDNA virus vCPc0 (Fig. 5). The average size of a plaque generated by vEL0-N/X was similar to that of the α0 deletion virus dl1403 (Fig. 5). This result suggested that deletion of the NH2-terminal acidic domain of ICP0 results in defects in the growth of HSV-1 in tissue culture. To further address the effect(s) of deleting this region of ICP0, the growth kinetics of vEL0-N/X relative to those of wild-type HSV-1, vCPc0, and the α0 null virus dl1403 were examined. Vero cells were infected at an MOI of 0.01 FFU per cell (see below), and virus yields were determined at the indicated times postinfection by titration on the ICP0-complementing cell line L7 (55). This analysis demonstrated that vEL0-N/X exhibits delayed growth kinetics and reduced virus yields relative to wild-type HSV-1 or the α0 cDNA virus vCPc0 (Fig. 6); however, these defects are not as severe as those exhibited by the α0 deletion virus dl1403 (Fig. 6). Because vEL0-N/X has a high FFU/PFU ratio (Table 1) and exhibits growth defects on both Vero and L7 cells, the number of virus particles yielded by cells infected with this virus may be underrepresented in Fig. 6 by approximately 8- to 10-fold. Consistent with the MOI-dependent growth of α0 mutant viruses (61), vEL0-N/X exhibited nearly wild-type growth kinetics and yields in high-MOI infections (data not shown). The growth kinetics of vEL0-TIF at low MOIs were intermediate between what was seen with vEL0-TIF and with wild-type HSV-1 (Fig. 6). Thus, the NH2 terminus of ICP0 is important for the growth of HSV-1 in low-MOI infections.
FIG. 5.
Wild-type and mutant herpesvirus plaque sizes. Vero cells were infected with the indicated wild-type and mutant herpesviruses at an MOI of 0.001. At 60 h postinfection, infected cell monolayers were fixed with 90% methanol and stained with 1% crystal violet. (A) Representative plaques photographed by standard techniques; (B) plaque sizes for wild-type HSV-1, the α0 cDNA virus vCPc0, and the α0 mutants vEL0-N/X and dl1403, expressed as the averages of 40 measurements per virus. The average plaque sizes are expressed relative to that of wild-type HSV-1 strain 17.
FIG. 6.
Growth kinetics of wild-type and α0 mutant herpesviruses. Vero cells were infected at an MOI of 0.01 FFU per cell with wild-type HSV-1, the α0 cDNA virus vCPc0, and the α0 mutant vEL0-N/X, vEL0-TIF, or dl1403. Virus yields at the indicated times postinfection were determined by titration on monolayers of the ICP0-expressing cell line L7 as described in Materials and Methods. Growth curves represent the average of two independent infections, each titrated in duplicate.
TABLE 1.
FFU/PFU ratios for wild-type and mutant viruses
| Virus | Avg FFU/PFU ratioa
|
|
|---|---|---|
| Vero cells | L7 cells | |
| HSV-1 strain 17 | 1.04 | 1.16 |
| vCPc0 | 1.16 | 1.04 |
| vEL0-N/X | 9.43 | 8.18 |
| vEL0-NXR | 1.4 | NDb |
| vEL0-TIF | 2.3 | 2 |
| dl1403 | 137.3 | 1.52 |
Determined in Vero and L7 cells as described in Materials and Methods.
ND, not determined.
To exclude the possibility that the growth defect(s) of vEL0-N/X resulted from extraneous mutations introduced during the construction of this recombinant, the N/X deletions within this virus were replaced with wild-type sequences from an α0 cDNA to create vEL0-NXR. This virus directs the synthesis of wild-type ICP0 from an α0 cDNA. The sequence arrangements of the α0 loci of vEL0-NXR were verified by Southern blot analysis, and restoration of the DNA encoding the wild-type acidic ICP0 NH2 terminus was confirmed by PCR and cycle sequencing (data not shown). Analysis of the growth of this recombinant in low-MOI infections demonstrated that this virus yielded wild-type levels of virus at 10 and 24 h postinfection (data not shown). Thus, the growth defects of vEL0-N/X arise only from deletion of ICP0 aa 3 to 104.
