Correction to: Scientific Reports 10.1038/s41598-023-50245-7, published online 04 January 2024
The original version of this Article contained an error in Figure 3 where the western blot decorated with the anti-Pgk1 antibody for loading control was incorrect in panel (g). The original Figure 3 and accompanying legend appear below.
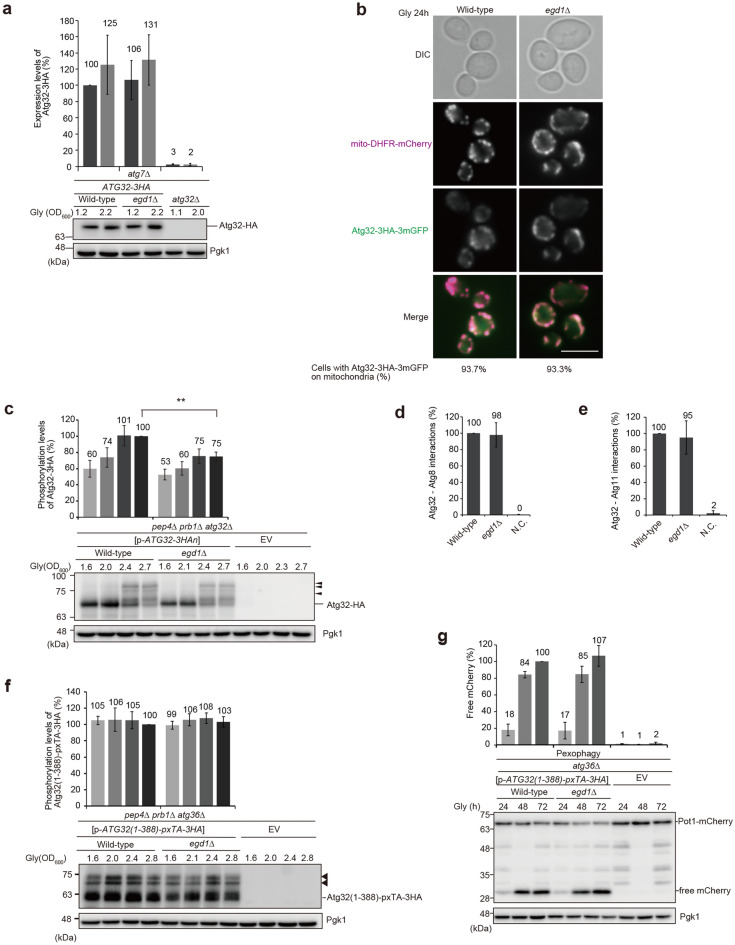
Figure 3.
Loss of Egd1 leads to a decrease in Atg32 phosphorylation. (a) Wild-type, egd1∆, and atg32∆ cells expressing Atg32-3HA were grown in glycerol medium (Gly), collected at the indicated OD600 points, and subjected to western blotting. All strains are derivatives lacking Atg7, a protein essential for all autophagy-related processes, to avoid degradation of Atg32-3HA via mitophagy. Atg32-3HA signals normalized with Pgk1 (loading control) signals were quantified more than three times in independent experiments. The expression level of Atg32-3HA in wild-type cells at the OD600 = 1.2 point was set to 100%. Data represent the averages of all experiments, with bars indicating standard deviations. (b) Wild-type and egd1∆ cells expressing mito-DHFR-mCherry and Atg32-3HA-3mGFP were grown in glycerol medium for 24 h and observed under a fluorescence microscope. Cells (n = 100) with mitochondria-localized Atg32-3HA-3mGFP signals were quantified in more than three experiments, and the average percentages were indicated on the bottom side of image panels. Scale bar, 5 µm. (c) Wild-type and egd1∆ cells transformed with a plasmid encoding Atg32-3HA (p-ATG32-3HA), or an empty vector (EV) were grown in non-fermentable glycerol medium (Gly), collected at the indicated OD600 points, and subjected to western blotting. All strains are pep4 prb1 atg32 triple-null derivatives defective in intravacuolar degradation. Arrowheads indicate putative phosphorylated Atg32. Phosphorylated Atg32-3HA signals normalized with all Atg32-3HA signals were quantified more than three times in independent experiments. The phosphorylation level of Atg32-3HA in wild-type cells at the OD600 = 2.7 point was set to 100%. Data represent the averages of all experiments, with bars indicating standard deviations. **P < 0.01 (unpaired two-tailed Student’s t-test). (d, e) Wild-type and egd1∆ cells expressing Atg32-3HA-3mGFP-3FLAG-LgBiT and SmBiT-3FLAG-8His-Atg8 or Atg11-HA-SmBiT, or wild-type cells expressing Atg32 and Atg11 (negative control, N.C.) were grown in glycerol medium (Gly), collected at the OD600 = 1.4 point, incubated with substrates, and subjected to measurements of GFP and luminescent signals in more than three experiments using a microplate reader. (f) Wild-type and egd1∆ cells transformed with a plasmid encoding the Atg32 cytoplasmic domain anchored to the peroxisome (p-Atg32(1–388)-pxTA-3HA), or an empty vector (EV) were grown in non-fermentable glycerol medium (Gly), collected at the indicated OD600 points, and subjected to western blotting. All strains are pep4 prb1 atg36 triple-null derivatives defective in intravacuolar degradation and endogenous pexophagy. Arrowheads indicate putative phosphorylated protein bands. Phosphorylated Atg32(1–388)-pxTA-3HA signals normalized with all Atg32(1–388)-pxTA-3HA signals were quantified more than three times in independent experiments. The phosphorylation level of Atg32(1–388)-pxTA-3HA in wild-type cells at the OD600 = 2.8 point was set to 100%. Data represent the averages of all experiments, with bars indicating standard deviations. (g) Wild-type and egd1∆ cells transformed with p-Atg32(1–388)-pxTA-3HA, or an empty vector (EV) were grown in non-fermentable glycerol medium (Gly), collected at the indicated time points, and subjected to western blotting. All strains are atg36-null derivatives expressing Pot1-mCherry (a peroxisome marker). The amounts of free mCherry in cells under respiratory conditions for 24 h, 48 h, and 72 h were quantified in three experiments. The signal intensity value of free mCherry in wild-type cells at the 72 h time point was set to 100%. Data represent the averages of all experiments, with bars indicating standard deviations.
The original Article has been corrected.