Significance
We used a genome-wide CRISPR screen to identify regulators of NK cell cytotoxicity. Deletion of TMEM30A was shown to provide protection to cancer cells from NK cell killing. The encoded protein is a subunit of a flippase that transports phospholipids in the plasma membrane and we show that disruption of the gene caused phosphatidylserine accumulation on the outside of the cells. TMEM30A-knock-out cells from various hematological malignancies were more resistant to NK cell killing and we demonstrate that this is due to the interaction between phosphatidylserine and the inhibitory NK cell receptor, TIM-3. TMEM30A mutations are recurrent in lymphomas, and our findings suggest that this may serve as an immuno-evasive strategy in these malignancies.
Keywords: natural killer cells, CRISPR screen, TMEM30A, phosphatidylserine, TIM-3
Abstract
Natural killer (NK) cell immunotherapy has gained attention as a promising strategy for treatment of various malignancies. In this study, we used a genome-wide CRISPR screen to identify genes that provide protection or susceptibility to NK cell cytotoxicity. The screen confirmed the role of several genes in NK cell regulation, such as genes involved in interferon-γ signaling and antigen presentation, as well as genes encoding the NK cell receptor ligands B7-H6 and CD58. Notably, the gene TMEM30A, encoding CDC50A–beta-subunit of the flippase shuttling phospholipids in the plasma membrane, emerged as crucial for NK cell killing. Accordingly, a broad range of TMEM30A knock-out (KO) leukemia and lymphoma cells displayed increased surface levels of phosphatidylserine (PtdSer). TMEM30A KO cells triggered less NK cell degranulation, cytokine production and displayed lower susceptibility to NK cell cytotoxicity. Blockade of PtdSer or the inhibitory receptor TIM-3, restored the NK cell ability to eliminate TMEM30A-mutated cells. The key role of the TIM-3 – PtdSer interaction for NK cell regulation was further substantiated by disruption of the receptor gene in primary NK cells, which significantly reduced the impact of elevated PtdSer in TMEM30A KO leukemic cells. Our study underscores the potential significance of agents targeting the interaction between PtdSer and TIM-3 in the realm of cancer immunotherapy.
Natural killer cells (NK cells) are cytotoxic lymphocytes that are important in response to viral infections and in cancer immunosurveillance. In contrast to cytotoxic T 0cells, circulating NK cells are able to kill susceptible target cells without prior sensitization. NK cell effector functions are regulated by a complex integration of signals from inhibitory and activating receptors. The activating NK cell receptors (aNKR) comprise the natural cytotoxicity receptors NKp46, NKp44, and NKp30; NKG2D; DNAM-1, and the Fc-receptor CD16 (1). The main inhibitory NK cell receptors (iNKR), killer immunoglobulin-like receptors (KIRs) and NKG2A-CD94 bind different specific HLA-A, B, C and HLA-E, respectively. As these proteins are expressed by all nucleated cells in the body, the inhibitory receptors generally prevent NK cells from killing healthy cells. NK cells also express other inhibitory receptors, such as LIR-1, TIGIT, TIM-3, and IRp60/CD300a, that can also influence the activity of NK cells (2). For example, both TIM-3 and IRp60 have been described to recognize phosphatidylserine (PtdSer) on the surface of cells (3–8). Several strategies have been developed to target interactions between PtdSer and its receptors, and such agents have shown efficacy in mouse tumor models (9–12).
Despite the importance of NK cells in disease, much is still unknown about what influences NK cell activity toward malignant cells. A growing number of studies have used targeted or genome-wide CRISPR screens to identify cancer cell genes that enable or prevent NK cell killing (13–17). In these screens, genes are stochastically targeted and deleted by a library of guide RNAs in the presence of a Cas protein. These cells are then cultured with or without NK cells and the relative abundance of edited cells can be determined. To obtain maximal resolution, CRISPR screens are commonly performed at high selection pressures (18). We speculated that less harsh selection conditions may be advantageous for identification of more structures involved in NK cell cytotoxicity. Thus, in the present study, we performed a genome-wide CRISPR screen using the K562 leukemic cancer cell line under carefully pretitrated conditions to allow the selection to be completed after overnight culture and to enable identification of structures that are involved in NK cell cytotoxicity. The screen identified CDC50A as a regulator of NK cell activity via its shuttling of PtdSer from the outer surface to the inner leaflet of the cell membrane. Deletion of the corresponding gene, TMEM30A, resulted in accumulation of PtdSer on the extracellular side of the membrane which inhibited NK cells via the inhibitory receptor TIM-3, providing a possible escape mechanism for cancer cells with TMEM30A mutations.
Results
Genes Involved in Interferon-γ Signaling Pathways and Antigen Presentation Provide Protection against NK Cell Cytotoxicity.
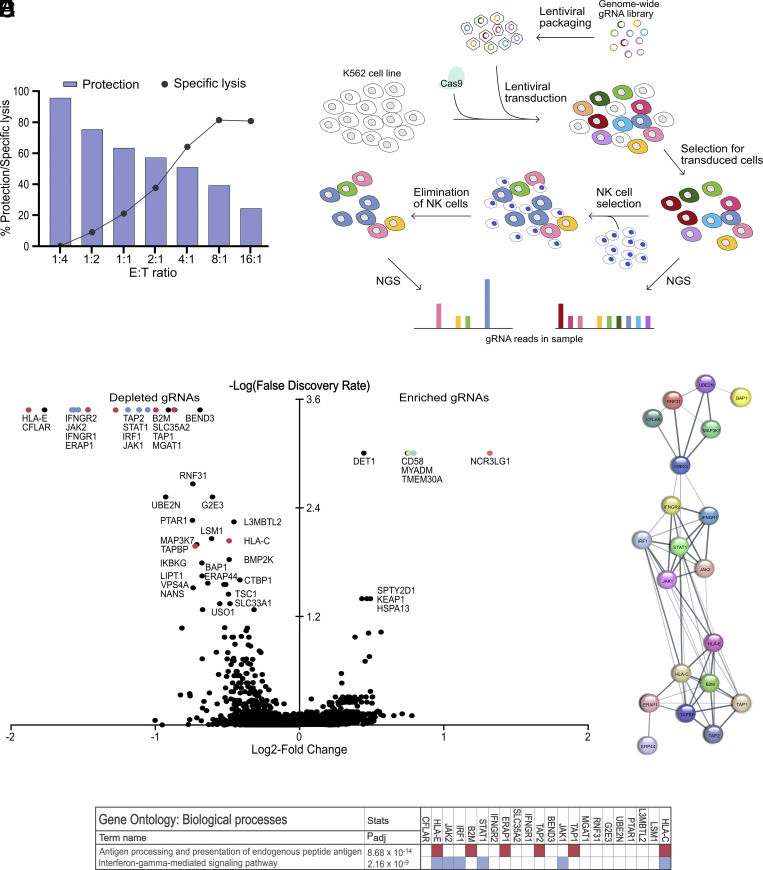
CRISPR screens are commonly performed at high selection pressures to maximize the difference between exposed cells and control cells (18). Although important biological insights have been generated from such screens, we speculated that such setups may fail to identify other physiologically relevant structures that determine whether a tumor cell escapes or is eliminated after an NK cell encounter. To test this hypothesis, we performed NK cell cytotoxicity experiments at different E:T ratios and evaluated to what extent blockade of one activating receptor–ligand interaction could prevent NK cell killing. As shown in Fig. 1A, aNKR blockade was very effective when the natural cytotoxicity was moderate. However, under harsher conditions, target cells still succumbed despite blockade of a major receptor–ligand interaction. Since these experiments reflect the situation in a CRISPR screen where a single receptor–ligand interaction is abrogated by a gene deletion, we decided to perform the screen under conditions where only approximately 60% of target cells died, which allowed NKp30 blockade to protect approximately 50% of otherwise killed K562 cells (Fig. 1A). Thus, Cas9-expressing K562 cells that had been transduced with a genome-wide library of gRNAs to stochastically delete genes, were exposed to NK cells in a genome-wide CRISPR loss-of-function screen (Fig. 1B). Compared to K562 control cells, cells with a gene knock-out resulting in increased sensitivity to NK cells were depleted to a higher extent in cultures with NK cells. The data obtained in the screen confirm and extend the findings of previous reports that genes encoding proteins involved in interferon-γ (IFNγ) signaling pathways and antigen presentation dominated the resulting list of most depleted gRNAs (Fig. 1C and Dataset S1) (13–15). Thus, among the top-depleted genes, the corresponding Gene Ontology biological processes, antigen presentation and IFNγ-mediated signaling, were highly overrepresented (P = 8.68 × 10−14 and P = 2.16 × 10−9, respectively) which was also seen in analyses of protein interactions (Fig. 1 D and E). The results obtained for top-deleted genes were also confirmed in analyses of individual gRNAs by use of specific UMIs (Dataset S2).
