Abstract
The Paramyxoviridae family includes established human pathogens such as measles virus, mumps virus, and the human parainfluenza viruses; highly lethal zoonotic pathogens such as Nipah virus; and a number of recently identified agents, such as Sosuga virus, which remain poorly understood. The high human-to-human transmission rate of paramyxoviruses such as measles virus, high case fatality rate associated with other family members such as Nipah virus, and the existence of poorly characterized zoonotic pathogens raise concern that known and unknown paramyxoviruses have significant pandemic potential. In this review, the general life cycle, taxonomic relationships, and viral pathogenesis are described for paramyxoviruses that cause both systemic and respiratory system–restricted infections. Next, key gaps in critical areas are presented, following detailed conversations with subject matter experts and based on the current literature. Finally, we present an assessment of potential prototype pathogen candidates that could be used as models to study this important virus family, including assessment of the strengths and weaknesses of each potential prototype.
Keywords: pandemic preparedness, paramyxovirus, prototype pathogens, zoonosis
Paramyxoviruses include important human pathogens such as measles virus (MV) and mumps virus (MuV), zoonotic agents such as Hendra virus (HeV) and Nipah virus (NiV) that emerge from bats [1], impactful animal viruses with a very wide host range such as canine distemper virus (CDV) and the eradicated rinderpest virus (RPV) [2], and a swathe of recently identified viruses and sequences with unknown zoonotic and pandemic potential [3]. Effective live-attenuated vaccines and a subunit vaccine have been developed for a small subset of paramyxoviruses, and these have been used to limit geographical distribution and eradicate important human and veterinary diseases. However, no effective approved antiviral treatments or vaccines exist to mitigate the majority of paramyxoviral diseases and we are woefully unprepared should a respiratory paramyxovirus emerge and spread in humans with no preexisting immunity. Paramyxoviruses are members of the order Mononegavirales and have nonsegmented, negative-sense RNA genomes encapsidated into a ribonucleoprotein (RNP) complex within an enveloped virion (Figure 1). All paramyxovirus genomes encode for a nucleocapsid (N) protein, a phospho- (P) protein, a matrix (M) protein, a fusion (F) glycoprotein, an attachment hemagglutinin (H)/hemagglutinin-neuraminidase (HN)/glyco- (G) protein, and an RNA-dependent RNA polymerase, or large (L) protein. In addition, some paramyxoviruses encode a small hydrophobic protein and RNA editing of the P protein gene leads to expression of additional nonstructural proteins that play key roles in the antiviral response.
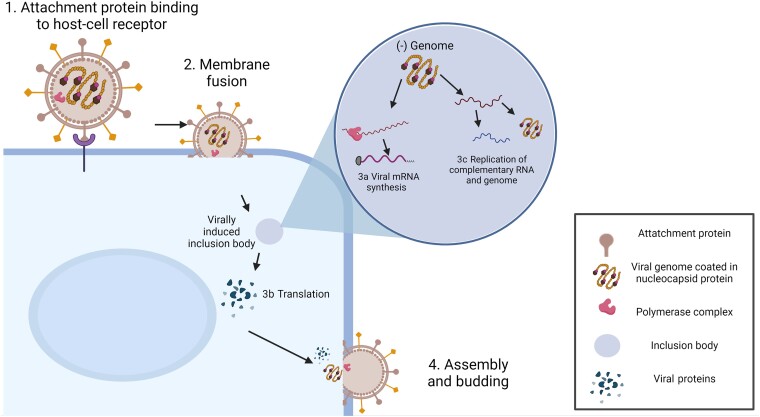
Figure 1.
Generalized paramyxovirus life cycle. (1) Attachment of the viral particle, mediated by the attachment protein. (2) Fusion between the viral and a cellular membrane to release the ribonucleoprotein complex containing the negative sensed genome into the cytosol. Transcription (3a) of viral messenger RNA (mRNA) is promoted by the viral polymerase complex within viral-induced inclusion bodies, followed by translation (3b) in the cytosol, and replication of viral genomes within intracytoplasmic inclusion bodies (3c). (4) Viral proteins and encapsidated genome are transported to sites of assembly and budding occurs from the plasma membrane.
