Abstract
This review focuses on discussing key mechanisms in disease pathogenesis mediated by the protein post-translational modification citrullination. These processes are discussed in depth in the context of complex diseases such as rheumatoid arthritis, cancer, central nervous system disorders, and cardiovascular disease. Additionally, a critical evaluation of challenges in laboratory detection of citrullination sites is also outlined. In this context, the role of mass spectrometry is discussed with a focus on contemporary techniques that offer promising options to detect the exact site of protein citrullination. Novel methods described in the paper have the potential to detect and quantify the occurrence of post-translational modification sites for diagnosis and therapeutic purposes with a high degree of specificity and sensitivity. Furthermore, they offer a much faster performance than traditional techniques making them ideal for large-scale experimentation.
Keywords: citrullination, rheumatoid arthritis, cancer, atherosclerosis, cardiac disease, CNS, mass spectrometry
1. INTRODUCTION
Citrullination is a protein post-translational modification (PTM) that has a role in the pathogenesis of human diseases. It can be a key mediator of multifactorial conditions such as rheumatic arthritis, cancer, multiple sclerosis, and cardiovascular disease. However, the low cellular abundance of citrullinated proteins, as well as the low stoichiometry of citrullinated residues within modified proteins (among other factors), makes it difficult to identify and quantify the extent of citrullination in samples through traditional laboratory methods. The focus of this paper is to investigate and determine the role of citrullination in the pathogenesis of disease and explain novel citrullination-specific mass spectrometry (MS) approaches. The reviewed literature presented in this paper will provide clarity in the current state of understanding of citrullination-mediated mechanisms in disease onset and its progression, and summarizes MS methods that can isolate specific sites of modification that may serve as probable targets for diagnosis and treatment.
2. CITRULLINATION AS A PROTEIN POST-TRANSLATIONAL MODIFICATION
Modification of proteins following the translation of mRNA (mRNA) is a normal physiological process. Scientists have been able to identify more than 200 PTMs in eukaryotes.1 PTMs can influence protein cellular localization, turnover, and interaction with substrates and/or proteins. PTMs may also affect their inherent function or activity state.2 The main PTM addressed in the context of disease pathogenesis in the rest of the paper is citrullination.
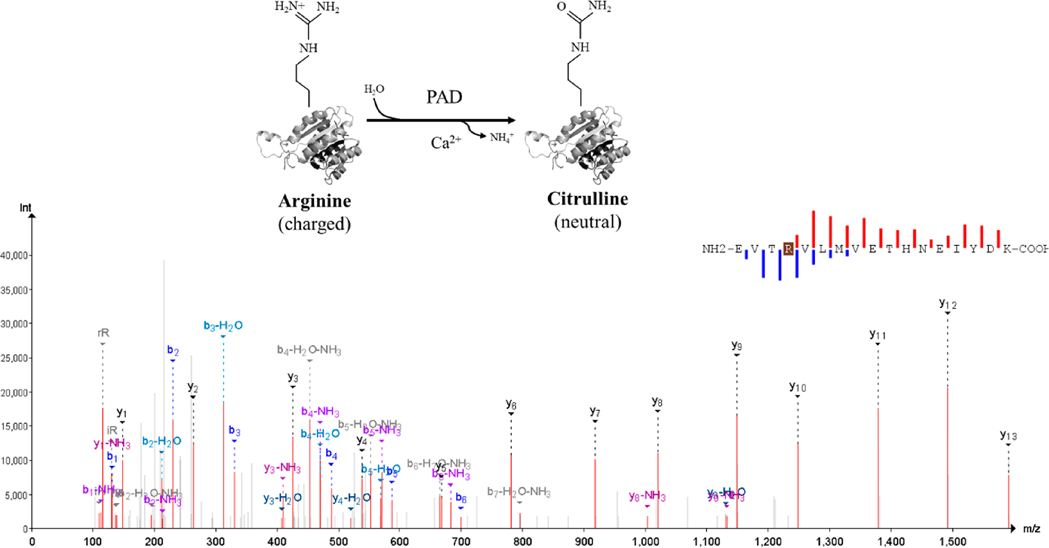
Citrullination is the deimination of the amino acid arginine characterized by the hydrolysis of nitrogen atoms of one of its side chains. This irreversible PTM converts the guanidium group of the arginine residue to an ureido group. The reaction leads to the production of the nonstandard amino acid citrulline, ammonia release, and the loss of a positive charge with a monoisotopic mass contrast of +0.984016 (Figure 1), lowering the overall charge of the protein each time the reaction takes place. Citrullination, as an enzymatic reaction, can take place in alkaline solutions at ambient temperatures. Peptidylarginine deiminases (PADs), enzymes dependent on Ca2+, act as catalysts for protein citrullination.3 The five PAD enzymes expressed in the human body are PAD 1–4 and PAD 6.4 The guanidine group of arginine provides a positive charge at physiological pH, while citrulline has a neutral charge (Figure 1). The charge difference has a direct impact on the isoelectric point of the protein, its ability to form hydrogen bonds, and its charge distribution. Citrullination can also increase the local and global hydrophobicity of the modified peptide,5 which in turn indicates that citrullination can modify protein folding, change its polarity, or trigger denaturation, thus making citrullinated proteins potentially more susceptible to degradation by enzymatic activity.6 Research evidence suggests that cellular protein such as histones, vimentin, and filaggrin and extracellular matrix proteins such as collagens may be citrullinated under normal and/or disease conditions.7
Figure 1.
Citrullination as the protein arginine deiminases-catalyzed reaction80 with an annotated spectrum of an illustrative citrullinated peptide.
