Significance
Severe acute respiratory syndrome coronavirus (SARS-CoV)-2 causes a spectrum of respiratory disease ranging from asymptomatic infections to severe pneumonia and death. Innate immune responses during SARS-CoV-2 infection have been associated with clinical disease severity, with robust early interferon responses in the nasal epithelium reported to be protective. Thus, elucidating mechanisms through which SARS-CoV-2 induces and antagonizes host innate immune responses is crucial to understanding viral pathogenesis. CoVs encode various innate immune antagonists, including the conserved nonstructural protein 15 (nsp15) which contains an endoribonuclease (EndoU) domain. We demonstrate that SARS-CoV-2 EndoU is a crucial interferon antagonist, by providing further evidence for the role of the conserved CoV nsp15 in antagonizing innate immune activation, thereby optimizing CoV replication.
Keywords: SARS-CoV-2, endoribonuclease, nsp15, interferon, dsRNA-induced pathways
Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV)-2 has caused millions of deaths since its emergence in 2019. Innate immune antagonism by lethal CoVs such as SARS-CoV-2 is crucial for optimal replication and pathogenesis. The conserved nonstructural protein 15 (nsp15) endoribonuclease (EndoU) limits activation of double-stranded (ds)RNA-induced pathways, including interferon (IFN) signaling, protein kinase R (PKR), and oligoadenylate synthetase/ribonuclease L (OAS/RNase L) during diverse CoV infections including murine coronavirus and Middle East respiratory syndrome (MERS)-CoV. To determine how nsp15 functions during SARS-CoV-2 infection, we constructed a recombinant SARS-CoV-2 (nsp15mut) expressing catalytically inactivated nsp15, which we show promoted increased dsRNA accumulation. Infection with SARS-CoV-2 nsp15mut led to increased activation of the IFN signaling and PKR pathways in lung-derived epithelial cell lines and primary nasal epithelial air–liquid interface (ALI) cultures as well as significant attenuation of replication in ALI cultures compared to wild-type virus. This replication defect was rescued when IFN signaling was inhibited with the Janus activated kinase (JAK) inhibitor ruxolitinib. Finally, to assess nsp15 function in the context of minimal (MERS-CoV) or moderate (SARS-CoV-2) innate immune induction, we compared infections with SARS-CoV-2 nsp15mut and previously described MERS-CoV nsp15 mutants. Inactivation of nsp15 had a more dramatic impact on MERS-CoV replication than SARS-CoV-2 in both Calu3 cells and nasal ALI cultures suggesting that SARS-CoV-2 can better tolerate innate immune responses. Taken together, SARS-CoV-2 nsp15 is a potent inhibitor of dsRNA-induced innate immune response and its antagonism of IFN signaling is necessary for optimal viral replication in primary nasal ALI cultures.
There are currently seven known human coronaviruses (HCoVs), three of which emerged in the past 20 y and cause severe disease [severe acute respiratory syndrome (SARS)-CoV, Middle East respiratory syndrome (MERS)-CoV, and SARS-CoV-2]. SARS-CoV-2 is the causative agent of COVID-19 and the ongoing pandemic that has claimed millions of lives worldwide (1).
CoV genomes are positive-sense, single-stranded (ss)RNAs of approximately 30 kilobases in length. The 5′ proximal two-thirds of the CoV genome is composed of two open reading frames (ORFs), ORF1a and ORF1b, that encode 16 nonstructural proteins (nsps), while the 3′ proximal third encodes structural and accessory proteins. Following viral entry, ORF1a and ORF1b are translated from genomic RNA and processed into nsps that form replication–transcription complexes (RTCs) localized to double-membrane vesicles formed from rearranged endoplasmic reticulum. Nsps also function as RNA-processing enzymes, proteases, and protein modifiers (2). Here, we focused on nsp15, which contains an endoribonuclease (EndoU) that is localized to RTCs during infection. We previously reported that, during infection of bone marrow–derived macrophages (BMMs), murine coronavirus nsp15 EndoU cleaves genomic RNA with a preference for U↓A and C↓A sequences (3). Alternatively, it has been proposed that nsp15 EndoU cleaves polyU sequences on the 5′-end of negative-sense, antigenome RNA (4). Both studies concluded that EndoU limits activation of double-stranded (ds)RNA-induced antiviral responses, including interferon (IFN) signaling, the oligoadenylate synthetase/ribonuclease L (OAS/RNase L) pathway, and the protein kinase R (PKR) pathway (2, 3, 5).
DsRNA is a by-product of viral replication that can be detected by host cell sensors. Melanoma differentiation-associated protein 5 (MDA5) detects dsRNA and triggers the production of type I and type III IFNs (6–9). Once secreted, IFN binds to its receptor on both infected and uninfected cells, which results in Janus kinase (JAK) activation and phosphorylation of signal transducer and activator of transcription (STAT)1 and STAT2. STAT phosphorylation and subsequent nuclear translocation induces transcription from promoters that regulate a coordinated antiviral response involving hundreds of interferon-stimulated genes (ISGs) (10–12). Upon sensing of dsRNA, OASs (isoforms 1-3) produce 2′-5′-oligoadenylates (2-5A), which activate RNase L. RNase L cleaves both viral and cellular RNAs, thereby suppressing viral replication and protein synthesis as well as inducing inflammation and apoptosis (13, 14). PKR autophosphorylates upon sensing of dsRNA and then phosphorylates eukaryotic initiation factor 2 (eIF2)α, leading to activation of the integrated stress response and inhibition of protein synthesis (15). Both PKR and OASs are ISGs and, due to the interconnected nature of these pathways, dsRNA is a key regulator of many antiviral host cell responses.
Previous studies have characterized the endoribonuclease activity of nsp15 expressed by murine coronavirus, mouse hepatitis virus (MHV) (3, 5, 16, 17). Mutation of either catalytic histidine (H262A or H277A) within the EndoU domain of MHV nsp15 promotes increased induction of type I IFN, activation of the OAS/RNase L and PKR pathways, and significant attenuation of replication in both BMMs and in mice (16, 17). These findings suggest that inactivation of EndoU results in increased sensing of dsRNA by host sensors which was concluded to occur due to either an increase in total dsRNA or a redistribution of dsRNA away from RTCs (3, 16). Similar findings were reported with mutant viruses expressing catalytically inactive nsp15 of HCoV-229E, avian infectious bronchitis virus (IBV), porcine epidemic diarrhea virus (PEDV), and MERS-CoV (5, 16–20). We have previously characterized multiple recombinant MERS-CoV mutant viruses with catalytic inactivation of nsp15, deletion of accessory protein NS4a (a dsRNA-binding protein), inactivation of accessory protein NS4b (which contains a phosphodiesterase that specifically antagonizes RNase L activation and also functions as an IFN signaling inhibitor via inhibition of the NF-κB pathway), or double mutants of nsp15/NS4a or nsp15/NS4b (19, 21–24). We found that although the MERS-CoV nsp15 mutant virus had a minor replication defect in respiratory epithelial cell lines, the double mutants were significantly more attenuated and elicited increased expression of IFN and ISG mRNAs (19). These data suggest that nsp15 is crucial to CoV evasion of host detection due to its ability to antagonize dsRNA-induced innate immune responses.
Published reports on SARS-CoV-2 nsp15 have utilized either in vitro biochemical assays, ectopic overexpression, or bioinformatic approaches to characterize the structural and RNA-binding/cleavage capacity of SARS-CoV-2 nsp15 (25–34). However, these studies were carried out in the absence of other viral proteins and moreover did not address the role of nsp15 in SARS-CoV-2 infection. Therefore, to understand the activity of nsp15 EndoU in the context of authentic infection, we generated and characterized a recombinant SARS-CoV-2 expressing a catalytically inactive nsp15 (SARS-CoV-2 nsp15mut). We assessed the impact of nsp15 on viral replication and host responses in lung-derived epithelial cell lines as well as primary nasal epithelial air–liquid interface (ALI) cultures, which recapitulate many features of the in vivo upper airway, including its heterogeneous cellular population (primarily ciliated epithelial cells and mucus-producing goblet cells) and mucociliary functions (35, 36). These nasal ALI cultures mimic the initial site of infection, where innate immune responses are crucial for early control of viral infections (37, 38). Our data indicate that SARS-CoV-2 nsp15 contributes to innate immune evasion by reducing dsRNA accumulation, dampening IFN induction, ISG expression, and PKR pathway activation in all cell types examined, and that inactivation of nsp15 results in attenuation of replication in primary cell ALI cultures.
Results
Generation of Recombinant SARS-CoV-2 nsp15mut Expressing an Inactive Endoribonuclease.
