Abstract
Microbial communities associated with plants growing in harsh conditions, including salinity and water deficiency, have developed adaptive features which permit them to grow and survive under extreme environmental conditions. In the present study, an ex-situ plant trapping method has been applied to collect the culturable microbial diversity associated with the soil from harsh and remote areas. Oryza sativa cv. Baldo and Triticum durum Primadur plants were used as recruiters, while the soil surrounding the roots of Oryza glaberrima plants from remote regions of Mali (West Africa) was used as substrate for their growth. The endophytic communities recruited by the two plant species belonged to Proteobacteria and Firmicutes, and the dominant genera were Bacillus, Kosakonia, and Enterobacter. These endophytes were characterized by analyzing some of the most common plant growth promoting traits. Halotolerant, inorganic phosphate-solubilizing and N-fixing strains were found, and some of them simultaneously showing these three traits. We verified that ‘Baldo’ recruited mostly halotolerant and P-solubilizers endophytes, while the endophytes selected by ‘Primadur’ were mainly N-fixers. The applied ex-situ plant trapping method allowed to isolate endophytes with potential beneficial traits that could be applied for the improvement of rice and wheat growth under adverse environmental conditions.
Keywords: endophytes, Ex-situ plant trapping, harsh environments, microbial diversity, rice, wheat
Ex-situ plant trapping for successful isolation of endophytes with potential beneficial features.
Introduction
Microbes, including bacteria, fungi, viruses, and other microorganisms, play a fundamental role in agricultural ecosystems, impacting plant health, nutrient cycling, soil fertility, and overall crop productivity (Compant et al. 2019; Compant et al. 2021). The thin film of soil surrounding the roots, with its richness in low-molecular weight organic substances, such as carbohydrates, amino acids, fatty acids, organic acids, vitamins, and secondary metabolites secreted by plant roots, creates a unique space for microbial activity (Radhakrishnan et al. 2017; Pathan et al. 2020, Chepsergon and Moleleki 2023).
All plants exert selective effects on soil microbial communities aimed at the acquisition of specific functional traits necessary for their growth and fitness, regardless of their geographical origin or recent location (; Fu et al. 2023). In addition, the soil microbiome represents a hotspot of microbial competition. To colonize host plants, soil microorganisms must possess the ability to enhance plant fitness and survival (plant selection) and be able to successfully compete against other microorganisms (microbial competition) (Marasco et al. 2022). Therefore, the soil microbiome greatly expands the functional repertoire of plants through different mechanisms, which remain largely unexplored. However, this information is crucial in the understanding and management of microbial functions in ecosystems and in the support of future plant growth in a rapidly changing environment. Moreover, since the diversity and composition of soil bacterial communities is also function of soil properties, an alteration of the latter due to climate change, along with artificial irrigation and unbalanced use of chemicals, are leading to the destruction of microbial communities in arable soils (Ray et al. 2020).
Despite the recognized importance of root-associated microorganisms for plant growth and health, few studies are available on how the diversity of the microbiome associated with plants growing in hostile environments can be exploited to support plant development under adverse environmental conditions, such as water scarcity (Marasco et al. 2022; Alsharif et al. 2020, Marasco et al. 2023).
Soil-inhabiting bacteria enabling plant growth and health belong to the class of bacteria collectively defined as plant growth promoting bacteria (PGPB) (Santoyo et al. 2016).
The endophytes belong to the PGPB class and live within plants for at least part of their life cycle with no apparent disease. They promote plant growth through nitrogen fixation, phytohormone production, nutrient acquisition, beside conferring tolerance to abiotic and biotic stresses (Kandasamy and Kathirvel 2023).
Plant-endophyte communication during the early stages of the colonization process is likely to facilitate access of endophytes to the inside of the host plant tissues (Mushtaq et al. 2023).