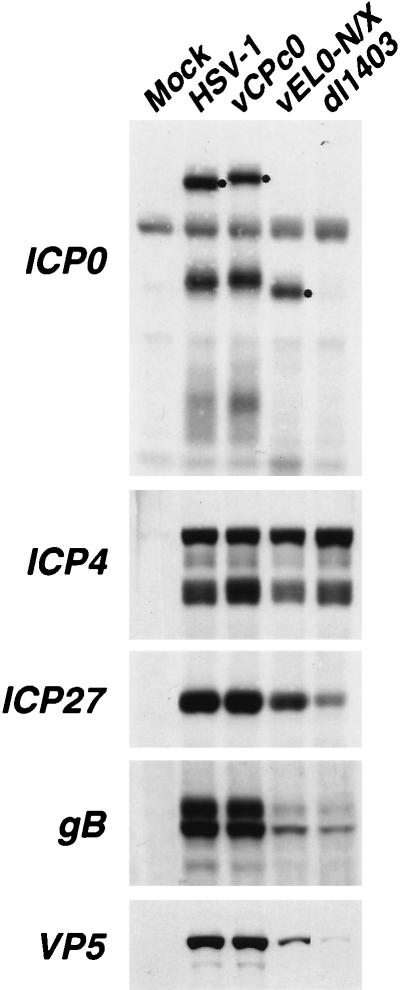
To further address the growth defects of vEL0-N/X, Western blot analysis was performed to determine the abundance of ICP0-N/X in infected cells (Fig. 7). ICP0-N/X accumulated to wild-type levels in cells infected with vEL0-N/X (Fig. 7). Thus, the delayed growth kinetics and reduced virus yields of vEL0-N/X cannot be attributed to defects in the accumulation of this protein. Subsequent analysis of the abundance of representative proteins from each of the three major temporal classes of HSV genes (α, β, and γ) demonstrated that the essential α regulatory protein ICP4 accumulates to wild-type levels in cells infected with vEL0-N/X. However, the abundance of ICP27 and β/γ proteins gB and VP5 was reduced (Fig. 7). These findings are consistent with the intermediate growth defects of vEL0-N/X relative to wild-type HSV-1 and the α0 deletion virus dl1403, as the levels of ICP27, gB, and VP5 in cells infected with dl1403 were lower than those in cells infected with vEL0-N/X (Fig. 7).
FIG. 7.
Accumulation of HSV-1-specified proteins in infected cells. Vero cells were infected with the indicated viruses at an MOI of 1 FFU per cell, and infected cell extracts were prepared at 8 h postinfection as described in Materials and Methods. The relative levels of accumulation of the immediate-early proteins ICP0, ICP4, and ICP27 and the early-late proteins gB and VP5 were determined by Western blot analysis using the mouse monoclonal antibody 58S (anti-ICP4) or the rabbit polyclonal antibody CLU7 (anti-ICP0), CLU38 (anti-ICP27), R69 (anti-gB), or NC-1 (anti-VP5). Secondary antibodies were conjugated to horseradish peroxidase, and immunoblots were developed with the chemiluminescent substrate LumiGLO. ICP0 is marked (•).
As described above, fusion of the αTIF-AD to ICP0 aa 105 to 775 (ICP0-TIF) partially compensated for deletion of the ICP0 acidic NH2 terminus (Fig. 3 and 6). As another measure of ICP0-TIF function, the FFU/PFU ratio of this virus was determined because the ICP0 NH2-terminal deletion virus vEL0-N/X exhibited high FFU/PFU ratios (see below). vEL0-TIF exhibited an intermediate FFU/PFU ratio relative to those of wild-type HSV-1 and vEL0-N/X (Table 1). Thus, the αTIF-AD can partially compensate for deletion of the NH2 terminus in both transfected and infected cells (Fig. 3 and 6; Table 1).
