Abstract
Latent membrane protein 2A (LMP2A) of Epstein-Barr virus (EBV) is expressed on the plasma membrane of B lymphocytes latently infected with EBV and blocks B-cell receptor (BCR) signal transduction in EBV-immortalized B cells in vitro. The LMP2A amino-terminal domain that is essential for the LMP2A-mediated block on BCR signal transduction contains eight tyrosine residues. Association of Syk protein tyrosine kinase (PTK) with LMP2A occurs at the two tyrosines of the LMP2A immunoreceptor tyrosine-based activation motif, and it is hypothesized that Lyn PTK associates with the YEEA amino acid motif at LMP2A tyrosine 112 (Y112). To examine the specific association of Lyn PTK to LMP2A, a panel of LMP2A cDNA expression vectors containing LMP2A mutations were transfected into an EBV-negative B-cell line and analyzed for Lyn and LMP2A coimmunoprecipitation. Lyn associates with wild-type LMP2A and other LMP2A mutant constructs, but Lyn association is lost in the LMP2A construct containing a tyrosine (Y)-to-phenylalanine (F) mutation at LMP2A residue Y112 (LMP2AY112F). Next, the LMP2AY112F mutation was recombined into the EBV genome to generate stable lymphoblastoid cell lines (LCLs) transformed with the LMP2AY112F mutant virus. Analysis of BCR-mediated signal transduction in the LMP2AY112F LCLs revealed loss of the LMP2A-mediated block in BCR signal transduction. In addition, LMP2A was not tyrosine phosphorylated in LMP2AY112F LCLs. Together these data indicate the importance of the LMP2A Y112 residue in the ability of LMP2A to block BCR-mediated signal transduction and place the role of this residue and its interaction with Lyn PTK as essential to LMP2A phosphorylation, PTK loading, and down-modulation of PTKs involved in BCR-mediated signal transduction.
In primary B lymphocytes, cross-linking the B-cell receptor (BCR) leads to an intricate signal cascade including the recruitment and activation of the Src family protein tyrosine kinases (PTKs); subsequent activation and recruitment of other kinases, phosphatases, or adaptor proteins; the hydrolysis of phospholipids; mobilization of intracellular calcium; activation of protein kinase C; activation of nuclear transcription factors; and transcription of BCR signal-specific genes (1, 5, 10, 13, 44). These signal cascades also occur in Epstein-Barr virus (EBV)-negative transformed B-cell lines in vitro, but B-cell lines transformed with EBV are blocked in the ability to transduce signals through the BCR. One EBV gene product, the latent membrane protein 2A (LMP2A), has been demonstrated to be responsible for this phenotype (26–28).
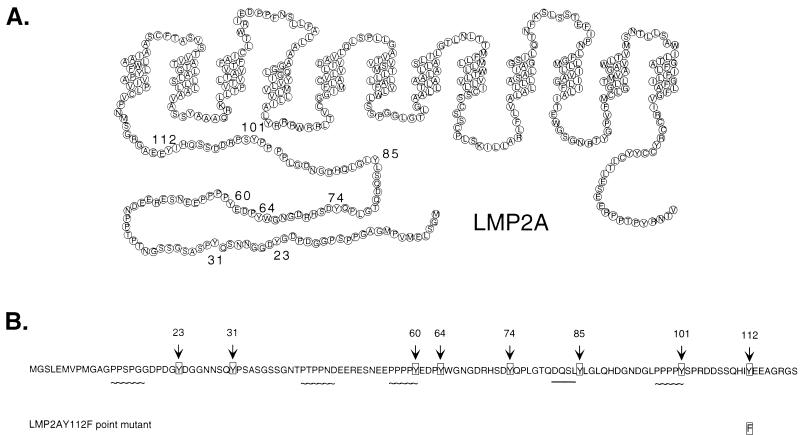
LMP2A is one of nine viral proteins expressed in B cells latently infected with EBV in vitro. LMP2A contains a 119-amino-acid amino-terminal cytoplasmic domain, 12 hydrophobic transmembrane domains, and a 27-amino-acid cytoplasmic carboxyl domain and is expressed in aggregates in the plasma membranes of latently infected B cells (Fig. 1) (21). The amino-terminal domain of LMP2A includes eight tyrosine residues (20), two of which form an immunoreceptor tyrosine-based activation motif (ITAM) (4, 37). This amino-terminal domain has been shown to be tyrosine phosphorylated and is necessary for LMP2A association with the Src family PTKs and the Syk PTK (3, 20, 26). Each phosphorylated tyrosine residue provides a potential binding site for cellular proteins containing Src homology 2 (SH2) domains. SH2 domains are noncatalytic domains conserved among cytoplasmic signaling proteins which bind tyrosine-phosphorylated proteins (34, 38).
FIG. 1.
LMP2A structure and amino acid sequence indicating important motifs of the LMP2A amino-terminal domain. (A) Schematic of the predicted structure of LMP2A in the B-cell plasma membrane. LMP2A contains a 119-amino-acid amino-terminal cytoplasmic domain, 12 hydrophobic transmembrane domains, and a 27-amino-acid cytoplasmic carboxyl domain and is expressed in aggregates in the plasma membranes of latently infected B cells. Numbers denote the locations of the eight tyrosine residues in the LMP2A amino-terminal domain. (B) Amino acid sequence of the LMP2A amino-terminal domain, with the eight tyrosine residues (arrows), four proline-rich motifs (∼∼∼∼∼), and DQSL motif (▪▪▪▪▪) indicated. Each of the eight LMP2A amino-terminal Y residues was mutated to an F residue in pLMP2A cDNA expression vector constructs, including a double Y-to-F mutation at LMP2A residues Y74 and Y85. Four LMP2A deletion mutations, lacking LMP2A residues 21 to 36, 21 to 64, 21 to 85, and 80 to 112, were also incorporated into pLMP2A cDNA expression vector constructs. In addition, multiple LCLs incorporating a Y-to-F mutation at LMP2A Y112 were generated.
LMP2A was first shown to block normal BCR signal transduction in the EBV-negative B-cell line BJAB. In BJAB cells expressing LMP2A and no other EBV gene products, intracellular calcium was not mobilized following BCR cross-linking (28). Studies using EBV-transformed lymphoblastoid cell lines (LCLs), referred to as EBV+LMP2A+ LCLs, demonstrate a similar block in calcium mobilization after BCR cross-linking as well as a block in protein tyrosine phosphorylation and nuclear gene transcription following BCR cross-linking (26–28). In addition, BCR cross-linking failed to activate cellular signal transducers such as Lyn, Syk, phosphatidylinositol 3-kinase (PI3-kinase), phospholipase Cγ2, (PLCγ2), Vav, mitogen-activated protein kinase (MAPK), and Shc in the EBV+LMP2A+ LCLs (26). In contrast, parallel BCR cross-linking studies of LCLs with null mutations in LMP2A, referred to as EBV+LMP2A− LCLs, resulted in normal BCR signal transduction, as measured by the induction of tyrosine phosphorylation, mobilization of intracellular calcium, and induction of lytic viral replication (26, 27). These results indicated that the amino-terminal domain of LMP2A was sufficient for the block in BCR-mediated signal transduction observed in EBV+LMP2A+ LCLs.
The importance of the LMP2A amino-terminal domain in blocking BCR signal transduction was confirmed by the analysis of EBV-infected LCLs with deletion mutations within the LMP2A amino-terminal domain. We constructed three deletions which removed amino acids 21 to 36, 21 to 64, or 21 to 85 from the LMP2A amino-terminal domain. Amino acids 21 to 36 were dispensable, whereas amino acids 37 to 85 were essential for full LMP2A function (11). The importance of the LMP2A ITAM in the LMP2A-mediated block in BCR signal transduction was confirmed by the analysis of LCLs with tyrosine-to-phenylalanine point mutations in the LMP2A ITAM. Mutation of either tyrosine restored normal BCR signal transduction and prevented the binding of the Syk PTK to LMP2A (12).
In this study, a tyrosine (Y)-to-phenylalanine (F) mutation was engineered into the LMP2A gene at amino acid 112 (Y112F) and incorporated into both LMP2A cDNA expression vectors as well as the viral genome to generate LCLs infected with LMP2AY112F recombinant virus (11, 12). The LMP2AY112F LCLs were analyzed for the ability to transduce signals through the BCR and the effect of the LMP2AY112F mutation on the Lyn and Syk PTKs.
MATERIALS AND METHODS
Cell lines and cell culture.
All cell lines were maintained in RPMI 1640 medium containing 10% inactivated fetal bovine serum, 1,000 U of penicillin per ml, and 1,000 μg of streptomycin (Sigma, St. Louis, Mo.) per ml (complete RPMI). BJAB is an EBV-negative B-lymphoma cell line (24). B95-8 is an EBV-infected marmoset cell line that is partially permissive for viral replication (29, 30). HPB.ALL is an acute lymphoblastic leukemia T-cell line. The HH514-16 subclone of the original EBV-infected P3HR1 (P3JHR1) Burkitt’s lymphoma cell line (a gift of George Miller) has EBV genomes with only a single type of DNA, from which the segment encoding EBNA2 and the last two exons of EBNALP are deleted (14). WT1 and WT4 are EBV+LMP2A+ LCLs, and ES1 and ES4 are EBV+LMP2A− LCLs (27). Primary human mononuclear cells were obtained from blood samples of healthy donors by centrifugation over a cushion of Ficoll-Paque (Pharmacia, Piscataway, N.J.). For EBV-positive donors, T lymphocytes were removed as previously described (23).
Antibodies.
Whole goat anti-human immunoglobulin (Ig) for cross-linking surface Ig (sIg) experiments was purchased from Southern Biotechnical Associates (Santa Cruz, Calif.), and goat anti-human F(ab′)2 was purchased from Jackson ImmunoResearch Laboratories (West Grove, Pa.). The rat monoclonal anti-human LMP2A antibody (14B7) has been previously described (11). The mouse monoclonal antihemagglutinin (HA1) epitope antibody (12CA5) was obtained from BAbCO (Richmond, Calif.). The anti-Lyn mouse monoclonal antibody was purchased from Transduction Laboratories (Lexington, Ky.), and the anti-Lyn rabbit polyclonal antibody was purchased from Upstate Biotechnology Inc. (Lake Placid, N.Y.). The mouse monoclonal antibody to Syk (4D10) and the antiphosphotyrosine (APT) antibody (PY20) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). The mouse monoclonal antibody to the BZLF1 protein (BZ1) has been described previously (48). All horseradish peroxidase (HRP)-linked secondary antibodies were purchased from Amersham (Arlington Heights, Ill.) except for the NeutrAvidin-HRP, purchased from Pierce (Rockford, Ill.). The donkey anti-rat secondary antibody Cy3 used for immunofluorescence was purchased from Jackson.