Fig. 1.
Genes involved in antigen presentation and IFNγ pathways provide protection against NK cell cytotoxicity. (A) Cytotoxicity assay with WT K562 at various E:T, combined with blocking antibody against NKp30, showing the percentage protection (bars) and the specific lysis (dots). (B) Overview of the experimental design. Cas9-expressing K562 cell clones were established and transduced with gRNA library and cocultured together with polyclonally activated NK cells. Next-generation sequencing revealed the difference in distribution of gRNAs between surviving cells and unexposed control culture. (C) Depleted and enriched gRNAs from one replicate in the genome-wide CRISPR screen through analysis using MaGeCK software. Depleted genes involved in IFNγ signaling or antigen presentation are highlighted in blue or red, respectively. (D) Protein interaction analysis of significantly depleted gRNAs, displaying clusters of genes involved in IFNγ signaling and antigen presentation. (E) Gene Ontology biological processes results of significantly depleted gRNAs.
Depletion of B7-H6 Provides Striking Survival Benefit against NK Cell Cytotoxicity.
Previous studies have identified the interaction between the activating NK cell receptor, NKp30, and its ligand B7-H6 to be critical for NK cell recognition of various malignant cells (13, 14, 17). Accordingly, the top enriched gRNAs (Fig. 1C) targeted the B7-H6-encoding gene, NCR3LG1. Another top-enriched gene in the screen was CD58–encoding the ligand for the costimulatory receptor CD2, which was also observed in a recent study (19). Collectively, these results suggest that ligation of NKp30 and CD2 play a key role for NK cell recognition of K562 cells. We verified these results in NK cell cytotoxicity experiments where blocking antibodies for NKp30 and/or CD2 partially inhibited NK cell killing, with a similar nonsignificant trend for DNAM-1 (Fig. 2A). However, the genes encoding DNAM-1 ligands, PVR/CD155 and Nectin-2/CD112 did not show up as significant hits in the CRISPR screen.
Fig. 2.
NCR3LG1 knock-out provides striking survival benefit against NK cell cytotoxicity. (A) Cytotoxicity assay with WT K562 cells at E:T ratio 4:1, combined with blocking antibodies against indicated activating NK cell receptors (n = 8). (B) Staining of CD58 and B7-H6 for confirmation of KO cell lines. (C) Degranulation assay with WT K562 (blue), CD58 sKO (green), NCR3LG1 sKO (red) and NCR3LG1/CD58 dKO (orange) cells, showing the percentage of CD107+ cells among CD56dim NK cells (n = 9). (D and E) Intracellular cytokine staining assays with indicated target cell lines, showing the percentage of IFNγ+ cells among CD56 dim NK cells (D) or TNFα+ cells among CD56 dim NK cells (E) (n = 9). (F) Cytotoxicity assay with indicated target cell lines at E:T ratio 2:1 (n = 6). (G) Cytotoxicity assay using blocking antibodies with NCR3LG1/CD58 dKO cells at E:T ratio 8:1 (n = 8). n indicates experiments with NK cells from different donors and error bars represent SEM. One-way ANOVA with Dunnett’s multiple comparisons test for (A) and (G) and Šidák’s multiple comparisons test was used for statistical analyses of (C–F).
To further validate the role of these receptor–ligand interactions, we generated KO cell lines with disrupted expression of B7-H6, CD58, or both (henceforth referred to as NCR3LG1 sKO, CD58 sKO, and NCR3LG1/CD58 dKO; Fig. 2B and SI Appendix, Fig. S1A). Both CD58 sKO and NCR3LG1 sKO K562 cells triggered a significant reduction of NK cell degranulation and IFNγ- and TNFα production compared to wild-type (WT) K562 cells, which was further pronounced by the combined NCR3LG1/CD58 dKO K562 cell line (Fig. 2 C–E with example plots in SI Appendix, Fig. S1B). In cytotoxicity experiments, the NCR3LG1 sKO and NCR3LG1/CD58 dKO cells displayed high resistance to lysis at low E:T ratios (Fig. 2F). However, these cells were readily killed at higher E:T ratios (SI Appendix, Fig. S1C). Notably, in cytotoxicity experiments with blocking antibodies, a shift from NKp30 dependence in WT cells to dependence on NKG2D and DNAM-1 in NCR3LG1/CD58 dKO K562 cells was observed (Fig. 2 A and G; sKOs in SI Appendix, Fig. S1 D and E).
TMEM30A Deletion Protects K562 Cells from NK Cell Cytotoxicity by Enhanced Inhibition through Inhibitory PtdSer Receptors.