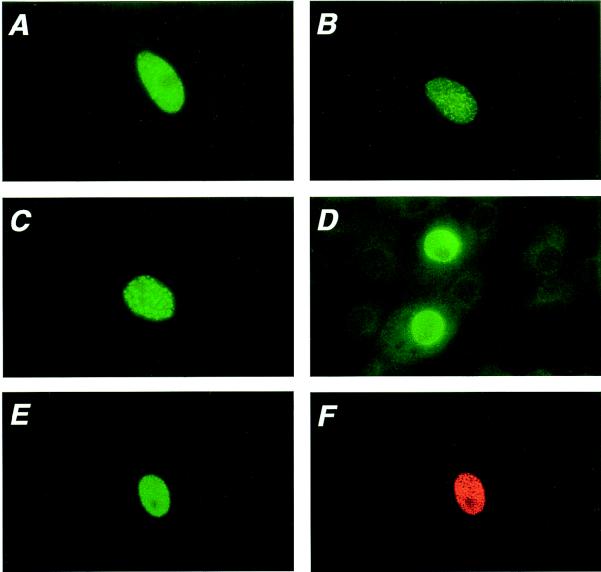
Analysis of the distribution pattern of ICP0-N/X in infected cell nuclei demonstrated that it differed from that of wild-type ICP0 (Fig. 8). Vero cells were infected with wild-type HSV-1 or vEL0-N/X at a low MOI, and ICP0 was detected by indirect immunofluorescence at 8 h postinfection as described in Materials and Methods. Unlike wild-type ICP0, which accumulates as large punctate bodies, the immunofluorescence pattern of ICP0-N/X was more finely dispersed in the nuclei of infected cells (Fig. 8). Next, we asked whether the altered immunofluorescence pattern of ICP0-N/X represented an intrinsic property of this protein, or if it was a result of altered protein-protein interaction(s) between ICP0-N/X and an infected cell protein(s). To address this question, the distribution patterns of ICP0 and ICP0-N/X, in the absence of other virus-specified proteins, were determined in transfected Vero cells (Fig. 8). In transfected cells, ICP0 is localized to characteristic doughnut-like structures whereas ICP0-N/X is more finely dispersed in smaller intranuclear bodies (Fig. 8C and D).
FIG. 8.
Intranuclear distribution of ICP0 and ICP0-N/X in infected and transfected cells. Vero cells were infected with wild-type HSV-1 strain 17 (A) or the α0 mutant vEL0-N/X (B) or were transfected with plasmids encoding HSV-1 ICP0 (C), ICP0-N/X (D), or both proteins (E and F). At 7 h postinfection or 48 h posttransfection, wild-type ICP0 and the NH2-terminal deletion mutant ICP0-N/X were detected by indirect immunofluorescence using rabbit polyclonal antibody CLU7, which recognizes epitopes between aa 312 and 400 of HSV-1 ICP0 (A to D), the affinity-purified rabbit polyclonal antibody α0-N18, which recognizes only the NH2-terminal 18 aa acids of HSV-1 ICP0 and thus fails to recognize ICP0-N/X (E), or the anti-ICP0 mouse monoclonal antibody H1083, which recognizes both ICP0 and ICP0-N/X (F). Panels E and F show the same field of cells. The secondary antibodies used in this analysis were coupled to either fluorescein isothiocyanate (A to E) or rhodamine (F).
ICP0-N/X is a dominant negative protein.
The growth of recombinant herpesviruses with deleterious mutations in the α0 gene can be complemented in L7 cells (55), which express ICP0 under the control of its endogenous promoter/regulatory sequences. However, vEL0-N/X exhibited a small-plaque phenotype when grown on either Vero (Fig. 5) or L7 (data not shown) cells. This finding suggested that ICP0-N/X may be a dominant negative inhibitor of wild-type ICP0. To test this hypothesis, the FFU/PFU ratios for wild-type HSV-1, vCPc0, vEL0-N/X, vEL0-NXR, and the α0 deletion virus dl1403 were determined on Vero and L7 cells. As determined here, the FFU/PFU ratio is a measure of the efficiency with which a virus can infect a cell and initiate the synthesis of immediate-early proteins relative to its ability to complete the lytic virus life cycle. α0 mutant viruses have previously been shown to have high FFU/PFU ratios in noncomplementing cell lines (61). Consistent with these findings, our analysis demonstrated that the ICP0 deletion virus dl1403 has a very high FFU/PFU ratio in Vero cells and a nearly wild-type FFU/PFU ratio in L7 cells (Table 1). The FFU/PFU ratio of vEL0-N/X, however, was found to be significantly higher than those of wild-type HSV-1, vCPc0, and the repaired control virus vEL0-N/X in both Vero and L7 cells (Table 1). These data suggest a dominant negative phenotype for ICP0-N/X.