Microbial communities in soil can exhibit high taxonomic and functional diversity and contribute to several ecosystem services that can improve plant growth, nutrient assimilation, disease suppression, and overall plant health (Schlaeppi and Bulgarelli 2015, Singh et al. 2020, Aguilar-Paredes et al. 2023, Fagorzi et al. 2023).
It has been suggested that the synthetic microbial communities (SynComs), which help simplify the complexity of soil microbiome, can be exploited to promote plant productivity across contrasting climate and in response to extreme events (Delgado‐Baquerizo 2022, Sáez-Sandino et al. 2023). SynComs derived from harsh environments might further promote plant productivity and improve stress resilience in agricultural systems (Schmitz et al. 2022, Gonçalves et al. 2023).
Culturing and establishing microbial culture collections is essential: (i) to study and preserve the microbial diversity; (ii) provide the basis for microbiota manipulation; (iii) to produce bioinoculants and synthetic communities for the enhancement of plant growth and health (Qiu et al. 2019, Mapelli et al. 2022, Riva et al. 2022). Neglected crops, landraces, non-model plants, and progenitors of modern crop varieties are generally more resilient to environmental stresses but suffer from the lack of information on associated microbial diversity. These types of plants can be valuable sources of novel microorganisms (Vaccaro et al. 2022); indeed, they have been found to harbour a higher diversity of relevant plant growth-promoting bacteria as compared to modern crops varieties (Bulgarelli et al. 2015, Escudero-Martinez and Bulgarelli 2019, Cangioli et al. 2022, Massa et al. 2022, Bianco et al. 2021). The presence of these bacteria in the progenitors of modern crop varieties suggests that they may have played a role in the domestication and improvement of these crops and that modern crops could benefit from the application of such bacteria as bioinoculants (Fagorzi et al. 2023).
Moreover, wild ancestors harbor microbial genera with traits which are depleted in the microbiome of modern crops, though it is unknown to what extent rewilding could enhance the growth and health of modern crops (Raaijmakers and Kiers 2022, Rodrigues and Melotto 2023).
The isolation and characterization of microbial communities from plants grown in remote areas (as in case of many landraces, neglected crops, and non-model plants) is often challenging due to a lack of suitable laboratory facilities and resources. This difficulty limits our understanding of the role of microorganisms in these ecosystems, thus hindering the possibility of exploiting the microbial diversity for the growth of cultivated crops.
Ex-situ plant trapping is a useful strategy for the collection of endophytes associated with a particular plant species (Guimarãesa et al. 2012, Favero et al. 2021). This method involves the collection of soils from natural habitat and their transfer to laboratory environment, where they can be used as substrate for the growth of cultivated crops under controlled conditions. Of course, ex-situ trapping cannot be used to assess the diversity of the microbiome from native plants and infer ecological parameters of native soils. However, even if local plant genotypes might harbor different microbiomes because of a genotype-specificity with local microbial strains, the use of modern crops in trapping of the microbial diversity present in a particular habitat allows the isolation and direct assessment of microbes colonizing and enhancing growth and the health of the modern crop of interest (Petipas et al. 2021, Xiong et al. 2021). Therefore, thanks to the established agronomic practices used for modern crops, ex-situ crop trapping allows exploiting the biodiversity of plant-associated microbiomes for practical applications (Sessitsch et al. 2019, Fagorzi et al. 2023).
By applying the ex-situ plant trapping on soils collected in harsh environments, it is possible to isolate microorganisms well adapted to those environmental conditions (Tian et al. 2022; Wang et al. 2020; Mohanram and Kumar 2019). Plant growth in regions characterized by extreme conditions, such as those of the Mali (West Africa), is challenged by strong abiotic stresses such as extreme temperature fluctuations, high radiation, water scarcity, low nitrogen, organic matter, and soil salinity (Van Andel 2010, Sie et al. 2012). However, plants of this region have acquired adaptive mechanisms to thrive on soils with extremely low water content, salinity, and high temperatures. The diverse microbial communities residing within the tissues of these plants (endophytes) contributed to the adaptation of host plants.