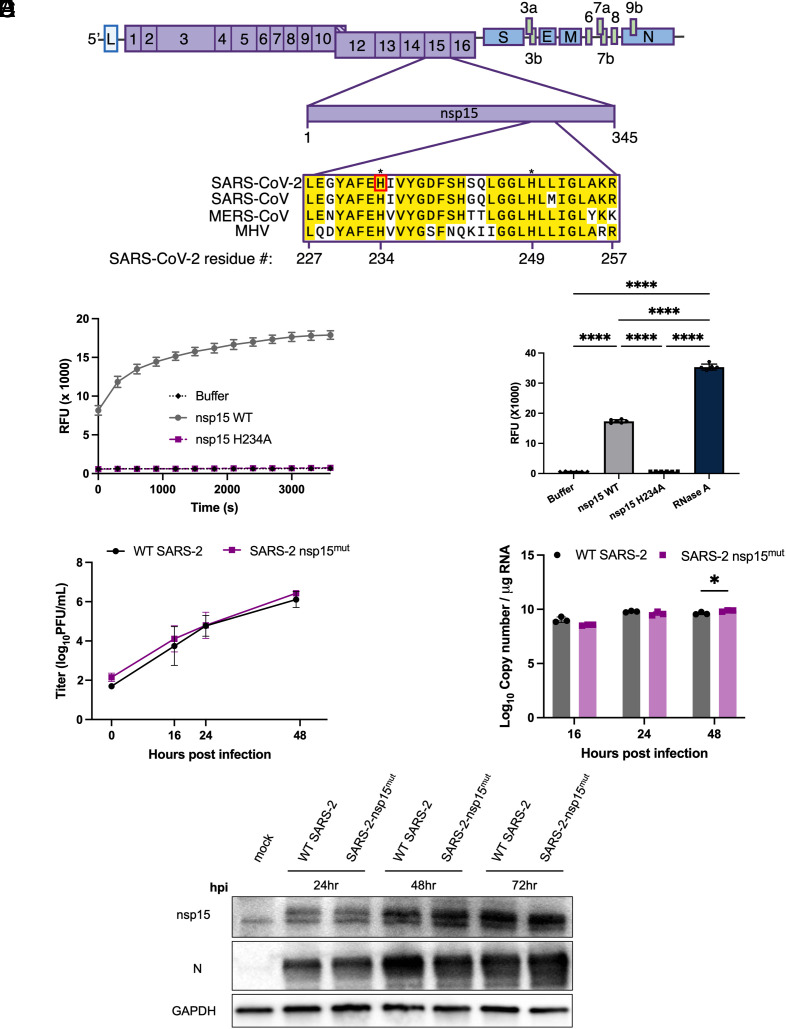
To determine the role of nsp15 in antagonizing dsRNA-induced innate immune responses during SARS-CoV-2 infection, we generated a recombinant SARS-CoV-2 expressing a catalytically inactive nsp15 (denoted SARS-CoV-2 nsp15mut) using a bacterial artificial chromosome (BAC)-based reverse genetic system (39). SARS-CoV-2 nsp15 contains two catalytic histidines at amino acid residue positions 234 and 249, which are conserved among coronaviruses, as illustrated for a subset of betacoronaviruses in Fig. 1A (40). We generated a catalytically inactive nsp15 via a histidine (H) to alanine (A) substitution at amino acid position 234 (nsp15H234A). To validate that the H234A substitution was sufficient to abrogate endoribonuclease activity, wild-type (WT) nsp15 and the H234A mutant proteins were expressed in Escherichia coli and purified to homogeneity (SI Appendix, Fig. S1). Endoribonuclease activity of both nsp15 proteins was tested using a fluorescence-based assay in which a synthetic ssRNA substrate containing a Förster resonance energy transfer pair was incubated with each recombinant nsp15. Endoribonuclease activity results in cleavage of ssRNA substrate and fluorescence detection (Fig. 1 B and C). While nuclease activity was observed for WT nsp15, the activity of nsp15H234A was significantly lower and comparable to the negative control. These results suggest that the H234A substitution was sufficient to abolish the endoribonuclease activity of SARS-CoV-2 nsp15.
Fig. 1.
Construction of recombinant SARS-CoV-2 with inactive nsp15mut endoribonuclease. (A) Diagram of the SARS-CoV-2 genome. Full-length nsp15 of SARS-CoV-2 is shown in the center. Sequence alignment among betacoronaviruses, from Top to Bottom: SARS-CoV-2 USA-WA1/2020, SARS-CoV, MERS-CoV EMC/2012, and MHV strain A59. Conserved residues are shown in yellow with the catalytic histidine residues of SARS-CoV-2, H234, and H249, designated with asterisks. Nsp15 mutation site for SARS-CoV-2 nsp15mut generated in this study, H234, shown in red. (B) SARS-CoV-2 WT, nsp15mut, and buffer alone catalytic activity measured by dequenching of 6-FAM upon ssRNA cleavage measured in relative fluorescence units recorded at 5-min intervals over a 60-min period. (C) Total quantified ssRNA cleavage measured for WT nsp15, nsp15mut, buffer alone, and RNase A at the end of 1 h. (D–F) VeroE6 cells were infected with either WT or nsp15mut SARS-CoV-2 (MOI 0.1 in D and MOI 1 in E and F). (D) Supernatants were collected at indicated times post infection and titered via plaque assay. Data shown are the average of two independent experiments. (E) Intracellular RNA was collected at the indicated time points post infection and genome copy number per μg RNA was quantified by RT-qPCR using a standard curve and primers directed against SARS-CoV-2 RdRp (nsp12). (F) Cells were lysed at indicated time points using protein lysis buffer. Samples were separated via SDS-PAGE and transferred to a PVDF membrane for immune detection with antibodies against SARS-CoV-2 nsp15, SARS-CoV-2 N, and GAPDH.
To determine whether the H234A substitution was associated with an inherent viral growth defect, we compared the kinetics of replication of SARS-CoV-2 nsp15mut with that of WT SARS-CoV-2 in VeroE6 cells, an IFN-deficient cell line (41, 42). Viral growth curves quantifying infectious virus in the supernatant of infected cells at various time points post infection revealed no significant difference between WT and nsp15mut SARS-CoV-2 (Fig. 1D). Corroborating this finding, no significant differences in viral genome copy number (quantified via RT-qPCR from intracellular RNA) were detected (Fig. 1E). Additionally, western blots probed for nsp15 and nucleocapsid (N) protein revealed no detectable differences in viral protein expression between WT and nsp15mut SARS-CoV-2 (Fig. 1F). Thus, the H234A catalytic mutation in SARS-CoV-2 nsp15mut does not directly impact SARS-CoV-2 replication in IFN-incompetent VeroE6 cells.
Inactivation of SARS-CoV-2 nsp15 Results in Increased dsRNA Accumulation.
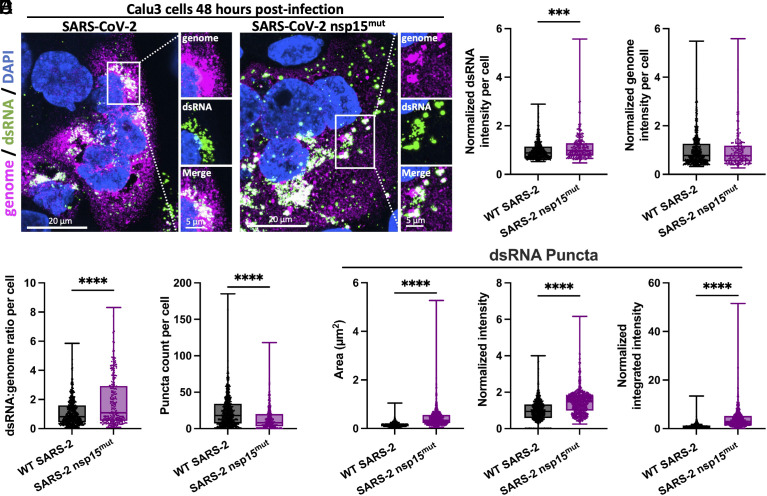
Given the in vitro data confirming that catalytic inactivation of SARS-CoV-2 nsp15 resulted in reduced endoribonuclease activity, we infected the human lung-derived Calu3 cell line with either WT or nsp15mut SARS-CoV-2 and evaluated viral genome expression (ORF1a oligonucleotide probes) and dsRNA (K1 antibody) abundance using single-molecule fluorescent in situ hybridization (smFISH) and immunofluorescence; representative images are shown in Fig. 2A and SI Appendix, Fig. S2. No significant difference in average viral genome intensity between WT and nsp15mut SARS-CoV-2 infected cells was detected, however, SARS-CoV-2 nsp15mut infected cells had greater total dsRNA intensity per cell and more dsRNA per genome (Fig. 2 B–D). Interestingly, SARS-CoV-2 nsp15mut infected cells contained on average fewer dsRNA puncta than WT SARS-CoV-2 infected cells (Fig. 2E). However, further analysis of dsRNA puncta revealed that the average size, intensity, and integrated intensity of dsRNA puncta was increased during nsp15mut infection (Fig. 2F). These data are consistent with a role for SARS-CoV-2 nsp15 EndoU in limiting dsRNA accumulation during infection.
Fig. 2.
SARS-2 nsp15mut induces elevated levels of dsRNA. (A) Immunofluorescence assay for dsRNA using K1 antibody (green) and smFISH using fluorescent probes targeting ORF1a of the SARS-CoV-2 genome (magenta) in Calu3 cells. Regions positive for both genome and dsRNA are shown in white in merged panels. Nuclei were stained with DAPI (blue). High-resolution imaging of SARS-CoV-2 genome and dsRNA using 100× objective. (B) Intensity of dsRNA in individual cells. (C) Intensity of the viral genome in individual cells. (D) Ratio of dsRNA:genome in individual cells. (E) Number of dsRNA puncta per cell. (B–E) Data represents individual cells (dots) from three fields of view from two independent experiments. 652 cells for WT SARS-CoV-2 and 239 cells for SARS-CoV-2 nsp15mut were quantified in B–D. 848 cells for WT SARS-CoV-2 and 354 cells for SARS-CoV-2 nsp15mut were quantified in E. (F) Quantification of the area, mean intensity, and integrated intensity (total intensity) of dsRNA. 975 puncta from 19 cells in six fields of view were quantified for WT SARS-CoV-2. 719 puncta from 22 cells were quantified in six fields of view for SARS-CoV-2 nsp15mut. All intensity data (B–D and F) are shown as normalized values relative to the average intensity for WT SARS-CoV-2 in each experiment. Statistical significance was determined using Student’s t-test, P < 0.0001 (****).