Citrullination has physiological significance as well as a role in disease pathogenesis. Physiological processes manifesting citrullination include gene regulation, apoptosis, and epidermal terminal differentiation.8,9 It also plays a part in immune system functions, apoptosis, and skin physiology.6,10 There is also an ever growing number of pathological circumstances where citrullinated proteins are linked to multiple sclerosis, rheumatoid arthritis, Alzheimer’s disease, cancer, central nervous system disorders, and cardiovascular disease.4,11–14 This is illustrated in Supplemental Table 1, which further summarizes the ongoing information regarding citrullinated proteins, their biological role, associated pathologies, and the methodology employed for enrichment and/or detection. Recently, there is indication of a role for citrullination in epigenetic regulation, as citrullination can alter histones directly or through the interaction of citrullinated histone-modifying enzymes and coactivators. Furthermore, citrullination can alter transcription by potentially antagonizing the methylation of splicing factors such as SFPF and its association with mRNA.15
3. POST TRANSLATIONAL MODIFICATIONS AND PATHOGENESIS OF DISEASE
3.1. . Rheumatoid Arthritis (RA)
In RA, protein and peptide citrullination contribute to the inflammation of the synovial membrane’s joints. The research community is largely interested in understanding the significance of anti-citrullinated protein antibodies (ACPAs) found in the sera of affected individuals.16 Dysregulation of PAD enzyme, citrullination, and its contribution to the formation of neutrophil extracellular traps (NETs) containing DNA and protein from neutrophils, as a result of inflammation, is characteristic of the pathogenesis of RA. RA synovium contains high levels of citrullinated proteins.17 In fact, anticyclic citrullinated proteins (anti-CCPs) are an important type of ACPAs and are used to diagnose RA in its early, as well as, advanced stages.18 As already described, filaggrin, fibronectin, fibrinogen alpha and beta chains, vimentin, and collagen are autoantigens for the ACPAs and are present in the synovial fluid.19–21 Tutturen and team applied MS to determine the degree of citrullination and frequency of citrullinated peptides present in the synovial fluid obtained from patients with RA. They identified new autoantigens, PAD4 and alpha-enolase, as well as known proteins fibrinogen alpha chain, fibrinogen beta chain, and vimentin. Interestingly, different citrullination protein patterns existed in different patients with RA.11
Shelef and team linked the presence of citrullinated fibronectin with synovial fibroblasts in RA.22 Under normal physiological conditions, fibronectin acts as the mediator for several synovial fibroblast functions. However, when fibronectin is citrullinated, fibroblast adhesion and migration on the modified fibronectin are impaired. The authors elucidated that citrullinated fibronectin in RA can modify the behavior of both rheumatoid and normal synovial fibroblasts. They stated that much deeper research is required to discover the role of citrullinated extracellular matrix proteins and citrullinated fibronectin in RA.22
Yu and team suggested that citrullinated proteins may be generated in human fibroblast-like synoviocytes by PAD2, an event induced by hypoxia in RA synovium.36 Exposure to hypoxia increased the levels of both PAD2, as well as citrullinated proteins, while PAD4 levels were decreased. Hypoxia also increased the levels of IL-6 and IL-8.23
Sohn et al. found that citrullinated histones from neutrophils were targeted by ACPAs during an autoimmune response,12 and increased histone citrullination could be linked to an increased innate immunostimulatory capacity.24 They proposed that specific citrullinated histone proteins activated cytokine production by macrophages, which in turn activated neutrophils. Furthermore, histones cH2A and cH2B also acted as autoantigens in stimulating the innate immune response.12
RA patients are known to experience coronary heart disease and atherosclerosis to a greater degree than normal subjects do.25 Spinelli and researchers further explored the role of anti-citrullinated protein antibodies (ACPAs) in their review of atherosclerosis.26 The studies also included that RA patients with comorbid Porphyromonas gingivalis could also produce PAD enzymes leading to the development of RA in these susceptible individuals. Linking this evidence together with an ACPA-specific response to citrullination revealed the link between RA and citrullination.27,28 Other studies demonstrated a correlation between the coronary artery calcium score and citrullinated fibrinogen and vimentin. PAD enzymes and citrullinated proteins were also detected in the perivascular myocardial interstitium of patients diagnosed with RA.29,30 Moreover, the proper diagnosis and identification of early RA patients should be based on the presence of ACPAs together with rheumatoid factor (RF) and anti-carbamylated protein (anti-CarP) antibodies.31 Carbamylated proteins have been positively associated with the erosive form of RA. Moreover, antibodies to carbamylated proteins can be detected before the onset of RA, similar to the ACPA and rheumatoid factor.32,33 Carbamylation of low-density lipoprotein (LDL) leads to endothelial dysfunction through the lectin-type oxidized LDL receptor 1 that is a proposed biomarker for RA. Carbamylated LDL may have several other adverse reactions, such as uncoupling of endothelial nitric oxide synthase that reduces nitric oxide bioavailability and impairs endothelium vasodilation. Many more mechanisms of carbamylated LDL in the pathogenesis of atherosclerosis are still under study.34,35
3.2. Cardiovascular Disease and Heart Failure
Fert-Bober and colleagues performed a comprehensive experiment on human and mouse heart tissue using the citrullination-targeted proteomic strategy through a data-independent acquisition method (discussed in section 3). They were able to identify and validate 304 citrullinated sites mapping 145 proteins in human myocardium, from healthy individuals and also with two different forms of heart failure.36 Through RT-PCR, they confirmed PAD2 as the main isoform found in both cardiac fibroblasts and cardiomyocytes, while PAD1 and PAD4 were only expressed in cardiac fibroblasts. Interestingly, protein citrullination was shown to be dominant in sacromeric and mitochondrial subproteome. Furthermore, increased citrullination of enzymes involved in metabolic pathways was observed and found in ischemia cases, but not in idiopathic dilated cardiomyopathy (IDCM) cases. The sacromeric proteins are involved in muscle contraction via ATP-dependent interaction between F-actin and myosin and are regulated by tropomyosin. Among significant findings was the ability of citrullinated F-actin to modulate the enzymatic activity of biologically active fragment of myosin (heavy meromyosin, HMM). Citrullinated F-actin also modulated citrullinated tropomyosin by blocking the ability of myosin to bind to the actin filament. Citrullination in sacromeric proteins is suggestive of PTM regulation for cardiac contractility.36
Furthermore, apolipoprotein A-1 (apoA-1), present in high-density lipoproteins (HDLs), offers protective effects against atherosclerosis by suppressing low-density lipoprotein (LDL) oxidation. Dysfunctional HDL is characteristic of cardiovascular disease (CVD). Dysfunctional HDL is the result of PTM modification of apoA-1 that triggers inflammatory processes and promotes the development of CVD in individuals with atherosclerosis. Loss of function of HDL is the result of oxidized phospholipid accumulation in HDL, with the potential to mediate inflammation and oxidation.37,38 As suggested, a higher risk of CVD in RA populations is, in part, the result of shared inflammatory events between the two processes.39
Anti-citrullinated protein antibodies (ACPA) are associated with adverse outcomes in cardiovascular disease.40 The presence of these anti-citrullinated antibodies that are not specific to RA may account for a possible autoimmune component mediating inflammation in other pathological conditions such as cardiovascular disease.41 Westerlind and researchers further elaborated on the role of ACPA from their experimental testing of antibody load with a follow-up period of 12 years. Results indicated that ACPA has a clear link with cardiovascular disease as a comorbid condition in RA individuals. The individual ACPAs, more so, the total number of ACPAs found (antibody load), raised the risk of cardiovascular disease.42 Hermans and colleagues also worked on a similar research initiative to identify the long-term outcomes associated with ACPA in patients diagnosed with ST-elevation myocardial infarction (STEMI) and without RA as a concomitant condition. On testing the association, they found an increased “cumulative cardiac mortality” and an increased incidence of reinfarction and mortality. The study established ACPA as a potential risk factor for cardiovascular disease.43