Construction of plasmids.
The LMP2A cDNA expression vectors were generated by a PCR-based strategy and incorporated into pLMP2A (20). All LMP2A mutations were sequenced to confirm the presence of the desired mutation and the absence of any mutations introduced by PCR. pSVNaeZ, used to induce lytic EBV replication, and EcoRI-A, used to rescue the EBNA2 and EBNALP mutations contained in the P3HR1 cell line, have been described previously (23, 40). The genomic LMP2AY112F mutation was generated by using a PCR-based strategy, and a unique PmlI restriction site adjacent to the Y112 codon was also generated. The recombinant LMP2A fragment containing both the tyrosine mutation and the PmlI site was incorporated into pRH6, which contains a KpnI (B95-8, bp 161097)-to-EcoRI (B95-8, bp 1) fragment from B95-8 EBV DNA. All recombinant LMP2A constructs were sequenced to confirm the presence of the desired tyrosine-to-phenylalanine mutation, the incorporation of the PmlI site, and the absence of any mutations introduced by PCR.
Transfection.
DNA used for transfections was banded twice on CsCl density gradients. Recombinant cell lines were generated by transfecting 107 P3HR1 cells in 0.4 ml of complete RPMI with 15 μg of pSVNaeZ, 7 μg of EcoRI-A, and 25 μg of recombinant LMP2A DNA. BJAB cells (107) were transfected similarly with 50 μg of each pLMP2A cDNA and labeled with [35S]methionine as described previously (20). Cells were pulsed once with a Gene Pulser (Bio-Rad, Hercules, Calif.) at 200 V and 960-μF capacitance in a 0.4-cm electrode gap cuvette (Bio-Rad) and immediately diluted in 10 ml of complete RPMI. The induction of lytic EBV replication in LCLs was performed similarly except that these cells were pulsed at 230 V with 25 μg of pSVNaeZ and diluted in complete RPMI containing 20 ng of 12-O-tetradecanolylphorbol-13-acetate per ml.
Generation of LMP2AY112F LCLs.
PCR-mediated mutagenesis was used to create an EBV DNA fragment encoding LMP2A with a tyrosine-to-phenylalanine point mutation at amino acid 112 of LMP2A. The recombinant LMP2A construct was verified by sequencing and then incorporated into virus through a strategy using the transformation-incompetent, replication-competent EBV deletion mutant, P3HR1. Recombinant virus was generated by the cotransfection of the P3HR1 cell line with pSVNaeZ DNA, to induce lytic viral replication; EcoRI-A DNA, to rescue the EBNA2 and EBNALP deletions in the P3HR1 genome; and the recombinant LMP2A DNA fragment (22). The recombinant P3HR1 virus was used to infect primary blood mononuclear cells (PBMCs) or purified B lymphocytes in culture as previously described (11). Clones emerged 3 to 5 weeks after initial plating and were screened by PCR for incorporation of the LMP2AY112F point mutation at 6 to 8 weeks.
PCR and Southern blot verification of LMP2AY112F mutant LCLs.
Genomic DNA from LCLs was prepared (40) and amplified in a 25-μl volume for 40 cycles as previously described (22). Primers 5′LMP1 (CTAGGCGCACCTGGAGGTGG) and 3′LMP1 (AGTCAGTCAGGCAAGCCTAT) were used to screen LCLs containing the 81-bp smaller LMP1 gene present in the EBV genomic DNA fragment in which the LMP2A mutation was incorporated. The LMP1 gene is adjacent to the LMP2A gene within the EBV genome. After this initial screen, primers 5′LMP2A (CTGCTGCAGCTATGGGGTCC) and 3′112-LMP2A (TCCTCTGCCCGCTTCTTCGA) were used to specifically amplify DNA from LCLs incorporating the LMP2AY112F point mutation. Ethidium bromide-stained PCR products were viewed after electrophoresis through 1.5% EEO agarose (Fisher, Pittsburgh, Pa.) to determine the sizes of the amplified products.
PCR-identified LMP2AY112F mutant LCLs and wild-type controls were further verified by Southern blot hybridization. A 1,872-bp fragment, generated from a BglI digest of pRL49 (22), was isolated and used as a 32P-labeled hybridization probe. Genomic DNA was prepared by cell lysis, proteinase K treatment, phenol extraction, and ethanol precipitation (43). Prepared DNAs from representative LCLs were digested with BglI alone or with both BglI and PmlI. The digested DNAs were separated by electrophoresis on a 1% agarose gel, transferred to a nylon membrane (GeneScreen; NEN, Boston, Mass.), and probed as previously described (43). Both digests verified the correct restriction sites present in the LMP2A wild-type and mutant LCLs, and the BglI-PmlI double digest verified the unique PmlI site present only in the LMP2AY112F mutants.
The verified LMP2AY112F mutant LCLs and wild-type controls were further characterized for sIg expression by flow cytometry using a fluorescein isothiocyanate-linked goat anti-human Ig diluted 1:50 (Southern Biotech, Birmingham, Ala.) as previously described (28).
Cellular activation and preparation of lysates.
Tissue culture cells (cell numbers are indicated in figure legends) were resuspended and equilibrated in serum-free RPMI for 15 min at 37°C. Cells were stimulated with 25 μg of goat anti-human Ig (Southern Biotech) per ml for the indicated times, and cells were immediately lysed in 1% Nonidet P-40 (NP-40) lysis buffer (1% NP-40, 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 10 μg each of pepstatin and leupeptin per mL, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate). The insoluble materials were removed by centrifugation at 4°C, and cleared supernatants were processed for immunoprecipitations or immunoblotting.
Immunoprecipitations and immunoblotting.
Cleared lysates were incubated with the appropriate antisera for 1 h at 4°C; immune complexes were captured with either protein A- or protein G-Sepharose (Pharmacia) for 1 h as indicated in the figure legends and then washed four times with 1% NP-40 lysis buffer. Immunoprecipitated proteins were then resuspended in 2× sodium dodecyl sulfate (SDS) sample buffer, heated at 70°C for 5 min, and separated by electrophoresis through SDS-polyacrylamide gels (percentages are indicated in the figure legends) (SDS-PAGE).
For immunoblotting, immune complex materials were electrophoresed and transferred to either nitrocellulose or Immobilon, as indicated in the figure legends. Membranes were blocked in 3 to 5% milk for 1 h at room temperature and then incubated in primary antibody, diluted in Tris-buffered saline-Tween (TBST), for 1 h at room temperature. Membranes were washed three times in TBST, incubated with a HRP- or biotin-linked secondary antibody for 30 min at room temperature, and washed four times in TBST, and proteins were detected by enhanced chemiluminescence (ECL; Pierce, Rockford, Ill.).
In vitro kinase assays.
Lyn and Syk immunoprecipitation reactions were done as described above, and the immune complexes were washed four times with 1% Triton X-100 lysis buffer. Immune complexes were further washed twice with kinase buffer (50 mM Tris [pH 7.4], 10 mM MgCl2, 10 mM MnCl2). After washing, the immune complexes were resuspended in a 25-μl volume of kinase buffer labeled with 10 μCi of [γ-32P]ATP per sample and incubated at room temperature for 20 min. Reactions were stopped with the addition of 2× SDS sample buffer; the samples were heated at 70°C for 5 min and loaded onto SDS–6% polyacrylamide gels. The 32P-labeled products were visualized by autoradiography of the dried gels.
Calcium mobilization.
Tissue culture cells (3 × 106) were resuspended in loading buffer (145 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM glucose, 10 mM HEPES, 1% bovine serum albumin, 2.5 mM probenicid) and loaded with 2 mM calcium-binding dye (fluo-3-acetoxymethyl ester [fluo-3]; Molecular Probes, Eugene, Oreg.) for 30 min at room temperature. After loading, the cells were washed twice and resuspended in loading buffer. The prepared cells were then measured for fluorescence after BCR cross-linking with goat anti-human Ig F(ab′)2 antibody (Jackson) in a Perkin-Elmer (Norwalk, Conn.) LS-5B luminescence spectrometer. Excitation and emission values for fluo-3 are 505 and 530 nm, respectively. Autofluorescence (or Fmin [minimal fluorescence]) for each LCL was also determined by performing the above procedure without adding fluo-3 dye. Baseline fluorescence (F) was recorded for 1 min, and maximal fluorescence (Fmax) was measured by addition of digitonin (40 μM). Calcium concentration (nanomolar) was calculated according to the formula [Ca2+] = 400 (F − Fmin)/(Fmax − F) (18, 25).
Immunofluorescence.
Immunofluorescence of LCLs was performed as previously described (21). In brief, LCLs were fixed to glass slides with acetone at −20°C for 5 min, blocked with 20% normal goat serum for 10 min at room temperature, incubated with the anti-LMP2A rat monoclonal antibody 14B7 diluted 1:1,000, incubated with a goat anti-rat secondary antibody Cy3 diluted 1:1,000 (Jackson) for 30 min, washed in phosphate-buffered saline and viewed with a Zeiss Axioscope fluorescence microscope.
RESULTS
Site-specific cDNA mutation of the LMP2A amino-terminal domain.
PCR-mediated mutagenesis was used to create LMP2A cDNA expression vector constructs with point mutations and deletion mutations in the LMP2A amino-terminal domain. Each of the eight Y residues located within this 119-amino-acid domain was changed to an F residue, including a double Y-to-F mutation at LMP2A residues Y74 and Y85. The LMP2A deletion mutations eliminated LMP2A residues 21 to 36, 21 to 64, 21 to 85, and 80 to 112. In addition, an 11-amino-acid peptide from the influenza virus HA1 protein that is reactive with monoclonal antibody 12CA5 was added to the carboxyl terminus of each LMP2A cDNA construct to facilitate analysis of the cDNA mutants. All of the recombinant LMP2A constructs were verified by sequencing (data not shown) and then incorporated into the expression vector pLMP2A (20). Figure 1 shows the amino acid sequence of the LMP2A amino-terminal domain, with the eight tyrosine residues highlighted.