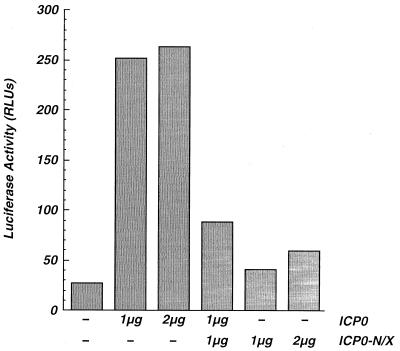
Next, we sought to determine whether ICP0-N/X would act as a dominant negative inhibitor of ICP0-mediated transcriptional activation in transient expression assays. Vero cells were cotransfected with plasmids encoding ICP0, ICP0-N/X, or both ICP0 and ICP0-N/X and with reporter plasmids directing luciferase expression under the control of the HSV-1 α4 promoter. Consistent with previous results, ICP0 activated reporter gene expression whereas ICP0-N/X was less effective at activation of the α-gene promoter reporter (Fig. 9). Coexpression of ICP0 and ICP0-N/X led to a significant reduction in reporter gene expression (Fig. 9). This effect did not result from promoter competition, as no evidence of competition was observed in cells transfected with equivalent amounts (2 μg) of plasmids directing the expression of wild-type ICP0 (Fig. 9). Thus, ICP0-N/X is a dominant negative inhibitor of ICP0 in transfected cells.
FIG. 9.
Effect of ICP0-N/X on transcriptional activation by ICP0. Vero cells were cotransfected with the indicated amounts of plasmids directing the expression of wild-type HSV-1 ICP0, ICP0-N/X, or both proteins and 1 μg of the pCPC-4P-LUC reporter plasmid. Total cell extracts were prepared at 48 h posttransfection, and luciferase activities (relative light units [RLUs]) were quantified as described in Materials and Methods. Cells transfected with reporter plasmids only are indicated (−). The data represent the averages of three independent experiments, each done in quadruplicate.
To further characterize the dominance of ICP0-N/X over ICP0, we examined the immunofluorescence patterns of these two proteins in transfected and infected Vero cells (Fig. 8). Unlike wild-type ICP0, which is located in large discrete punctate structures in the nuclei of transfected cells (Fig. 8C), ICP0-N/X exhibited a heterogeneous micropunctate distribution (Fig. 8D). Cotransfection of Vero cells with constructs directing the expression of ICP0 and ICP0-N/X and subsequent immunodetection of both proteins with monoclonal antibody H1083 suggested that ICP0-N/X altered the immunofluorescence pattern of ICP0, as no large discrete nuclear structures were observed (Fig. 8F). Staining of these cells with an affinity-purified polyclonal antibody that recognizes only aa 1 to 18 of ICP0 (α0-N18) confirmed that the pattern of ICP0 was altered in the presence of ICP0-N/X (compare Fig. 8C and E).
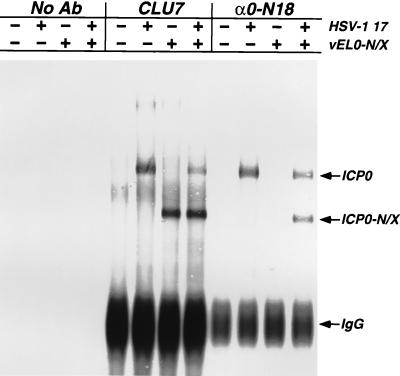
Previous evidence suggests that ICP0 forms oligomers in transfected and infected cells (5, 42). Thus, the formation of oligomers containing mutant and wild-type forms of ICP0 could account for the dominance of ICP0-N/X. This hypothesis is supported by the existence of at least one oligomerization domain within ICP0 (10, 42, 67) and our observation that ICP0-N/X appears to alter the intranuclear distribution of wild-type ICP0 (Fig. 8). Therefore, we determined whether wild-type ICP0 and ICP0-N/X form hetero-oligomers in infected cells (Fig. 10). Vero cells were infected at an MOI of 5 with wild-type HSV-1, vEL0-N/X, or both viruses. At 7 h postinfection, lysates were prepared, ICP0 and ICP0-N/X were immunoprecipitated with antibodies that recognize both proteins (CLU7) or only ICP0 (α0-N18), and the immunoprecipitates were subjected to Western blot analysis using the rabbit polyclonal antibody CLU7. As predicted, the α0-N18 antibody immunoprecipitated wild-type ICP0 exclusively, as this antibody failed to recognize ICP0-N/X (because the interacting epitope was deleted) in cell extracts that did not contain wild-type ICP0 (Fig. 10). However, antibody α0-N18 immunoprecipitated ICP0-N/X from infected-cell lysates containing both ICP0 and ICP0-N/X (Fig. 10). These data reveal that ICP0-N/X oligomerizes with wild-type ICP0 and that deletion of the ICP0 NH2 terminus does not impair oligomerization of the protein. This finding further demonstrates that ICP0 forms oligomers in infected cells and supports the conclusion that ICP0 function is dependent on oligomerization during the lytic life cycle.