Entry of paramyxoviruses into target cells initiates with binding of the attachment glycoprotein (H/HN/G) to a cell surface receptor. These can be proteins on the plasma membrane; for example, CD150 is the primary morbillivirus receptor [4] and ephrin B2 is used by HeV and NiV [5] or moieties such as sialic acid, present on cell surface glycoproteins or glycolipids [6] (Figure 1). The attachment protein then triggers the F glycoprotein to initiate the membrane fusion process and the negative-sense (−)RNP is liberated into the cytoplasm [7]. The incoming (−)RNP is initially used by the polymerase complex as the template for primary transcription of viral messenger RNA (mRNA). It is the basic unit of infectivity and is comprised of multiple copies of the N protein, which encapsidate the (−)RNA, and a number of P and L proteins in complex, which initially function as a transcriptase. Subsequently the (−)RNP is utilized for the replication of full-length complementary RNA by the same polymerase functioning as a replicase. Resulting (+)RNPs are in turn replicated into nascent (−)RNPs that are incorporated into progeny virions. Transcription and replication occur within virally induced inclusion bodies [8–10]. In the later stages of infection, the viral (−)RNP complex and viral proteins are targeted to specific sites on the plasma membrane for assembly and budding of new viral particles [11].
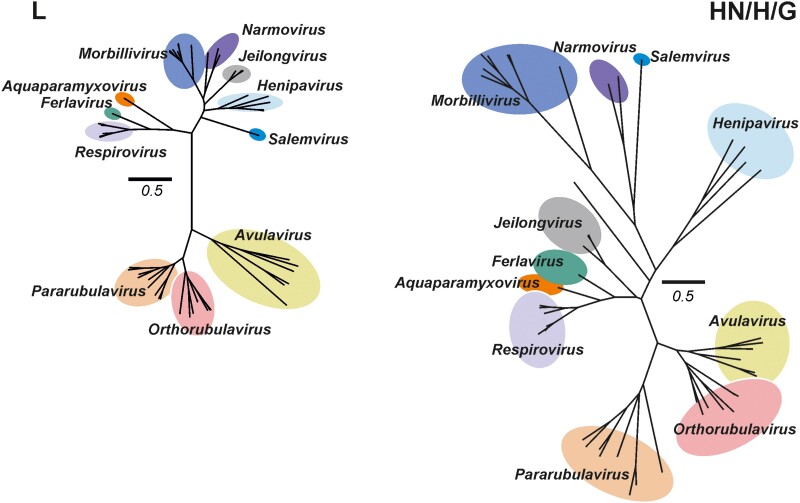
Taxonomically the family is divided into 4 subfamilies (Orthoparamyxovirinae, Metaparamyxovirinae, Rubulavirinae, and Avulavirinae) based on phylogenetic analysis of the L gene. This genotypic subdivision is consistent with the phenotypic classification based on biological, biochemical, and host range [12]. Phylogenetic analysis of the L and receptor-binding proteins currently describe 20 genera, although not all have been assigned to a subfamily (Figure 2). Avulaviruses infect birds and are not zoonotic; therefore, these were triaged from discussion as potential prototype pathogens. Orthoviruses and pararubulaviruses originate from animals, primarily bats, and were therefore considered to pose somewhat of a pandemic risk. A metaparamyxovirus has yet to be isolated and exists only as sequence information, making metaparamyxoviruses unsuitable as prototype pathogens. Orthoparamyxoviruses represent the largest grouping in the family and based on their biological properties, the Morbillivirus, Respirovirus, and Henipavirus genera were prioritized for closer consideration over the other genera (Figure 2).
Figure 2.
Maximum likelihood phylogenetic tree of L and attachment (HN/H/G) proteins of recognized Paramyxoviridae families. Colored circles correspond to characterized genera by the International Committee on Taxonomy of Viruses. Avulavirinae subfamily members (genera Metaavulavirus, Orthoavulavirus, and Paraavulavirus) are not shown for clarity of presentation. Scale bar indicates 0.5 amino acid substitutions per site. Genera prioritized for discussion by the Paramyxovirus Expert Review Group were the rubulaviruses, respiroviruses, morbilliviruses and Henipaviruses. Trees were constructed using Mega 6 using a complete deletion option and WAG substitution model. Trees constructed by Andres Moreira-Soto and Jan Felix Drexler, Charité, Berlin, Germany.
PARAMYXOVIRUSES PATHOGENESIS: SYSTEMIC AND RESPIRATORY
The pathogenesis of paramyxoviruses varies from the extreme of systemic infection to localized infection in the respiratory tract. These boundaries are well illustrated by the morbillivirus MV (more similar to the highly virulent systemic henipaviruses) and the Respirovirus human parainfluenza viruses (HPIVs) (more similar to the less pathogenic but still clinically relevant rubulaviruses).