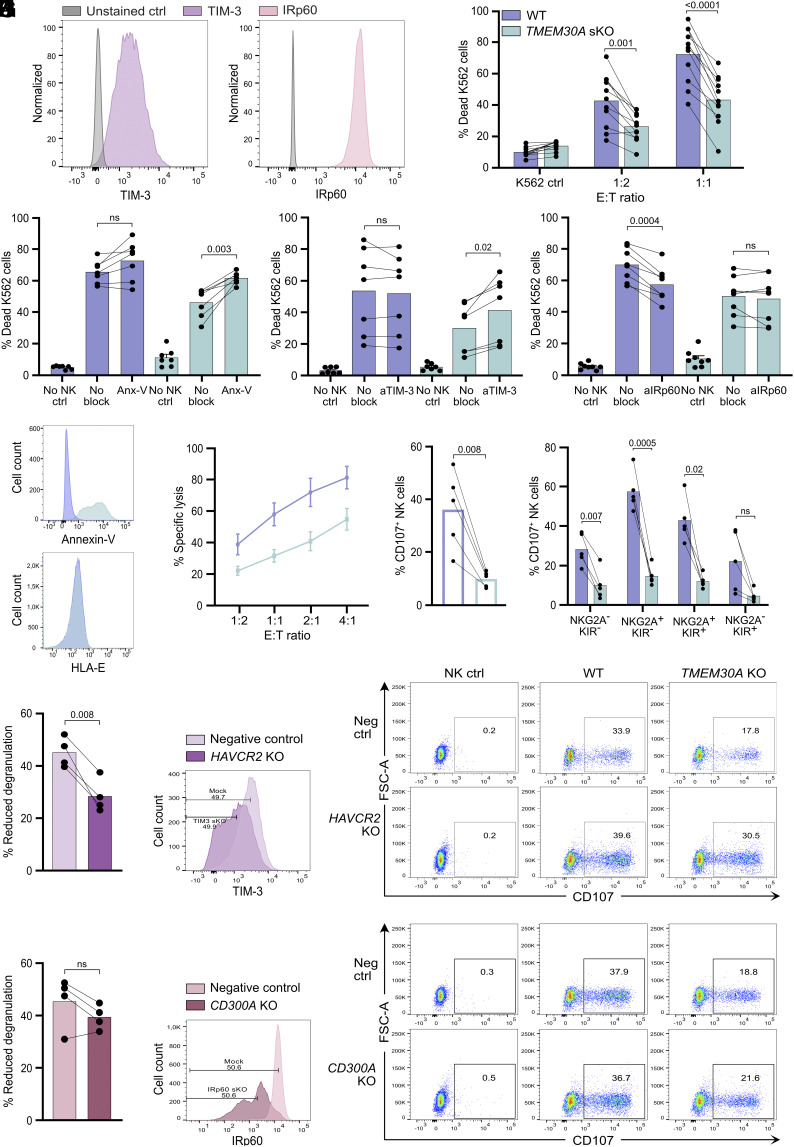
Another potentially important gene for NK cell cytotoxicity identified in the screen was TMEM30A (Fig. 1C). The gene encodes CDC50A, which is one of the β-subunits of the phospholipid flippase, P4-ATPase, that maintains leaflet asymmetry in the cellular plasma membrane. This gene was recently reported to be recurrently mutated in diffuse large B cell lymphoma (DLBCL) (20) and intravascular large B cell lymphoma (IVLBCL) (21, 22) and the occurrence of this gene in the screen suggested that this may be an immune evasion mechanism exploited by lymphoma cells to avoid NK cell immunosurveillance. To address this possibility, we generated TMEM30A KO K562 cells (SI Appendix, Fig. S2A). The expression of ligands for most NK cell receptors remained unchanged (Fig. 3A and SI Appendix, Fig. S3A) in two independent clones of TMEM30A KO cells. An increase in HLA-E expression was observed in TMEM30A KO cells and the expression of CD58 was marginally reduced. The discrepancy in the CD58 staining disappeared when the slight variation in cell size was accounted for (SI Appendix, Fig. S3B). In accordance with the screen results, TMEM30A KO cells triggered less NK cell activation, both in terms of NK cell degranulation and production of IFNγ and TNFα (Fig. 3 B–F). Importantly, the reduced NK cell degranulation to TMEM30A KO K562 cells was not dependent on the increased HLA-E expression, as reduced NK cell degranulation toward TMEM30A KO cells was also observed among NKG2A- NK cell subsets (Fig. 3C).
Fig. 3.
TMEM30A KO cells display extracellular PtdSer expression and trigger lower NK cell degranulation and cytokine production. (A) Staining for NK cell receptor ligands of WT K562 cells (blue) and TMEM30A KO cell line (green). (B and C) Degranulation assay with WT K562 cells and TMEM30A KO cell line, showing the percentage of CD107+ cells among CD56dim NK cells (B) or among CD56dim NKG2A− NK cells (C) (n = 12). (D–F) Intracellular cytokine staining assays with WT K562 cells and TMEM30A KO cells, showing the percentage of IFNγ+ cells among CD56 dim NK cells (D) or TNFα+ cells among CD56 dim NK cells (E) (n = 12), with representative density plots (F). (G) Annexin-V staining of live TMEM30A KO K562 cells, compared to WT K562 cells. n indicates experiments with NK cells from different donors. Error bars represent SEM, and the paired t-test was used for statistical analyses.
Notably, established TMEM30A KO clones displayed elevated levels of extracellularly exposed PtdSer as determined by fluorescently labeled Annexin-V (Fig. 3G; additional clones in SI Appendix, Fig. S3C and using additional TMEM30A CRISPR /Cas9 gRNAs in SI Appendix, Fig. S3D). Two immune cell receptors that have been described to bind PtdSer are TIM-3 and IRp60 (6–8). In accordance with previous studies, we observed both IRp60 and TIM-3 to be expressed by NK cells (Fig. 4A) (4, 7). We speculated that exposure of PtdSer in TMEM30A KO K562 cells could lead to increased NK cell inhibition through engagement of these inhibitory receptors. Accordingly, cytotoxicity assays confirmed that the TMEM30A KO cell line was killed by NK cells to a significantly lower extent compared to WT cells, but by blocking PtdSer on the surface of the K562 cells, the killing of TMEM30A KO cell line was largely restored (Fig. 4 B and C; additional clone in SI Appendix, Fig. S3E and using additional TMEM30A guides in SI Appendix, Fig. S3 F–H). Next, attempts were made to restore cytotoxicity by blocking PtdSer receptors on NK cells. In a first series of experiments, we exposed NK cells to WT or TMEM30A KO K562 cells in the presence or absence of an antibody to TIM-3. As shown in Fig. 4D, TIM-3 blockade had no effect on NK cell cytotoxicity against WT K562 cells, while it enhanced the cytotoxicity exerted by NK cells against TMEM30A KO K562 cells. A fab fragment to IRp60 had an agonistic activity on the receptor, thereby reducing NK cell effector function against WT K562 cells. By contrast, NK-cell mediated killing of TMEM30A KO cells was largely unaffected by anti-IRp60 Fab fragments (Fig. 4E), possibly indicating that the agonistic effect of the antibody was equivalent to the inhibition triggered by PtdSer. To further validate the role of these receptors, we used CRISPR/Cas9 to knock out the genes encoding TIM-3 or IRp60 in primary polyclonally activated NK cells. These activated NK cells killed the K562 cells to a too high degree to distinguish any differences (SI Appendix, Fig. S3I). Instead, we generated TMEM30A KO Jurkat (T-ALL) cells that are less sensitive to NK cells. In accordance with K562 cells, the TMEM30A KO Jurkat cells displayed increased exposure of PtdSer, also seen previously (20), but no change in HLA-E expression (Fig. 4F). Similar to the TMEM30A KO K562 cells, the TMEM30A KO Jurkat cells demonstrated a reduced sensitivity to NK cell cytotoxicity and triggered lower NK cell degranulation, which was not dependent on one particular NK cell subset (Fig. 4 G–I). The depletion of TIM-3 and IRp60 was not complete, but degranulation experiments further supported the role of TIM-3 for the reduced susceptibility of TMEM30A KO cells. Thus, the difference in NK cell degranulation after coculture with WT as compared to TMEM30A KO cells was significantly reduced with the TIM-3-depleted NK cells (Fig. 4J). A similar nonsignificant trend could also be seen for IRp60-depleted NK cells (Fig. 4K). Taken together, these results suggest that engagement of TIM-3 by surface-exposed PtdSer leads to TMEM30A KO-dependent inhibition of NK cells, while further experiments are warranted to determine the role of IRp60 in this context.