Infection with SARS-CoV-2 nsp15mut Promotes Increased IFN Signaling and PKR Pathway Activation Compared to WT SARS-CoV-2 in Respiratory Epithelial Cell Lines.
To understand the role that SARS-CoV-2 nsp15 plays in evading dsRNA-induced innate immunity, we utilized two lung-derived epithelial cell lines, Calu3 cells and A549 cells transduced to stably express the SARS-CoV-2 receptor, human angiotensin converting-enzyme 2 (ACE2) (A549-ACE2). Both of these cell lines have intact IFN signaling, PKR pathway, and OAS/RNase L pathway responses. We and others have previously reported that WT SARS-CoV-2 induces all of these pathways during infection of both Calu3 and A549-ACE2 cells (9, 43–45).
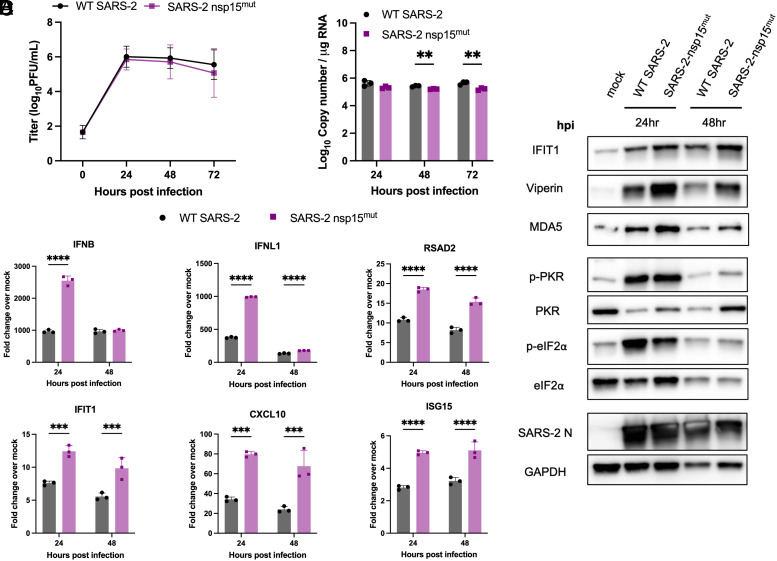
Infection of either Calu3 (Fig. 3A) or A549-ACE2 (Fig. 4A) cells with SARS-CoV-2 nsp15mut revealed no significant difference in replication kinetics compared to WT SARS-CoV-2 (Figs. 3A and 4A). However, a slight but significant decrease in intracellular viral genome copies was detected at 48 and 72 hours post infection (hpi) during SARS-CoV-2 nsp15mut infection compared to WT in Calu3 cells (Fig. 3B).
Fig. 3.
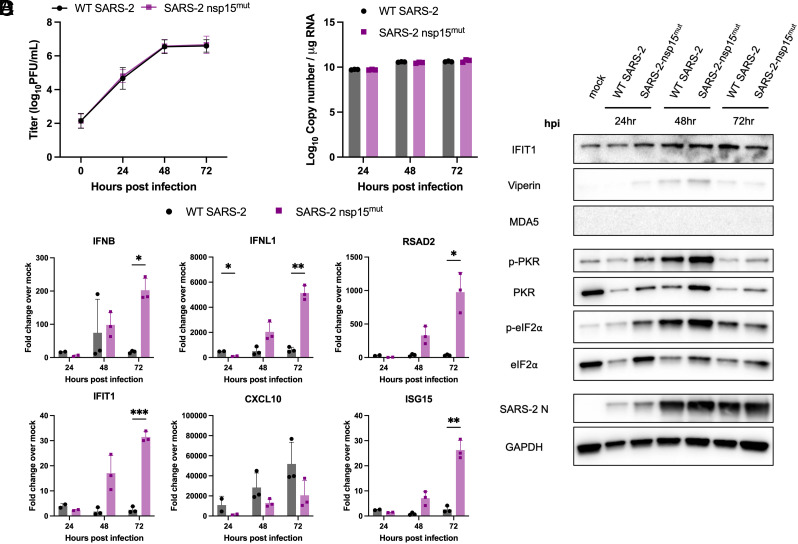
SARS-2 nsp15mut induces increased IFN mRNA and ISG expression in Calu3 cells compared to WT SARS-CoV-2. Calu3 cells were infected with either WT or nsp15mut SARS-CoV-2 at MOI 0.1 in (A) and MOI 1 in (B–D). (A) Supernatants were collected at 0, 24, 48, and 72 hpi and viral titers were measured by plaque assay. Data shown are the average of three independent experiments. (B and C) At the indicated time points post infection, intracellular RNA was extracted and analyzed by RT-qPCR. (B) Genome copy number was quantified using primers against SARS-CoV-2 nsp12 and a standard curve. (C) Relative mRNA expression of IFNB, IFNL1, and four representative ISGs: RSAD2, IFIT1, CXCL10, and ISG15. Fold change is expressed as 2−Δ(ΔCt). (D) Whole cell lysates were resolved using SDS-PAGE and analyzed via western blot analysis using antibodies against indicated proteins. Data in B–D are from one representative experiment of three total experiments.
Fig. 4.
SARS-2 nsp15mut induces increased IFN mRNA and ISG expression as well as PKR activation in A549-ACE cells compared to WT SARS-CoV-2. A549-ACE2 cells were infected with either WT or nsp15mut SARS-CoV-2 at MOI 0.1 in (A) or MOI 1 in (B–D). (A) At indicated time points, supernatants were collected and infectious virus was measured by plaque assay. Data shown are the average of three independent experiments. (B–D) At 24, 48, and 72 hpi either intracellular RNA was extracted and analyzed by RT-qPCR or whole cell lysates were collected. (B) Genome copy number was calculated using a standard curve and primers targeting SARS-CoV-2 RdRp. (C) Relative mRNA expression of IFNB, IFNL1, and four representative ISGs: RSAD2, IFIT1 CXCL10, and ISG15 were measured. Fold change is expressed as 2−Δ(ΔCt). (D) Whole cell lysates were separated via SDS-PAGE and analyzed by western blot analysis with antibodies against indicated proteins. Data in B–D are from one representative experiment of three total experiments.
Expression of type I (IFNB) and type III IFN (IFNL1) mRNAs, as well as mRNA expression of four representative ISGs [radical S-adenosyl methionine domain containing 2 (RSAD2), IFN-induced protein with tetratricopeptide repeats 1 (IFIT1), C-X-C motif chemokine ligand 10 (CXCL10), and ISG15] were quantified by RT-qPCR to detect activation of the IFN signaling pathway (Figs. 3C and 4C). Despite the lack of viral attenuation, we observed increased mRNA induction of both type I and type III IFNs upon infection with SARS-CoV-2 nsp15mut compared to WT in both cell lines. mRNA expression of representative ISGs was also upregulated in A549-ACE2 and Calu3 cells with SARS-CoV-2 nsp15mut.
To confirm our observations of increased IFN and ISG mRNA induction, western blots were performed to assess protein expression of IFIT1, viperin, MDA5, and PKR in lysates collected from cells either mock-infected or infected with WT or nsp15mut SARS-CoV-2. A trend toward increased IFIT1, viperin, MDA5, and PKR expression was observed during nsp15mut infection relative to WT in Calu3 cells (Fig. 3D). Similar analysis of infected A549-ACE2 cells revealed that while levels of IFIT1 and viperin were moderately increased during infection with nsp15mut compared to WT early (24 hpi) during infection, MDA5 was not detected (Fig. 4D).
Activation of the PKR pathway was also evaluated via western blot to assess phosphorylation of PKR and its downstream substrate eIF2α. In Calu3 cells, clear phosphorylation of PKR and eIF2α was observed early (24 hpi) during infection, however, phosphorylation levels were reduced at 48 hpi compared to 24 hpi in both WT and nsp15mut infected cells (Fig. 3D). Similar analysis in A549-ACE2 cells revealed that both p-PKR and p-eIF2α levels were increased during SARS-CoV-2 nsp15mut infection relative to WT (Fig. 4D) indicating both an earlier (24 hpi) and more robust activation of the PKR pathway during SARS-CoV-2 nsp15mut infection. Though increased PKR pathway activation (above WT levels) was detected in A549-ACE2 cells, we hypothesize that robust activation of the PKR pathway by WT SARS-CoV-2 in Calu3 cells renders any additional PKR activation by SARS-CoV-2 nsp15mut difficult to detect. It is important to note that in both cell lines, comparable levels of SARS-CoV-2 N were observed following infection with either nsp15mut or WT SARS-CoV-2, consistent with the lack of replication defect (Figs. 3D and 4D). Finally, we compared OAS/RNase L activation in Calu3 and A549-ACE2 cells infected with WT and nsp15mut SARS-CoV-2 by assessing ribosomal RNA (rRNA) degradation using a bioanalyzer. No increase in RNase L activity was observed during SARS-CoV-2 nsp15mut infection relative to activation observed by WT SARS-CoV-2 in either cell type (SI Appendix, Fig. S3 A and B).