Following transmission in respiratory droplets, MV initially targets alveolar macrophages and dendritic cells in the deep lung [13]. It is here the virus begins its cell-associated, lymphotropic existence during systemic disease. Migration of these infected cells to bronchus-associated lymphoid tissue brings the viruses into close proximity to other dendritic cells, as well as T and B lymphocytes, which express high levels of the primary MV receptor human CD150 when activated. Intimate cell-to-cell contacts and microfusion events are critical in seeding the infection and amplifying the virus in these tertiary lymphoid tissues. Migration of infected cells to tracheo-bronchial lymph nodes ensures that cell-to-cell transmission continues in the more organized secondary lymphoid tissues. It is from here that the MV becomes viremic and the virus disseminates in immune cells throughout the body. Finally, it is within the epithelium that immune cells make the initial contact with the adherens junction protein nectin-4, transferring infectivity to the basolateral surface of epithelial cells, leading to virus release from the apical surface into the air [14–16]. Absence of both CD150 and nectin-4 from the apical surface leads to unimpeded shedding. This likely explains why MV is the most transmissible human virus known. Morbillivirus infection leads to lifelong immunity. Highly efficacious, live-attenuated vaccines are available that are also considered to protect from clinical disease for life.
Primary pathogenesis and cell-to-cell spread of the HPIVs is less well understood. The primary target cells are ciliated epithelial cells and infection is generally restricted to the upper respiratory tract and lung epithelium. Infection causes croup, bronchiolitis, or pneumonia depending on the location [17]. Repeat infections are frequent [18], and no licensed vaccine is available. There are similarities to the more intensely studied human respiratory syncytial virus (HRSV) which, although it is now classified in the Pneumoviridae, is closely related to the respiroviruses biologically. Transmissibility and transmission of HPIV has been compared to other common respiratory pathogens (adenoviruses, rhinoviruses, coronaviruses, pneumoviruses, influenza viruses, etc) and this informs vaccine, antiviral, and nonpharmaceutical intervention strategies [19].
KEY GAPS
Basic Biology
While considerable research on paramyxoviruses has been undertaken over many decades, a series of key gaps in our fundamental understanding of their biology were identified that should be addressed if a future paramyxovirus pandemic is to be rapidly and effectively handled. First, the majority of research has been conducted with laboratory-adapted viral strains, which grow efficiently within the tissue culture models that have been utilized for decades [20]. However, research shows that viral adaptations can occur under these circumstances [21], such that these laboratory-adapted strains are imperfect models for the clinical strains from which a pandemic virus might arise. For example, vaccine and laboratory-adapted strains of MV use CD46, which appears to be quite irrelevant in vivo [22]. The development of sequence-verified clinical strains of known provenance that can be widely distributed is needed to address this. These strains should be augmented by the generation of molecular clones from the unpassaged clinical isolates from which they were derived and concomitant rescue of stable, well-characterized recombinant paramyxoviruses [13, 23, 24].
Second, a number of fundamental issues related to paramyxovirus replication during infection remain unexplained. Neurotropism and endotheliotropism are observed for some, but not all, paramyxoviruses, but the molecular basis for this differential within the family has yet to be deciphered. Similarly, factors that restrict some paramyxoviruses to primarily respiratory while other family members produce systemic infection remain to be fully elucidated. In addition, viral persistence after the initial infection has been documented, but the molecular basis for paramyxovirus persistence has only recently begun to be explored [24, 25]. For example, MV represents the paradigm for the persistence of an RNA virus in humans. Novel morbilliviruses that cause chronic infections have been identified [26], demonstrating that persistence has potential ramifications for pathogen emergence and possible eradication, making this a critical area for further research [27].
While the overall life cycle of paramyxoviruses is well conserved within the family (Figure 1), there are variations in critical areas that could impact understanding of an emerging paramyxovirus. First, the cellular receptor utilized by paramyxoviruses differs and how glycoprotein evolution drives these changes is poorly understood. It will be important to understand factors that facilitate or restrict cross-species infection and host-to-host transmission. As the glycoproteins are the major paramyxovirus antigens, a more detailed understanding of their evolution and structural diversity is also critical for predicting cross-reaction and responding to novel paramyxoviruses. Next, paramyxovirus nonstructural proteins are crucial in combating the host antiviral response, but there is considerable variation among family members in both which nonstructural proteins are produced and in the sequence of those that are synthesized. A more detailed analysis of how variation within the nonstructural proteins impacts viral–host interactions would provide an important framework for understanding the risks of novel paramyxoviruses as they emerge.