3.3. CNS Disorders: Multiple Sclerosis, Alzheimer’s, Stroke, and Injury
Increased levels of MBP and GFAP found in the cells, tissues, serum, cerebral spinal fluid, and other body fluids of spinal cord and brain occur in patients diagnosed with multiple sclerosis. Neither the exact modification site nor the PAD enzyme isoform has been identified, although the PAD2 isoform is predominant in the central nervous system. GFAP is an intermediate filament protein found in the cytoskeleton and serves as an important biomarker for brain injury and stroke. GFAP is also citrullinated in multiple sclerosis.44 Patients suffering from brain injury also had circulating MBP and GFAP. In fact, increased circulating GFAP and UCH1 (ubiquitin carboxyl-terminal hydrolase) are FDA-approved for diagnosis of traumatic brain injury.45,46
Another brain protein neurogranin (NRGN) responsible for learning was found in the cerebral spinal fluid of patients diagnosed with Alzheimer’s disease. Researchers used direct MS approach to identify 12 sites of endogenous citrullination in the MBP protein, 4 endogenous citrullinated residues in the GFAP protein, and 1 in NRGN protein. Their findings also suggested that methylation of arginine prevented citrullination catalyzed by PAD2 at the site of the same residue. Researchers concluded that citrullination of arginine in CNS-specific proteins increases the immunogenicity of proteins. Autoantibodies against the citrullinated proteins act as autoantigens after brain injury and may exacerbate it. Detecting and blocking antibodies may serve as an important step for a better prognosis, stratification of risk, and therapy.4,47
3.4. Cancer
Citrullination may have a significant role in the pathogenesis of cancer. Limited evidence exists on the influence of citrullination in the progression of tumors.48 In an attempt to study the citrullination-cancer link, Yuzhalin identified increased PAD4 expression in tumors and peripheral blood of patients diagnosed with lung cancer.49–51 Benign tumors and nontumor-inflamed tissues did not show any such characteristic.49 In the case of metastasis, PAD4 levels were even higher than primary tumors.52 These findings are suggestive of the role of citrullination in disease progression, from benign tissue overgrowth to an invasive form of cancer. Citrullination also affected cell signaling in cancer cells such as removing the methylation tag from histones, PAD4 and p53 interactions with implications in apoptosis, PAD4 mediated p53-to-ING4 binding leading to repression of p53 acetylation and inhibition of downstream p21 expression, inhibition of Wnt/beta-catenin signaling, and PAD2-mediated activation of androgen signaling. So far, research evidence appears to point to a potential link between the formation of NETs and citrullination, and its potential role in inducing antitumor immunity.14
4. METHODS FOR PROTEIN CITRULLINATED ANALYSIS
4.1. Pitfalls of Existing Approaches for Laboratory Detection of Protein Citrullination
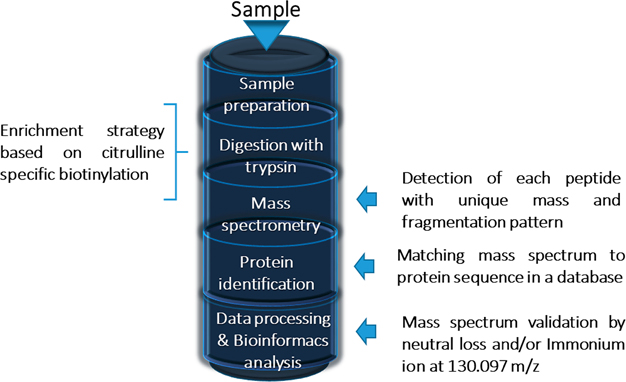
A classical method for detection of protein citrullination is the Color Development Reagent (COLDER assay), which relies on the chemical derivatization of the urea group but cannot detect citrullinated proteins in low concentration nor provide information about which amino acid residue is modified. Regarding citrullinated, antibody-based detection system for proteins offer greater sensitivity, but do not exhibit the required sensitivity. Traditional MS approaches are challenging and arduous, as a high level of mass accuracy is required to detect the small change in mass among other reasons but provide both protein and site information (and quantification). The MS approach has been optimized through the incorporation of additional processes such as collision-induced dissociation (CID), neutral loss, and enriched citrullination-specific peptide libraries that can be used alone or in combination with protein enrichment approaches53 (as illustrated in Figure 2), these approaches being discussed in the rest of this section.
Figure 2.
Mass spectrometry approach to analyze citrullinated peptides.
A common approach in detecting protein citrullination is to chemically modify the citrulline ureido group, through the use of antipyrine and diacetylmonoxime, and staining the polyclonal antibodies with the ability to detect modified citrulline. The same technique also helps visualize citrullinated proteins in tissues with immunohistochemistry or using the combination of gel electrophoresis and Western blotting and afterward identifying the protein by peptide mass fingerprinting. The method has limitations in terms of only being able to detect whether protein citrullination has taken place and cannot identify the exact sites of citrullination.54–57
One way to get around this problem is the use of diacetylmonoxime–antipyrine derivatization for targeting citrullinated peptides, themselves. This method uses liquid chromatography-tandem MS, and the detection of a signature ion that is generated by CID fragmentation. In an analogous approach, protein citrulline residues can be modified using 2,3-butandione.58 Another MS signature ion for citrullination with the CID fragmentation method is the neutral loss of isocyanic acid of the citrullinated peptides. Scientists also presented a technique to visualize protein citrullination by using a rhodamine-phenylglyoxal probe before performing SDS-PAGE and fluorescent analysis or cutting out the gel spot and analysis by MS.59
4.2. Mass Spectrometry Approaches for Identifying Citrullinated Proteins
The identification of citrullinated residues cannot be detected directly using immunostaining or standard chemical derivatization approaches.4 Therefore, mass spectrometry appears as the technique of choice. In traditional MS approaches, a protease digests proteins before analysis by MS. A common protease used in MS analysis is trypsin, which cleaves the C-terminal of arginine or lysine unless it is succeeded by proline. Trypsin may not cleave after citrulline as it lacks the positive charge.3 Although Bennike and colleagues have reported the inability of trypsin to cleave citrulline residue by using 24 synthetic peptide sets with arginine and citrulline containing sequences from citrullinated proteins reported in the literature,3 others have shown that trypsin is able to cleave citrullinated residues but probably at a different/lower rate.4 These authors also suggest that it is probably protein specific. Regardless, these data suggest that when searching large MS generated peptide data sets that identify an extreme C-terminal citrullinated residue, it should be further investigated and the spectra manually validated.
An interesting study conducted by Bennike et al. demonstrated that a synthetic peptide pair SP10 and SP10CE, with Arg or citrulline residue within the sequence 480-PAPDRKGFRLLLASPR/CitSCYK-499, was only cleaved at arginine residues.60 Preceding digestion, SP10 was detected by extracted ion chromatography (XIC). After digestion, three peptide fragments PAPDR, LLLASPR, and SCYK were identified, relating to a total cleavage after 495Arg. However, in SP10CE, PAPDR, and LLLASPCitSCYK peptide fragments were distinguished after digestion.
As inferred above, MS is the preferred method for the identification and quantification of proteins and peptides with PTMs. Validation of these identifications, i.e., peptides carrying PTMs, is a challenge as heterogeneous protein mixtures as they may only contain a small number of modified peptides. Targeted analysis of PTM-peptides generally includes an enrichment step such as detecting phosphorylation of serine and threonine by neutral loss scans. However, selective enrichment has not been carried out for citrullinated proteins and collision-induced dissociation (CID) that refers to the neutral loss of isocyanic acid from polypeptide-bound citrulline serves as a marker for the diagnosis of protein citrullination.61
Lee and colleagues performed data mining from mass-spectrometry-based deep proteomic profiling to identify citrullinated residues on endogenous proteins.62 Their sample consisted of 30 different human tissues. The methods used in the experiment were a neutral loss of isocyanic acid with the manual interpretation of the spectrum and the use of reference spectra of deiminated peptides or synthetically citrullinated peptides. They validated 375 sites of citrullination on 209 human proteins. Their research identified a high level of novel modification sites (more than 80% of the identified citrullinated sites were validated). Furthermore, citrullination was detected for the first time in 56% of the citrullinated proteins. Gly and Asp residues were largely found among citrullinated sites. Twenty-six human tissues were characterized by the modification; however, it is not clear if there is a linear or conformational consensus sequence for citrullination. Interestingly, there was increased levels of citrullination in the brain and lung tissues, suggestive of a level of tissue-specificity. However, researchers were unable to identify a relationship between the citrullinated proteins abundance and PAD enzymes activity. Researchers indicated that it was challenging to distinguish citrullination from deamidation of Gln and Asn residues, which occurred frequently. Lee and team admitted that a comprehensive study of cellular protein citrullination would only be possible through methods that involve protein enrichment.15