FIG. 10.
Coimmunoprecipitation of ICP0 and ICP0-N/X. Vero cells were infected at an MOI of 5 FFU per cell with either wild-type HSV-1, vEL0-N/X, or both viruses. At 7 h postinfection, cell lysates were prepared and proteins were immunoprecipitated as described in Materials and Methods. The antibodies (Ab) used in these immunoprecipitations were rabbit polyclonal antibody CLU7, which recognizes epitopes between aa 312 and 400 of ICP0, and the affinity-purified rabbit polyclonal antibody α0-N18, which recognizes only the NH2-terminal 18 aa of wild-type ICP0. ICP0 and ICP0-N/X were detected by Western blotting using CLU7. IgG, immunoglobulin G.
DISCUSSION
ICP0 is an immediate-early virus regulatory protein that functions as a promiscuous activator of gene expression during the HSV-1 life cycle (4, 6, 7, 15, 17, 19). This report reveals a role for the acidic NH2 terminus of ICP0 in the activation of gene expression from HSV-1 α-gene promoters. The data presented here also suggest that ICP0 possesses more than one transcriptional activation domain. They also demonstrate that the NH2-terminal deletion mutant ICP0-N/X is a dominant negative inhibitor of ICP0’s activation function and that dominance results from the formation of hetero-oligomers. These data support the premise that ICP0 functions as an oligomer in infected cells.
The mechanism(s) underlying ICP0 function remains undefined. Recent data, however, suggest that it acts at the level of transcription (32). ICP0 does not appear to possess any sequence-specific DNA binding activity and thus is unlikely to recognize specific elements within virus gene regulatory sequences. The possibility does exist, however, that ICP0 acts as a coactivator or adapter (1) by interacting with other sequence-specific DNA binding proteins and/or ubiquitous components of the basal transcription machinery. ICP0 interacts with and acts synergistically with ICP4, the major HSV-1 regulatory protein, to mediate the activation of virus gene expression (6, 7, 15, 17, 19, 46, 67, 69).
Transient expression analysis of ICP0 mutants indicates that the carboxy terminus contains a transcriptional activation domain (4, 6, 7, 15, 17, 19). However, the possibility that mutations within this region of ICP0 result in the loss of reporter gene expression by altering the protein’s ability to oligomerize and/or interact with another protein(s) is suggested by the proximity of the major ICP0 oligomerization domain to the carboxy terminus (42). Mutations that disrupt the three-dimensional structure of ICP0, or its intracellular localization, could also result in the loss of ICP0 function. To exclude these possibilities, we performed experiments to identify regions of ICP0 capable of activating gene expression in S. cerevisiae.
These studies demonstrated that the acidic NH2 terminus of ICP0 can act as a transcriptional activation domain. Our results were generated by using proteins containing the NH2 terminus of ICP0 fused to either the GAL4-BD or the bacterial LexA protein and yeast strains containing the appropriate target promoters. Thus, the possibility that our findings represent artifacts resulting from the generation of artificial transcriptional activators or the use of a specific promoter context is unlikely. Moreover, overproduction of fusion proteins containing the ICP0 NH2-terminal region resulted in squelching (25) (data not shown), a finding consistent with data obtained using other transcriptional activators (reference 25 and references therein).
The approach taken here to identify transcriptional activation domains within ICP0 is not without complications. For example, many of the GAL4-BD–ICP0 fusion proteins that contained the carboxy terminus of ICP0 failed to accumulate to detectable levels in yeast. Another complication is that fusion with a DNA binding domain might alter the structure of ICP0 in a way that inactivates it. This possibility became apparent when we attempted to perform similar experiments with mammalian cells. None of the GAL4-BD–ICP0 fusion proteins was able to activate a target promoter containing GAL4 DNA binding sites. A third complication involved the failure of GAL4-BD–ICP0 fusion proteins containing aa 1 to 105 or 1 to 212 of ICP0 to localize to the nucleus despite the presence of a functional nuclear localization signal within the GAL4-BD. This aberrant localization could explain the inability of these proteins to activate transcription from the G5-TK promoter in mammalian cells (Fig. 2).