SARS-CoV-2 nsp15mut Is Attenuated for Replication and Induces Increased IFN Signaling and PKR Pathway Activation in Primary Nasal Epithelial ALI Cultures.
We sought to investigate the role of nsp15 in a primary nasal epithelial cell culture system which models the initial site of viral replication and the primary barrier to respiratory virus infections. Thus, nasal epithelial cells derived from four to six donors were pooled prior to growth and differentiation at an air–liquid interface (ALI) to recreate the cell types and functions present in the nasal airway. Infections comparing WT and nsp15mut SARS-CoV-2 in nasal ALI cultures were conducted at 33 °C (approximate nasal airway temperature) to optimize replication of SARS-CoV-2 and model the in vivo nasal temperature dynamics (46–48).
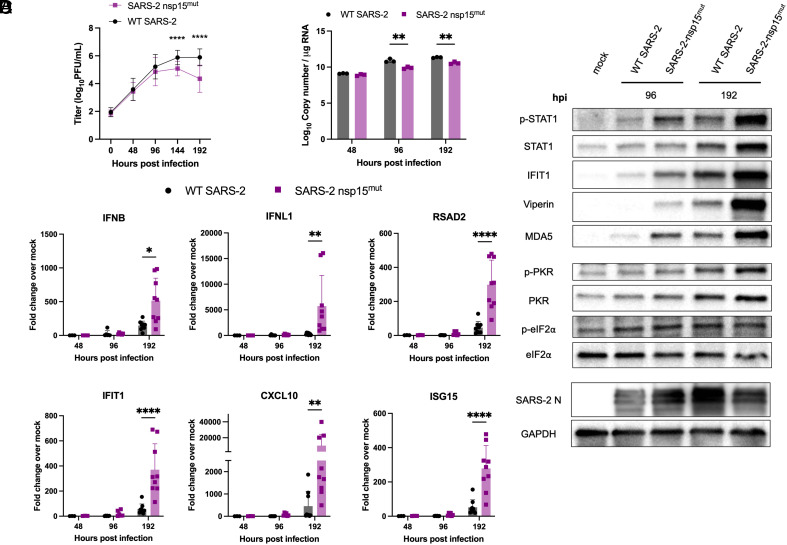
To investigate the kinetics of viral replication, nasal ALI cultures were infected at the apical surface with WT or nsp15mut SARS-CoV-2. Apical surface liquid (ASL) was collected at 48-h intervals from 0 to 192 hpi and quantified via plaque assay. Average viral titers are shown in Fig. 5A. WT SARS-CoV-2 reached peak titers at 144 hpi and then plateaued, as we have previously reported (46). In contrast to growth curves in both A549-ACE2 and Calu3 cells, SARS-CoV-2 nsp15mut replication was significantly attenuated compared to WT by approximately 10-fold PFU/mL at both 144 and 192 hpi. In addition, a significant reduction in intracellular genome copy number was detected at both 96 and 192 hpi during SARS-CoV-2 nsp15mut infection compared to WT as quantified by RT-qPCR (Fig. 5B).
Fig. 5.
SARS-2 nsp15mut is attenuated for replication compared to WT SARS-CoV-2 and induces increased IFN signaling and PKR pathway activation in primary nasal epithelial ALI cultures. Primary nasal ALI cultures were infected at MOI 1 with either WT or nsp15mut SARS-CoV-2. (A) ASL was collected every 48 h and infectious virus was titered by plaque assay. Growth curves shown are the average titers from five independent experiments, each performed in triplicate with a different set of four to six pooled nasal cell donors. (B and C) Intracellular RNA was extracted and analyzed by RT-qPCR at 48, 96 and 192 hpi. (B) Genome copy number was quantified using primers against nsp12 and a standard curve. (C) Relative mRNA expression of IFNB and IFNL1 as well as four representative ISGs: RSAD2, IFIT1 CXCL10, and ISG15. Fold change is expressed as 2–Δ(ΔCt). (D) Western blot analysis of whole cell lysates collected at 96 and 192 hpi was performed using antibodies against indicated proteins. Data in B–D are from one representative experiment of three total experiments.
Next, we compared dsRNA-induced innate immune responses in nasal ALI cultures infected with either WT or nsp15mut SARS-CoV-2. We observed delayed IFN and ISG responses during both WT and nsp15 mutant virus infections. At 48 and 96 hpi, there was minimal induction of IFN and ISG mRNA in cultures infected with either virus (Fig. 5C). However, both IFN and the representative ISG mRNAs were significantly (~10-fold) upregulated at 192 hpi during SARS-CoV-2 nsp15mut infection compared to WT. The kinetics of IFN and ISG mRNA induction in nasal ALI cultures were consistent with the timing of the replication defect observed for SARS-CoV-2 nsp15mut (Fig. 5A).
Given the growth defect and increased expression of IFN and ISG mRNA during infection with SARS-CoV-2 nsp15mut compared to WT in nasal ALI cultures, we examined STAT1 phosphorylation (p-STAT1) and ISG induction by western blot. At 96 hpi, we observed an increase in p-STAT1, as well as increased levels of ISG protein expression (IFIT1, Viperin, MDA5, and PKR) in samples infected with nsp15mut compared to WT SARS-CoV-2 (Fig. 5D). This increase in p-STAT1 and upregulation of ISGs during SARS-CoV-2 nsp15mut infection was even more robust at 192 hpi.
Protein lysates from infected nasal ALI cultures were further evaluated to assess PKR pathway activation (Fig. 5D). A clear increase in p-PKR as well as total PKR was observed during infection with both viruses at 192 hpi. Despite a clear increase in p-PKR, we have not detected a consistent increase in p-eIF2α above mock levels in nasal ALI cultures from five independent sets of pooled donors infected with either WT or nsp15mut SARS-CoV-2 at any time point assayed. Western blotting for SARS-CoV-2 N levels showed decreased expression at 192 hpi during nsp15mut infection compared to WT (Fig. 5D), consistent with nsp15mut attenuation in primary nasal cells. Additionally, we compared OAS/RNase L activation in nasal ALI cultures but did not detect activation of RNase L as indicated by the lack of detectable rRNA degradation during either WT or nsp15mut infection (SI Appendix, Fig. S3C).
Finally, to determine whether SARS-CoV-2 nsp15 EndoU contributes to suppressing inflammation and/or cytotoxicity as well as IFN signaling pathways, we quantified cytotoxicity and expression of inflammatory mediators (TNF-α, IL-6). Infection with SARS-CoV-2 nsp15mut did not result in increased cytotoxicity as measured via lactate dehydrogenase assay relative to WT in nasal ALI cultures (SI Appendix, Fig. S4A). However, a trend toward increased expression of inflammatory cytokines TNF-α and IL-6 was observed during SARS-CoV-2 nsp15mut infection (SI Appendix, Fig. S4B).
Attenuation of SARS-CoV-2 nsp15mut in Primary Nasal Epithelial Cells Is IFN-mediated.
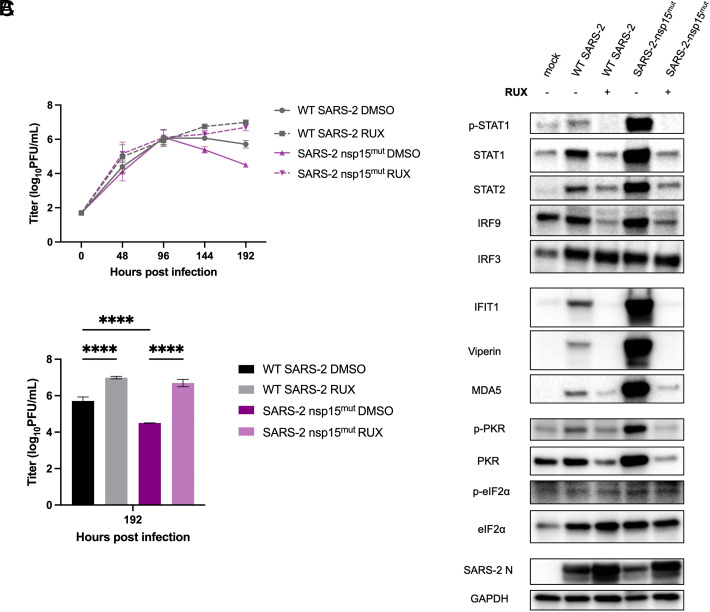
To determine the extent to which the attenuation of SARS-CoV-2 nsp15mut is due to increased IFN signaling, we infected nasal ALI cultures in the presence of the small molecule JAK1/2 inhibitor, ruxolitinib (RUX). RUX treatment prevents phosphorylation of STATs by JAK1/2 and subsequent nuclear translocation; thus RUX-treated nasal cells can produce and release IFN, but cells are unable to respond to IFN and induce ISG transcription (49). Nasal ALI cultures pretreated with RUX, at a noncytotoxic concentration (SI Appendix, Fig. S5), were infected with either WT or nsp15mut SARS-CoV-2. ASL was collected every 48 h and infectious virus was quantified via plaque assay. As expected, in control DMSO-treated cultures, replication of SARS-CoV-2 nsp15mut was attenuated compared to WT at late times post infection (144 and 192 hpi) (Fig. 6A). While RUX treatment led to a significant increase in replication of WT SARS-CoV-2 (10-fold at 192 hpi), RUX treatment of SARS-CoV-2 nsp15mut infected cultures resulted in a larger increase in viral titers (100-fold). No significant difference in replication was observed between the RUX-treated cultures infected with either WT or nsp15mut SARS-CoV-2 at 192 hpi. Fig. 6B highlights the viral titers from Fig. 6A for each virus and treatment condition at 192 hpi plotted as a bar graph, illustrating complete rescue of the nsp15mut growth defect to WT SARS-CoV-2 levels with RUX treatment. As might be expected, RUX treatment significantly impacts viral titers at late time points post infection, which coincides with the timing of IFN signaling induction during SARS-CoV-2 infection of nasal cells (Fig. 5 C and D).