4.3. Novel Probes to Quantify Citrullination Peptides and Proteins
A significant area of research in the context of understanding citrullination in disease pathogenesis is the elucidation of PAD biology and the effects of its dysregulation in the manifestation of the disease. Toward this end, Lewallen and colleagues synthesized phenylglyoxal-based probes and demonstrated the use of biotin-PG in detecting protein citrullination and monitoring the activity of PAD inhibitor BB-Cl-amidine. They also indicated that RNA splicing apparatus components could be substrates for PAD enzymes. Another important discovery was that citrullination could possibly antagonize arginine methylation. A key advantage of their approach is the previous protein enrichment by enhancing peptide coverage. Their method is capable of uncovering a full range of substrates linked to PAD enzyme activity in a range of cell signaling mechanisms. This process is capable of detecting citrullinated proteins with a high level of confidence.63 Researchers used streptavidin–biotin beads with BB-CL to capture protein and perform on-bead tryptic digestion. After digestion, only unmodified or non-citrullinated peptides are released. A cleavable biotin linker may be the final step to release citrullinated peptides into the mixture for further analysis by MS.63 Therefore, their enrichment boosts citrullinated proteome coverage by reducing sample complexity before its analysis using MS.57 Tutturen et al. used citrulline specific biotinylation as the chosen strategy for protein enrichment. Their objective was to detect citrullinated peptides in the synovial fluid for patients diagnosed with rheumatoid arthritis. The enrichment technique was highly effective, and the estimated number of citrullinated peptides evident from the presence of a modification-specific signature ion was 13 times greater than nonenriched protein.11
Another plausible protein enrichment technique is dependent on the reaction between the ureido residue of citrulline and glyoxal derivative glyoxal benzoic acid (GBA) in a strong acidic medium.64 To do so, the GBA molecule has been coupled with a biotin moiety, which resulted in biotinylation of citrulline peptides when modified by biotin-PEG-GBA (BPG). This process allowed their selective enrichment using streptavidin beads. To calculate the sensitivity and specificity of the protein enrichment, enzymatically deiminated peptides or enriched synthetic citrulline peptides were spiked on a heterogeneous biological digest. Their improved method of streptavidin enrichment and BPG labeling resulted in a 20-fold increase in sensitivity. The tool is highly sensitive and specific and may be used in biological processes with a high degree of confidence.57
4.4. Fragmentation Characteristics of Citrullinated Peptides
Stensland and colleagues suggested a targeted MS-based method to detect citrullinated peptides postchemical modification. Their approach combined liquid chromatography with tandem MS analysis, where collision-induced fragmentation (CID) and electron-transfer dissociation (ETD) were performed alternately. The citrullinated residues of a model protein deiminated in vitro were identified using CID for specific and signature ion-based detection and ETD for peptide sequence determination.16
Jin and team identified five citrullination sites of glial fibrillary acid protein (GFAP), two sites on neurogranin (NRGN) and 14 sites of myelin basic protein (MBP) in a group of patients diagnosed with Alzheimer’s disease.4 Since the identification of citrullinated proteins in complex mixtures poses significant challenges, researchers recommended higher-energy collisional dissociation (HCD) triggered by CID for optimal MS-based identification of proteins to identify MBP, GFAP, and NRGN in vivo and in vitro. Results indicated that CID-triggered-HCD along with high-resolution MS coupled to reversed-phase HPLC (RP-HPLC) separation served as an ideal approach to identify citrullinated peptides. Their method was effective for complex protein matrices and had a high confidence level.
Steckel and colleagues optimized MS measurements by focusing on fragmentation of deiminated proteins.65 They analyzed a solution of purified nonmodified and citrullinated peptides and used low-energy collision-induced detection method of tandem MS. Their research successfully elucidated the “citrulline effect” and suggested its use as a complementary tool for the modification site confirmation, through the simultaneous identification of specific fragmentation ions along with the losses of isocyanic acid from fragment ions or protonated molecules.58 This group specifically focused on features of fragmentation in citrullinated peptides using protein epitopes that are significant in the pathogenesis of rheumatoid arthritis (RA).
RA is characterized by the generation of citrullinated autoantigens which are recognized by antibodies and trigger chronic inflammation. To further study this mechanism, Steckel and colleagues selected model peptides from epitope regions of proteins that act as substrates for PAD enzymes.58 The selected proteins were collagen, filaggrin, and fibrin. The advantage of using model peptides resided in their ability to present basic, acidic, neutral, and aromatic side chains, in order to apply the mobile proton theory and to find the fragmentation characteristics of peptides. They classified the peptides as those containing no mobile protons, partially mobile protons, and mobile protons. Their experiment identified the major fragmentation pathways of deiminated peptides. These pathways were conventional sequential fragmentation, specific sequential fragmentation pathways such as citrulline effect, and the neutral loss of isocyanic acid. These insights could provide identification and sequencing of deiminated peptides with greater reliability, specificity, and speed.58
4.5. Overcoming Misidentification of Citrullination
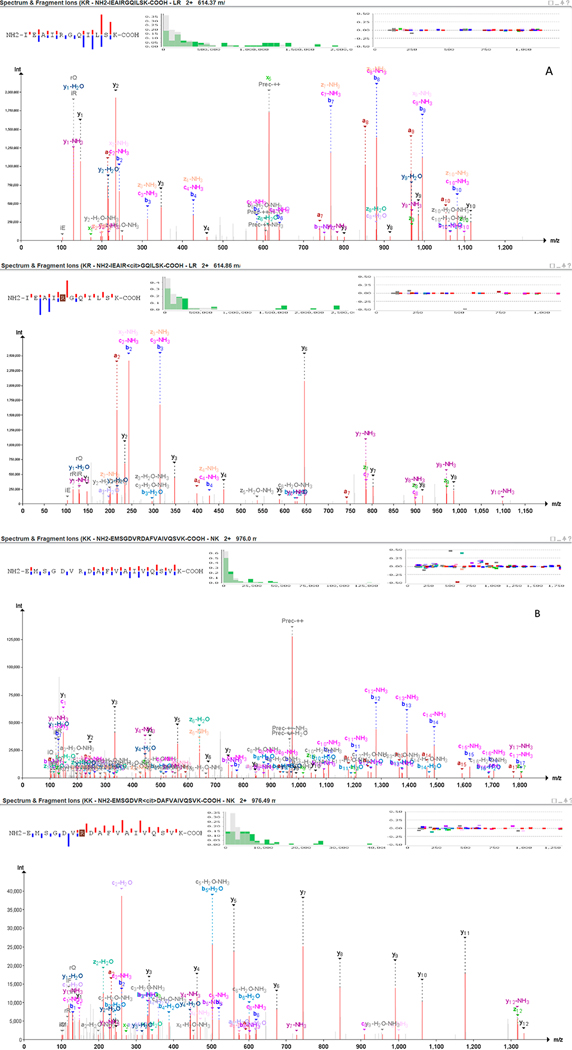
De Ceuleneer and his research group proposed to explore an isotope strategy for the detection of citrullinated peptides.66 They streamlined their general strategy utilizing isotope-marked synthetic peptides by RP-HLPC. Their strategy depended on comparing the isotopic sites of citrullinated and noncitrullinated PADs which partially overlap if the modified form is present, since it will cause a deviation from the original nonmodified isotopic distribution pattern. This approach was successfully applied to synovial fluid samples obtained from RA patients, even though an additional step was required to differentiate between citrullinated and deamidation of Gln and Asp compared to the unmodified peptide.67 In a study conducted by Lee et al., on high energy collision dissociation (HCD), spectrum of E(R+1)YFD(R+1)INEND-PEYIR peptide showed a loss of 43 Da on the modification site. It suggested the process of citrullination,15 as the peptide comprised Asn or Gln residues and gave a final peptide fragment ALE(R+1)GLQDEDGYR. As always, spectra validation is mandatory as the loss of isocyanic acid is not altogether specific for citrulline. Pair mass spectra of carbamylated Lys (i.e., homocitrulline) deposits that can be framed when utilizing urea containing lysis buffers show a similar loss of isocyanic acid inferable from fundamentally the same side chains of citrulline and homocitrulline. Citrullination and homocitrullination (mass distinction of a 14 Da for the extra CH2 bunch in homocitrulline) can be confounded in cases in which a peptide contains at any rate two missed cleavage destinations of which one speaks to a potential citrulline and the other a homocitrulline or the other way around. Thus, we recommend manual validation of MS spectra to ensure citrullination is correctly assigned. Figure 3 shows two pairs of modified versus nonmodified peptides spectra from two distinct proteins, respectively, transforming growth factor beta-1 proprotein (TGFB1_HUMAN) (Figure 3A) and annexin (E5RK69_HUMAN) (Figure 3B) (data were available through PRIDE PXD003132 and reprocessed using Peptide-Shaker).68 Both proteins were identified as citrullinated in the context of RA.69 Modified peptides spectra clearly evidence the citrulline effect characterized by an enhanced cleavage preference at the C-terminus of citrullinated residues upon collision-induced dissociation (CID) tandem mass spectrometry, which results in a marked abundance of series y ions for tryptic peptides.70
Figure 3.