While there is solid understanding of the developing immune response to some paramyxoviruses, a better picture of how immune response varies over time between family members should be developed, including analysis of potential conserved epitopes that would facilitate response to a novel family member. In addition, while all human paramyxovirus vaccines to date have been live-attenuated, the factors leading to attenuation and how conserved these are should be more deeply explored. There is an urgent need to understand the good vaccines we have developed if we are to move toward rational attenuation of novel agents. Some licensed paramyxovirus vaccines have been under- or overattenuated, further highlighting the importance of dissecting the molecular basis governing disease.
Finally, few data exist on the transmission and stability of paramyxoviruses within the environment and how this might vary between family members and under different circumstances. The effectiveness of various types of decontamination methods and how this varies among the family is also unclear, and thus remains an important area of study.
Models of Disease
It is critical to have a battery of in vitro, ex vivo, and in vivo models that recapitulate aspects of the infection in the natural host to be fully prepared to develop and test vaccines, antivirals, and other interventions when pathogens emerge. This is exemplified in the recent coronavirus disease 2019 (COVID-19) pandemic, which required the rapid and costly establishment of a range of animal models of disease [28]. First, given the fact that paramyxoviruses are largely lymphotropic, epitheliotropic, neurotropic, and endotheliotropic, efforts should be made to establish in vitro models using primary or inducible pluripotent stem cells, which are more disease-relevant. Given that enveloped viruses are the product of the cell in which they are grown, more attention needs to be paid to cell provenance since this governs lipid bilayer composition and glycosylation patterns. Cellular substrate may have significant impact on the effectiveness of antivirals, and the use of transformed cell lines generates spurious results. An impediment is the significant costs associated with the maintenance and differentiation of primary cells. Second, ex vivo models—for example, lung slices from a range of species—have been used to investigate cell-to-cell spread and tropism of paramyxoviruses [29]. These, along with optimal in vitro models, can be used to inform the design of in vivo experiments and assist in the reduction of animals required. Third, a weakness in the field in general is the focus on using human viruses in suboptimal animal species. The ubiquitous mouse is often the first port of call and when standard models fail to deliver, virologists often turn to transgenic or outbred animals [30]. Even these may require the use of rodent-adapted viruses, which in turn bring their own challenges. It is clear infection biology needs to move beyond the mouse and this major gap. For some paramyxoviruses there is a need to develop better animal models (eg, MuV); others (eg, MV) that only infect humans must be studied in nonhuman primates (NHPs), which are costly and can only be used in small numbers. The use of animal viruses in naturally infected species (eg, CDV in ferrets) provides an excellent means to study disease progression, cross-species infection, and respiratory transmission of paramyxoviruses. More efforts need to be made to champion the use of veterinary pathogens as acceptable surrogates to study key elements of pathogen emergence. This in turn will require the paucity of immunological and cell biological reagents for commonly used species like ferrets and hamsters to be developed. While there are tractable animal models for henipaviruses, the viruses are primarily (except for Cedar virus) Biosafety Level (BSL) 4, making these challenging to work with for most researchers and less suitable as prototypes. Key questions remain: Can we improve the predictive power of animal models for humans, and can human challenge studies [31] augment or replace animal models for some viruses?
Medical Countermeasures
The primary medical countermeasure for human and animal paramyxoviruses has been live-attenuated vaccines, and the measles and mumps vaccines have been in widespread use for decades [32]. Live-attenuated vaccines for CDV and parainfluenza virus 5 have also been developed, and extensive work on the immune response in animals has been performed [33]. While these vaccines are highly effective, rapid creation of a new live-attenuated vaccine is slow and challenging, especially as the molecular determinants of attenuation remain unclear. How these vary across the family remain to be deciphered. New subunit vaccine candidates for HRSV and NiV based on F-G glycoproteins and stabilized by a molecular clamp have also been reported [34]. Piloting the rapid development of an mRNA or subunit vaccine and demonstrating equivalence to the live-attenuated licensed product should be a high priority for at least 1 member of the paramyxoviruses. A protein-based subunit vaccine for HeV has been successfully deployed in horses [35, 36], providing proof of principle for paramyxovirus protein-based vaccines. Cross-protection to NiV has been observed, and correlates of protection for this vaccine are currently being investigated. There remains a need to develop pan-paramyxovirus antivirals. However, this has been challenging for a number of reasons. Virus resurgence, reemergence in highly vaccinated populations, and vaccine hesitancy alongside the global efforts to eradicate MV suggest that some priority needs to be given to remove this roadblock [37].