Fig. 6.
The attenuated replication of SARS-2 nsp15mut relative to WT in primary nasal ALI cultures is IFN-mediated. Primary nasal ALI cultures were treated with either DMSO or RUX at a concentration of 10 µM for 48 h prior to infection. Cultures were then infected with either WT or nsp15mut SARS-CoV-2. (A) ASL was collected every 48 h and titered via plaque assay. (B) Represents the 192 hpi data from (A) presented as a bar graph with significance comparisons shown. (C) Whole cell lysates were collected at 192 hpi for separation via SDS-PAGE and western blot analysis using antibodies against indicated proteins. All data shown are from one experiment representative of two total experiments, and each set of pooled nasal cell cultures is derived from four to six individual donors.
Protein lysates from control- or RUX-pretreated infected nasal ALI cultures were analyzed via western blot to confirm the inhibitory activity of RUX and to further investigate the relationships between the PKR pathway and IFN signaling during SARS-CoV-2 infection (Fig. 6C). At 192 hpi in DMSO-treated control samples, increased ISG expression was observed following SARS-CoV-2 nsp15mut infection compared to WT, as described earlier. RUX treatment completely inhibited STAT1 phosphorylation, as well as upregulation of both STAT1 and STAT2 expression during either WT or nsp15mut SARS-CoV-2 infection. Downstream upregulation of ISG expression was also inhibited in RUX-treated cultures. Interestingly, no increase in p-PKR and total PKR signal was detected in RUX-treated cultures, suggesting that IFN-mediated upregulation of PKR is necessary for PKR pathway activation in nasal ALI cultures. Finally, consistent with viral attenuation, SARS-CoV-2 N levels were decreased during SARS-CoV-2 nsp15mut infection compared to WT in DMSO-treated cultures, but this defect was rescued with RUX treatment. These data indicate that the growth defect observed for SARS-CoV-2 nsp15mut in nasal ALI cultures is IFN-mediated.
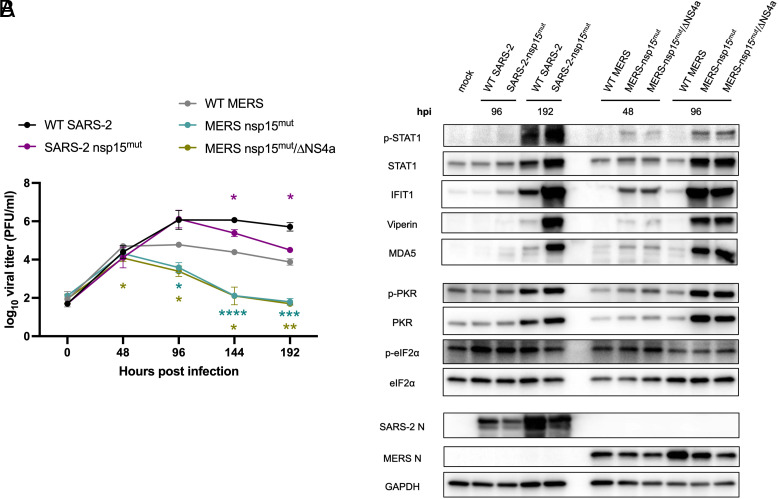
Nsp15 Endoribonuclease Activity Is More Crucial in Promoting Viral Replication during MERS-CoV Infection Compared to SARS-CoV-2 Infection.
We sought to evaluate the role of nsp15 in innate immune antagonism and optimization of viral replication during SARS-CoV-2 infection compared with another lethal CoV, MERS-CoV. Since these viruses utilize different cellular receptors, we chose two cell types in which both viruses can efficiently replicate for these comparisons (Calu3 cells and nasal ALI cultures) (50). For these comparisons, we used previously characterized MERS-CoV mutants, MERS-CoV nsp15mut and a MERS-CoV nsp15mut/ΔNS4a double mutant, which also lacks expression of dsRNA-binding protein NS4a. Previously, we found that both of these MERS-CoV mutant viruses were attenuated in A549 cells expressing the MERS-CoV receptor, but the deletion of NS4a in addition to the inactivation of nsp15 conferred a more dramatic replication defect and greater stimulation of IFN signaling and PKR activation (19). Thus, along with WT SARS-CoV-2 and SARS-CoV-2 nsp15mut, we infected Calu3 and nasal ALI cultures with WT MERS-CoV, MERS-CoV nsp15mut and MERS-CoV nsp15mut/ΔNS4a.
In Calu3 cells, both MERS-CoV nsp15mut and MERS-CoV nsp15mut/ΔNS4a were attenuated compared to WT MERS-CoV; this contrasts with the lack of attenuation observed for SARS-CoV-2 nsp15mut (Fig. 7 A and B). Additionally, MERS-CoV nsp15mut/ΔNS4a exhibited a larger growth defect than MERS-CoV nsp15mut in Calu3 cells. In nasal ALI cultures, WT SARS-CoV-2 replicated to higher titers than WT MERS-CoV (Fig. 8A), which is consistent with our previous findings (19, 43). A significant growth defect for SARS-CoV-2 nsp15mut was observed at 144 and 192 hpi, while both MERS-CoV mutants were attenuated relative to WT MERS-CoV beginning earlier, at 96 hpi. The magnitude of the growth defect was also larger for both MERS-CoV mutants (~100-fold) compared to SARS-CoV-2 nsp15mut (~10-fold). Thus, nsp15 activity appears to play a more crucial role in viral replication during MERS-CoV compared to SARS-CoV-2 infection, as MERS-CoV nsp15mut and MERS-CoV nsp15mut/ΔNS4a exhibited an earlier and larger replication defect in both cell types.
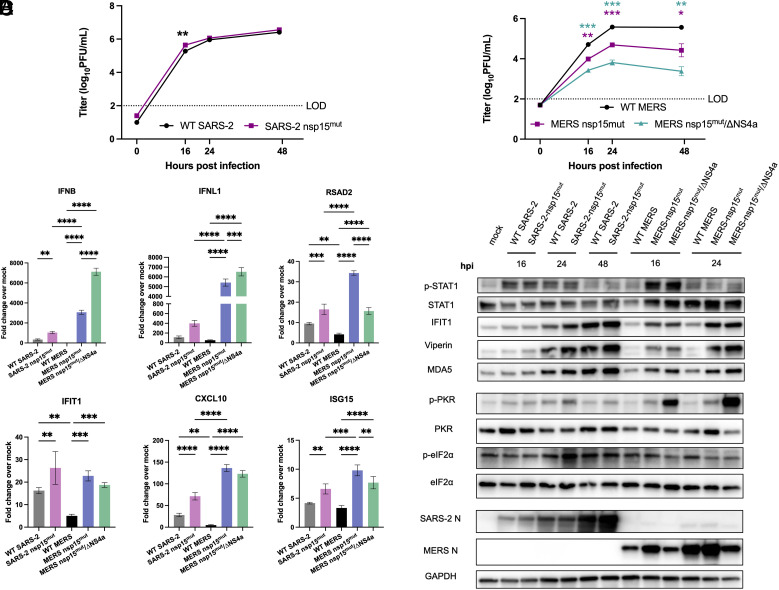
Fig. 7.
Inactivation of nsp15 endoribonuclease has more impact on viral replication during MERS-CoV infection in Calu3 cells compared to SARS-CoV-2. Calu3 cells were infected with either WT SARS-CoV-2, SARS-CoV-2 nsp15mut, WT MERS-CoV, MERS-CoV nsp15mut, or MERS-CoV nsp15mut/ΔNS4a at MOI 0.1. (A and B) Supernatants were collected at the indicated time points post infection and infectious virus was quantified by plaque assay. (C) Intracellular RNA was extracted at 24 hpi and relative mRNA expression of IFNB and IFNL1 as well as four representative ISGs: RSAD2, IFIT1, CXCL10, and ISG15 was determined by RT-qPCR. Fold change is expressed as 2−Δ(ΔCt). (D) Western blot analysis of whole cell lysates was performed at indicated time points using antibodies against indicated proteins. Data shown are from one representative experiment of three total experiments.
Fig. 8.
Inactivation of nsp15 endoribonuclease significantly impacts viral replication and IFN signaling of MERS-CoV as well as SARS-CoV-2 in nasal ALI cultures. Nasal ALI cultures were infected with either WT SARS-CoV-2, SARS-CoV-2 nsp15mut, WT MERS-CoV, MERS-CoV nsp15mut, or MERS-CoV nsp15mut/ΔNS4a at MOI 1. (A) ASL was collected every 48 h and titered via plaque assay. (B) Whole cell lysates were collected at the indicated time point post infection for western blot analysis. Blots were probed with antibodies against indicated proteins. All data shown are from one experiment representative of three total experiments, and each set of pooled nasal cell cultures is derived from four to six individual donors.