Tandem mass spectra of the modified versus nonmodified doubly protonated peptide precursors. Panel A shows the modified versus nonmodified peptide NH2-IEAIRGQILSK-COOH originated from transforming growth factor beta-1 proprotein (TGFB1_HUMAN). Panel B shows the modified versus nonmodified peptide NH2-EMSGDVRDAFVAIVQSVK-COOH originated from annexin (E5RK69_HUMAN).
Fert-Bober and team assumed a different approach to identify, quantify, and validate citrullination residues while circumventing the challenges associated with current methods in preparing samples and analyzing data.71 Their research was in response to the widespread false positive results through MS in the PTMs domain. Identifying the extent and degree of citrullination is a significant challenge due to the inherently limited capability of existing techniques in detecting and localizing the modification even after applying the required enrichment. The result is evident in the form of only a small fraction of citrullination available and a mandatory high degree of confidence for further exploration.15,47,72,73 Their method was more efficient than the current citrullination quantification methods, as scientists were able to achieve a 4-fold increase in the coverage of the citrullinated proteome of their analysis carried out across six mouse organs.71
To prepare the hyper-citrullinated library, researchers isolated six tissue samples from three wild-type mouse models. They split the samples into two parts and treated one of them with a PAD cocktail, in order to allow for an afterward determination of all possible citrullination targets. They treated the second sample with water to act as a control sample. Lys-C was employed to digest samples while removing any potential issues that can arise due to differences in trypsin digestion of a modified versus nonmodified Lys. Data-independent acquisition-MS was used for quantification. Finally, they validated the accuracy of the site-specific citrullination based on a number of rules which they provide as an algorithm for others researchers to employ.71 Data acquired for the various mouse tissue showed that citrullinated proteins belonged to different functional groups, but their distribution was tissue specific. Data also indicated involvement of multiple physiological processes not pointed out by previous research studies. Brain tissue had the highest percentage of citrullinated protein, followed by lung and kidney tissues. Comparison of this mouse data with a collection of human citrullinated proteins indicated that 34 sites overlapped in both models.
5. CONCLUSION
The identification and quantification of protein citrullination is a particularly challenging area, and more rigor is required in the field to draw plausible conclusions. With respect to laboratory detection of these PTM sites, meaningful conclusions will require additional research to further enlighten the ongoing technical specifications of proteomics laboratory tools and approaches, as well as to standardize them. So far, the most widely studied PTMs are Ser/Thr phosphorylation, Lys methylation, ubiquitination, acetylation, and N-linked glycosylation.74 Although MS technique optimizations allow researchers the freedom to quantify thousands of proteins, liberating them from focusing on only a few specific pathways, a high level of sensitivity, specificity, accuracy, resolution, and robustness in the technique is still required. Advancements in MS approaches improves “stability and uniqueness” in the peptides identified in this process. The next challenge relies on improving the right statistical techniques and considering possible bias in clinical studies to open up new avenues in the practice of “personalized medicine”. Prujin sums up the research works undertaken so far and hints at the need to differentiate between the physiological and pathological significance of citrullination in experiments targeted at RA pathogenesis. Highly specific conclusions are required to elucidate the role of specific citrullination sites, extracellular traps, enzymatic responses, ACPAs, and epitopes.75 To achieve a high level of accuracy, laboratory approaches must be supported by online analysis and prediction tools that combine available data and machine learning algorithms to identify sequence motifs and enzymes. They offer visualization techniques and assist in the discovery of functional proteins. However, considerable research is still required to figure out complex biological events in vivo such as PTM crosstalk, and enzymatic activity. The later, regarding PADs activity and consensus site specificity, represents a very important topic for researchers to address. Recent studies have aimed at discovering tissue-specific preferences of citrullinated sites in order to further correlate with different PAD isoforms and tissue-based consensus sequence motifs.76 Stensland and team have determined that approximately one-fifth of PAD4 substrates contained an RG/RGG motif in human tissue and plasma.77 Instead, it pointed to an enrichment of glutamic acid residues in the N-terminus of citrullinated sites in heart and lung tissue versus a preference for lysine residues in the N-terminus of citrullinated site found in muscle tissue.8,78,79 Identification and correlation of compartment-specific sequence motifs of citrullinated proteins/peptides to specific PAD isoforms should be performed with the combination of MS-based approaches combined with knock down mutants or inhibitors, as performed for kinases (S/T/Y), as a way to further characterize PADs site interaction or specificity to citrullinated residues/motifs in either a physiological or a pathological context.
This paper presents a review of the role of citrullination concerning the pathogenesis of RA, cancer, CNS disorders, and cardiac disease. It also emphasizes novel techniques in MS and optimizations that would help detect precise citrullination sites of diagnostic and therapeutic significance. Future research efforts must aim at studying mainly relevant citrullinated epitopes, environmental factors, and enzyme isotopes to identify viable therapeutic targets.
Supplementary Material
Table S1: List of citrullinated proteins and their UniProt ID, origin, biological significance/role, associated pathology and applied methodology for identification/detection (XLSX)
ACKNOWLEDGMENTS
This work was supported by Portuguese Foundation for Science and Technology (FCT), European Union, QREN, FEDER, and COMPETE for funding the Unidade de Investigação Cardiovascular (UID/IC/00051/2019), iBiMED (UIDB/04501/2020, POCI-01–0145-FEDER-007628), and FCT QOPNA ((FCT UID/QUI/00062/2019) and LAQV/REQUIMTE (UIDB/50006/2020) research units and the research project. R.V. is supported by individual fellowship grants (IF/00286/2015). C.V. also acknowledges the Coimbra Chemistry Centre, supported by FCT, through the Project-UID/QUI/00313/2020. J.V.E. is the Erika Glazer Endowed Chair in Women’s Heart Health.
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00474.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.0c00474
Contributor Information
Rui Vitorino, QOPNA & LAQV-REQUIMTE, Departamento de Qúimica, Universidade de Aveiro, Aveiro, Portugal; iBiMED, Department of Medical Sciences, University of Aveiro, Aveiro, Portugal; Unidade de Investigação Cardiovascular, Departamento de Cirurgia e Fisiologia, Faculdade de Medicina, Universidade do Porto, Porto, Portugal.
Sofia Guedes, QOPNA & LAQV-REQUIMTE, Departamento de Qúimica, Universidade de Aveiro, Aveiro, Portugal.
Carla Vitorino, Faculty of Pharmacy, University of Coimbra, Coimbra, Portugal.
Rita Ferreira, QOPNA & LAQV-REQUIMTE, Departamento de Qúimica, Universidade de Aveiro, Aveiro, Portugal.
Francisco Amado, QOPNA & LAQV-REQUIMTE, Departamento de Qúimica, Universidade de Aveiro, Aveiro, Portugal.