While the data described above suggest a role for the acidic NH2 terminus of ICP0 in the activation of transcription, they also underscore the potential drawbacks in studying fusion proteins. Therefore, we deleted the NH2 terminus of ICP0 and tested the effects of this change to the protein in transient expression assays and in the context of the virus genome. Our findings demonstrate that this domain specifically enhances the activation of immediate-early virus gene promoters by ICP0. This effect did not result from reduced accumulation of ICP0-N/X (Fig. 4), nor did it result from a gross structural alteration in ICP0-N/X, as it directed wild-type levels of reporter gene expression from the HSV-1 TK and gC promoters (Fig. 3).
Postulating that the acidic nature of the NH2 terminus was responsible for activation, we asked whether the αTIF-AD could functionally substitute for this domain of ICP0. Our results suggest that the αTIF-AD can substitute for the ICP0 NH2 terminus, as the ICP0-TIF fusion protein activated all of the HSV-1 α-gene promoters examined in this study to nearly wild-type levels and a virus expressing this fusion protein replicated with near-wild-type kinetics (Fig. 3 and 6). Restoration of activation did not result just from the presence of the additional amino acids of the αTIF-AD, as a stuffer peptide containing aa 761 to 861 of E. coli β-galactosidase failed to restore activity. Collectively, these data support the following conclusions. (i) The NH2 terminus of ICP0 is a transcriptional activation domain. (ii) ICP0 contains more than one transcriptional activation domain, as ICP0-N/X activates at least one β and one γ HSV-1 promoter, yet is defective in the activation of transcription from four α promoters. (iii) The acidic NH2 terminus of ICP0 is involved in the activation of transcription in a promoter-specific fashion.
To address the role of the ICP0 NH2 terminus in the context of the virus life cycle, a recombinant herpesvirus which directs the synthesis of ICP0-N/X was generated. This virus exhibited growth defects characteristic of other mutant viruses lacking functional ICP0. Specifically, vEL0-N/X formed small plaques in Vero cell monolayers, exhibited MOI-dependent growth, and had a high FFU/PFU ratio. However, these defects were less severe than those of the α0 deletion virus dl1403. The growth and replication defects of vEL0-N/X were not caused by low abundance of ICP0-N/X, as this protein accumulated to wild-type levels in infected cells (Fig. 7), although the intranuclear localization of this mutant form of ICP0 was frequently aberrant. The salient difference between ICP0 and ICP0-N/X was in the distribution patterns of these two proteins: the ICP0-NX protein was more diffuse in its distribution within the nuclei of transfected and infected cells.
The growth and replication defects of α0 mutant viruses, such as dl1403, are complemented in L7 cells (55). However, unlike dl1403 and several other α0 mutants (38), vEL0-N/X exhibits a small-plaque phenotype and high FFU/PFU ratios in L7 cells (Fig. 5 and Table 1). These data suggest that ICP0-N/X is a dominant negative mutant, a conclusion supported by the following observations: (i) in transient expression assays, ICP0-N/X inhibited the activation of reporter gene expression by wild-type ICP0 (Fig. 9), and (ii) ICP0-N/X altered the intranuclear localization of wild-type ICP0 (Fig. 8). Formation of hetero-oligomers between the two proteins might result in the observed effects. In cells coinfected with HSV-1 strain 17 and vEL0-N/X, both proteins were immunoprecipitated with an antibody that recognizes ICP0 but fails to bind ICP0-N/X (Fig. 10). This result was also obtained in cotransfection experiments (data not shown). These data provide the first clear evidence suggesting that (i) no other virus-specified protein(s) is required for ICP0 oligomerization and (ii) ICP0 functions as an oligomer in infected cells.
The results presented here define the acidic NH2-terminal region of ICP0 as a transcriptional activation domain. They also suggest that ICP0 must contain other transcriptional activation domains. Our findings do not define whether ICP0 activates transcription directly or indirectly. However, the observation that ICP0 interacts with and/or alters the activity of cyclin D3 (33), eEF1Bα (32), a DNA-dependent protein kinase (35), and a ubiquitin-specific protease (21) may suggest that ICP0 activates transcription indirectly. Our results indicate that ICP0 can also affect transcription through an acidic transcriptional activation domain. The function of this activation domain is not redundant, as defects resulting from its deletion were not compensated for by other transactivation domains within this protein. Future experiments will be required to more finely map this activation domain and to identify the other transactivation domains within ICP0 as well as the factors that bring ICP0 to gene regulatory regions or otherwise allow it to function as an activator.