Loss of LMP2A phosphorylation and Src family PTK association in the Y112 LMP2A mutant.
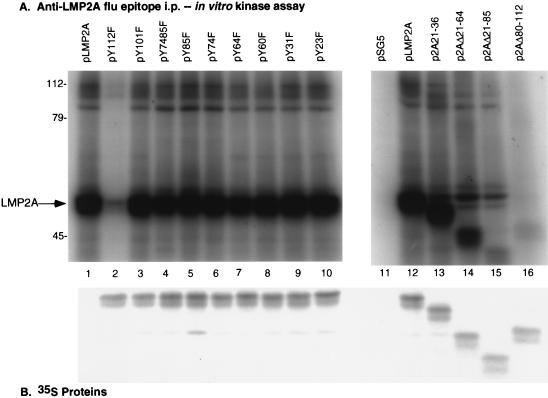
To determine which sequences of the LMP2A amino-terminal domain were responsible for LMP2A associations with cellular proteins and necessary for LMP2A phosphorylation, the epitope-tagged wild-type and mutant LMP2A cDNA expression vectors were transfected into the EBV-negative B-lymphoma cell line BJAB, labeled with [35S]methionine, lysed in NP-40, and immunoprecipitated with the anti-HA1 antibody 12CA5. One-half of the immunoprecipitated proteins were separated by SDS-PAGE, and the immunoprecipitated 35S-labeled LMP2A proteins were visualized by autoradiography (Fig. 2B). As expected, no 35S-labeled LMP2A was immunoprecipitated in vector control (pSG5)-transfected cells (Fig. 2B, lane 11), while approximately equal amounts of LMP2A were detected in each of the LMP2A transfections (Fig. 2B, lanes 2 to 10 and 12 to 16). The LMP2A protein in the deletion mutants migrated more rapidly than wild-type LMP2A, reflecting the size of each deletion.
FIG. 2.
LMP2A expression and LMP2A phosphoprotein immune complexes in transfected BJABs. Results of in vitro immune complex kinase reactions and 35S-labeled proteins from BJAB cells transfected with wild-type pLMP2A and pLMP2A deletion and point mutants are shown. BJABs cells (107) were transfected with the influenza virus (Flu) HA1 epitope-tagged LMP2A expression vectors, labeled with [35S]methionine, lysed in 1% NP-40, immunoprecipitated (i.p.) with anti-HA1 antibody 12CA5, and captured with protein G-Sepharose, and the immunoprecipitates were divided equally. The first half was separated by SDS-PAGE (7% gel) and transferred to nitrocellulose, and the 35S-labeled LMP2A protein was visualized by autoradiography (B). LMP2A expression was detected in all of the LMP2A-transfected cells (B, lanes 2 to 10 and 12 to 16), while no LMP2A was detected in the control pSG5 transfected cells (B, lane 11). The other half of each immune complex was 32P labeled in vitro by an immune complex kinase reaction and separated by SDS-PAGE (7% gel), and the resulting 32P-labeled proteins were visualized by autoradiography (A) after blocking of the 35S-labeled proteins by two sheets of film. A number of phosphorylated proteins complex with both wild-type and mutant pLMP2A proteins (A, lanes 1 to 10 and 12 to 16), including LMP2A (arrow), 72-kDa Syk, and 53- to 60-kDa Src family PTKs, as well as unidentified proteins of 96, 104, 110, and 112 kDa. The pY112 and p2AΔ80-112 mutants demonstrate a complete reduction of total phosphorylation (A, lanes 2 and 16). No proteins were detected in the control pSG5-transfected cells (A, lane 11). Protein standards are indicated at the left in kilodaltons.
In vitro immune complex kinase reactions were performed with the other half of the LMP2A immunoprecipitated proteins, and the resulting 32P-labeled proteins were separated by SDS-PAGE and visualized by autoradiography (Fig. 2A). Only the 32P-labeled proteins were visualized, because the 35S signal was blocked by two sheets of film. As expected, no phosphoproteins were detected in the in vitro kinase reaction of the vector control (pSG5)-transfected cells (Fig. 2A, lane 11). Multiple phosphorylated proteins coimmunoprecipitated with pLMP2A and mutant pLMP2A proteins (Fig. 2A), including 72-kDa Syk and the 53- to 60-kDa Src family PTKs, as well as unidentified proteins of 96, 104, 110, and 112 kDa. In addition, phosphorylated LMP2A was clearly evident migrating at 54 kDa (Fig. 2A, arrow) or decreasingly less according to the size of each LMP2A deletion. The level of LMP2A phosphorylation decreased in the LMP2A deletion mutant constructs: p2AΔ21-64 showed reduced LMP2A phosphorylation, p2AΔ21-85 showed even less phosphorylation, and LMP2A phosphorylation in p2AΔ80-112 was nearly undetectable (Fig. 2A, lanes 14 to 16). The pY112F mutant also had nearly undetectable levels of LMP2A phosphorylation, as seen in the p2AΔ80-112 mutant (Fig. 2A, lanes 2 and 16). In addition, fewer phosphorylated proteins were immunoprecipitated in the pY112F-transfected cells, and the level of phosphorylation was less than in the other in vitro kinase reactions (Fig. 2A), although all reactions were carried out with similar 35S-labeled LMP2A protein levels (Fig. 2B). The Src family PTKs, which migrate just above LMP2A, and a protein with a molecular mass of approximately 96 kDa appeared to be reduced or absent from the pY112F LMP2A immune complex. The in vitro kinase reactions of the cells transfected with pY74F, pY85F, and pY7485F did not contain the 72-kDa Syk PTK. These results are in agreement with previous work that identified the LMP2A ITAM, LMP2A residues Y74 and Y85, as the site of Syk binding to LMP2A (12). The in vitro kinase reaction of the pY60F-transfected cells appeared to be missing a protein of approximately 112 kDa (Fig. 2A, lane 8).
The in vitro kinase reactions of the four LMP2A deletion mutants supported the associations of the same specific proteins with regions of LMP2A as was seen with the LMP2A tyrosine point mutants. The p2AΔ21-36 deletion mutant, which lacks LMP2A residues Y23 and Y31, appeared to contain the same immunoprecipitating proteins as the pY23F and pY31F point mutant immune complexes, and all three mutants appeared to contain the same proteins as seen in immunoprecipitates with wild-type LMP2A (Fig. 2A, lane 13). Therefore, the loss of residues 21 to 36, or specifically Y23 or Y31, did not seem to affect LMP2A associations with cellular proteins. The in vitro kinase reaction of the p2AΔ21-64 deletion mutant, which lacks Y23, Y31, Y60, and Y64, was missing the same 112-kDa protein as observed in the pY60F immunoprecipitate (Fig. 2A, lane 14). The p2AΔ21-85 deletion mutant, which lacks Y23, Y31, Y60, Y64, Y74, and Y85, lost the associations of both the 72-kDa (Syk) and 112-kDa proteins (Fig. 2A, lane 15). The lost LMP2A association with the 112-kDa protein was consistent with the loss of LMP2A residue Y60, and the missing 72-kDa (Syk) protein was consistent with the loss of LMP2A residues Y74 and Y85, as was demonstrated in the in vitro kinase reactions of the pY60F, pY74F, and pY85F LMP2A point mutant-transfected cells. The in vitro kinase reaction of the p2AΔ80-112 deletion mutant, which lacks Y112, Y101, and Y85, contained fewer 32P-labeled proteins than observed in wild-type LMP2A immunoprecipitates (Fig. 2A, lanes 12 and 16), although both samples contained similar levels of LMP2A protein in the immunoprecipitates (Fig. 2B, lanes 12 and 16). The Src family PTKs, which migrated as a three bands of approximately 53, 56, and 60 kDa, were clearly absent from both p2AΔ80-112 and pY112F in vitro kinase reaction (Fig. 2A, lanes 2 and 16). The presence of the Src family PTKs was clearly evident in the LMP2A deletion mutant constructs p2AΔ21-64 and p2AΔ21-85, where the smaller LMP2A protein no longer comigrated with the Src PTKs (Fig. 2A, lanes 14 and 15). In summary, these data indicate that LMP2A residue Y60 is important for LMP2A association with a 112-kDa phosphorylated protein, Y74 and Y85 are important for 72-kDa Syk association with LMP2A as previously shown (12), and Y112 is critical for the 53- to 60-kDa Src family PTK association with LMP2A.
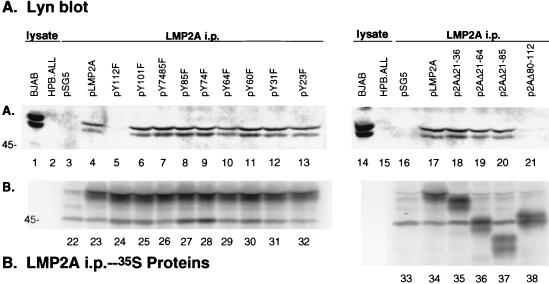
Mutation of Y112 in the LMP2A amino-terminal domain prevents the association of the Lyn PTK with LMP2A.