We further compared activation of the IFN and PKR pathways in Calu3 cells. WT MERS-CoV infection resulted in nearly undetectable IFN and ISG mRNA expression whereas WT SARS-CoV-2 infection promoted mild induction of the IFN signaling pathway, consistent with prior experiments (Fig. 7C) (19, 43). Infection with SARS-CoV-2 nsp15mut promoted an increase in IFN induction compared to WT SARS-CoV-2 while infection with either MERS-CoV nsp15mut or MERS-CoV nsp15mut/ΔNS4a induced a more robust increase in IFNB and IFNL1 mRNA expression compared SARS-CoV-2 nsp15mut (>threefold and >10-fold, respectively). Interestingly, differences in ISG induction were more nuanced. MERS-CoV mutants induced some ISGs more robustly than SARS-CoV-2 nsp15mut (CXCL10 and RSAD2), while other ISGs were induced to similar degrees by SARS-CoV-2 nsp15mut and both MERS-CoV mutants (IFIT1 and ISG15). At the protein level, the SARS-CoV-2 nsp15 mutant as well as both MERS-CoV mutants exhibited peak STAT1 phosphorylation at 16 hpi, although p-STAT1 levels were higher during infection with both MERS-CoV mutants (Fig. 7D). Both MERS-CoV mutants induced ISGs (IFIT1, viperin, MDA5) earlier (beginning at 16 hpi) than SARS-CoV-2 nsp15mut (first induction at 24 hpi). The SARS-CoV-2 nsp15mut did not reach peak ISG induction levels until 48 hpi. This suggests that nsp15 antagonizes IFN responses earlier during MERS-CoV infections compared with SARS-CoV-2. Regarding the PKR pathway, neither SARS-CoV-2 nsp15mut nor MERS-CoV nsp15mut induced PKR phosphorylation as robustly as MERS-CoV nsp15mut/ΔNS4a. This may partially explain why MERS-CoV nsp15mut/ΔNS4a is more attenuated in Calu3 cells.
In nasal ALI cultures, given the delayed IFN induction observed during SARS-CoV-2 infection, we first evaluated IFN and ISG induction for SARS-CoV-2 and MERS-CoV at a late time point (192 hpi). We found that IFN and ISG mRNA expression had returned to mock levels for both MERS-CoV mutants at 192 hpi, suggesting that this time point was too late to detect IFN induction for these viruses (SI Appendix, Fig. S6). Therefore, protein expression of ISGs and PKR pathway activation were examined via western blot at relevant time points for each virus (96, 192 hpi for SARS-CoV-2 and 48, 96 hpi for MERS-CoV) (Fig. 8B). Consistent with our observations in Calu3 cells, increased phosphorylation of STAT1 as well as ISG expression occurred at earlier times post infection in nasal ALI cultures with either of the MERS-CoV mutants compared to SARS-CoV-2 nsp15mut. Interestingly, SARS-CoV-2 nsp15mut stimulated the highest accumulation of p-STAT1 at 192 hpi. However, induction of each of the ISGs analyzed occurred to a similar extent with MERS-CoV nsp15mut and MERS-CoV nsp15mut/ΔNS4a at 96 hpi and SARS-CoV-2 nsp15mut at 192 hpi. Comparable levels of PKR phosphorylation were detected at 48 and 96 hpi with the MERS-CoV mutants and at 192 hpi with SARS-CoV-2 nsp15mut. Overall, the kinetics of IFN induction and ISG expression occur at earlier times post infection following inactivation of MERS-CoV nsp15 compared to SARS-CoV-2 nsp15. This kinetic signature correlates with an earlier and more dramatic impact on viral replication during infection with either of the MERS-CoV nsp15 mutants compared to SARS-CoV-2 nsp15mut in nasal ALI cultures. These data indicate that other viral antagonists in addition to nsp15 may be contributing to innate immune antagonism by SARS-CoV-2.
Discussion
The conserved CoV nsp15 endoribonuclease has been shown to be a potent inhibitor of host innate immunity during infection with multiple CoVs. Infections with viruses expressing nsp15 with an inactive endoribonuclease have resulted in significant attenuation of viral replication compared to their WT counterparts. Increased dsRNA-induced responses, including IFN production and signaling, as well as PKR and OAS/RNase L pathway activation were also observed (5, 16–19). To build upon previous literature that utilized overexpression systems to identify SARS-CoV-2 nsp15 EndoU as an inhibitor of IFN signaling, we generated a recombinant SARS-CoV-2 expressing a catalytically inactive nsp15 EndoU to investigate the role of nsp15 in limiting dsRNA-induced pathway activation and thereby optimizing viral replication. We found that inactivation of nsp15 EndoU promotes increased dsRNA accumulation during SARS-CoV-2 infection. DsRNA accumulation was increased as quantified by total dsRNA intensity per infected cell as well as size and intensity of individual dsRNA puncta (Fig. 2).
Despite evidence of robust IFN signatures as well as increased PKR activation in lung-derived epithelial cell lines and primary nasal ALI cultures, replication of SARS-CoV-2 nsp15mut was attenuated only in primary nasal ALI cultures (Figs. 3–5). These data suggest a role for SARS-CoV-2 nsp15 as an IFN antagonist whose function is essential for optimizing viral replication in certain cellular contexts. Various studies have demonstrated that SARS-CoV-2 is highly sensitive to IFN pretreatments (51). Thus, its ability to evade or antagonize IFN and ISG signaling impacts its ability to replicate.
We have previously reported that SARS-CoV-2 infections of nasal ALI cultures produce a relatively low percentage of infected cells (~10%) compared to cell lines (46). We hypothesize that this low percentage of infected cells, which may more closely reflect in vivo infections, may accentuate the impact that inactivation of nsp15 has on SARS-CoV-2 infection. Secreted IFN from a relatively small population of infected cells provides a signal to neighboring cells to create an antiviral state to limit viral spread (52, 53). Additionally, nasal ALI cultures are more robust in terms of overall IFN and ISG signaling when compared to transformed epithelial cell lines. We compared the magnitude of IFN and ISG mRNA induction during infection of A549-ACE2, Calu3, and nasal ALI cultures and found that nasal cultures consistently exhibited the strongest induction of IFIT1 and ISG15 mRNAs (SI Appendix, Fig. S7). Both IFIT1 and ISG15 have been previously characterized as authentic anti-SARS-CoV-2 ISGs (54–56). It is also important to note that we observed some variability in the magnitude of IFN and ISG induction within these primary nasal ALI cultures. There is a level of donor-dependent variability in the susceptibility to infection as well as magnitude of immune responses that we and other groups have previously observed (Fig. 5C) (46, 57). To help mitigate this variability, four to six individual donors were pooled prior to seeding nasal cultures to model average host responses, and thus we cannot make specific conclusions regarding individual donors’ innate immune responses to infection.
Given that SARS-CoV-2 nsp15mut replication is attenuated in nasal cells, we used a JAK1/2 inhibitor, RUX, to test to what extent the observed growth defect is IFN-mediated. While RUX treatment led to an increase in WT SARS-CoV-2 titers, it had a more robust impact on SARS-CoV-2 nsp15mut titers, such that its replication was rescued to WT SARS-CoV-2 levels in the presence of RUX (Fig. 6). The correlation between SARS-CoV-2 attenuation and IFN induction is consistent with our previous findings that characterized multiple MERS-CoV nsp15 mutant viruses in A549 cells. We found that growth defects in these mutant viruses were similarly IFN-mediated, as infections conducted in mitochondrial antiviral-signaling protein (MAVS) knockout cells resulted in complete rescue of viral titers to WT MERS-CoV levels in the absence of IFN signaling (19). Similar findings have been observed during MHV infection, whereby nsp15 mutant viruses can only replicate efficiently in BMMs deficient in either IFNAR or both PKR and RNase L (17). Our data further illustrate that nsp15 is an IFN antagonist since SARS-CoV-2 nsp15mut is able to replicate to WT levels when IFN signaling is abrogated by RUX treatment in nasal ALI cultures.
RUX treatment of nasal cells also highlighted the interconnected nature of dsRNA-induced pathways. PKR pathway activation indicated by increased p-PKR occurred in nasal cells infected with either WT or nsp15mut SARS-CoV-2. However, RUX treatment resulted in p-PKR levels that were comparable to mock levels during infection by both viruses (Fig. 6). Since PKR itself is an ISG, this suggested that IFN-mediated upregulation may be necessary for sufficient PKR expression that allows for its activation and autophosphorylation in nasal cell cultures (58, 59). Parallel observations have been made during infection of BMMs with MHV, whereby low basal expression of OASs resulted in undetectable RNase L activation (60). We have been unable to detect OAS/RNase L pathway activation in nasal ALI cultures, either during WT or nsp15mut SARS-CoV-2 infection or following transfection with varying concentrations of the dsRNA mimetic, poly(I:C), (SI Appendix, Fig. S3C), suggesting that cell type-dependent differences in dsRNA-induced pathway activation exist.
In contrast to both PKR and OAS proteins, the downstream target of PKR, eIF2α, is not an ISG and thus would not be upregulated in the context of increased IFN signaling during SARS-CoV-2 nsp15mut infection. Detection of increased phosphorylated eIF2α proved difficult in both Calu3 and nasal ALI cultures, which may be linked to its lack of IFN-mediated upregulation. It is important to note that PKR is not the only kinase responsible for eIF2α phosphorylation. Three other kinases [PKR-like ER Kinase (PERK), general control nonderepressible 2 (GCN2), and Heme-regulated eIF2α kinase (HRI)] have the capacity to phosphorylate eIF2α, but only PKR is activated following dsRNA detection. Instead, accumulation of unfolded proteins, amino acid starvation, or heme deficiency respectively activate these kinases (61, 62). Additionally, we hypothesize that due to the robust activation of both the PKR and RNase L pathways during WT SARS-CoV-2 infection in the epithelial-derived cell lines, any further activation by the nsp15 mutant was difficult to detect.