These microorganisms, being evolutionarily well adapted to extreme environmental conditions, can promote plant growth, and enhance soil fertility more than microbes found in plants grown in non-degraded soils (Alsharif et al. 2020, Bouri et al. 2022).
As previously demonstrated, endophytes from a culture collection established from O. glaberrina plants collected in the Inner Delta region of Mali could efficiently colonize O. sativa rice (Bianco et al. 2021). This collection included several strains whose plant-growth promoting abilities were effective for commercial varieties of O. sativa, indicating that the microbial diversity, both associated with and isolated from plants grown in harsh environment, is a valuable resource for conventional crops.
In the present study, ex-situ plant trapping experiments were performed on soil surrounding O. glaberrima plants collected in Mali to exploit the microbial diversity associated with plants adapted to marginal environments and, at the same time, overcome the difficulty of accessing remote areas of Mali. Using two cereal crops (O. sativa cv. Baldo and Triticum durum Primadur) as trapping plants, a collection of bacterial endophytes was established. This experimental design allowed to evaluate and compare the composition and main physiological features of endophytes trapped by the two cereal crops and analyze the main differences with the native endophytic bacterial communities of O. glaberrima. Results indicated that the trapped communities were dependent on the host crop and were mainly composed by strains of Bacillales and Enterobacteriales order, in agreement with the previous isolation of endophytes from O. glaberrina (Bianco et al. 2021).
Additionally, host-dependency was found also at the physiological level, with the ‘Baldo’ plants trapping many halotolerant bacteria, whereas ‘Primadur’ plants trapped mainly N-fixing ones.
Our findings foster the use of ex-situ plant trapping method for the isolation of plant-associated microbial diversity and reinforce the concept that plant microbiome is mainly shaped by host species selecting endophytic bacteria with different characteristics.
Materials and methods
Seeds and soil sources
Seeds of Oryza sativa L. cv. Baldo were obtained from the Breeder Ente Nazionale Risi (Milan, Italy), whereas seeds of Triticum durum Primadur were obtained from Consiglio per la Ricerca in Agricoltura e l'Analisi dell'Economia Agraria (CREA, Foggia, Italy).
The portion of soil surrounding the roots of O. glaberrima plants grown in the region of Bamako (Mali) (12°39′0.00“ N -8°00′0.00″ W) were collected one month after sowing and transferred to laboratory conditions.
Plant growth
Dehulled seeds of rice (‘Baldo’) and wheat (‘Primadur’) plants were surface sterilized as follows: incubation in 70% EtOH, washing several times with sterile distilled water, incubation in 5% sodium hypochlorite solution containing tween 20, washing several times with sterile distilled water. The detailed protocol was described by Bianco et al. (2021). Sterilized seeds were positioned onto the surface of 0.8% water-agar plates and incubated at 21°C in the dark for germination. After 5 days, germinated seeds were transferred into plastic pots units (7 cm in length and 10 cm in diameter) containing soil (200 g) from Mali. The soil pH was measured from 1 : 1 soil/water slurry equilibrated for 30 min (Hue and Evans 1986). This analysis showed that the soil was weakly acid (pH = 6.0). Measurement of %C and %N of soil were made using an elemental analyzer NA1500 (CARLO ERBA, Milan, Italy) coupled to a mass spectrometer (Isoprime GV, Elementar Gmbh, Isoprime Ltd, Germany). The measured %C and %N values (0.611 ± 0.058 and 0.090 ± 0.004, respectively) were used to calculate the C/N ratio (C/N = 6.8).
Plants grown in pots containing sterilized sand (1.0 mm granule size) and perlite (3-4 mm granule size) in 1 : 1 ratio (rice) (Andreozzi et al. 2019) or 3 : 1 ratio (wheat) (Guizani et al. 2023) were used as reference of endophytic colonization. For both rice and wheat plants, 3 pots containing 2 germinated seeds were prepared. Each planting unit was kept in the growth chamber under long daylight (16 h), 19-°C temperature and 75% relative humidity and watered daily. Once a week a nitrogen-free nutrient medium (Bianco et al. 2021) was added to the plants.