To investigate the tyrosine-specific association of the Lyn PTK with LMP2A, HA1 epitope-tagged LMP2A cDNA expression vectors were transfected into BJAB cells, labeled with [35S]methionine, lysed in NP-40, and immunoprecipitated with the anti-HA1 antibody 12CA5. Autoradiography of the transferred proteins confirmed that similar amounts of LMP2A immunoprecipitated from each LMP2A-expressing transfection (Fig. 3B, lanes 23 to 32 and 34 to 38), and no LMP2A expression was detected in the control pSG5-transfected BJABs (Fig. 3B, lanes 22 and 33). The membranes were then immunoblotted with an anti-Lyn polyclonal antiserum to detect Lyn coimmunoprecipitation with LMP2A (Fig. 3A). Lyn is the most abundant Src family PTK expressed in BJAB cells (2). Both p56 and p53 forms of Lyn readily coimmunoprecipitated with wild-type LMP2A in BJABs transfected with a wild-type pLMP2A cDNA construct (Fig. 3A, lanes 4 and 17). The two forms of Lyn are derived from alternative splicing by utilizing an internal splice donor site in the Lyn mRNA (47). Lyn was not detected in pSG5 control immunoprecipitates (Fig. 3A, lanes 3 and 16). Lyn also coimmunoprecipitated with pLMP2A constructs containing LMP2A tyrosine point mutations (pY101F, pY7485F, pY85F, pY74F, pY64F, pY60F, pY31F, and pY23F [Fig. 3A, lanes 6 to 13]), as well as with pLMP2A deletion mutants (p2AΔ21-36, p2AΔ21-64, and p2AΔ21-85 [Fig. 3A, lanes 18 to 20]). However, Lyn coimmunoprecipitation with LMP2A was barely detectable in pY112F-transfected BJABs (Fig. 3A, lane 5) or the p2AΔ80-112-transfected BJABs (Fig. 3A, lane 21). Both forms of Lyn were readily detected in control cellular lysates from BJAB cells (Fig. 3A, lanes 1 and 14), while no Lyn expression was detected in cellular lysates from the T-cell line HPB.ALL (Fig. 3A, lanes 2 and 15). A more detailed analysis, performed by titrating BJAB cell lysates and comparing levels of immunoprecipitating p56 and p53 with levels of Lyn bound to LMP2A in immune complexes, demonstrated that the two forms of Lyn bound equally well to LMP2A in amounts similar to their relative abundance in BJAB cells (data not shown). These data indicate that LMP2A tyrosine 112 is the location of Lyn PTK association with LMP2A. Without the tyrosine residue at 112, Lyn does not associate with LMP2A.
FIG. 3.
Lyn binds to Y112 of LMP2A. BJAB cells (107) were transfected with HA1 epitope-tagged LMP2A expression vectors, labeled with [35S]methionine, lysed in 1% NP-40, immunoprecipitated (i.p.) with the anti-HA1 antibody 12CA5, captured with protein G-Sepharose, separated by SDS-PAGE (7% gel), and transferred to nitrocellulose, and the immunoprecipitated LMP2A protein was visualized by autoradiography (B). LMP2A expression within the transfected BJAB cells was detected in all of the LMP2A-transfected cells (B, lanes 23 to 32 and 34 to 38), while no LMP2A was detected in the control pSG5-transfected cells (B, lanes 22 and 33). The membrane was then immunoblotted with a polyclonal Lyn antiserum and incubated with an HRP-conjugated secondary antibody, and proteins were detected by ECL (A). Lyn coimmunoprecipitated with LMP2A in BJAB cells transfected with the wild-type pLMP2A expression vector (A, lanes 4 and 17) and not with control pSG5 vector (A, lanes 3 and 16). Lyn also coimmunoprecipitated with pLMP2A constructs containing tyrosine-to-phenylalanine point mutations at LMP2A tyrosine residues (Y101F, Y7485F, Y85F, Y74F, Y64F, Y60F, Y31F, and Y23F [A, lanes 6 to 13]), as well as pLMP2A deletion mutants that lack LMP2A residues 21 to 36, 21 to 64, and 21 to 85 (A, lanes 18 to 20). However, Lyn did not coimmunoprecipitate with LMP2A in the Y112F point mutant (A, lane 5) or the deletion mutant lacking residues 80 to 112 (lane 21). BJAB and HPB.ALL cellular lysates were included as positive and negative controls for Lyn expression, respectively (A, lanes 1, 2, 14, and 15). Protein standards are indicated at the left in kilodaltons.
Generation of LCLs infected with LMP2AY112F EBV.
The LMP2AY112F mutation was incorporated into the EBV genome through a strategy previously described (22). The generated LMP2AY112F recombinant virus was used to infect PBMCs to generate LCLs as previously described (11). The generated LCLs were screened by PCR to identify the presence of the LMP2AY112F mutation within the infecting EBV genome, and each LMP2AY112F LCL was further verified for correct incorporation of the recombinant LMP2A DNA fragment by Southern blot hybridization (data not shown). Multiple cell lines were transformed by the LMP2AY112F recombinant virus, and these EBV+LMP2AY112F LCLs are characterized in this study.
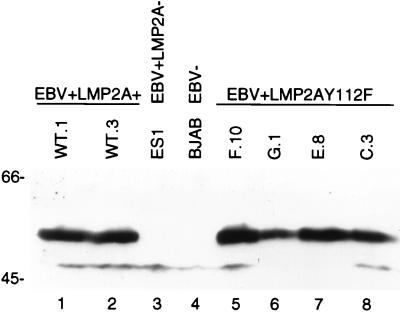
LMP2A protein expression in EBV+LMP2AY112F LCLs.
Expression of LMP2A in the generated EBV+LMP2AY112F LCLs was confirmed by immunoblot analyses using the LMP2A-specific antibody 14B7. Representative data are shown in Fig. 4. LMP2A expression in two EBV+LMP2A+ LCLs was clearly evident (Fig. 4, lanes 1 and 2), while no LMP2A was detected in the negative control EBV+LMP2A− LCL, ES1 (Fig. 4, lane 3), or the EBV− cell line BJAB (Fig. 4, lane 4). LMP2A expression in four representative EBV+LMP2AY112F LCLs (Fig. 4, lanes 5 to 8) was similar to the LMP2A expression in EBV+LMP2A+ LCLs. Immunofluorescence microscopy using anti-LMP2A antibodies verified that the LMP2A subcellular protein localization within the EBV+LMP2AY112F LCLs was similar to LMP2A subcellular localization in EBV+LMP2A+ LCLs (data not shown). These data demonstrate the similar expression levels and subcellular localization of LMP2A protein in EBV+LMP2A+ and EBV+LMP2AY112F LCLs.
FIG. 4.
LMP2A expression in EBV+LMP2A+ and EBV+LMP2AY112F LCLs. NP-40 lysates of LCLs (5 × 106 cells/lane) were separated by SDS-PAGE (8% gel), transferred to Immobilon, immunoblotted with anti-LMP2A antiserum 14B7, and incubated with an HRP-conjugated secondary antibody, and proteins were detected by ECL. The levels of LMP2A expression were similar in two wild-type EBV+LMP2A+ LCLs (lanes 1 and 2) and four EBV+LMP2AY112F LCLs (lanes 5 to 8). LMP2A expression was not detected in an EBV+LMP2A− LCL (lane 3) or the EBV− cell line BJAB (lane 4). LCL clone numbers are indicated above each lane, and protein standards are indicated at the left in kilodaltons.
LMP2A is not phosphorylated in EBV+LMP2AY112F LCLs.
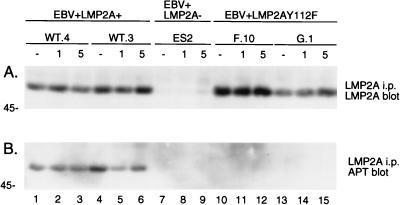
To verify the phosphorylation state of LMP2A in response to BCR cross-linking, unstimulated and stimulated cellular lysates of EBV+LMP2A+, EBV+LMP2A−, and EBV+LMP2AY112F LCLs were immunoprecipitated with an anti-LMP2A antibody, divided equally, and separated by duplicate SDS-PAGE. Two parallel immunoblot analyses were performed to detect immunoprecipitated LMP2A (Fig. 5A) and tyrosine-phosphorylated LMP2A (Fig. 5B). Equivalent levels of LMP2A were immunoprecipitated from all LMP2A-expressing LCLs: the two EBV+LMP2A+ LCLs (Fig. 5A, lanes 1 to 6) and two EBV+LMP2AY112F LCLs (Fig. 5A, lanes 10 to 15), while no LMP2A was detected in the control EBV+LMP2A− LCL (Fig. 5A, lanes 7 to 9). In the APT blot, LMP2A was constitutively phosphorylated in the two EBV+LMP2A+ LCLs (Fig. 5B, lanes 1 to 6), while LMP2A in the two EBV+LMP2AY112F LCLs was not phosphorylated before or after BCR cross-linking (Fig. 5B, lanes 10 to 15). As expected, no phosphorylated LMP2A was detected in the control EBV+LMP2A− LCL (Fig. 5B, lanes 7 to 9). These data demonstrate the complete loss of LMP2A tyrosine phosphorylation in EBV+LMP2AY112 LCLs.
FIG. 5.
LMP2A is not phosphorylated in EBV+LMP2AY112F LCLs. LCLs were untreated (−) or treated at 3 × 107 cells per ml with anti-BCR antibodies for the indicated times (1 or 5 min), lysed in NP-40, immunoprecipitated (i.p.) with anti-LMP2A 14B7 antiserum, captured with protein G-Sepharose, separated by duplicate SDS-PAGE (6% gel), and transferred to Immobilon. Parallel immunoblots were probed with anti-LMP2A (14B7-biotin) antiserum (A) or APT antibody PY20 (B), incubated with HRP-conjugated secondary antibodies, and detected by ECL. (A) LMP2A expression levels in two EBV+LMP2A+ LCLs (A, lanes 1 to 6) and two EBV+LMP2AY112F LCLs (A, lanes 10 to 15) were similar in all four LCLs during the time course. No LMP2A was detected in the EBV+LMP2A− LCL included as a negative control (A, lanes 7 to 9). (B) LMP2A was constitutively phosphorylated in two EBV+LMP2A+ LCLs which did not change after BCR cross-linking (B, lanes 1 to 6). In contrast, LMP2A was not phosphorylated in the two EBV+LMP2AY112F LCLs even after BCR cross-linking (B, lanes 10 to 15). No phosphorylated LMP2A was detected in the negative control EBV+LMP2A− LCL (B, lanes 7 to 9). LCL clone numbers are indicated above each group of lanes, and protein standards are indicated at the left in kilodaltons.