PROTOTYPE PATHOGENS: KEY CONSIDERATIONS AND RECOMMENDATIONS
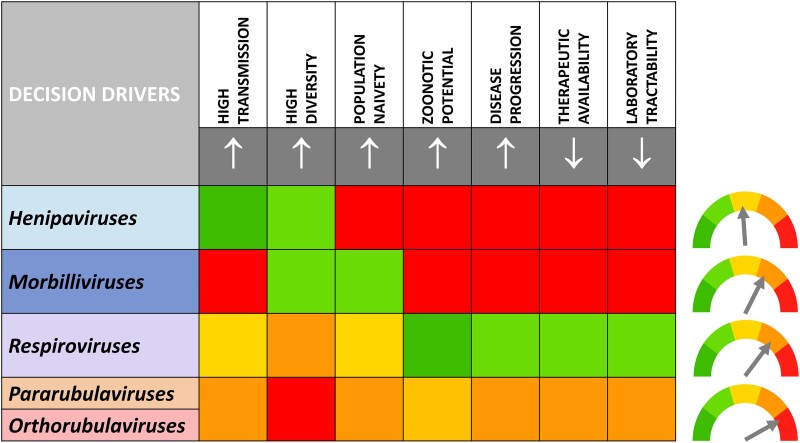
Consideration of the optimal prototype pathogens and which viral subfamily they should come from was driven by a variety of factors, transmissibility, genetic diversity, population naivety to subfamily members, zoonotic potential, morbidity and mortality, availability of therapeutics to current subfamily members, and the tractability of working with the pathogen in a laboratory setting. With this in mind, we developed a paramyxovirus pandemic pathogen prioritization matrix (Figure 3) to facilitate and focus a collective, objective assessment. Our goal was to formulate a general approach to prototype prioritization that could be used to select viruses from any family by focusing the 7 key decision drivers.
Figure 3.
Pandemic pathogen prioritization matrix. Five decision drivers considered to increase the likelihood of selection of a prototypic virus from the selected genera (↑) and 2 decision drivers considered to decrease the likelihood of selection of a prototypic virus from the selected genera (↓) were prioritized by the Paramyxovirus Expert Group. These were assigned a number from 1 (dark green) to 5 (red) by the group with of goal of objective ranking. This evaluative tool proved to be useful in helping the group to prioritize prototypes iteratively. This led to a ranking of henipaviruses < morbilliviruses < respiroviruses < pararubulaviruses/orthorubulaviruses as indicated by the arrows.
Henipaviruses
NiV and HeV represent a considerable current threat, given the high mortality rates (>40% for NiV) and zoonotic transmission [38]. However, while human-to-human transmission has been documented, the low R0 value for these viruses reduces the concern about evolution to a highly transmissible agent. Recognition of the importance of these pathogens has resulted in significant funding for vaccine and antiviral research, including through the Coalition for Epidemic Preparedness Innovations, though to date only a subunit equine vaccine has been approved (see Medical Countermeasures). Use of either HeV or NiV as a prototype pathogen is significantly hampered by the need to work with these select agents at BSL-4. However, recent identification of the nonpathogenic henipavirus Cedar virus may provide a potential prototype from this subfamily that could be more widely worked on due to its BSL-2+ designation [39]. Of the 4 subfamilies considered, selection of a prototypic paramyxovirus from the currently known henipaviruses was considered the lowest priority (Figure 3).
Morbilliviruses
Several characteristics of morbilliviruses make them potential pandemic threats. They are among the most highly transmissible viruses known [40]; a number of emerging morbilliviruses have been recently identified in cats, pigs, and bats [41–43]. They have shown a significant propensity to jump species, best exemplified by reverse zoonotic outbreaks caused by CDV in macaques [44, 45]. This represents a significant concern for vaccination strategies post–MV eradication. At present, given the widespread use and efficacy of the live-attenuated MV vaccine, population naivety in the developed world is not a concern. However, vaccine hesitancy, political conflicts, and the COVID-19 pandemic are leading to a lower number of children who have been fully or partially vaccinated against MV. This is reflected by the increase in deaths due to measles from 2016 to 2019 by 50%. The current dogma is that morbillivirus vaccines are cross-protective. However, understanding the level of cross-genus protection to novel morbilliviruses that is provided by measles vaccination is important. Likewise, dissecting barriers that restrict cross-species infection of these highly transmissible pathogens using structural biology to map evolutionary trajectories was considered a priority. All known morbilliviruses are BSL-2 agents; therefore, they can be studied by the largest number of researchers. Potential prototype pathogens are CDV or a novel bat morbillivirus. Of the 4 subfamilies considered, selection of a prototypic paramyxovirus from the currently known morbillivirus was considered a midlevel priority (Figure 3).