Jennifer E. Van Eyk, Advanced Clinical Biosystems Research Institute, The Smidt Heart Institute, Cedars-Sinia Medical Center, Los Angeles, California, United States
REFERENCES
- (1).Tak I.-u.-R.; Ali F; Dar JS; Magray AR; Ganai BA; Chishti MZ Chapter 1 - Posttranslational Modifications of Proteins and Their Role in Biological Processes and Associated Diseases. In Protein Modificomics; Dar TA.; Singh LR., Eds.; Academic Press, 2019; pp 1–35. [Google Scholar]
- (2).Bennike T; Lauridsen KB; Olesen MK; Andersen V; Birkelund S; Stensballe A Optimizing the Identification of Citrullinated Peptides by Mass Spectrometry: Utilizing the Inability of Trypsin to Cleave after Citrullinated Amino Acids. J. Proteomics Bioinf. 2013, 6, 12. [Google Scholar]
- (3).Bennike T; Lauridsen K; Olesen M; Andersen V; Birkelund S; Stensballe A Optimizing the Identification of Citrullinated Peptides by Mass Spectrometry: Utilizing the Inability of Trypsin to Cleave after Citrullinated Amino Acids. J. Proteomics Bioinf. 2013, 6, 12. [Google Scholar]
- (4).Jin Z; Fu Z; Yang J; Troncosco J; Everett AD; Van Eyk JE Identification and characterization of citrulline-modified brain proteins by combining HCD and CID fragmentation. Proteomics 2013, 13, 2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Raijmakers R; van Beers JJ; El-Azzouny M; Visser NF; Bozic B; Pruijn GJ; Heck AJ Elevated levels of fibrinogen-derived endogenous citrullinated peptides in synovial fluid of rheumatoid arthritis patients. Arthritis Res. Ther. 2012, 14, R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Witalison EE; Thompson PR; Hofseth LJ Protein Arginine Deiminases and Associated Citrullination: Physiological Functions and Diseases Associated with Dysregulation. Curr. Drug Targets 2015, 16, 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Baka Z; Gyorgy B; Geher P; Buzas EI; Falus A; Nagy G Citrullination under physiological and pathological conditions. Jt., Bone, Spine 2012, 79, 431–436. [DOI] [PubMed] [Google Scholar]
- (8).György B; Tóth E; Tarcsa E; Falus A; Buzás EI Citrullination: a posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 2006, 38, 1662–1677. [DOI] [PubMed] [Google Scholar]
- (9).Koziel J; Mydel P; Potempa J The link between periodontal disease and rheumatoid arthritis: an updated review. Curr. Rheumatol. Rep. 2014, 16, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Alghamdi M; Alasmari D; Assiri A; Mattar E; Aljaddawi AA; Alattas SG; Redwan EM An Overview of the Intrinsic Role of Citrullination in Autoimmune Disorders. J. Immunol. Res. 2019, 2019, 7592851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Tutturen AE; Fleckenstein B; de Souza GA Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J. Proteome Res. 2014, 13, 2867–2873. [DOI] [PubMed] [Google Scholar]
- (12).Sohn DH; Rhodes C; Onuma K; Zhao X; Sharpe O; Gazitt T; Shiao R; Fert-Bober J; Cheng D; Lahey LJ; Wong HH; Van Eyk J; Robinson WH; Sokolove J Local Joint inflammation and histone citrullination in a murine model of the transition from preclinical autoimmunity to inflammatory arthritis. Arthritis Rheumatol. 2015, 67, 2877–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tonelli M; Karumanchi SA; Thadhani R Epidemiology and Mechanisms of Uremia-Related Cardiovascular Disease. Circulation 2016, 133, 518–536. [DOI] [PubMed] [Google Scholar]
- (14).Yuzhalin AE Citrullination in Cancer. Cancer Res. 2019, 79, 1274–1284. [DOI] [PubMed] [Google Scholar]
- (15).Lee CY; Wang D; Wilhelm M; Zolg DP; Schmidt T; Schnatbaum K; Reimer U; Ponten F; Uhlen M; Hahne H; Kuster B Mining the Human Tissue Proteome for Protein Citrullination. Mol. Cell. Proteomics 2018, 17, 1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Stensland M; Holm A; Kiehne A; Fleckenstein B Targeted analysis of protein citrullination using chemical modification and tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 2754–2762. [DOI] [PubMed] [Google Scholar]
- (17).Kim S; Hwang J; Xuan J; Jung YH; Cha HS; Kim KH Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One 2014, 9, No. e97501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Vos I; Van Mol C; Trouw LA; Mahler M; Bakker JA; Van Offel J; De Clerck L; Huizinga TW Anti-citrullinated protein antibodies in the diagnosis of rheumatoid arthritis (RA): diagnostic performance of automated anti-CCP-2 and anti-CCP-3 antibodies assays. Clin. Rheumatol. 2017, 36, 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sakkas LI; Bogdanos DP; Katsiari C; Platsoucas CD Anti-citrullinated peptides as autoantigens in rheumatoid arthritis-relevance to treatment. Autoimmun. Rev. 2014, 13, 1114–1120. [DOI] [PubMed] [Google Scholar]
- (20).Farid S; Azizi G; Mirshafiey A Anti-citrullinated protein antibodies and their clinical utility in rheumatoid arthritis. Int. J. Rheum. Dis. 2013, 16, 379–386. [DOI] [PubMed] [Google Scholar]
- (21).Kimura E; Kanzaki T; Tahara K; Hayashi H; Hashimoto S; Suzuki A; Yamada R; Yamamoto K; Sawada T Identification of citrullinated cellular fibronectin in synovial fluid from patients with rheumatoid arthritis. Mod. Rheumatol. 2014, 24, 766–769. [DOI] [PubMed] [Google Scholar]
- (22).Shelef MA; Bennin DA; Mosher DF; Huttenlocher A Citrullination of fibronectin modulates synovial fibroblast behavior. Arthritis Res. Ther. 2012, 14, R240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yu R; Li C; Sun L; Jian L; Ma Z; Zhao J; Liu X Hypoxia induces production of citrullinated proteins in human fibroblast-like synoviocytes through regulating HIF1alpha. Scand. J. Immunol 2018, 87, No. e12654. [DOI] [PubMed] [Google Scholar]
- (24).Sohn DH; Rhodes C; Onuma K; Zhao X; Sharpe O; Gazitt T; Shiao R; Fert-Bober J; Cheng D; Lahey LJ; Wong HH; Van Eyk J; Robinson WH; Sokolove J Local Joint inflammation and histone citrullination in a murine model of the transition from preclinical autoimmunity to inflammatory arthritis. Arthritis Rheumatol. 2015, 67, 2877–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Skeoch S; Bruce IN Atherosclerosis in rheumatoid arthritis: is it all about inflammation? Nat. Rev. Rheumatol. 2015, 11, 390–400. [DOI] [PubMed] [Google Scholar]