Fig. 4.
TMEM30A knock-out protects K562 cells from NK cell cytotoxicity correlating with enhanced extracellular PtdSer expression, interacting with inhibitory receptors. (A) Staining of TIM-3 (purple) and IRp60 (pink) of activated NK cells. (B) Cytotoxicity assay with WT K562 cells (blue) and TMEM30A KO cell line (green) at E:T ratios 1:2 and 1:1 (n = 11). (C–E) Cytotoxicity assay with K562 WT and TMEM30A KO cells at E:T ratio 1:1, combined with Annexin-V (Anx-V) for blockade of PtdSer (n = 7) (C), combined with TIM-3 blockade (n = 7) (D) or combined with IRp60 blockade (n = 8) (E). (F) Annexin-V and HLA-E staining of live Jurkat WT and TMEM30A KO cells. (G) Cytotoxicity assay with Jurkat WT and TMEM30A KO cells at various E:T ratios (n = 5). (H and I) Degranulation assay with Jurkat WT and TMEM30A KO cells, showing the percentage of CD107+ cells among CD56dim NK cells (H), separated based on NK cell NKG2A/KIR subtype (I) (n = 5). n indicates experiments with NK cells from different donors. (J and K) Degranulation assay with Jurkat WT and TMEM30A KO together with CRISPR/Cas9-engineered NK cells with modified expression of TIM-3 (HAVCR2 KO) (J) and IRp60 (CD300A KO) (K), showing percentage reduced degranulation against TMEM30A KO cells, compared to WT cells and representative dot plots where indicated gating strategy of 50% TIM-3low or IRp60low cells is shown (n = 4; representing separate KO-transfections). ns not significant, error bars represent SEM and one-way ANOVA with Šidák’s multiple comparisons test was used for (B–E), and (I) and the paired t-test was used for statistical analyses of (H), (J), and (K).
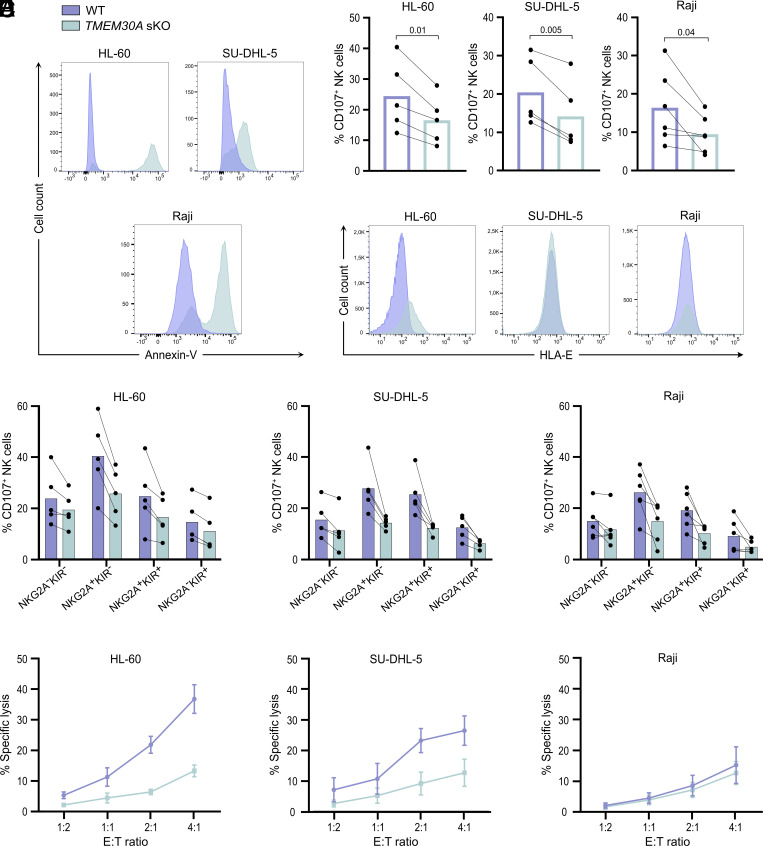
TMEM30A Deletions Allow Multiple Leukemia and Lymphoma Cell Lines to Evade NK Cell Cytotoxicity.
Next, we investigated whether the immunoevasive property of TMEM30A mutations was shared among other hematological malignancies. To this end, we generated TMEM30A KO versions of HL-60 (AML), Raji (Burkitt lymphoma), and SU-DHL-5 (DLBCL) cells. In accordance with K562 and Jurkat cells, CRISPR/Cas9 editing of all tested cell lines resulted in enhanced Annexin-V staining (Fig. 5A). Furthermore, in all cell lines tested, TMEM30A KO cells triggered less NK cell degranulation than WT cells (Fig. 5B). Notably, TMEM30A deletion resulted in enhanced expression of HLA-E also in the other myeloid cell line, HL-60, but this was not observed in the lymphoid cell lines (Fig. 5C). However, the elevated HLA-E expression did not explain the reduced NK cell degranulation toward TMEM30A KO cells, as both NKG2A+ and NKG2A− subsets displayed reduced degranulation to the KO cell line (Fig. 5D). Despite the fact that WT Raji cells triggered NK cell degranulation, Raji cells were largely resistant to NK cell cytotoxicity, which is in line with previous reports (23, 24). All other cell lines tested became more resistant to NK cell cytotoxicity after TMEM30A KO (Fig. 5E).
Fig. 5.
TMEM30A aberration in other hematological malignancies results in reduced sensitivity to NK cells, correlated by enhanced extracellular PtdSer expression. (A) Annexin-V staining of live TMEM30A KO (green) cells, compared to WT (blue) cells. (B) Degranulation assay with WT and TMEM30A KO cell line, showing the percentage of CD107+ cells among CD56dim NK cells (n = 5, Raji n = 6). (C) Staining for HLA-E on WT and TMEM30A KO cells. (D) Degranulation assay with WT and TMEM30A KO cells, showing the percentage of CD107+ cells among CD56dim NK cells, separated based on NK cell NKG2A/KIR subtype (n = 5, Raji n = 6). (E) Cytotoxicity assay with WT and TMEM30A KO cells at various E:T ratios (n = 5). n indicates experiments with NK cells from different donors. Error bars represent SEM, and the paired t-test was used for statistical analyses.