Respiroviruses
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has demonstrated the disastrous consequences of a moderately transmissible respiratory pathogen infecting a naive human population in the absence of vaccines, antivirals, or biopharmaceutical interventions. This should increase the concern that members of the respirovirus subfamily could emerge and spread rapidly across the globe. Novel respiroviruses have recently been identified, including from pangolins and squirrels, though the zoonotic nature remains to be clarified. All know respiroviruses are BSL-2 agents; therefore, they can be studied by the largest number of researchers. Established respiroviruses such as HPIV-1 or -3 or emerging pathogens such as the bat respiroviruses could serve as prototypes for this subfamily. Of the 4 subfamilies considered, selection of a prototypic paramyxovirus from the currently known morbilliviruses was considered a midlevel priority (Figure 3).
Rubulaviruses
There are key similarities between these and the respiroviruses, and many equivalent arguments for prioritization apply. Factors that decrease concern that a rubulavirus with pandemic potential might emerge stem primarily from the availability of a safe and efficacious live-attenuated vaccine for MuV. However, in the past 12 years, multiple mumps outbreaks have occurred in vaccinated young adults and concerns have been raised about waning immunity [46]. This, coupled with greater genetic diversity in circulating MuV strains, has led some to suggest that an updated MuV or a third dose might be needed in doubly vaccinated adolescents. If the current mumps vaccine fails to induce durable immunity, this in turn may reduce potential cross-protection to new members of this genus that can infect humans. Other factors that increase concerns about an emerging rubulavirus include ability to infect numerous cell types due to the use of sialic acid and the fact that novel members have been found in animal reservoirs and zoonotic transmission has been observed [47, 48]. All known rubulaviruses are BSL-2 agents, so they can be studied by the largest number of researchers. Potential prototypes include the recently identified Menangle, Sosuga, Tioman, and Achimota viruses or a bat rubulavirus. Either HPIV-2 or parainfluenza virus 5 could also serve as a prototype. Given the fact MuV is a human pathogen that has been understudied and there are questions about the longevity of immune responses provided by the existing vaccine that may be overattenuated, there are compelling arguments to use human MuV as the prototypic pathogen. Of the 4 subfamilies considered, selection of a prototypic paramyxovirus from the currently known rubulaviruses was considered a top priority (Figure 3).
CONCLUSIONS
Prototype pathogen prioritization and selection is an important but challenging task. However, its success has been demonstrated by focused, fundamental research into Middle East respiratory syndrome coronavirus vaccine development through structure-guided, antigen design. This knowledge was elegantly leveraged into SARS-CoV-2 spike glycoprotein stabilization during the COVID-19 pandemic during the development of mRNA vaccines [49]. We took an agnostic approach to prototype choice and developed an evaluative tool that should be used iteratively for downstream selection. This was used to rank subfamilies within the Paramyxoviridae and suggest tractable prototypes that should be studied in greater depth with a view to understanding virus emergence and evolution.
Contributor Information
W Paul Duprex, Center for Vaccine Research; Department of Microbiology and Molecular Genetics, University of Pittsburgh, Pennsylvania.
Rebecca Ellis Dutch, Department of Molecular and Cellular Biochemistry, University of Kentucky College of Medicine, Lexington.
Notes
Acknowledgments. We are extremely grateful to the outstanding group of virologists whose lively and wisdom-filled discussion during several focus groups formed the basis for this review. This Paramyxovirus Expert Group was comprised of Christopher Broder, Uniformed Services University; Emmie De Wit, National Institute of Allergy and Infectious Diseases; Jan Felix Drexler, Charité–Universitätsmedizin Berlin; Benhur Lee, Icahn School of Medicine at Mount Sinai; Anne Moscona, Columbia University; Richard Plemper, Georgia State University; Veronika von Messling, German Federal Ministry of Education and Research; and Linfa Wang, Duke University–National University of Singapore Medical School. We thank Kearstin Edmonds for drawing the generalized viral life cycle; Andres Moreira-Soto and Jan Felix Drexler for generating the paramyxovirus phylogenetic tree; and Linda Rennick for careful review and critical input into the manuscript.