Isolation of endophytic bacteria from rice and wheat tissues
Endophytic bacteria were isolated from whole plants of rice ‘Baldo’ and wheat ‘Primadur’ grown up to 7 days in sterilized sand-perlite substrate and in soil from Mali. Plants were carefully removed from the pots, washed with running water to clean the soil off. Plants (three replicates each containing two plants) were surface sterilized through 1 min incubation in 5% sodium hypochlorite solution, washing with sterile distilled water, 1 min incubation with 70% EtOH, and washing several times with sterile distilled water as described in detail by Bianco et al. (2021). To assess the absence of microbial growth around the plant tissues, sterilized plants were placed on LB agar plates and incubated at 30 °C for 3 days. Sterilized samples (whole plants) were ground with 5 ml of 1× PBS buffer using a sterile mortar and pestle. To isolate the endophytic bacteria, favoring the slow-growing ones, the tissue extracts were serially diluted (1 : 10, 1 : 100, 1 : 1000) in 1× PBS solution and plated onto R-2A plates (Sigma-Aldrich, St. Louis, Missouri, USA). The plates were incubated for up to 5 days at 30 °C for bacterial growth. Forty colonies, randomly selected from the plates showing the best bacterial colony separation, were streaked onto new R2A plates. This procedure was repeated several times to gain pure isolates. All purified colonies were grown in R-2A broth (HiMedia, VWR, Radnor, Pennsylvania, USA) at 30°C for 2 days. The bacterial cultures in exponential growth phase (OD600 ± 0.7) were stored at −80°C in a solution containing 16% (v/v) dimethyl sulfoxide (DMSO) and 10% (v/v) glycerol.
Dereplication of the culture collection and taxonomic assignment
Dereplication of the collections was done by performing Random Amplified Polymorphic DNA (RAPD) analysis. Lysates from each isolate were used as templates for PCR amplification using previously optimized conditions with primer 1253 (5′-GTTTCCGCCC-3′) (Mengoni et al. 2014).
Reactions were assembled in 25 µl total volume containing 1X GoTaq® Master Mix (Promega Corporate, WI, USA) with 3 mM MgCl2, 500 ng of primer. Reactions were performed on a T100 thermal cycler (Bio-Rad Corporate, CA, USA) programmed for an initial melting step at 94°C for 5 min followed by 45 cycles each at 94°C for 30 sec, 36°C for 30 sec and 72°C for 2 min. A final extension step at 72°C for 10 min was performed.
After agarose gel electrophoresis (2.5% in 40 mM Tris-acetate, 1 mM EDTA buffer) for 1.5 h at 10 V/cm, band profiles were analysed by GelComparII ver. 2.5 software (Applied Maths, Sint-Martens-Laten, Belgium) to identify distinct band patterns and assign them to isolates. Band patterns are indicated as “groups” and correspond to distinct haplotypes. The amplification and sequencing of 16S rRNA gene for each isolate were carried out as previously reported (Bianco et al. 2021). Taxonomic assignment was performed with SILVA database v. 138.1 (Quast et al. 2013). The matrix of occurrence of each RAPD haplotype in the collections was used to compute alpha diversity values (Shannon H and Eveness), multivariate and univariate analyses (Øyvind et al. 2001). Biplots and PCA were performed using the function ‘prcomp()’ in R. Plots were visualized using the package ‘ggplot2’ (version 3.3.3) and the function ‘autoplot ()’ (Wickham 2009).
SIMPER (Similarity Percentage) (Clarke 1993) was used for assessing which RAPD group are primarily responsible for observed differences between trapped and reference plant collection and was performed with Past 4 software (Hammer et al. 2001).