- (26).Spinelli FR; Pecani A; Conti F; Mancini R; Alessandri C; Valesini G Post-translational modifications in rheumatoid arthritis and atherosclerosis: Focus on citrullination and carbamylation. J. Int. Med. Res. 2016, 44, 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Aggarwal R; Liao K; Nair R; Ringold S; Costenbander KH Anti-citrullinated peptide antibody assays and their role in the diagnosis of rheumatoid arthritis. Arthritis Rheum. 2009, 61, 1472–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Modi S; Soejima M; Levesque MC The effect of targeted rheumatoid arthritis therapies on anti-citrullinated protein autoantibody levels and B cell responses. Clin. Exp. Immunol. 2013, 173, 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sokolove J; Brennan MJ; Sharpe O; Lahey LJ; Kao AH; Krishnan E; Edmundowicz D; Lepus CM; Wasko MC; Robinson WH Brief report: citrullination within the atherosclerotic plaque: a potential target for the anti-citrullinated protein antibody response in rheumatoid arthritis. Arthritis Rheum. 2013, 65, 1719–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Giles JT; Fert-Bober J; Park JK; Bingham CO 3rd; Andrade F; Fox-Talbot K; Pappas D; Rosen A; van Eyk J; Bathon JM; Halushka MK Myocardial citrullination in rheumatoid arthritis: a correlative histopathologic study. Arthritis Res. Ther. 2012, 14, R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Verheul MK; Böhringer S.; van Delft MAM.; Jones JD; Rigby WFC; Gan RW; Holers VM; Edison JD; Deane KD; Janssen KMJ; Westra J; Brink M; Rantapää-Dahlqvist S; Huizinga TWJ; van der Helm-van Mil AHM.; van der Woude D.; Toes REM.; Trouw LA. Triple Positivity for Anti-Citrullinated Protein Autoantibodies, Rheumatoid Factor, and Anti-Carbamylated Protein Antibodies Conferring High Specificity for Rheumatoid Arthritis: Implications for Very Early Identification of At-Risk Individuals. Arthritis Rheumatol. 2018, 70, 1721–1731. [DOI] [PubMed] [Google Scholar]
- (32).Shi J; Knevel R; Suwannalai P; van der Linden MP; Janssen GM; van Veelen PA; Levarht NE; van der Helm-van Mil AH.; Cerami A.; Huizinga TW.; Toes RE.; Trouw LA. Autoantibodies recognizing carbamylated proteins are present in sera of patients with rheumatoid arthritis and predict joint damage. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 17372–17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Shi J; van de Stadt LA; Levarht EWN; Huizinga TWJ; Toes REM; Trouw LA; van Schaardenburg D Brief Report: Anti–Carbamylated Protein Antibodies Are Present in Arthralgia Patients and Predict the Development of Rheumatoid Arthritis. Arthritis Rheum. 2013, 65, 911–915. [DOI] [PubMed] [Google Scholar]
- (34).Jaisson S; Desmons A; Doue M; Gorisse L; Pietrement C; Gillery P Measurement of Homocitrulline, A Carbamylation-derived Product, in Serum and Tissues by LC-MS/MS. Curr. Protoc. Protein Sci. 2018, 92, No. e56. [DOI] [PubMed] [Google Scholar]
- (35).Speer T; Owala FO; Holy EW; Zewinger S; Frenzel FL; Stahli BE; Razavi M; Triem S; Cvija H; Rohrer L; Seiler S; Heine GH; Jankowski V; Jankowski J; Camici GG; Akhmedov A; Fliser D; Luscher TF; Tanner FC Carbamylated low-density lipoprotein induces endothelial dysfunction. Eur. Heart J. 2014, 35, 3021–3032. [DOI] [PubMed] [Google Scholar]
- (36).Fert-Bober J; Giles JT; Holewinski RJ; Kirk JA; Uhrigshardt H; Crowgey EL; Andrade F; Bingham CO 3rd; Park JK; Halushka MK; Kass DA; Bathon JM; Van Eyk JE Citrullination of myofilament proteins in heart failure. Cardiovasc. Res. 2015, 108, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Rosenson RS; Brewer HB Jr.; Ansell BJ; Barter P; Chapman MJ; Heinecke JW; Kontush A; Tall AR; Webb NR Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat. Rev. Cardiol. 2016, 13, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Demoruelle MK; Bowers E; Lahey LJ; Sokolove J; Purmalek M Antibody Responses to Citrullinated and Non-citrullinated Antigens in the Sputum of Subjects With Rheumatoid Arthritis and Subjects at Risk for Development of Rheumatoid Arthritis. Arthritis Rheumatol. 2018, 4, 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ferri L,CG.; Kuang Y; Breznen B; Fazeli M. Cardiovascular Disease in Rheumatoid Arthritis: Risk Factors and the Role of Autoantibodies Arthritis Rheumatol. 2019, 71, Suppl 10. [Google Scholar]
- (40).Hermans MP; Volkov M; Velden D v. d.; Cabezas JMM; Huizinga TW; Kuiper J; Toes RE; Schalij MJ; Jukema JW; van der Woude D 08.18 Anti-citrullinated protein antibodies: a marker of cardiovascular disease and mortality in patients without rheumatoid arthritis. Ann. Rheumatic Dis. 2017, No. Suppl 1, A82–A82. [Google Scholar]
- (41).van der Woude D; Catrina AI HLA and anti-citrullinated protein antibodies: Building blocks in RA. Best Pract Res. Clin. Rheumatol. 2015, 29, 692–705. [DOI] [PubMed] [Google Scholar]
- (42).Westerlind H; Holmqvist M; Ljung L; Frisell T; Askling J Siblings of patients with rheumatoid arthritis are at increased risk of acute coronary syndrome. Ann. Rheum. Dis. 2019, 78, 683. [DOI] [PubMed] [Google Scholar]
- (43).Hermans MPJ; van der Velden D; Montero Cabezas JM; Putter H; Huizinga TWJ; Kuiper J; Toes REM; Schalij MJ; Wouter Jukema J; van der Woude D Long-term mortality in patients with ST-segment elevation myocardial infarction is associated with anti-citrullinated protein antibodies. Int. J. Cardiol. 2017, 240, 20–24. [DOI] [PubMed] [Google Scholar]
- (44).Wizeman JW; Nicholas AP; Ishigami A; Mohan R Citrullination of glial intermediate filaments is an early response in retinal injury. Mol. Vis. 2016, 1137–1155. [PMC free article] [PubMed] [Google Scholar]
- (45).Bazarian JJ; Biberthaler P; Welch RD; Lewis LM; Barzo P; Bogner-Flatz V; Gunnar Brolinson P; Büki A.; Chen JY; Christenson RH; Hack D; Huff JS; Johar S; Jordan JD; Leidel BA; Lindner T; Ludington E; Okonkwo DO; Ornato J; Peacock WF; Schmidt K; Tyndall JA; Vossough A; Jagoda AS Serum GFAP and UCH-L1 for prediction of absence of intracranial injuries on head CT (ALERT-TBI): a multicentre observational study. Lancet Neurol. 2018, 17, 782–789. [DOI] [PubMed] [Google Scholar]
- (46).Bazarian JJ; Biberthaler P; Welch RD; Lewis LM; Barzo P; Bogner-Flatz V; Gunnar Brolinson P; Büki A.; Chen JY; Christenson RH; Hack D; Huff JS; Johar S; Jordan JD; Leidel BA; Lindner T; Ludington E; Okonkwo DO; Ornato J; Peacock WF; Schmidt K; Tyndall JA; Vossough A; Jagoda AS Serum GFAP and UCH-L1 for prediction of absence of intracranial injuries on head CT (ALERT-TBI): a multicentre observational study. Lancet Neurol. 2018, 17, 782–789. [DOI] [PubMed] [Google Scholar]
- (47).Lazarus RC; Buonora JE; Flora MN; Freedy JG; Holstein GR; Martinelli GP; Jacobowitz DM; Mueller GP Protein Citrullination: A Proposed Mechanism for Pathology in Traumatic Brain Injury. Front. Neurol. 2015, 6, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Hanahan D; Weinberg RA Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- (49).Chang X; Han J; Pang L; Zhao Y; Yang Y; Shen Z Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 2009, 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Chang X; Hou X; Pan J; Fang K; Wang L; Han J Investigating the pathogenic role of PADI4 in oesophageal cancer. Int. J. Biol. Sci. 2011, 7, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ulivi P; Mercatali L; Casoni GL; Scarpi E; Bucchi L; Silvestrini R; Sanna S; Monteverde M; Amadori D; Poletti V; Zoli W Multiple marker detection in peripheral blood for NSCLC diagnosis. PLoS One 2013, 8, No. e57401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Yuzhalin AE; Gordon-Weeks AN; Tognoli ML; Jones K; Markelc B; Konietzny R; Fischer R; Muth A; O’Neill E; Thompson PR; Venables PJ; Kessler BM; Lim SY; Muschel RJ Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat. Commun. 2018, 9, 4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Clancy KW; Weerapana E; Thompson PR Detection and identification of protein citrullination in complex biological systems. Curr. Opin. Chem. Biol. 2016, 30, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Holm A; Rise F; Sessler N; Sollid LM; Undheim K; Fleckenstein B Specific modification of peptide-bound citrulline residues. Anal. Biochem. 2006, 352, 68–76. [DOI] [PubMed] [Google Scholar]