Discussion
In the present study, we sought to identify protein-encoding genes that modulate the susceptibility of target cells to NK cell cytotoxicity using a genome-wide CRISPR screen in K562 cells. These datasets provide insight into proteins expressed on the surface of target cells, acting as ligands for NK cell receptors as well as intracellular proteins involved in cellular pathways that have direct or indirect impact on the interaction between the target and effector cells. Previously performed NK cell-based CRISPR screens have typically used high selection pressure (13, 16, 17, 25). This may limit the amount of identified genes as only the most protected cells stand a chance to survive. Furthermore, under harsh conditions, most cells succumb, and it can make it difficult to identify gene deletions that sensitize the cells further. We hypothesized that a lower selection level and a shorter coculture would enable enrichment of additional gRNA hits that target other important interactions. In this study, we thus obtained several additional hits aside from NCR3LG1. We could identify CD58, the gene encoding the ligand to the coreceptor CD2, as well as TMEM30A. Establishing KO cell lines of specific genes identified in the screen allowed us to verify the role of the genes in the interaction between NK cells and target cells. Deletion of the CD58 gene resulted in a marginal effect on NK cell effector function, while the effect of NCR3LG1 was larger. However, we observed an additive effect of CD58 and NCR3LG1 deletion on NK cell cytokine production. Interestingly, in experiments using blocking antibodies to various activating NK cell receptors, we observed a shift from NKp30 dependence for NK cell cytotoxicity against WT K562 cells to a more pronounced role of NKG2D and DNAM-1 in cytotoxic responses to CD58/NCR3LG1 dKO K562 cells (26). This demonstrates that it is possible to skew the receptor–ligand interactions between target and NK cell by knocking out ligand genes. Notably, genes encoding ligands for DNAM-1 were not among the top hits in the screen despite the contributing role of DNAM-1 for NK cell killing of K562 cells. This likely reflects the existence of inhibitory receptors, such as TIGIT, TACTILE, PVRIG, that also recognize these ligands. Thus, the net effect of deletion of the ligand genes may be neutral.
Previous CRISPR screen studies with NK cells have highlighted roles for IFNγ signaling and MHC class I (13–15, 25). Our screen extends these findings and demonstrates a striking depletion of gRNAs related to MHC class I expression and the IFNγ signaling pathway. Thus, the top 50 genes comprised 13 genes involved in antigen presentation or IFNγ signaling. Although the K562 cell line is commonly used due to its lack of MHC class-I expression, the top depleted gRNAs in fact targeted the nonclassical MHC class-I molecule HLA-E–the ligand to the inhibitory receptor heterodimer, CD94/NKG2A. This is probably due to upregulation of MHC class-I in response to IFNγ secretion of the activated NK cells in the coculture (13). Thus, a knock-out of any gene associated with IFNγ signaling pathway or antigen presentation would reduce MHC class-I upregulation and sensitize the target cells to NK cell killing (13). By contrast, in a CRISPR screen study where the NK-92 cell line was used to select for susceptibility in K562 cells, loss of genes belonging to the IFNγ pathway prevented killing (17). These contradictory results are probably related to the use of the NK-92 cell line that display lower expression of many key inhibitory receptors that interact with MHC class I (27). In this setting, the IFNγ pathway did instead promote increased sensitivity, for example by boosting the expression of the key adhesion molecule, ICAM-1 that enables efficient NK cell lysis of target cells (28).
A key finding in our screen was the selective enrichment of cells carrying a TMEM30A deletion. TMEM30A encodes the protein CDC50A, which forms a heterodimer with P4-ATPases to create flippases that shuttle phospholipids from the outer to the inner leaflet of the plasma membrane (29). This enzyme system maintains plasma membrane asymmetry, but in case of caspase activation, the flippase is cleaved and inactivated resulting in upregulation of PtdSer on the outside of the apoptotic cell. A previous study identified the gene ATP11C along with TMEM30A as two key genes for the shuttling of PtdSer to the inner leaflet in cells (30). ATP11C encodes a P4-ATPase which is the α-subunit of the functional flippase. Despite the functional importance of the flippase for NK cell recognition of K562 cells, neither ATP11C nor any other genes encoding P4-ATPases appeared among the top hits in the screen. This probably reflects a redundancy among the P4-ATPases. There are 14 known P4-ATPases but only three known CDC50 proteins. Coimmunoprecipitation studies have shown that CDC50A forms complexes with 11 different P4-ATPases, while CDC50B and C, which are not ubiquitously expressed, only form complexes with a limited number of P4-ATPases. Besides ATP11C, ATP11A and ATP8A2 have also been shown to act as plasma membrane flippases (31). Thus, deletion of one single P4-ATPase may not be enough to stop transport of PtdSer to the inner leaflet as there are other interaction partners for CDC50A to produce a functional flippase in the plasma membrane.
Apart from PtdSer induction, deletion of the TMEM30A gene did generally not induce an altered expression profile of NK cell receptor ligands in the hematological cell lines evaluated in this study. An important exception was HLA-E, which was selectively up-regulated in myeloid TMEM30A-deficient cells. This lineage-restricted phenomenon is intriguing and warrants further investigation. HLA-E induction may further enhance the immuno-evasive properties of TMEM30A-mutated cells as HLA-E triggers the inhibitory NKG2A receptor on subsets of NK cells and T cells. However, the HLA-E induction did not fully explain the NK cell-resistant phenotype of TMEM30A-deficient myeloid cells as also NKG2A- NK cell subsets displayed reduced degranulation toward TMEM30A-KO cells.
Apoptotic cells with high expression of PtdSer are rapidly cleared by myeloid cells in a phagocytic process known as efferocytosis. PtdSer is believed to be one of the key “eat-me signals” and there are several PtdSer receptors promoting the uptake of apoptotic cells. In NK cells, inhibitory PtdSer receptors may serve to limit NK cell cytotoxicity to avoid overwhelming the clearance capacity of myeloid cells and ensuing secondary necrosis that may damage the tissue. An interesting possibility is that malignant cells exploit this inhibitory interaction and up-regulate PtdSer to limit NK cell cytotoxicity. In fact, inactivating mutations in TMEM30A were reported to be recurrent in DLBCL and IVLBCL (20–22), but to what extent loss of TMEM30A results in enhanced PtdSer in DLBCL has hitherto been unclear (20, 32). Our study shows that loss of the TMEM30A gene results in enhanced expression of PtdSer and inhibition of NK cell effector functions, ranging from cytokine release to cytotoxicity. Similar results were seen in a broad range of cell lines from different hematological malignancies, including DLBCL, indicating that this may be a general strategy to evade NK cell recognition and elimination. It is conceivable that altered flippase activity may affect NK cell interactions with target cells in other ways. Accumulation of phospholipids in the outer leaflet affects the membrane curvature (33), which could potentially impact immune synapse formation. Nevertheless, the ability to reverse the immunoevasive phenotype of TMEM30A-deficient cells by blocking either PtdSer or TIM-3 provides support for the mechanism proposed in our study.