ACKNOWLEDGMENT
This study was supported by Public Health Service grant AI-33952 to Saul J. Silverstein.
REFERENCES
- 1.Barlev N, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 2.Bohenzky R A, Papavassiliou A G, Gelman I H, Silverstein S. Identification of a promoter mapping within the reiterated sequences that flank the herpes simplex virus type 1 UL region. J Virol. 1993;67:632–642. doi: 10.1128/jvi.67.2.632-642.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasier A R, Tate J E, Habener J F. Optimized use of the firefly luciferase assay as a reporter gene in mammalian cell lines. BioTechniques. 1989;7:1116–1122. [PubMed] [Google Scholar]
- 4.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63:4579–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Panagiotidis C, Silverstein S. Multimerization of ICP0, a herpes simplex virus immediate-early protein. J Virol. 1992;66:5598–5602. doi: 10.1128/jvi.66.9.5598-5602.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Silverstein S. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J Virol. 1992;66:2916–2927. doi: 10.1128/jvi.66.5.2916-2927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Zhu X, Silverstein S. Mutational analysis of the sequence encoding ICP0 from herpes simplex virus type-1. Virology. 1991;180:207–220. doi: 10.1016/0042-6822(91)90025-7. [DOI] [PubMed] [Google Scholar]
- 8.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien C T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciufo D M, Mullen M A, Hayward G S. Identification of a dimerization domain in the C-terminal segment of the IE110 transactivator protein from herpes simplex virus. J Virol. 1994;68:3267–3282. doi: 10.1128/jvi.68.5.3267-3282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon R F, Schaffer P A. Fine structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein VP175. J Virol. 1980;36:189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DuBridge R B, Tang P, Hsia H C, Phaik-Mooi L, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett R, O’Hare P, O’Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett R D. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol. 1988;202:87–96. doi: 10.1016/0022-2836(88)90521-9. [DOI] [PubMed] [Google Scholar]
- 16.Everett R D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 17.Everett R D. A detailed mutational analysis of VMW110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 1987;6:2069–2076. doi: 10.1002/j.1460-2075.1987.tb02472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett R D. The products of herpes simplex virus type 1 (HSV-1) immediate early genes 1, 2, and 3 can activate HSV-1 gene expression in trans. J Gen Virol. 1986;67:2507–2513. doi: 10.1099/0022-1317-67-11-2507. [DOI] [PubMed] [Google Scholar]
- 19.Everett R D. Trans-activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett R D, Barlow P, Milner A, Luisi B, Orr A, Hope G, Lyon D. A novel arrangement of zinc-binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J Mol Biol. 1993;234:1038–1047. doi: 10.1006/jmbi.1993.1657. [DOI] [PubMed] [Google Scholar]
- 21.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML domain and binds to a herpes virus regulatory protein. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelman I H, Silverstein S. Coordinate regulation of herpes simplex virus gene expression is mediated by the functional interaction of two immediate early gene products. J Mol Biol. 1986;191:395–409. doi: 10.1016/0022-2836(86)90135-x. [DOI] [PubMed] [Google Scholar]
- 23.Gelman I H, Silverstein S. Herpes simplex virus immediate-early promoters are responsive to virus and cell trans-acting factors. J Virol. 1987;61:2286–2296. doi: 10.1128/jvi.61.7.2286-2296.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelman I H, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;344:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 26.Goldin A, Sandri-Goldin R, Levine M, Glorioso J. Cloning of herpes simplex virus type 1 sequences representing the whole genome. J Virol. 1981;38:50–58. doi: 10.1128/jvi.38.1.50-58.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Vol. 194. Pasadena, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 28.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper J, Adami G, Wei N, Keyomarsi K, Elledge S. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 30.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan R, Schaffer P. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Lees-Miller S, Long M, Kilvert M, Lam V, Rice S, Spencer C. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leopardi R, Michael N, Roizman B. Repression of the herpes simplex virus type 1 α4 gene by its gene product (ICP4) within the context of the viral genome is conditioned by the distance and stereoaxial alignment of the ICP4 DNA binding site relative to the TATA box. J Virol. 1995;69:3042–3048. doi: 10.1128/jvi.69.5.3042-3048.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lium E K, Panagiotidis C A, Wen X, Silverstein S J. Repression of the α0 gene by ICP4 during a productive herpes simplex virus infection. J Virol. 