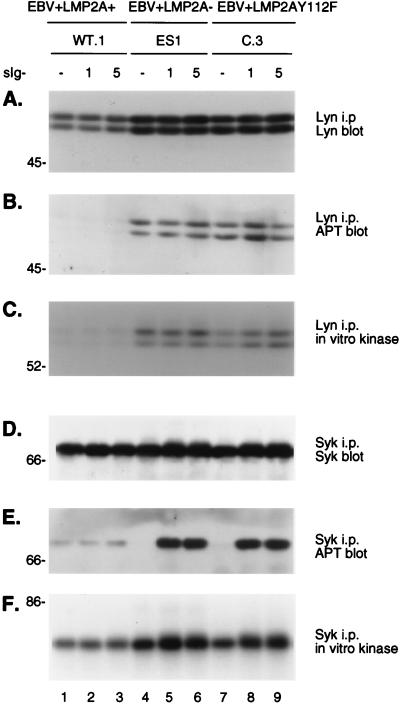
EBV+LMP2AY112F LCLs demonstrate high levels of constitutive Lyn phosphorylation and kinase activity and the induction of Syk phosphorylation and kinase activity similar to those of EBV+LMP2A− LCLs following BCR cross-linking.
To investigate the effect of the LMP2AY112F point mutation on the phosphorylation state and kinase activities of the Lyn and Syk PTKs, parallel anti-Lyn and anti-Syk immunoprecipitations were performed on cellular lysates from representative EBV+LMP2A+, EBV+LMP2A−, and EBV+LMP2AY112F LCLs. The Lyn and Syk immunoprecipitations were divided equally and were either analyzed for autokinase activities by in vitro immune complex kinase assays or probed for Lyn, Syk, or APT reactivities on immunoblots. The Lyn immunoprecipitation and Lyn immunoblot revealed similar Lyn protein expression levels in all LCLs, although the EBV+LMP2A+ LCL demonstrated slightly less protein than either the EBV+LMP2A− or EBV+LMP2AY112F LCLs (Fig. 6A, lanes 1 to 9). This observation was reproducible in other Lyn immunoprecipitation experiments of EBV+LMP2A+ LCLs (data not shown) and was consistent with observations previously described (26). The parallel APT immunoblot of the same Lyn immunoprecipitates revealed phosphorylation differences between the wild-type and mutant LCLs (Fig. 6B). The EBV+LMP2A+ LCL demonstrated undetectable Lyn phosphorylation levels both before or after BCR cross-linking (Fig. 6B, lanes 1 to 3), while constitutive Lyn phosphorylation was clearly evident in the EBV+LMP2A− and EBV+LMP2AY112F mutant LCLs both before and after BCR cross-linking (Fig. 6B, lanes 4 to 9). Finally, in vitro autophosphorylation kinase assays were performed on the same Lyn immunoprecipitates (Fig. 6C). The representative EBV+LMP2A+ LCL demonstrated low Lyn kinase activity (Fig. 6C, lanes 1 to 3), while the EBV+LMP2A− and EBV+LMP2AY112F LCLs demonstrated greater Lyn kinase activities (Fig. 6C, lanes 4 to 9). These data demonstrate that unlike the down-modulation of Lyn phosphorylation and kinase activity characteristic of EBV+LMP2A+ LCLs, the EBV+LMP2AY112 LCL demonstrated both the increased Lyn phosphorylation and kinase activity characteristic of an EBV+LMP2A− LCL.
FIG. 6.
Expression, phosphorylation, and kinase activities of Lyn and Syk in EBV+LMP2A+, EBV+LMP2A−, and EBV+LMP2AY112F LCLs following BCR cross-linking. LCLs were untreated (−) or treated at 5 × 107 cells per ml with anti-BCR antibodies for the indicated times (1 or 5 min) and lysed in 1% NP-40. Lysates were divided equally, immunoprecipitated (i.p.) in parallel with either a Lyn monoclonal antibody (A to C) or a Syk monoclonal antibody (D to F), and captured with protein A-Sepharose beads. One third of each immunoprecipitation was further subjected to an in vitro kinase assay (C and F). All immune complexes were separated in parallel by SDS-PAGE (6% gel). The 32P-labeled products were visualized by autoradiography of the dried gels (C and F). The nonradioactive proteins were transferred to Immobilon and immunoblotted in parallel with monoclonal APT (B and E), anti-Lyn (A), or anti-Syk (D) antibodies, followed by incubation with HRP-conjugated secondary antibodies and ECL detection. (A) The level of p56/p53 Lyn expression did not change following BCR cross-linking in the EBV+LMP2A+, EBV+LMP2A−, and EBV+LMP2AY112F LCLs (A, lanes 1 to 9). (B) Lyn remained constitutively phosphorylated before and after BCR cross-linking in the three LCLs, but the level of p56/p53 Lyn phosphorylation was elevated in the EBV+LMP2A− and EBV+LMP2AY112F LCLs (B, lanes 4 to 9) compared with the barely detectable Lyn phosphorylation in the EBV+LMP2A+ LCL (B, lanes 1 to 3). (C) Lyn remained constitutively active in autophosphorylation assays before and after BCR cross-linking in the three LCLs, but the level of p56/p53 Lyn activity was increased in the EBV+LMP2A− and EBV+LMP2AY112F LCLs (C, lanes 4 to 9) compared with the barely detectable Lyn activity in the EBV+LMP2A+ LCL (C, lanes 1 to 3). (D) The level of 72-kDa Syk expression was similar in the EBV+LMP2A+, EBV+LMP2A−, and EBV+LMP2AY112F LCLs following BCR cross-linking (D, lanes 1 to 9). (E) Syk remained constitutively phosphorylated before and after BCR cross-linking in the EBV+LMP2A+ LCL (E, lanes 1 to 3), while Syk demonstrated no constitutive phosphorylation before BCR cross-linking in the EBV+LMP2A− and EBV+LMP2AY112F LCLs (E, lanes 4 and 7) and became rapidly phosphorylated within 1 min after BCR cross-linking in both the EBV+LMP2A− and EBV+LMP2AY112F LCLs (E, lanes 5 to 6 and 8 to 9). (F) Syk demonstrated a low level of constitutive activity before and after BCR cross-linking in the EBV+LMP2A+ LCL (F, lanes 1 to 3), whereas Syk demonstrated a higher level of constitutive activity that induced to even greater levels following BCR cross-linking in the EBV+LMP2A− and EBV+LMP2AY112F LCLs (F, lanes 4 to 9). LCL clone numbers are indicated above each group of lanes, and protein standards are indicated at the left in kilodaltons.
The Syk immunoprecipitation and Syk immunoblot demonstrated similar Syk protein expression levels in all LCLs before and after BCR cross-linking (Fig. 6D, lanes 1 to 9). However, the parallel APT immunoblot of the same Syk immunoprecipitates revealed phosphorylation differences between the wild-type and mutant LCLs (Fig. 6E). The representative EBV+LMP2A+ LCL demonstrated a low level of constitutive Syk phosphorylation that was not induced by BCR cross-linking (Fig. 6E, lanes 1 to 3), while both the EBV+LMP2A− and EBV+LMP2AY112 LCLs demonstrated the absence of Syk phosphorylation before BCR cross-linking and the rapid induction of Syk phosphorylation within 1 min after BCR cross-linking (Fig. 6E, lanes 4 to 9). The APT studies were further supported by an in vitro autophosphorylation kinase assay performed on the same Syk immunoprecipitates (Fig. 6F). Syk demonstrated a low constitutive kinase activity in the EBV+LMP2A+ LCL that was not induced by BCR cross-linking (Fig. 6F, lanes 1 to 3), while the EBV+LMP2A− LCL and EBV+LMP2AY112 LCL demonstrated increased baseline Syk activities that were further induced to greater activities after BCR cross-linking (Fig. 6C, lanes 4 to 9). These data demonstrate that unlike the down-modulation of Syk phosphorylation and kinase activity characteristic of EBV+LMP2A+ LCLs, the EBV+LMP2AY112 LCL demonstrated the induction of both Syk phosphorylation and kinase activity as characteristic of an EBV+LMP2A− LCL. The results of this study are consistent with previous observations of Lyn and Syk protein expression, phosphorylation, and kinase activity levels in EBV+LMP2A+ and EBV+LMP2A− LCLs (26).
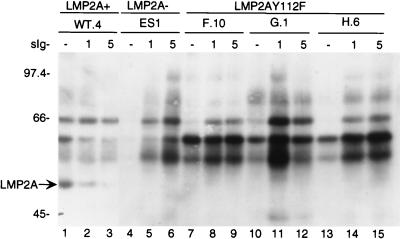
Mutation of LMP2AY112F restores BCR-activated tyrosine phosphorylation.
To investigate the effect of the LMP2AY112F mutation on the induction of APT activity, one of the earliest known biochemical events after BCR cross-linking, EBV+LMP2A+, EBV+LMP2A−, and EBV+LMP2AY112F LCLs, matched for BCR expression by flow cytometry (data not shown), were analyzed for induction of tyrosine phosphorylation following BCR cross-linking. An EBV+LMP2A+ LCL demonstrated the constitutive phosphorylation of a number of proteins in unstimulated LCLs (Fig. 7, lane 1), and this pattern remained relatively unchanged after BCR cross-linking (Fig. 7, lanes 2 to 3). As shown in Fig. 5, LMP2A was phosphorylated in EBV+LMP2A+ LCLs, and its level of phosphorylation did not change over time (Fig. 7, lanes 1 to 3, arrow). This phosphorylated protein was confirmed to be LMP2A by stripping the blot and reprobing it with the LMP2A-specific antibody 14B7 (data not shown). In contrast, there was little constitutive phosphorylation in untreated cellular lysates from EBV+LMP2AY112F LCLs (Fig. 7, lanes 7, 10, and 13) or the control EBV+LMP2A− LCL (Fig. 7, lane 4), but upon BCR cross-linking there was a dramatic increase in the number of proteins which were tyrosine phosphorylated in all mutant cell lines (Fig. 7, lanes 4 to 6, 7 to 9, 10 to 12, and 13 to 15). This increase in tyrosine phosphorylated proteins in the EBV+LMP2AY112F LCLs following BCR cross-linking was similar to the induction of protein tyrosine phosphorylation observed in either EBV+LMP2A− LCLs or an EBV− B-lymphoma cell line as previously described (26). As shown in Fig. 5, LMP2A was not phosphorylated in any of the EBV+LMP2AY112F point mutant LCLs either before or after BCR cross-linking, while LMP2A phosphorylation in the EBV+LMP2A+ LCL was clearly present before and after BCR cross-linking (Fig. 7, lanes 1 to 3, arrow). These data indicate that the induction of tyrosine phosphorylation following BCR cross-linking is not blocked in EBV+LMP2AY112F LCLs. Interestingly, two proteins with the approximate molecular masses of 60 and 57 kDa were constitutively phosphorylated in the unstimulated lysates of the LMP2AY112F LCLs and not present in the unstimulated lysate of the EBV+LMP2A− LCL (Fig. 7; compare lanes 4, 7, 10, and 13). Proteins of similar molecular masses are also constitutively phosphorylated in the unstimulated lysate of the EBV+LMP2A+ LCL (Fig. 7, lane 1).