Bacterial identification by 16S rRNA gene sequence analysis
Cells of each strain were grown aerobically in LB medium at 30 °C, on a shaker at 200 r/m, for 24 h. Aliquots (200 µl) of cultures were aseptically collected and centrifuged at 10 000 × g for 10 min, to obtain supernatant-free cells. Cells were then suspended in 200 μl TE 1x containing 1% Triton and incubated at 100 °C for 10 min. The bacterial suspension was centrifuged for 5 min at 10.000 x g and the resulting supernatant used as DNA source to amplify the 16S rRNA gene fragment (1.4 Kb). To amplify the 16S rRNA gene fragment (1.4 Kb), the universal primer pairs 8–27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1510–1492R (5′-ACG GCT ACC TTG TTA CGA CTT-3′), 100 ng of genomic DNA and 0.03 U of DreamTaq Hot Start DNA Polymerase (Thermo Scientific, Walthman, MA, USA) were used following the procedure reported in Bianco et al. (2021). Sanger (dideoxy chain terminator technology) sequencing was performed from 8–27F using the Brilliant Dye terminator 1.1 kit (NimaGen, Nijmegen, The Netherlands) by BMR Genomics (Padova, Italy). Sequences were manually checked using Bioedit (Hall et al. 2011) and assigned to bacterial taxonomy using SILVAngs Version: 1.9.10 / 1.4.9 run on SILVA database r138.1 (https://ngs.arb-silva.de/). Sequence data are deposited on GenBank database under the accessions OR186984-OR187002 and OR187006-OR187042.
Screening of plant growth-promoting (PGP) activities
Indole-3-acetic acid (IAA) production by Salkowski colorimetric assay
Bacterial cultures grown over-night in LB medium containing 100 μM L-tryptophan at 30 °C were centrifuged and the supernatant mixed with Salkowski reagent in a 1 : 1 ratio and incubated at 30 °C for 30 min in the dark (room temperature). The absorbance of the mixtures was estimated at 530 nm. The bacterial cells were used for data normalization (Bianco et al. 2021).
Phosphate solubilization in liquid medium
Cells of the selected strains were grown aerobically in LB medium at 30°C, on a shaker at 200 r/m for 24 h, washed three times with 0.9% NaCl and then resuspended in Pikovskaya's (PVK) medium containing insoluble phosphate substrate (Nautiyal 1999). The composition of the PVK medium was (g L−1): Glucose 10 g, Ca3(PO4)2 5 g, (NH4)2SO4 0.5 g, NaCl 0.2 g, KCl 0.2 g, MgSO4.7H2O 0.1 g, Yeast Extract 0.5 g, MnSO4.H2O 0.1 mg, FeSO4.7H2O 0.1 mg.
Bacterial cultures in PVK medium were incubated at 30°C on a shaker at 200 r/m. After 6 days of incubation, aliquots (250 µl) of cultures were aseptically collected and centrifuged at 10 000 × g for 10 min, to obtain a biomass-free supernatant. The soluble phosphate concentration in the supernatants was estimated using the molybdenum blue method (Saheki et al. 1985). The reaction mixtures containing 150 µl sample, 120 µl Reagent A and 480 µl Reagent B were incubated for 10 min a 30°C. The absorbance of the samples was measured at 850 nm using a DU 800 UV/Visible spectrophotomer (Beckman Coulter, Brea, CA, US). A mixture containing Reagent A, Reagent B and buffer was used as reference.
Halotolerance assay
The endophytic isolates were screened for salt-tolerance properties using LB media supplemented with various levels of NaCl (w/v) such as, 2% (342 mM), 4% (684 mM), 6% (1.03 M), 8% (1.37 M). Control plates were maintained with 1% (171 mM) NaCl (w/v). Fresh cultures of each strain were streaked on plates containing different levels of NaCl. After incubation for 24 h at 30◦C, the growth on the NaCl-supplemented plates was compared with that of the control ones.