The IFN-mediated growth defect of SARS-CoV-2 nsp15mut in nasal ALI cultures was only detected at late times post infection (144 and 192 hpi), concurrent with time points at which nsp15mut induced more IFN signaling than WT SARS-CoV-2 (Fig. 5). Similar findings of delayed kinetics of IFN and ISG signaling in primary nasal cells were previously reported based on comparisons with influenza which induces the IFN pathway much earlier than SARS-CoV-2 (63). The mechanism behind this delay in IFN signaling can likely be explained in part by the immune antagonist activities of accessory and additional nonstructural proteins encoded by SARS-CoV-2. Additional conserved CoV nsps likely contribute to SARS-CoV-2 immune evasion, including nsp1 (which selectively inhibits host protein translation), nsp3 (which modifies host proteins via its macrodomain and deubiquitinase domain to suppress antiviral responses), as well as nsp14 and nsp16 (which contribute to RNA modification important for CoV immune evasion) (56, 64–67). SARS-CoV-2 encodes multiple additional accessory proteins, encoded in ORF3b, ORF6, ORF7a, ORF7b, and ORF8, some of which may play roles in immune evasion (68). Specifically, the protein encoded by SARS-CoV-2 ORF6 has been reported to inhibit nuclear translocation of transcription factors STAT1/2, thus functioning as an IFN signaling inhibitor (69–72). Since SARS-CoV-2 nsp15mut retains other functional IFN antagonist activities in its accessory proteins, this may contribute to the delayed IFN responses observed in nasal ALI cultures. We previously reported that MERS-CoV encodes three potent IFN antagonists that together shut down dsRNA-induced pathways (the conserved CoV nsp15 EndoU and accessory proteins NS4a and NS4b), so it is likely that SARS-CoV-2 also encodes multiple strategies to evade host innate immunity (19, 21).
Another potential contributor to delayed IFN kinetics observed during SARS-CoV-2 infection and its nsp15 mutant counterpart is temperature. Infections in primary nasal cultures were conducted at 33 °C to model in vivo nasal airway temperatures, whereas infections in respiratory epithelial cell lines were conducted at 37 °C (47, 73). Temperatures in the nasal passages range from 32 to 35 °C, whereas temperatures in the lung are closer to ambient body temperature (37 °C). Temperature has been shown to modulate innate immune responses and replication of SARS-CoV-2 as well as other viruses such as human rhinovirus-16 (48, 74). Our prior work has indicated that while SARS-CoV-2 can replicate in nasal cultures incubated at either 33 °C or 37 °C, its replication is optimal at nasal airway temperature (33 °C) (46). This preference for replication of SARS-CoV-2 at 33 °C was corroborated in a lower airway ALI model (48). The delayed kinetics of activation of IFN and ISG signaling in nasal ALI cultures is likely multifactorial, contributed to by both temperature and immune antagonists encoded by SARS-CoV-2.
Having established that nsp15 EndoU activity contributes to IFN antagonism and promotes viral replication during SARS-CoV-2 infection, we directly compared the effects of an inactive nsp15 EndoU domain during either SARS-CoV-2 or MERS-CoV infection in Calu3 cells and nasal ALI cultures (Figs. 7 and 8). MERS-CoV nsp15 mutants exhibited an earlier and more robust IFN signature associated with attenuation in both Calu3 cells and nasal ALI cultures. While SARS-CoV-2 nsp15mut induced IFN and downstream ISG expression in all cellular systems analyzed, it was only associated with a growth defect in nasal ALI cultures (Figs. 3–5). WT MERS-CoV is particularly adept at shutting down dsRNA-induced pathway activation in contrast to WT SARS-CoV-2, which induces activation of IFN, PKR, and RNase L (43). Indeed, WT MERS-CoV induced ten-fold less IFN-β and two-fold less IFN-λ mRNA compared to WT SARS-CoV-2 in Calu3 cells (Fig. 7C). Additionally, although WT SARS-CoV-2 titers were relatively equal to WT MERS-CoV in Calu3 cells, the levels of IFN and ISG induction were all higher during WT SARS-CoV-2 infection (Fig. 7). We hypothesize that SARS-CoV-2 has evolved to replicate despite moderate innate immune induction, and thus its replication is not significantly impacted when IFN induction is increased above WT levels due to the inactivation of nsp15 EndoU. Although nsp15 serves as a strong IFN antagonist encoded by both viruses, our data indicate that nsp15 may play a more crucial role in optimizing MERS-CoV replication. It is not surprising that CoVs would have varying degrees of sensitivity to IFN. SARS-CoV-2, its variants, and SARS-CoV are all genetically similar to each other but demonstrate variability in their sensitivity to IFN (51, 75–78). Experiments directly comparing the sensitivity of SARS-CoV-2 and MERS-CoV, as well as their respective nsp15 mutants, to IFN would be useful in characterizing these two lethal coronaviruses.
Here, we have characterized a recombinant SARS-CoV-2 nsp15 mutant virus, demonstrating that nsp15 is a potent antagonist of IFN induction and ISG signaling in multiple cellular contexts. Abrogation of IFN antagonism by SARS-CoV-2 nsp15 can have a dramatic impact on viral replication, illustrated most clearly in primary nasal cells in which the SARS-CoV-2 nsp15 mutant exhibits IFN-mediated attenuation of replication. Future studies will investigate the combinatorial role of other SARS-CoV-2 accessory genes in antagonism of IFN signaling, such as the ORF6 encoded protein, as well as the downstream consequences of increased dsRNA-induced pathway activation, such as inflammatory cytokine production and cell death, and how these responses may contribute to overall pathogenesis. Additionally, although nasal ALI cultures possess a heterogenous cellular population of ciliated and mucus-secreting epithelial cells, they lack immune cells, which have been shown to play a prominent role in pathogen recognition as well as inflammatory cytokine production (79). The establishment of a co-culture nasal ALI system containing various innate immune cell populations would mitigate this pitfall present in all three cellular systems used in this study. It is imperative to characterize the mechanisms of innate immune evasion by SARS-CoV-2 and other HCoVs to understand how viruses interact with host cells to optimize replication and spread. Moreover, these findings will inform identification of effective antivirals and development of live-attenuated vaccines against SARS-CoV-2 and other respiratory coronaviruses.
Materials and Methods
Nasal epithelial cells were collected via cytologic brushing of patients’ nasal cavities after obtaining informed consent and then grown and differentiated on transwell inserts to establish ALI cultures. The full study protocol was approved by the University of Pennsylvania Institutional Review Board (protocol # 800614) and the Philadelphia VA Institutional Review Board (protocol #00781). A549-ACE2, Calu3, and primary nasal epithelial cells were infected with recombinant WT SARS-CoV-2, SARS-CoV-2 nsp15mut, WT MERS-CoV, MERS-CoV nsp15mut, or MERS-CoV nsp15mut/ΔNS4a at the indicated multiplicities of infection (19, 43). At various times post infection, infectious viruses in cellular supernatants or apical surface liquid collected via apical wash of nasal ALI cultures were quantified via plaque assay. Total RNA or protein was also collected from infected cells for analysis of dsRNA-induced pathway activation. All of these techniques are described in SI Appendix, Materials and Methods. Any materials or related protocols mentioned in this work can be obtained by contacting the corresponding author.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank members of the Weiss lab for feedback and discussion of this project and Dr. Volker Thiel (University of Bern, Switzerland) for discussion and early mutant construction. We thank Drs. David W. Kennedy, James N. Palmer, Nithin D. Adappa, and Michael A. Kohanski for aid in the collection of nasal tissue for establishing primary nasal epithelial cultures. This work was supported by NIH grants R01AI140442 (S.R.W.), R01AI169537 (N.A.C. and S.R.W.), R01AI104887 (R.H.S. and S.R.W.), R35GM151249 (J.M.B.), R35GM138029 (A.R.F.), P20GM113117 (A.R.F.); R01A1AI161175 (L.M.-S.); Department of Veterans Affairs Merit Review 1-I01-BX005432-01 (N.A.C. and S.R.W.); the Penn Center for Research on Coronaviruses and Other Emerging Pathogens (S.R.W.). C.J.O. was supported in part by F30AI172101 and T32AI055400 and N.B. in part by T32AI007324 and K12GM081259.
Author contributions
C.J.O., N.B., N.A.P., J.M.B., L.M.-S., and S.R.W. designed research; C.J.O., N.B., N.A.P., C.Y., A.A., E.K.B., L.H.T., N.J., and J.M.B. performed research; N.A.P., C.Y., J.J.P., A.R.F., J.M.B., and L.M.-S. contributed new reagents/analytic tools; C.J.O., N.B., A.A., R.H.S., J.M.B., N.A.C., and S.R.W. analyzed data; and C.J.O., N.B., N.A.P., A.R.F., R.H.S., N.A.C., L.M.-S., and S.R.W. wrote the paper.
Competing interests
S.R.W. consults for Powell Gilbert LLP. N.A.C. consults for GSK, AstraZeneca, Novartis, Sanofi/Regeneron. N.A.C. has US Patent “Therapy and Diagnostics for Respiratory Infection” (10,881,698 B2, WO20913112865) and a licensing agreement with GeneOne Life Sciences.