FIG. 7.
Tyrosine phosphorylation following BCR cross-linking in EBV+LMP2A+, EBV+LMP2A−, and EBV+LMP2AY112F LCLs. LCLs were untreated (−) or treated at 3 × 107 cells per ml for the indicated times (1 or 5 min) with anti-BCR antibodies, lysed in 1% NP-40, immunoprecipitated with APT antibody PY20, captured with protein A-Sepharose, separated by SDS-PAGE (6% gel), transferred to Immobilon, and immunoblotted with HRP-conjugated APT antibody PY20, and proteins were detected by ECL. There was no induction of tyrosine phosphorylation in an EBV+LMP2A+ LCL (lanes 1 to 3), in contrast to the dramatic increase in phosphorylation in an EBV+LMP2A− LCL (lanes 4 to 6) and three EBV+LMP2AY112F LCLs (lanes 7 to 15). LCL clone numbers are indicated above each group of lanes, and protein standards are indicated at the left in kilodaltons.
Calcium mobilization in EBV+LMP2A+ and EBV+LMP2AY112F LCLs following BCR cross-linking.
To assess the effect of the LMP2AY112F mutation on calcium mobilization, multiple EBV+LMP2A+ and EBV+LMP2AY112 LCLs, derived in parallel and matched for BCR expression by flow cytometry (data not shown), were loaded with the calcium-binding dye fluo-3 and monitored for fluorescence after BCR cross-linking. Previously described EBV+LMP2A+ (Table 1, WT4) and EBV+LMP2A− (Table 1, ES1) LCLs were included as controls (27). EBV+LMP2A+ LCLs demonstrated a minimal calcium flux after BCR cross-linking (5 to 13%) (Table 1). These calcium flux values are small compared to the mean calcium flux value of the control EBV+LMP2A− LCL (411%) (Table 1). Representative EBV+LMP2AY112F point mutant LCLs were also tested and had an observed mean calcium flux of 80 to 368%, a mean greater than that for EBV+LMP2A+ LCLs (5 to 13%) (Table 1). These data demonstrate that the LMP2AY112 LCLs readily mobilize calcium following BCR cross-linking, unlike EBV+LMP2A+ LCLs, which demonstrate a block in BCR cross-linking induced calcium mobilization.
TABLE 1.
Calcium mobilization in EBV+LMP2A+, EBV+LMP2A−, and LMP2AY112F LCLs after BCR cross-linkinga
| Cell line type and nameb | No.c | Mean % increase in calcium (range) |
|---|---|---|
| EBV+LMP2A+ LCLs | ||
| WT4 | 5 | 8 (3–11) |
| KC.WT.1 | 2 | 13 (10–16) |
| KC.WT.3 | 2 | 10 (7–12) |
| RL.WT.C.12 | 2 | 5 (0–10) |
| RL.WT.G.2 | 5 | 6 (0–11) |
| EBV+LMP2A− LCL | ||
| ES1 | 5 | 411 (270–560) |
| EBV+LMP2AY112F LCLs | ||
| LMP2AY112.C.3 | 5 | 192 (139–276) |
| LMP2AY112.E.8 | 3 | 80 (56–123) |
| LMP2AY112.A.6 | 4 | 103 (69–149) |
| LMP2AY112.D.2 | 2 | 124 (88–159) |
| LMP2AY112.G.10 | 3 | 90 (41–149) |
| LMP2AY112.A.10 | 3 | 79 (65–93) |
| LMP2AY112.G.5 | 2 | 368 (181–554) |
| LMP2AY112.A.3 | 2 | 102 (75–128) |
| LMP2AY112.E.6 | 4 | 89 (46–173) |
| LMP2AY112.B.4 | 1 | 132 |
| LMP2AY112.H.2 | 3 | 156 (92–222) |
| LMP2AY112.H.10 | 3 | 102 (69–137) |
LCLs (3 × 106) were resuspended in loading buffer and loaded with 2 mM fluo-3 for 30 min at room temperature. Following loading, LCLs were washed twice and resuspended in loading buffer. The prepared cells were then measured for fluorescence after sIg cross-linking in a Perkin-Elmer LS-5B luminescence spectrometer (see Materials and Methods).
WT4 and ES1 were previously described (27). All other LCLs were produced in this study.
Number of times calcium mobilization was determined for each LCL.
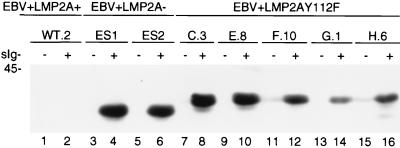
Induction of BZLF1 expression in EBV+LMP2AY112F mutant LCLs following BCR cross-linking.
The activation of lytic viral replication was investigated in EBV+LMP2A+, EBV+LMP2A−, and EBV+LMP2AY112 LCLs following BCR cross-linking by analyzing the expression of BZLF1, the immediate-early transactivator of EBV lytic replication. The BCR was either untreated or treated with goat anti-human Ig antibody for 48 h in representative EBV+LMP2A+, EBV+LMP2A−, or EBV+LMP2AY112F LCLs, and the cellular lysates were monitored for BCR-induced expression of BZLF1 by immunoblotting with monoclonal antibody BZ1 (27, 48). BZLF1 expression was evident after BCR cross-linking in the two control EBV+LMP2A− LCLs (Fig. 8, lanes 4 and 6) and the five representative EBV+LMP2AY112F LCLs (Fig. 8, lanes 8, 10, 12, 14, and 16) but was not detected in any of the EBV+LMP2AY112F LCLs before BCR cross-linking (Fig. 8, lanes 7, 9, 11, 13, and 15) or in the two EBV+LMP2A− LCLs (Fig. 8, lanes 3 and 5). BZLF1 expression was not detected in the representative EBV+LMP2A+ LCL either before or after BCR cross-linking (Fig. 8, lanes 1 and 2). The two different-sized BZLF1 proteins reflect a variable number of repeat sequences present within the BZLF1 gene of different viral isolates. The BZLF1 proteins of both molecular weights specifically react with the BZ1 monoclonal antibody. These data demonstrate that tyrosine 112 of LMP2A plays an important role in the LMP2A-mediated inhibition of viral nuclear gene transcription following BCR cross-linking and that loss of LMP2A tyrosine residue 112 results in the induction of BZLF1 expression, one of the first events leading to EBV lytic replication.
FIG. 8.
Induction of BZLF1 expression in EBV+LMP2A− and EBV+LMP2AY112F LCLs following BCR cross-linking. LCLs (4 × 106) were untreated (−) or treated (+) with anti-BCR antibodies for 48 h. Whole-cell lysates were separated by SDS-PAGE (8% gel), transferred to Immobilon, immunoblotted with anti-BZLF1 antibody BZ1, and incubated with an HRP-conjugated secondary antibody, and proteins were detected by ECL. In five EBV+LMP2AY112F LCLs, BZLF1 expression was induced after anti-BCR treatment (lanes 8, 10, 12, 14, and 16), similar to the result for two EBV+LMP2A− LCLs, included as positive controls (lanes 4 and 6). However, BZLF1 expression was not induced after anti-BCR treatment in an EBV+LMP2A+ LCL, included as a negative control (lane 2). Prior to BCR cross-linking, BZLF1 expression was not detected in any of the LCLs (lanes 1, 3, 5, 7, 9, 11, 13, and 15). LCL clone numbers are indicated above each pair of lanes, and protein standards are indicated at the left in kilodaltons.
DISCUSSION
The results reported in this study indicate that tyrosine 112 of LMP2A is essential for LMP2A-mediated down-modulation of BCR signal transduction. The LMP2AY112F mutation results in the restoration of normal BCR signal transduction, as demonstrated by the induction of tyrosine phosphorylation (Fig. 7), the mobilization of intracellular calcium (Table 1), the induction of nuclear gene transcription (Fig. 8), and the activation of cellular PTKs (Fig. 6) after BCR cross-linking. In addition, the LMP2AY112F mutation results in the loss of Lyn association with LMP2A (Fig. 3A) and the loss of LMP2A tyrosine phosphorylation (Fig. 5B).
Applying the data collected in this study with data from previously published studies of LMP2 and studies of signal transduction through the BCR, we have formulated a model of LMP2A function in latently infected B lymphocytes (Fig. 9). In this model, LMP2A is expressed in the plasma membranes of EBV latently infected B cells and forms aggregates resembling ligated BCRs (Fig. 9). Previously, studies have shown that the transmembrane domains of LMP2A are responsible for LMP2A aggregation. Truncation of LMP2A after two or five transmembrane domains result in the loss of LMP2A aggregation and a concomitant loss of LMP2A function (20, 27). Once LMP2A is aggregated in a newly infected B cell, the Lyn PTK may be recruited to the unphosphorylated LMP2A aggregates through strategies similar to those used for the recruitment of the nonactivated Src family PTKs to the unphosphorylated BCR. The amino-terminal unique region of Lyn plays an important role in the association of Lyn with the unphosphorylated Ig α chain (Igα). The unique region of Lyn binds the DCSM sequence located before the second conserved tyrosine of the Igα ITAM (8, 35). Interestingly, LMP2A has a homologous DQSL sequence located adjacent to the LMP2A ITAM which may play a similar role as the DCSM Igα sequence by providing a docking site for the Lyn PTK on unphosphorylated LMP2A (Fig. 9A). Alternatively, the Lyn SH3 domain may interact with one of the four proline-rich regions in the LMP2A amino-terminal domain (Fig. 9A). The small amount of the Lyn PTK detectable in the pY112F- and p2AΔ80-112-transfected immunoprecipitates is compatible with either of these hypotheses (Fig. 3A).