- (55).Makrygiannakis D; Hermansson M; Ulfgren AK; Nicholas AP; Zendman AJ; Eklund A; Grunewald J; Skold CM; Klareskog L; Catrina AI Smoking increases peptidylarginine deiminase 2 enzyme expression in human lungs and increases citrullination in BAL cells. Ann. Rheum. Dis. 2008, 67, 1488–1492. [DOI] [PubMed] [Google Scholar]
- (56).Senshu T; Sato T; Inoue T; Akiyama K; Asaga H Detection of citrulline residues in deiminated proteins on polyvinylidene difluoride membrane. Anal. Biochem. 1992, 203, 94–100. [DOI] [PubMed] [Google Scholar]
- (57).Tutturen AE; Holm A; Fleckenstein B Specific biotinylation and sensitive enrichment of citrullinated peptides. Anal. Bioanal. Chem. 2013, 405, 9321–9331. [DOI] [PubMed] [Google Scholar]
- (58).Steckel A; Uray K; Turiák L; Gömöry Á; Drahos L; Hudecz F; Schlosser G Mapping the tandem mass spectrometric characteristics of citrulline-containing peptides. Rapid Commun. Mass Spectrom. 2018, 32, 844–850. [DOI] [PubMed] [Google Scholar]
- (59).Choi M; Song JS; Kim HJ; Cha S; Lee EY Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of peptide citrullination site using Br signature. Anal. Biochem. 2013, 437, 62–67. [DOI] [PubMed] [Google Scholar]
- (60).Bennike T; Birkelund S; Stensballe A; Andersen V Biomarkers in inflammatory bowel diseases: current status and proteomics identification strategies. World J. Gastroenterol. 2014, 20, 3231–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Hao G; Wang D; Gu J; Shen Q; Gross SS; Wang Y Neutral loss of isocyanic acid in peptide CID spectra: a novel diagnostic marker for mass spectrometric identification of protein citrullination. J. Am. Soc. Mass Spectrom. 2009, 20, 723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Lee CY; Wang D; Wilhelm M Mining the Human Tissue Proteome for Protein Citrullination. Mol. Cell. Proteomics 2018, 7, 1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Lewallen DM; Bicker KL; Subramanian V; Clancy KW; Slade DJ; Martell J; Dreyton CJ; Sokolove J; Weerapana E; Thompson PR Chemical Proteomic Platform To Identify Citrullinated Proteins. ACS Chem. Biol. 2015, 10, 2520–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Tutturen AEV; Holm A; Fleckenstein B Specific biotinylation and sensitive enrichment of citrullinated peptides. Anal. Bioanal. Chem. 2013, 405, 9321–9331. [DOI] [PubMed] [Google Scholar]
- (65).Steckel A; Uray K; Turiák L; Gömöry Á; Drahos L; Hudecz F; Schlosser G Mapping the tandem mass spectrometric characteristics of citrulline-containing peptides. Rapid Commun. Mass Spectrom. 2018, 32, 844–850. [DOI] [PubMed] [Google Scholar]
- (66).De Ceuleneer M; Van Steendam K; Dhaenens M; Deforce D In vivo relevance of citrullinated proteins and the challenges in their detection. Proteomics 2012, 12, 752–760. [DOI] [PubMed] [Google Scholar]
- (67).De Ceuleneer M; Van Steendam K; Dhaenens M; Elewaut D; Deforce D Quantification of citrullination by means of skewed isotope distribution pattern. J. Proteome Res. 2012, 11, 5245–5251. [DOI] [PubMed] [Google Scholar]
- (68).Vaudel M; Burkhart JM; Zahedi RP; Oveland E; Berven FS; Sickmann A; Martens L; Barsnes H PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [DOI] [PubMed] [Google Scholar]
- (69).Sipilä KH; Ranga V; Rappu P; Torittu A; Pirilä L; Käpylä J; Johnson MS; Larjava H; Heino J Extracellular citrullination inhibits the function of matrix associated TGF-β. Matrix Biol. 2016, 55, 77–89. [DOI] [PubMed] [Google Scholar]
- (70).Steckel A; Schlosser G Citrulline Effect Is a Characteristic Feature of Deiminated Peptides in Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30, 1586–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Fert-Bober J; Venkatraman V; Hunter CL; Liu R; Crowgey EL; Pandey R; Holewinski RJ; Stotland A; Berman BP; Van Eyk JE Mapping Citrullinated Sites in Multiple Organs of Mice Using Hypercitrullinated Library. J. Proteome Res. 2019, 18, 2270–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Nemmara VV; Tilvawala R; Salinger AJ; Miller L; Nguyen SH; Weerapana E; Thompson PR Citrullination Inactivates Nicotinamide- N-methyltransferase. ACS Chem. Biol. 2018, 13, 2663–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Sipilä KH; Ranga V; Rappu P; Mali M; Pirilä L; Heino I; Jokinen J; Käpylä J; Johnson MS; Heino J Joint inflammation related citrullination of functional arginines in extracellular proteins. Sci. Rep. 2017, 7, 8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Liddy KA; White MY; Cordwell SJ Functional decorations: post-translational modifications and heart disease delineated by targeted proteomics. Genome Med. 2013, 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Pruijn GJM Citrullination and carbamylation in the pathophysiology of rheumatoid arthritis. Front. Immunol. 2015, 6, 192–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Tanikawa C; Ueda K; Suzuki A; Iida A; Nakamura R; Atsuta N; Tohnai G; Sobue G; Saichi N; Momozawa Y; Kamatani Y; Kubo M; Yamamoto K; Nakamura Y; Matsuda K Citrullination of RGG Motifs in FET Proteins by PAD4 Regulates Protein Aggregation and ALS Susceptibility. Cell Rep. 2018, 22, 1473–1483. [DOI] [PubMed] [Google Scholar]
- (77).Stensland ME; Pollmann S; Molberg Ø; Sollid LM; Fleckenstein B Primary sequence, together with other factors, influence peptide deimination by peptidylarginine deiminase-4. Biol. Chem. 2009, 390, 99–107. [DOI] [PubMed] [Google Scholar]
- (78).Stadler SC; Vincent CT; Fedorov VD; Patsialou A; Cherrington BD; Wakshlag JJ; Mohanan S; Zee BM; Zhang X; Garcia BA; Condeelis JS; Brown AM; Coonrod SA; Allis CD Dysregulation of PAD4-mediated citrullination of nuclear GSK3β activates TGF-β signaling and induces epithelial-to-mesenchymal transition in breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 11851–11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Tilvawala R; Nguyen SH; Maurais AJ; Nemmara VV; Nagar M; Salinger AJ; Nagpal S; Weerapana E; Thompson PR The Rheumatoid Arthritis-Associated Citrullinome. Cell Chem. Biol. 2018, 25, 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Bicker KL; Thompson PR The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers 2013, 99, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: List of citrullinated proteins and their UniProt ID, origin, biological significance/role, associated pathology and applied methodology for identification/detection (XLSX)