Targeting the interactions between PtdSer and its receptors may thus be a promising strategy in immunotherapy of cancer. It should be pointed out that PtdSer has broad effects on tumor immunity beyond its impact on NK cell function. It has been reported that the extracellular expression of PtdSer is commonly dysregulated and contributes to an immunosuppressive tumor microenvironment and that the elevated expression interferes with the development of efficacious antitumor immunity (34, 35). Blocking PtdSer can reportedly skew tumor-associated macrophages from myeloid-derived suppressor cell phenotypes to proinflammatory M1 macrophages (9, 35). An antibody targeting PtdSer, bavituximab is currently being evaluated in various combinations in different solid cancers. In addition, there are multiple companies developing TIM-3 blocking antibodies, which have shown promising antitumor efficacy in preclinical studies and some are currently being investigated in phase 1/2 clinical trials (36–39). Multiple ligands have been described to interact with TIM-3, such as PtdSer, Galectin 9, CEACAM1, and HMGB1, and it is still unknown which interaction partner that is the most important. We did not investigate whether the expression of the other TIM-3 ligands varied between WT and TMEM30A KO cells, but it seems reasonable to assume that the effect of TIM-3 blockade observed in our experiments was due to interactions with PtdSer. It was also reported that TIM-3 antibodies with antitumor activity all had in common that they inhibited TIM-3 binding of PtdSer and CEACAM-1 (40). In our study, we observed that TIM-3 blockade predominantly affected the NK cell killing of TMEM30A KO cells with elevated levels of PtdSer, and knockout of TIM-3 in primary NK cells partially restored the susceptibility to NK cell cytotoxicity in TMEM30A KO Jurkat cells. Along these lines, a study in mice showed that treatment of NK cells in vitro with PtdSer reduced cytokine release and degranulation in a TIM-3-dependent fashion, and this mechanism was also suggested to explain the efficiency of TIM-3 blockade in a liver tumor model (41).
In conclusion, we have demonstrated that disruption of the TMEM30A gene drives enhanced extracellular expression of PtdSer that inactivates NK cell cytotoxicity via inhibitory receptors in a broad range of hematological malignancies. The results provide insight into a potential immune evasion strategy for malignant cells to avoid NK cells and may be a contributing factor for the efficacy of TIM-3 blockade.
Materials and Methods
Cell lines.
The details of the culture conditions of the cell lines can be found in SI Appendix, Table S1. The WT K562 cell line was authenticated directly before the Cas9 transduction (MCA by Multiplexion). All cell lines were cultured at 37 °C with 5% CO2 and regularly tested for Mycoplasma [Eurofins Mycoplasmacheck (qPCR)].
Isolation of NK Cells.
Peripheral blood mononuclear cells (PBMCs) were obtained from deidentified buffy coats from the blood center at the Sahlgrenska University Hospital, through density gradient centrifugation with Lymphoprep (Stemcell technologies, cat# 07861). NK cells were isolated by negative selection using a human NK cell isolation kit (Miltenyi Biotec, cat# 130-092-657).
Generation of Polyclonally Activated NK Cells.
NK cells were cultured together with irradiated allogenic PBMCs and 221G feeder cells in medium containing PHA-M (Sigma, cat# L8902). After 7 d, the medium was gradually replaced with the same medium excluding PHA-M. Culture details can be found in SI Appendix, Table S1.
Genome-wide CRISPR Screen.
K562 cells were transduced with lentiCas9-Blast (42) (Feng Zhang, Addgene, Addgene_52962) by spinoculation in 5 µg/mL polybrene (Merck, cat# TR-1003-G) and exposed to blasticidin (30 µg/mL; InVivoGen, cat# ant-bl-1) for 20 d to select transduced cells. Clones were obtained by single-cell sorting using a FACSAria III (BD Biosciences, NJ, USA). Cas9 expression was confirmed by western blot (SI Appendix, Table S2).
The genome-wide Brunello single-guide RNA library (gRNA) (43) was resynthesized to include Unique Molecular Identifiers (UMI) (44). Guides were cloned in pool (oligos synthesized by CustomArray) and packaged into lentivirus. The lentiviral backbone was based on lentiGuide-Puro (Addgene_52963), with AU-flip as previously described (45).
Cas9-expressing K562 cells were transduced with the Brunello library (MOI of 0.4;1,000 cells/guide). Transduced cells were selected (4 µg/mL puromycin) and then split into two replicates and cultured independently before cryopreservation.
Before selection experiments with NK cells, transduced cells were thawed and maintained in K562 medium with puromycin (2 µg/mL) for a total of 9 d. Eighty million transduced K562 cells per replicate were cocultured with activated NK cells at effector:target (E:T) ratio 1:2 in flat-bottom 96-well plates for 20 h when approximately 60% of K562 cells had died. NK cells were depleted by puromycin treatment and surviving K562 cells were pelleted and frozen. Control cells were maintained under the same conditions except for the exposure to NK cells.
Genomic DNA was isolated using the QIAamp DNA Blood Maxi kit (Qiagen), and guide and UMI sequences were amplified by PCR as described elsewhere (44). The sequencing was performed using an Illumina Novaseq S1 flowcell with approximately 90 million reads allocated per sample. NGS data were analyzed with the MaGeCK (Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout) software (46) and by UMI lineage dropout analysis (44).
CRISPR/Cas9 Engineered Knock-out Cell Lines and NK Cells.
For knock-out in HL-60, Jurkat, Raji, SU-DHL-5, and primary NK cells, a ribonucleoprotein (RNP)-based method was used, whereas for Cas9-expressing K562 cells, both RNP and a plasmid-based method was utilized. The spacers for the genes NCR3LG1 (B7-H6), CD58, TMEM30A, HAVCR2 (TIM-3), and CD300A (IRp60) were chosen among Broad Institute validated guides or Integrated DNA technologies’ (IDT) predesigned and confirmed using Synthego’s guide design verification tool. For TMEM30A KO, two additional spacers (B and C) were used to confirm the results from the first spacer (A) (all sequences in SI Appendix, Table S3). For the plasmid-based method, a construct containing the scaffold tracrRNA and restriction sites for both incorporation of the construct into a plasmid as well as insertion of the gene-specific spacer was designed (sequence in SI Appendix, Table S3). The construct and the plasmids mEGFP-C1 and EBFP2-C1 (Addgene_54759 and Addgene_54665, both deposited to Addgene by Michael Davidson) were digested using FastDigest BamHI and XhoI (Thermo Scientific, cat# FD0054 and FD0694), gel purified using RECOCHIP (TaKaRa, (cat# 9039), ligated using T4 DNA ligase (NEB, cat# M0202S) followed by plasmid transformation in DH5α electroMAX cells (Invitrogen, cat# 11319019). The spacers were annealed before being subjected to combined digestion and ligation using BbsI-HF (NEB, cat# R3539L) and T4 DNA ligase with the mEGFP- or EBFP2-plasmid. After transformation, the plasmid was purified using NucleoBond Xtra Midi EF (Macherey-Nagel, cat# 740410.50).
For the RNP-based method, the gene-specific crRNAs were ordered as RNA and annealed to form a duplex with equimolar amount of Alt-R CRISPR-Cas9 tracrRNA (IDT, cat# 1072533) and then incubated with Alt-R S.p. HiFi Cas9 Nuclease V3 (IDT, cat# 1081059) for RNP formation.