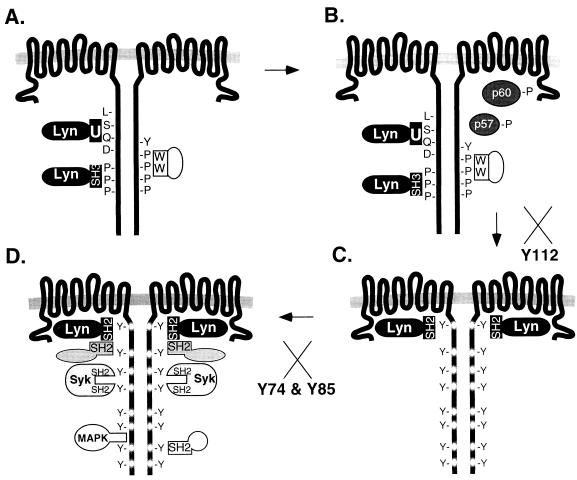
FIG. 9.
Model of LMP2A function. (A) The Src family PTK Lyn is recruited to LMP2A possibly by an interaction of the Lyn SH3 domain with the LMP2A proline-rich regions or an interaction of the Lyn unique region (U) with the LMP2A DQSL sequence. WW domain-containing proteins may also be recruited to the LMP2A nonphosphorylated PPPPY motifs. (B) LMP2A becomes phosphorylated at Y112 by the Lyn PTK or another unidentified cellular PTK, possibly the p57 or p60 protein whose phosphorylation is observed in both wild-type and LMP2AY112F LCLs prior to BCR cross-linking. (C) Once Y112 is phosphorylated, the Lyn SH2 domain binds. Following Lyn binding to LMP2A Y112, the remaining LMP2A tyrosines, including the LMP2A ITAM, become phosphorylated. Binding of Lyn to LMP2A and subsequent LMP2A phosphorylation are blocked in the Y112F mutants; thus, the Y112F mutants do not proceed to step C. (D) The presence of other phosphorylated LMP2A motifs allows the binding of the Syk PTK, other SH2-containing proteins, and MAPK to phosphorylated LMP2A. Once bound to LMP2A, their activities are reduced and they are no longer able to participate in BCR signal transduction. Binding of Syk to LMP2A is blocked in the LMP2A ITAM (Y74F and Y85F) mutants, although LMP2A still demonstrates the phosphorylation of other tyrosines. The LMP2A ITAM mutants do not proceed fully to step D, due to the loss of Syk binding to LMP2A.
Once Lyn is recruited to LMP2A by one or both of the mechanisms discussed above, Lyn phosphorylates LMP2A at Y112. Alternatively, an unknown kinase, which may be one of the proteins whose constitutive phosphorylation is induced in LMP2AY112F LCLs as well as EBV+LMP2A+ LCLs (Fig. 7), may phosphorylate LMP2A at Y112 (Fig. 9B). Once Y112 is phosphorylated, Lyn binds at Y112 and subsequently phosphorylates the remaining LMP2A tyrosines, in particular the two tyrosines of the LMP2A ITAM (Fig. 9C). The LMP2A phosphotyrosine motif at LMP2AY112 is YEEA, similar to the YEEI motif which is the SH2 domain binding motif preferred by the Src family PTKs (39). Interestingly, the BCR does not contain a similar motif. The specificity of Lyn for the LMP2A YEEA motif may confer preferential binding of Lyn to LMP2A and provide LMP2A with the ability to block normal BCR signal transduction. Data presented in this study provide evidence of the essential role of the Lyn PTK for LMP2A function. Without Lyn association with LMP2A at Y112 (Fig. 3A), LMP2A does not become phosphorylated (Fig. 5), and BCR-mediated signal transduction events occur, as evidenced by the induction of tyrosine phosphorylation (Fig. 7), mobilization of intracellular calcium (Table 1), induction of BZLF1 transcription (Fig. 8), and induction of PTK activities (Fig. 6) in the LMP2AY112F LCLs.
Following the phosphorylation of LMP2A, other SH2 domain-containing proteins may be recruited to LMP2A aggregates, just as SH2-containing proteins are recruited to the BCR upon BCR cross-linking. Specifically, the Syk PTK binds to the phosphorylated LMP2A ITAM (12) (Fig. 9D), similar to the binding of Syk to the ITAMs present in Igα and Igβ heterodimers. Binding of Syk to the Igα and Igβ heterodimer requires complete phosphorylation of the ITAMs (17), similar to the requirement of Syk binding only to a completely phosphorylated LMP2A ITAM. Mutation of either tyrosine in the LMP2A ITAM resulted in the loss of Syk binding to LMP2A (12). Although Syk is unable to bind to the LMP2A ITAM mutants, LMP2A is still phosphorylated at other LMP2A amino-terminal tyrosines (12). Once LMP2A is phosphorylated, other unidentified SH2 domain-containing proteins involved in normal BCR signal transduction may also be recruited to LMP2A phosphotyrosines. In immunoprecipitation studies, LMP2A associates with at least six unidentified cell proteins (2). Candidate proteins are Shc, PI3-K, PLCγ2, Abl, Crk, Nck, and Csk. In addition, other proteins such as MAPK or WW domain-containing proteins may also be recruited to LMP2A aggregates (Figs. 9A and D). MAPK has been shown to bind an LMP2A amino-terminal glutathione S-transferase fusion protein (33). Many of these same proteins (PI3-K, PLCγ2, Vav, Shc, and MAPK) failed to be activated by BCR cross-linking in LMP2A-expressing LCLs (26), thus demonstrating further evidence for LMP2A interaction with other cellular signal transducers and LMP2A down-modulation of their activities. Once bound to LMP2A, the functional activity of these proteins may be altered. Once recruited to LMP2A aggregates, the sequestered LMP2A-associated proteins would no longer be free to participate in normal BCR signal transduction. Alternatively, LMP2A may induce the desensitization of cellular signal transducers, thus preventing normal B-cell signal transduction. Desensitization may be mediated by Csk, a negative regulator of the Src family PTKs, or possibly by protein phosphatases recruited to LMP2A complexes. For example, SHP-1, when recruited to the BCR complex by the Fc receptor, results in down-modulation of BCR signal transduction (9). This type of functional inactivation of cellular proteins by viral proteins has been observed in other systems, such as the binding and inactivation of the tumor suppressor protein Rb by the human papillomavirus E7 oncoprotein (31).
Our studies of LMP2A function also highlight the central importance of the Lyn and Syk PTKs in BCR signal transduction. The Src family PTKs and Syk associate with the BCR and are activated following receptor activation (3, 6, 8, 16, 19, 36, 45, 46). A wealth of biochemical and genetic data has indicated the importance of these PTKs in B-cell signal transduction. Most relevant to studies of LMP2A function are the Lyn and Syk gene knockouts in murine and chicken B cells (7, 15, 32, 41, 42). EBV+LMP2A+ LCLs demonstrate a BCR-mediated signaling phenotype very similar to that of the Syk gene knockout in chicken B cells that demonstrate total disruption of BCR signal transduction (41). In the Lyn gene knockout in chicken B cells, there is only a modest alteration in B-cell signal transduction (41), presumably because of the replacement of Lyn function with other Src family PTKs. These observations further demonstrate the central importance of both the Lyn and Syk PTKs in the progression of normal B-cell signal transduction and how the loss or inactivation of these PTKs drastically alters the signaling pathways in B cells. The ability of LMP2A to bind and down-modulate the activities of both of these PTKs provides LMP2A with the means to efficiently block BCR-mediated signal transduction.
The role of other LMP2A-associated proteins remains to be determined. Proposed in our current model of LMP2A function is the binding of other cellular signaling proteins, containing modular SH2, SH3, or WW domains, to LMP2A phosphotyrosines or proline-rich regions. LMP2A may target proteins which are directly or indirectly activated by BCR-induced PTK phosphorylation events. Candidate proteins are the Shc, PI3-K, PLCγ2, Abl, Crk, Nck, and Csk proteins. Future functional analysis of the remaining LMP2A tyrosine residues and LMP2A proline-rich regions will determine the potential roles of these elements in the LMP2A-mediated block in BCR signal transduction.
Applying the knowledge gained in these studies to the role of LMP2A in vivo suggests an important role for LMP2A in maintaining a latent EBV infection within the peripheral blood B lymphocytes of an infected individual. In these cells, the blockade of BCR-mediated signal transduction may allow EBV-infected B cells to encounter BCR-specific signals without undergoing lytic replication, thus maintaining a reservoir of latent viral genomes within the infected host. However, lytic replication may occur in certain circumstances such as the interaction of an EBV-infected lymphocyte with epithelium-produced cytokines or with epithelial surfaces, using signal transduction pathways that are not efficiently blocked by LMP2A. This unique integration of a viral protein into the B-cell signal transduction pathway to block normal B-cell function offers an unusual opportunity to investigate the strategy that EBV has evolved to persist in the human population. Our investigations into this unusual interaction between LMP2A and cellular proteins may provide insights into the regulation of EBV infection, suggesting novel therapies for the control or prevention of EBV latency and latency-associated syndromes, and may further our understanding of B-cell signal transduction.
ACKNOWLEDGMENTS
R.L. is supported by Public Health Service grants CA62234 and CA73507 from the National Cancer Institute. R.L. is a Scholar of the Leukemia Society of America.