1996;70:3488–3496. doi: 10.1128/jvi.70.6.3488-3496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lium E K, Silverstein S J. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J Virol. 1997;71:8602–8614. doi: 10.1128/jvi.71.11.8602-8614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMahan L, Schaffer P A. Repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J Virol. 1990;64:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meredith M, Orr A, Elliott M, Everett R. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 43.Mullen M, Ciufo D, Hayward G. Mapping of intracellular localization domains and evidence for colocalization interactions between the IE110 and IE175 nuclear transactivator proteins of herpes simplex virus. J Virol. 1994;68:3250–3266. doi: 10.1128/jvi.68.5.3250-3266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Hare P, Hayward G S. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. J Virol. 1985;56:723–733. doi: 10.1128/jvi.56.3.723-733.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panagiotidis C A, Huang S C, Canellakis E S. Relationship of the expression of the S20 and L34 ribosomal proteins to polyamine biosynthesis in Escherichia coli. Int J Biochem Cell Biol. 1995;27:157–168. doi: 10.1016/1357-2725(94)00068-m. [DOI] [PubMed] [Google Scholar]
- 46.Panagiotidis C A, Lium E K, Silverstein S J. Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J Virol. 1997;71:1547–1557. doi: 10.1128/jvi.71.2.1547-1557.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phelan A, Carmo-Fonseca M, McLauchlan J, Lamond A I, Clements J B. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Preston C M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979;29:275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 50.Resnick J, Boyd B A, Haffey M L. DNA binding by the herpes simplex virus type 1 ICP4 protein is necessary for efficient down regulation of the ICP0 promoter. J Virol. 1989;63:2497–2503. doi: 10.1128/jvi.63.6.2497-2503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice S A, Knipe D M. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts M S, Boundy A, O’Hare P, Pizzorno M C, Ciufo D M, Hayward G S. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (α4) promoter and a specific binding site for the IE175 (ICP4) protein. J Virol. 1988;62:4307–4320. doi: 10.1128/jvi.62.11.4307-4320.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roizman B, Sears A. The human herpes viruses. New York, N.Y: Raven Press; 1993. [Google Scholar]
- 54.Sacks W R, Greene C C, Aschman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samaniego L, Wu N, DeLuca N A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samaniego L A, Webb A L, DeLuca N A. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J Virol. 1995;69:5705–5715. doi: 10.1128/jvi.69.9.5705-5715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandri-Goldin R M, Mendoza G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 58.Showalter S D, Zwieg M, Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smiley J. Construction in vitro and rescue of a thymidine kinase deficient deletion mutant of herpes simplex virus. Nature. 1980;285:333–335. doi: 10.1038/285333a0. [DOI] [PubMed] [Google Scholar]
- 60.Soliman T, Sandri-Goldin R, Silverstein S. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 62.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Zonneveld A J, Curriden S A, Luskutoff D J. Type 1 plasminogen activator inhibitor gene: functional analysis and glucocorticoid regulation of its promoter. Proc Natl Acad Sci USA. 1988;85:5525–5529. doi: 10.1073/pnas.85.15.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watson R J, Clements J B. A herpes simplex type 1 function continuously required for early and late virus RNA synthesis. Nature. 1980;285:329–330. doi: 10.1038/285329a0. [DOI] [PubMed] [Google Scholar]
- 65.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci USA. 1979;76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilcox K W, Kohn A, Sklyanskaya E, Roizman B. Herpes simplex virus phosphoproteins. I. Phosphate cycles on and off some viral polypeptides and can alter their affinity for DNA. J Virol. 1980;33:167–182. doi: 10.1128/jvi.33.1.167-182.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao F, Schaffer P A. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J Virol. 1994;68:8158–8168. doi: 10.1128/jvi.68.12.8158-8168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu X, Chen J, Silverstein S. Isolation and characterization of a functional cDNA encoding ICP0 from herpes simplex virus type 1. J Virol. 1991;65:957–960. doi: 10.1128/jvi.65.2.957-960.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu Z, Cai W, Schaffer P A. Cooperativity among herpes simplex virus type 1 immediate-early regulatory proteins: ICP4 and ICP27 affect the intracellular localization of ICP0. J Virol. 1994;68:3027–3040. doi: 10.1128/jvi.68.5.3027-3040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]