Footnotes
Reviewers: X.D., Oklahoma State University; and R.R., Rutgers Biomedical and Health Sciences.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.World Health Organization, WHO COVID-19 Dashboard 2020 (2023). https://covid19.who.int/. Accessed 2 November 2023.
- 2.V’Kovski P., et al. , Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ancar R., et al. , Physiologic RNA targets and refined sequence specificity of coronavirus EndoU. RNA 26, 1976–1999 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hackbart M., Deng X., Baker S. C., Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U.S.A. 117, 8094–8103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng X., Baker S. C., An "Old" protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology 517, 157–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth-Cross J. K., Bender S. J., Weiss S. R., Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 82, 9829–9838 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dias Junior A. G., Sampaio N. G., Rehwinkel J., A balancing act: MDA5 in antiviral immunity and autoinflammation. Trends Microbiol. 27, 75–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin X., et al. , MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 34, 108628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebendenne A., et al. , SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. 95, 02415-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Platanias L. C., Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Schoggins J. W., Rice C. M., Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1, 519–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Der S. D., et al. , Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U.S.A. 95, 15623–15628 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee S., et al. , OAS-RNase L innate immune pathway mediates the cytotoxicity of a DNA-demethylating drug. Proc. Natl. Acad. Sci. U.S.A. 116, 5071–5076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakrabarti A., Jha B. K., Silverman R. H., New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 31, 49–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García M. A., et al. , Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70, 1032–1060 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X., et al. , Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U.S.A. 114, E4251–E4260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kindler E., et al. , Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 13, e1006195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J., et al. , Coronavirus endoribonuclease ensures efficient viral replication and prevents protein kinase R activation. J. Virol. 95, 02103-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comar C. E., et al. , MERS-CoV endoribonuclease and accessory proteins jointly evade host innate immunity during infection of lung and nasal epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 119, e2123208119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y., et al. , Porcine epidemic diarrhea virus nsp15 antagonizes interferon signaling by RNA Degradation of TBK1 and IRF3. Viruses 12, 599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comar C. E., et al. , Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS Coronavirus infection. mBio 10, 00319-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemeyer D., et al. , Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 87, 12489–12495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornbrough J. M., et al. , Middle east respiratory syndrome Coronavirus NS4b protein inhibits host RNase L activation. mBio 7, e00258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabouw H. H., et al. , Middle east respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog. 12, e1005982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson I. M., et al. , Biochemical characterization of emerging SARS-CoV-2 Nsp15 endoribonuclease variants. J. Mol. Biol. 434, 167796 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Godoy A. S., et al. , Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease. Nucleic Acids Res. 51, 5255–5270 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D., et al. , SARS-CoV-2 Nsp15 suppresses type I interferon production by inhibiting IRF3 phosphorylation and nuclear translocation. iScience 26, 107705 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazier M. N., et al. , Characterization of SARS2 Nsp15 nuclease activity reveals it’s mad about U. Nucleic Acids Res. 49, 10136–10149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salukhe I., et al. , Regulation of coronavirus nsp15 cleavage specificity by RNA structure. PLoS One 18, e0290675 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pillon M. C., et al. , Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat. Commun. 12, 636 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhardwaj K., et al. , RNA recognition and cleavage by the SARS Coronavirus endoribonuclease. J. Mol. Biol. 361, 243–256 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhardwaj K., Guarino L., Kao C. C., The severe acute respiratory syndrome Coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78, 12218–12224 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuen C.-K., et al. , SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 9, 1418–1428 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boodhoo N., et al. , The severe acute respiratory syndrome coronavirus 2 non-structural proteins 1 and 15 proteins mediate antiviral immune evasion. Curr. Res. Virolog. Sci. 3, 100021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee R. J., et al. , Bacterial d-amino acids suppress sinonasal innate immunity through sweet taste receptors in solitary chemosensory cells. Sci. Signal 10, 7703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee R. J., et al. , Fungal aflatoxins reduce respiratory mucosal ciliary function. Sci. Rep. 6, 33221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallo O., et al. , The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 14, 305–316 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mifsud E. J., Kuba M., Barr I. G., Innate immune responses to influenza virus infections in the upper respiratory tract. Viruses 13, 2090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye C., et al. , Rescue of SARS-CoV-2 from a single bacterial artificial chromosome. mBio 11, 02168-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fehr A. R., Perlman S., Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chew T., et al. , Characterization of the interferon regulatory factor 3-mediated antiviral response in a cell line deficient for IFN production. Mol. Immunol. 46, 393–399 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Emeny J. M., Morgan M. J., Regulation of the interferon system: Evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43, 247–252 (1979). [DOI] [PubMed] [Google Scholar]
- 43.Li Y., et al. , SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 118, e2022643118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M., et al. , Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci. Immunol. 6, 9007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei X., et al. , Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11, 3810 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otter Clayton J., et al. , Infection of primary nasal epithelial cells differentiates among lethal and seasonal human coronaviruses. Proc. Natl. Acad. Sci. U.S.A. 120, e2218083120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keck T., et al. , Temperature profile in the nasal cavity. Laryngoscope 110, 651–654 (2000). [DOI] [PubMed] [Google Scholar]
- 48.V’Kovski P., et al. , Disparate temperature-dependent virus-host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium. PLoS Biol. 19, e3001158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mesa R. A., Yasothan U., Kirkpatrick P., Ruxolitinib. Nat. Rev. Drug Discovery 11, 103–104 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Nassar A., et al. , A review of human Coronaviruses’ receptors: The host-cell targets for the crown bearing viruses. Molecules 26, 6455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lokugamage Kumari G., et al. , Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 94, 01410-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sen G. C., Viruses and interferons. Annu. Rev. Microbiol. 55, 255–281 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Stanifer M. L., Pervolaraki K., Boulant S., Differential regulation of type I and type III interferon signaling. Int. J. Mol. Sci. 20, 1445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindewolf C., et al. , SARS-CoV-2 uses nonstructural protein 16 to evade restriction by IFIT1 and IFIT3. J. Virol. 97, e01532-22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu G., et al. , ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 6, 467–478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russ A., et al. , Nsp16 shields SARS-CoV-2 from efficient MDA5 sensing and IFIT1-mediated restriction. EMBO Rep. 23, e55648 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rijsbergen L. C., et al. , In vitro modelling of respiratory virus infections in human airway epithelial cells–A systematic review. Front. Immunol. 12, 683002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gal-Ben-Ari S., et al. , PKR: A kinase to remember. Front. Mol. Neurosci. 11, 480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pindel A., Sadler A., The role of protein kinase R in the interferon response. J. Interf. Cytokine Res. 31, 59–70 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Birdwell L. D., et al. , Activation of RNase L by murine Coronavirus in myeloid cells is dependent on basal oas gene expression and independent of virus-induced interferon. J. Virol. 90, 3160–3172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wek R. C., Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 10, a032870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pakos-Zebrucka K., et al. , The integrated stress response. EMBO Rep. 17, 1374–1395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hatton C. F., et al. , Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2. Nat. Commun. 12, 7092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakagawa K., Makino S., Mechanisms of Coronavirus Nsp1-mediated control of host and viral gene expression. Cells 10, 300 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clementz M. A., et al. , Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84, 4619–4629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alhammad Y. M., et al. , SARS-CoV-2 Mac1 is required for IFN antagonism and efficient virus replication in cell culture and in mice. Proc. Natl. Acad. Sci. U.S.A. 120, e2302083120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Case J. B., et al. , Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J. Virol. 92, e01531-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Redondo N., et al. , SARS-CoV-2 accessory proteins in viral pathogenesis: Knowns and unknowns. Front. Immunol. 12, 708264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kehrer T., et al. , Impact of SARS-CoV-2 ORF6 and its variant polymorphisms on host responses and viral pathogenesis. Cell Host Microbe 31, 1668–1684.e12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miorin L., et al. , SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 28344–28354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li T., et al. , Molecular mechanism of SARS-CoVs Orf6 targeting the Rae1–Nup98 complex to compete with mRNA nuclear export. Front. Mol. Biosci. 8, 813248 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Addetia A., et al. , SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. mBio 12, e00065-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lindemann J., et al. , Nasal mucosal temperature during respiration. Clin. Otolaryngol. Allied Sci. 27, 135–139 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Foxman E. F., et al. , Temperature-dependent innate defense against the common cold virus limits viral replication at warm temperature in mouse airway cells. Proc. Natl. Acad. Sci. U.S.A. 112, 827–832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rayhane N., et al. , Reduced replication but increased interferon resistance of SARS-CoV-2 Omicron BA.1. Life Sci. Alliance 6, e202201745 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo K., et al. , Interferon resistance of emerging SARS-CoV-2 variants. Proc. Natl. Acad. Sci. U.S.A. 119, e2203760119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shalamova L., et al. , Omicron variant of SARS-CoV-2 exhibits an increased resilience to the antiviral type I interferon response. PNAS Nexus 1, pgac067 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacob R. A., et al. , Sensitivity to neutralizing antibodies and resistance to type I interferons in SARS-CoV-2 R.1 Lineage Variants, Canada. Emerg. Infect. Dis. 29, 1386–1396 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akira S., Uematsu S., Takeuchi O., Pathogen recognition and innate immunity. Cell 124, 783–801 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.