Financial support. This work was supported by the National Institutes of Health (grant numbers 5R01 AI40758 and 1R01 AI051517 to R. E. D.) and the Coalition for Epidemic Preparedness Innovations and the Henry Hillman Family Foundation (to W. P. D.).
Supplement sponsorship. This article appears as part of the supplement “Pandemic Preparedness at NIAID: Prototype Pathogen Approach to Accelerate Medical Countermeasures—Vaccines and Monoclonal Antibodies,” sponsored by the National Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD.
References
- 1. Plemper RK, Lamb RA, Knipe DM, Howley PM. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, eds. Fields virology. 7th ed. Vol 1. Philadelphia, PA: Wolters Kluwer, 2021:504–58. [Google Scholar]
- 2. Hamilton K, Baron MD, Matsuo K, Visser D. Rinderpest eradication: challenges for remaining disease free and implications for future eradication efforts. Rev Sci Tech 2017; 36:579–88. [DOI] [PubMed] [Google Scholar]
- 3. Thibault PA, Watkinson RE, Moreira-Soto A, Drexler JF, Lee B. Zoonotic potential of emerging paramyxoviruses: knowns and unknowns. Adv Virus Res 2017; 98:1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature 2000; 406:893–7. [DOI] [PubMed] [Google Scholar]
- 5. Negrete OA, Levroney EL, Aguilar HC, et al. Ephrinb2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 2005; 436:401–5. [DOI] [PubMed] [Google Scholar]
- 6. Navaratnarajah CK, Generous AR, Yousaf I, Cattaneo R. Receptor-mediated cell entry of paramyxoviruses: mechanisms, and consequences for tropism and pathogenesis. J Biol Chem 2020; 295:2771–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azarm KD, Lee B. Differential features of fusion activation within the Paramyxoviridae. Viruses 2020; 12:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carlos TS, Young DF, Schneider M, Simas JP, Randall RE. Parainfluenza virus 5 genomes are located in viral cytoplasmic bodies whilst the virus dismantles the interferon-induced antiviral state of cells. J Gen Virol 2009; 90:2147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang S, Jiang Y, Cheng Q, Zhong Y, Qin Y, Chen M. Inclusion body fusion of human parainfluenza virus type 3 regulated by acetylated alpha-tubulin enhances viral replication. J Virol 2017; 91:01802–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma D, George CX, Nomburg JL, Pfaller CK, Cattaneo R, Samuel CE. Upon infection, cellular WD repeat-containing protein 5 (WDR5) localizes to cytoplasmic inclusion bodies and enhances measles virus replication. J Virol 2018; 92:e01726–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Najjar F E, Schmitt AP, Dutch RE. Paramyxovirus glycoprotein incorporation, assembly and budding: a three way dance for infectious particle production. Viruses 2014; 6:3019–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rima B, Balkema-Buschmann A, Dundon WG, et al. ICTV virus taxonomy profile: Paramyxoviridae. J Gen Virol 2019; 100:1593–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemon K, de Vries RD, Mesman AW, et al. Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathog 2011; 7:e1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muhlebach MD, Mateo M, Sinn PL, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 2011; 480:530–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ludlow M, Lemon K, de Vries RD, et al. Measles virus infection of epithelial cells in the macaque upper respiratory tract is mediated by subepithelial immune cells. J Virol 2013; 87:4033–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh BK, Li N, Mark AC, Mateo M, Cattaneo R, Sinn PL. Cell-to-cell contact and nectin-4 govern spread of measles virus from primary human myeloid cells to primary human airway epithelial cells. J Virol 2016; 90:6808–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moscona A. Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J Clin Invest 2005; 115:1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henrickson KJ. Parainfluenza viruses. Clin Microbiol Rev 2003; 16:242–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leung NHL. Transmissibility and transmission of respiratory viruses. Nat Rev Microbiol 2021; 19:528–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rima BK, Duprex WP. Morbilliviruses and human disease. J Pathol 2006; 208:199–214. [DOI] [PubMed] [Google Scholar]
- 21. Iketani S, Shean RC, Ferren M, et al. Viral entry properties required for fitness in humans are lost through rapid genomic change during viral isolation. mBio 2018; 9:e00898–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rennick LJ, de Vries RD, Carsillo TJ, et al. Live-attenuated measles virus vaccine targets dendritic cells and macrophages in muscle of nonhuman primates. J Virol 2015; 89:2192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rennick LJ, Nambulli S, Lemon K, et al. Recombinant subtype A and B human respiratory syncytial virus clinical isolates co-infect the respiratory tract of cotton rats. J Gen Virol 2020; 101:1056–68. [DOI] [PubMed] [Google Scholar]