Transfection of the cell lines was performed using the Neon electroporation system (100 µL tips, for settings, see SI Appendix, Table S4; Invitrogen). Cas9-expressing WT K562 cells were used as basis for knock-out (KO) of NCR3LG1, CD58 and TMEM30A, whereas a NCR3LG1/CD58 double KO (dKO) was created using NCR3LG1 single KO (sKO) K562 cells. After 2 d, the cells were single cell-sorted, based on GFP- or BFP-expression for NCR3LG1 and Annexin-V+ expression for TMEM30A KO cells, using a 3-laser FACSAria III and propagated to create clones. Promising clones were expanded and the genomic region covering the cut site was amplified (primers in SI Appendix, Table S5) and sent for Sanger sequencing (Eurofins Genomics) to verify double allelic KO of the gene. Synthego's ICE and CRISPR-ID tools were used for analysis of the sequencing results. CD58- K562 cells and TMEM30A KO HL-60, Jurkat, SU-DHL-5, and Raji cells were sorted in bulk and used polyclonally. TMEM30A KO K562 cells generated using additional guides B and C were also sorted in bulk and used polyclonally for confirmational experiments. Mock-transfected cells were generated using Alt-R Negative control crRNA (IDT, cat# 1072544) and were confirmed to behave similar to WT cells (SI Appendix, Fig. S1F).
Transfection of polyclonally activated NK cells was performed using 4D-nucleofector X Unit (Lonza) with P3 Primary Cell 4D-Nucleofector X Kit S (Lonza, cat# V4XP-3032) and the program DN-100. The RNP complex was prepared as for the cell lines. One million polyclonally activated NK cells were used per sample and 40 pmol RNP per transfection. Two guides for CD300A were used in combination. Mock-transfected cells were generated using Alt-R Negative control crRNA (IDT, cat# 1072544).
Flow Cytometry.
Details about antibodies and related reagents can be found in SI Appendix, Table S2. In general, cells were washed in buffered sodium chloride and stained for 30 min at 4 °C. TMEM30A KO K562 cells were washed and stained in Annexin-V buffer with Annexin-V and ToPro3 (invitrogen) for 15 min at room temperature. Data acquisition was performed on a 5-laser BD LSRFortessa (BD Biosciences, NJ, USA) and data analyzed using BD FACSDIVA version 9.0 or FlowJo software version 10.8.1.
Functional Assays.
Details on the antibodies and related reagents can be found in SI Appendix, Table S2. For the cytotoxicity assays, prestained target cells (Celltrace CFSE or violet, Invitrogen, cat# C34554 and C34557) were cocultured together with NK cells before staining with ToPro3. NK cell cytotoxicity against WT K562, NCR3LG1 KO and CD58 KO cells was determined after 3 h of coincubation at different effector/target ratios (1:2, 1:1, 2:1, 4:1 and 8:1). NK cell cytotoxicity responses to WT and TMEM30A KO K562, Jurkat, HL60, Raji, SU-DHL-5 cells were assayed after overnight incubation at effector/target ratios ranging from of 1:2 to 4:1 in the presence of 5 ng/mL IL-15 (PeproTech, cat# 200-15). To study the role of the different activating and inhibitory receptors involved in the interaction between the cells, blocking antibodies were used for the cytotoxicity assays. The antibody targeting NKp44 was used as a control antibody as the receptor is not expressed by resting NK cells. For blockade of IRp60, Fab fragments were generated in order to avoid receptor crosslinking using the Pierce Mouse IgG1 Fab and F(ab’)2 Micro Preparation Kit (Thermo Scientific, cat# 44680). The purification buffer was replaced by sodium chloride using Amicon Ultra-15 Centrifugal Filter Unit (Merck Millipore, cat# UFC903008). Blockade was performed either by preincubating NK cells with receptor antibodies for 15 min at room temperature or by preincubating K562 cells with Annexin-V for 1 h at 37 °C before coculture.
For degranulation and intracellular staining (ICS) assays, thawed PBMCs were cultured overnight with 500 IU/mL IL-2 before addition of target cells at PBMC:target ratio of 10:1 and the CD107a antibody (degranulation), followed by 1 or 5 h incubation respectively, with Brefeldin A (GolgiPlug, BD Biosciences, cat# 555029) added to ICS after 1 h. The cells were then stained with live/dead near-IR (Invitrogen, cat# L10119), followed by a mix of antibodies. For ICS, the cells were finally fixated and permeabilized with BD cytofix/cytoperm (BD biosciences, cat# 554722) and stained for IFNγ and TNFα.
Engineered KO NK cells were used together with WT and TMEM30A KO Jurkat cells in degranulation assays and stained for IRp60 and TIM-3 after 1 h coincubation. The experiments represent separate KO-transfections. In the gating strategy of both mock-transfected cells and KO cells, 50% TIM-3low or IRp60low NK cells were included. Percentage reduced degranulation against TMEM30A KO cells compared to WT cells was calculated as follows: 1 − (% degranulation against TMEM30A KO cells/% degranulation against WT cells).
Statistical Analyses.
The statistical analyses were performed using GraphPad Prism version 9.4. Data were represented as mean and comparison was performed using the paired t-test or one-way ANOVA followed by Šidák’s multiple comparison test for multiple group comparisons for preselected pairs or Dunnett’s multiple comparison test for comparisons to a control. P-values <0.05 were considered statistically significant.
Study Approval.
Blood samples were collected with written consent from donors.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Acknowledgments
Part of this work was carried out at the CRISPR Functional Genomics unit at Karolinska Institutet funded by Science for Life Laboratory. [We acknowledge support from the National Genomics Infrastructure. Computations were performed on resources provided by SNIC (project SNIC 2017-7-265) at the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX)]. This study was supported by the Swedish Research Council (2020-02783), the Swedish Cancer Society (19-0449), Region Västra Götaland, BioCARE, Wilhelm and Martina Lundgren foundation (2019-3167), Assar Gabrielsson’s foundation (FB21-76), the Roche foundation (per la Ricerca 2017), the Lerici foundation (2018), the Umberto Veronesi foundation (2018-1914), Leonardo da Vinci fellowship (2019) and the Sahlgrenska Academy at the University of Gothenburg.