REFERENCES
- 1.Bolen J B. Nonreceptor tyrosine protein kinases. Oncogene. 1993;8:2025–2031. [PubMed] [Google Scholar]
- 2.Burkhardt A L, Bolen J B, Kieff E, Longnecker R. An Epstein-Barr virus transformation-associated membrane protein interacts with Src family tyrosine kinases. J Virol. 1992;66:5161–5167. doi: 10.1128/jvi.66.8.5161-5167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkhardt A L, Brunswick M, Bolen J B, Mond J J. Anti-immunoglobulin stimulation of B lymphocytes activates src-related protein-tyrosine kinases. Proc Natl Acad Sci USA. 1991;88:7410–7414. doi: 10.1073/pnas.88.16.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cambier J C. New nomenclature for the Reth motif (or ARH1/TAM/ARAM/YXXL) Immunol Today. 1995;16:110. doi: 10.1016/0167-5699(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 5.Cambier J C, Pleiman C M, Clark M R. Signal transduction by the B cell antigen receptor and its coreceptors. Annu Rev Immunol. 1994;12:457–486. doi: 10.1146/annurev.iy.12.040194.002325. [DOI] [PubMed] [Google Scholar]
- 6.Campbell M A, Sefton B M. Association between B-lymphocyte membrane immunoglobulin and multiple members of the Src family of protein tyrosine kinases. Mol Cell Biol. 1992;12:2315–2321. doi: 10.1128/mcb.12.5.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng A M, Rowley B, Pao W, Hayday A, Bolen J B, Pawson T. Syk tyrosine kinase required for mouse viability and B-cell development. Nature. 1995;378:303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- 8.Clark M R, Johnson S A, Cambier J C. Analysis of Ig-alpha-tyrosine kinase interaction reveals two levels of binding specificity and tyrosine phosphorylated Ig-alpha stimulation of Fyn activity. EMBO J. 1994;13:1911–1919. doi: 10.1002/j.1460-2075.1994.tb06460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Ambrosio D, Hippen K L, Minskoff S A, Mellman I, Pani G, Siminovitch K A, Cambier J C. Recruitment and activation of PTP1C in negative regulation of antigen receptor signaling by FcgRIIB1. Science. 1995;268:293–297. doi: 10.1126/science.7716523. [DOI] [PubMed] [Google Scholar]
- 10.DeFranco A L. Transmembrane signaling by antigen receptors of B and T lymphocytes. Curr Opin Cell Biol. 1995;7:163–175. doi: 10.1016/0955-0674(95)80024-7. [DOI] [PubMed] [Google Scholar]
- 11.Fruehling S, Lee S, Herrold R, Frech B, Laux G, Kremmer E, Grässer F A, Longnecker R. Identification of latent membrane protein 2A (LMP2A) domains essential for the LMP2A dominant-negative effect on B-lymphocyte surface immunoglobulin signal transduction. J Virol. 1996;70:6216–6226. doi: 10.1128/jvi.70.9.6216-6226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fruehling S, Longnecker R. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology. 1997;235:241–251. doi: 10.1006/viro.1997.8690. [DOI] [PubMed] [Google Scholar]
- 13.Gold M R, Matsuuchi L. Signal transduction by the antigen receptors of B and T lymphocytes. Int Rev Cytol. 1995;157:181–276. doi: 10.1016/s0074-7696(08)62159-2. [DOI] [PubMed] [Google Scholar]
- 14.Heston L, Rabson M, Brown N, Miller G. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature. 1982;295:160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- 15.Hibbs M L, Tarlinton D M, Armes J, Grail D, Hodson G, Maglitto R, Stacker S A, Dunn A R. Multiple defects in the immune system of lyn-deficient mice, culminating in autoimmune disease. Cell. 1995;83:301–311. doi: 10.1016/0092-8674(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 16.Hutchcroft J E, Harrison M L, Geahlen R L. Association of the 72-kDa protein-tyrosine kinase PTK72 with the B cell antigen receptor. J Biol Chem. 1992;267:8613–8619. [PubMed] [Google Scholar]
- 17.Kimura T, Sakamoto H, Appella E, Siraganian R P. Conformational changes induced in the protein tyrosine kinase p72syk by tyrosine phosphorylation or by binding of phosphorylated immunoreceptor tyrosine-based activation motif peptides. Mol Cell Biol. 1996;16:1471–1478. doi: 10.1128/mcb.16.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S K, Stern P H. Studies on the mechanism of desensitization of the parathyroid hormone-stimulated calcium signal in UMR-106 cells: reversal of desensitization by alkaline phosphatase but not by protein kinase C downregulation. J Bone Miner Res. 1994;9:781–789. doi: 10.1002/jbmr.5650090604. [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Justement L B. The MB-1/B29 heterodimer couples the B cell antigen receptor to multiple src family protein tyrosine kinases. J Immunol. 1992;149:1548–1555. [PubMed] [Google Scholar]
- 20.Longnecker R, Druker B, Roberts T M, Kieff E. An Epstein-Barr virus protein associated with cell growth transformation interacts with a tyrosine kinase. J Virol. 1991;65:3681–3692. doi: 10.1128/jvi.65.7.3681-3692.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longnecker R, Kieff E. A second Epstein-Barr virus membrane protein (LMP2) is expressed in latent infection and colocalizes with LMP1. J Virol. 1990;64:2319–2326. doi: 10.1128/jvi.64.5.2319-2326.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longnecker R, Miller C L, Miao X-Q, Marchini A, Kieff E. The only domain which distinguishes Epstein-Barr virus latent membrane 2A (LMP2A) from LMP2B is dispensible for lymphocyte infection and growth transformation in vitro, and LMP2A is therefore nonessential. J Virol. 1992;66:6461–6469. doi: 10.1128/jvi.66.11.6461-6469.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini A, Tomkinson B, Cohen J I, Kieff E. BHRF1, the Epstein-Barr virus gene with homology to Bcl2, is dispensable for B-lymphocyte transformation and virus replication. J Virol. 1991;65:5991–6000. doi: 10.1128/jvi.65.11.5991-6000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menezes J, Leibold W, Klein G, Clements G. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt’s lymphoma. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- 25.Merrit J E, McCarthy S A, Davies M P A, Moores K E. Use of fluo-3 to measure cytosolic Ca2+ in platelets and neutrophils. Biochem J. 1990;269:513–519. doi: 10.1042/bj2690513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller C L, Burkhardt A L, Lee J H, Stealey B, Longnecker R, Bolen J B, Kieff E. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity. 1995;2:155–166. doi: 10.1016/s1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 27.Miller C L, Lee J H, Kieff E, Longnecker R. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc Natl Acad Sci USA. 1994;91:772–776. doi: 10.1073/pnas.91.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller C L, Longnecker R, Kieff E. Epstein-Barr virus latent membrane protein 2A blocks calcium mobilization in B lymphocytes. J Virol. 1993;67:3087–3094. doi: 10.1128/jvi.67.6.3087-3094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller G, Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci USA. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller G, Shope T, Lisco H, Stitt D, Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishizumi H, Taniuchi I, Yamanashi Y, Kitamura D, Ilic D, Mori S, Watanabe T, Yamamoto T. Impaired proliferation of peripheral B cells and indication of autoimmune disease in lyn-deficient mice. Immunity. 1995;3:549–560. doi: 10.1016/1074-7613(95)90126-4. [DOI] [PubMed] [Google Scholar]
- 33.Panousis C G, Rowe D T. Epstein-Barr virus latent membrane protein 2 associates with and is a substrate for mitogen-activated protein kinase. J Virol. 1997;71:4752–4760. doi: 10.1128/jvi.71.6.4752-4760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pawson T, Gish G D. SH2 and SH3 domains: from structure to function. Cell. 1992;71:359–362. doi: 10.1016/0092-8674(92)90504-6. [DOI] [PubMed] [Google Scholar]
- 35.Pleiman C M, Abrams C, Gauen L T, Bedzyk W, Jongstra J, Shaw A S, Cambier J C. Distinct p53/56lyn and p59fyn domains associate with nonphosphorylated and phosphorylated Ig-alpha. Proc Natl Acad Sci USA. 1994;91:4268–4272. doi: 10.1073/pnas.91.10.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pleiman C M, D’Ambrosio D, Cambier J C. The B-cell antigen receptor complex: structure and signal transduction. Immunol Today. 1994;15:393–399. doi: 10.1016/0167-5699(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 37.Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- 38.Schaffhausen B. SH2 domain structure and function. Biochim Biophys Acta. 1995;1242:61–75. doi: 10.1016/0304-419x(95)00004-y. [DOI] [PubMed] [Google Scholar]
- 39.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 40.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takata M, Sabe H, Hata A, Inazu T, Homma Y, Nukada T, Yamamura H, Kurosaki T. Tyrosine kinases Lyn and Syk regulate B cell receptor-coupled Ca2+ mobilization through distinct pathways. EMBO J. 1994;13:1341–1349. doi: 10.1002/j.1460-2075.1994.tb06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner M, Mee P J, Costello P S, Williams O, Price A A, Duddy L P, Furlong M T, Geahlen R L, Tybulewicz V L J. Perinatal lethality and blocked B-cell development in mice lacking the tyrosine kinase Syk. Nature. 1995;378:298–302. doi: 10.1038/378298a0. [DOI] [PubMed] [Google Scholar]
- 43.Wang F, Marchini A, Kieff E. Epstein-Barr virus (EBV) recombinants: use of positive selection markers to rescue mutants in EBV-negative B-lymphoma cells. J Virol. 1991;65:1701–1709. doi: 10.1128/jvi.65.4.1701-1709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss A, Littman D R. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 45.Yamada T, Tanaguchi T, Yang C, Yasue S, Saito H, Yamamura H. Association with B-cell-antigen receptor with protein-tyrosine kinase p72syk and activation by engagement of membrane IgM. Eur J Biochem. 1993;213:455. doi: 10.1111/j.1432-1033.1993.tb17781.x. [DOI] [PubMed] [Google Scholar]
- 46.Yamanashi Y, Fukui Y, Wongsasant W, Kinoshita Y, Ichimori Y, Toyoshima K, Yamamoto T. Activation of src-like protein-tyrosine kinase lyn and its association with PI(3)kinase upon B-cell antigen receptor-mediated signalling. Proc Natl Acad Sci USA. 1992;89:1118–1122. doi: 10.1073/pnas.89.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi T L, Bolen J B, Ihle J N. Hematopoietic cells express two forms of Lyn kinase differing by 21 amino acids in the amino terminus. Mol Cell Biol. 1991;11:2391–2398. doi: 10.1128/mcb.11.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young L S, Lau R, Rowe M, Niedobitek G, Packham G, Shanahan F, Rowe D T, Greenspan D, Greenspan J S, Rickinson A B, Farrell P J. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J Virol. 1991;65:2868–2874. doi: 10.1128/jvi.65.6.2868-2874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]