- 24. Greninger AL, Rybkina K, Lin MJ, et al. Human parainfluenza virus evolution during lung infection of immunocompromised individuals promotes viral persistence. J Clin Invest 2021; 131:e150506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Young DF, Wignall-Fleming EB, Busse DC, et al. The switch between acute and persistent paramyxovirus infection caused by single amino acid substitutions in the RNA polymerase P subunit. PLoS Pathog 2019; 15:e1007561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharp CR, Nambulli S, Acciardo AS, et al. Chronic infection of domestic cats with feline morbillivirus, United States. Emerg Infect Dis 2016; 22:760–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rima BK, Duprex WP. Molecular mechanisms of measles virus persistence. Virus Res 2005; 111:132–47. [DOI] [PubMed] [Google Scholar]
- 28. Munoz-Fontela C, Dowling WE, Funnell SGP, et al. Animal models for COVID-19. Nature 2020; 586:509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nguyen DT, de Vries RD, Ludlow M, et al. Paramyxovirus infections in ex vivo lung slice cultures of different host species. J Virol Methods 2013; 193:159–65. [DOI] [PubMed] [Google Scholar]
- 30. Noll KE, Ferris MT, Heise MT. The collaborative cross: a systems genetics resource for studying host-pathogen interactions. Cell Host Microbe 2019; 25:484–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahmad A, Eze K, Noulin N, et al. EDP-938, a respiratory syncytial virus inhibitor, in a human virus challenge. N Engl J Med 2022; 386:655–66. [DOI] [PubMed] [Google Scholar]
- 32. Buczkowski H, Muniraju M, Parida S, Banyard AC. Morbillivirus vaccines: recent successes and future hopes. Vaccine 2014; 32:3155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. da Fontoura Budaszewski R, von Messling V. Morbillivirus experimental animal models: measles virus pathogenesis insights from canine distemper virus. Viruses 2016; 8:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Isaacs A, Cheung STM, Thakur N, et al. Combinatorial F-G immunogens as Nipah and respiratory syncytial virus vaccine candidates. Viruses 2021; 13:1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Broder CC, Weir DL, Reid PA. Hendra virus and Nipah virus animal vaccines. Vaccine 2016; 34:3525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amaya M, Broder CC. Vaccines to emerging viruses: Nipah and Hendra. Annu Rev Virol 2020; 7:447–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Plemper RK. Measles resurgence and drug development. Curr Opin Virol 2020; 41:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ang BSP, Lim TCC, Wang L. Nipah virus infection. J Clin Microbiol 2018; 56:e01875–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marsh GA, de Jong C, Barr JA, et al. Cedar virus: a novel henipavirus isolated from Australian bats. PLoS Pathog 2012; 8:e1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roberts L. Is measles next? Science 2015; 348:958–61, 963. [DOI] [PubMed] [Google Scholar]
- 41. Woo PC, Lau SK, Wong BH, et al. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc Natl Acad Sci U S A 2012; 109:5435–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arruda B, Shen H, Zheng Y, Li G. Novel morbillivirus as putative cause of fetal death and encephalitis among swine. Emerg Infect Dis 2021; 27:1858–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Drexler JF, Corman VM, Muller MA, et al. Bats host major mammalian paramyxoviruses. Nat Commun 2012; 3:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qiu W, Zheng Y, Zhang S, et al. Canine distemper outbreak in rhesus monkeys, China. Emerg Infect Dis 2011; 17:1541–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sakai K, Nagata N, Ami Y, et al. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J Virol 2013; 87:1105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rasheed MAU, Hickman CJ, McGrew M, et al. Decreased humoral immunity to mumps in young adults immunized with MMR vaccine in childhood. Proc Natl Acad Sci U S A 2019; 116:19071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barr JA, Smith C, Marsh GA, Field H, Wang LF. Evidence of bat origin for Menangle virus, a zoonotic paramyxovirus first isolated from diseased pigs. J Gen Virol 2012; 93:2590–4. [DOI] [PubMed] [Google Scholar]
- 48. Albarino CG, Foltzer M, Towner JS, et al. Novel paramyxovirus associated with severe acute febrile disease, South Sudan and Uganda, 2012. Emerg Infect Dis 2014; 20:211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Corbett KS, Edwards DK, Leist SR, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020; 586:567–71. [DOI] [PMC free article] [PubMed] [Google Scholar]