Author contributions
L.K., C.B., Y.T., G.X., S.P., K.-W.T., and F.B.T. designed research; L.K., C.B., A.L., E.I., B.A.H., and S.P. performed research; L.K., C.B., A.L., E.I., and S.P. analyzed data; L.K., K.-W.T., and F.B.T. supervised research; and L.K. and F.B.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The data for this study were generated at CRISPR Functional Genomics (CFG) at Karolinska Institutet funded by Science for Life Laboratory. All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Bryceson Y. T., March M. E., Ljunggren H. G., Long E. O., Activation, coactivation, and costimulation of resting human natural killer cells. Immunol. Rev. 214, 73–91 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quatrini L., et al. , Human NK cells, their receptors and function. Eur. J. Immunol. 51, 1566–1579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeKruyff R. H., et al. , T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J. Immunol. 184, 1918–1930 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ndhlovu L. C., et al. , Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood 119, 3734–3743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakahashi-Oda C., Tahara-Hanaoka S., Honda S., Shibuya K., Shibuya A., Identification of phosphatidylserine as a ligand for the CD300a immunoreceptor. Biochem. Biophys. Res. Commun. 417, 646–650 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Simhadri V. R., et al. , Human CD300a binds to phosphatidylethanolamine and phosphatidylserine, and modulates the phagocytosis of dead cells. Blood 119, 2799–2809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lankry D., Rovis T. L., Jonjic S., Mandelboim O., The interaction between CD300a and phosphatidylserine inhibits tumor cell killing by NK cells. Eur. J. Immunol. 43, 2151–2161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith C. M., Li A., Krishnamurthy N., Lemmon M. A., Phosphatidylserine binding directly regulates TIM-3 function. Biochem. J. 478, 3331–3349 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Y., Huang X., Lynn K. D., Thorpe P. E., Phosphatidylserine-targeting antibody induces M1 macrophage polarization and promotes myeloid-derived suppressor cell differentiation. Cancer Immunol. Res. 1, 256–268 (2013). [DOI] [PubMed] [Google Scholar]
- 10.He J., Yin Y., Luster T. A., Watkins L., Thorpe P. E., Antiphosphatidylserine antibody combined with irradiation damages tumor blood vessels and induces tumor immunity in a rat model of glioblastoma. Clin. Cancer Res. 15, 6871–6880 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Gray M. J., et al. , Phosphatidylserine-targeting antibodies augment the anti-tumorigenic activity of anti-PD-1 therapy by enhancing immune activation and downregulating pro-oncogenic factors induced by T-cell checkpoint inhibition in murine triple-negative breast cancers. Breast Cancer Res. 18, 50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budhu S., et al. , Targeting phosphatidylserine enhances the anti-tumor response to tumor-directed radiation therapy in a preclinical model of melanoma. Cell Rep. 34, 108620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang X., Veltri D. P., Long E. O., Genome-wide CRISPR screen reveals cancer cell resistance to NK cells induced by NK-derived IFN-gamma. Front. Immunol. 10, 2879 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheffer M., et al. , Genome-scale screens identify factors regulating tumor cell responses to natural killer cells. Nat. Genet. 53, 1196–1206 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Bernareggi D., et al. , CHMP2A regulates tumor sensitivity to natural killer cell-mediated cytotoxicity. Nat. Commun. 13, 1899 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiba M., et al. , Genome-wide CRISPR screens identify CD48 defining susceptibility to NK cytotoxicity in peripheral T-cell lymphomas. Blood 140, 1951–1963 (2022), 10.1182/blood.2022015646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pech M. F., et al. , Systematic identification of cancer cell vulnerabilities to natural killer cell-mediated immune surveillance. Elife 8, e47362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bock C., et al. , High-content CRISPR screening. Nat. Rev. Method Prime 2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufva O., et al. , Single-cell functional genomics reveals determinants of sensitivity and resistance to natural killer cells in blood cancers. Immunity 56, 2816–2835.e3 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Ennishi D., et al. , TMEM30A loss-of-function mutations drive lymphomagenesis and confer therapeutically exploitable vulnerability in B-cell lymphoma. Nat. Med. 26, 577–588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada K., et al. , Frequent genetic alterations in immune checkpoint-related genes in intravascular large B-cell lymphoma. Blood 137, 1491–1502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Farre B., et al. , Intravascular large B-cell lymphoma genomic profile is characterized by alterations in genes regulating NF-κB and immune checkpoints. Am. J. Surg. Pathol. 47, 202–211 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roozemond R. C., van der Geer P., Bonavida B., Effect of altered membrane structure on NK cell-mediated cytotoxicity. II. Conversion of NK-resistant tumor cells into NK-sensitive targets upon fusion with liposomes containing NK-sensitive membranes. J. Immunol. 136, 3921–3929 (1986). [PubMed] [Google Scholar]
- 24.Kim J. H., Lee J. K., Naringenin enhances NK cell lysis activity by increasing the expression of NKG2D ligands on Burkitt’s lymphoma cells. Arch. Pharm. Res. 38, 2042–2048 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Dufva O., et al. , Single-cell functional genomics of natural killer cell evasion in blood cancers. bioRxiv [Preprint] (2022). 10.1101/2022.08.22.504722 (23 August 2022) [DOI] [PubMed]
- 26.Hussein B. A., et al. , NKG2A gene variant predicts outcome of immunotherapy in AML and modulates the repertoire and function of NK cells. J. Immunother Cancer 11, e007202 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunesch J. T., et al. , Genome-wide analyses and functional profiling of human NK cell lines. Mol. Immunol. 115, 64–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryceson Y. T., March M. E., Barber D. F., Ljunggren H. G., Long E. O., Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J. Exp. Med. 202, 1001–1012 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Mark V. A., Elferink R. P., Paulusma C. C., P4 ATPases: Flippases in health and disease. Int. J. Mol. Sci. 14, 7897–7922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segawa K., et al. , Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Segawa K., Kurata S., Nagata S., Human type IV P-type ATPAses that work as plasma membrane phospholipid flippases and their regulation by caspase and calcium. J. Biol. Chem. 291, 762–772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ennishi D., The biology of the tumor microenvironment in DLBCL: Targeting the “don’t eat me” signal. J. Clin. Exp. Hematop. 61, 210–215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke R. J., Hossain K. R., Cao K., Physiological roles of transverse lipid asymmetry of animal membranes. Biochim. Biophys. Acta Biomembr 1862, 183382 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Birge R. B., et al. , Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 23, 962–978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W., et al. , Mobilizing phospholipids on tumor plasma membrane implicates phosphatidylserine externalization blockade for cancer immunotherapy. Cell Rep. 41, 111582 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakuishi K., et al. , Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 207, 2187–2194 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ngiow S. F., et al. , Anti-TIM3 antibody promotes T cell IFN-gamma-mediated antitumor immunity and suppresses established tumors. Cancer Res. 71, 3540–3551 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Zhou Q., et al. , Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood 117, 4501–4510 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acharya N., Sabatos-Peyton C., Anderson A. C., Tim-3 finds its place in the cancer immunotherapy landscape. J. Immunother Cancer 8, e000911 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabatos-Peyton C. A., et al. , Blockade of Tim-3 binding to phosphatidylserine and CEACAM1 is a shared feature of anti-Tim-3 antibodies that have functional efficacy. Oncoimmunology 7, e1385690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan S., et al. , Tim-3 hampers tumor surveillance of liver-resident and conventional NK cells by disrupting PI3K signaling. Cancer Res. 80, 1130–1142 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmierer B., et al. , CRISPR/Cas9 screening using unique molecular identifiers. Mol. Syst. Biol. 13, 945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cross B. C., et al. , Increasing the performance of pooled CRISPR-Cas9 drop-out screening. Sci. Rep. 6, 31782 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W., et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Data Availability Statement
The data for this study were generated at CRISPR Functional Genomics (CFG) at Karolinska Institutet funded by Science for Life Laboratory. All other data are included in the manuscript and/or supporting information.