PCR amplification of the nitrogenase iron protein gene (nifH)
To identify nitrogen-fixing (diazotrophic) bacteria, a PCR amplification of the nifH gene was performed by using the primer pairs 19F (5′-GCI WTY TAY GGI AAR GGI GG-3′) and 407R (5′-AAI CCR CCR CAI ACI ACR TC-3′), which amplified a 388 bp fragment of the nifH gene. The amplification reactions were carried out by using 100 ng of genomic DNA and 0.03 U of DreamTaq Hot Start DNA Polymerase (Thermo Scientific, Walthman, MA, USA) as described in Bianco et al. (2021). The DNA of Sinorhizobium meliloti 1021 was used as positive control, while the DNA of the Escherichia coli MG1655 was used as negative control.
Nitrogenase activity by acetylene reduction assay (ARA) in inoculated plants
Dehulled seeds of O. sativa cv. Baldo (CREA-CI, Research Centre for Cereal and Industrial Crops, Vercelli, Italy) and Primadur (CREA-CI, Foggia, Italy) were surface sterilized and germinated as described above. After 5 days, germinated seeds were incubated in Petri dishes with 50 ml of 1× PBS solution containing each strain to a final concentration of 107 cells mL−1 for 4 h at room temperature. Seeds incubated in 1× PBS were used as control. Inoculated seeds were transferred into plastic pots ((7 cm in length and 10 cm in diameter) containing sand (1.0 mm granule size) and perlite (3–4 mm granule size) in 1 : 1 ratio (rice) or 3 : 1 ratio (wheat). One-week-old plants (‘Baldo’ and ‘Primadur’), grown as previously described, were carefully removed from the pots and the roots rinsed with water. The plants were transferred into 20 ml glass tubes containing 2 ml of minimal medium free of nitrogen sources, sealed with rubber serum stopper, incubated under a hypoxic atmosphere in a greenhouse for 20 h and analysed for the ARA assay as described by Defez et al. (2017). The amount of ethylene produced was measured by gas chromatography by using a TG-IBOND Alumina (Na2SO4 deactivate) column (Thermo Scientific) as previously described (Andreozzi et al. 2019). Data are expressed as nmol ethylene plant−1 min−1 and are the mean ± SD of at least six independent replicates. Tubes containing plants without injected acetylene were used as negative control. Six biological replicates were carried out for the measurement of nitrogenase activity by ARA test and the resulting data were subjected to Student's t-test. The results were considered statistically significant when P ≤ 0.05.
Results
Trapped biodiversity of the culture collection
The 462 random isolates obtained from tissues of rice (Baldo) and wheat (Primadur) plants grown in soil from Mali and in the reference soil, were dereplicated into 97 haplotypic groups by RAPD screening (Table S1). The diversity and differences of the two host plant species collections from the two different soils were evaluated. Table 1 reports the values of diversity (Richness, Shannon, Evenness) of the collections.
Table 1.
Values of diversity and number of isolates typed by RAPD.
| Baldo A | Baldo B | Baldo C | Baldo CtrL A | Baldo CtrL B | Baldo CtrL C | Primadur CtrL A | Primadur CtrL B | Primadur CtrL C | Primadur A | Primadur B | Primadur C | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotypes | 21 | 16 | 9 | 9 | 5 | 8 | 13 | 10 | 17 | 18 | 14 | 19 |
| Isolates | 40 | 36 | 40 | 41 | 37 | 40 | 39 | 39 | 39 | 36 | 38 | 37 |
| Shannon_H | 2.605 | 2.413 | 1.597 | 1.46 | 1.00 | 0.804 | 1.51 | 1.95 | 2.39 | 2.58 | 2.44 | 2.77 |
| Evenness | 0.644 | 0.697 | 0.548 | 0.482 | 0.546 | 0.279 | 0.350 | 0.707 | 0.648 | 0.736 | 0.820 | 0.845 |
A, B, and C refer to isolates from the three biological replicates. Ctrl, control plants.
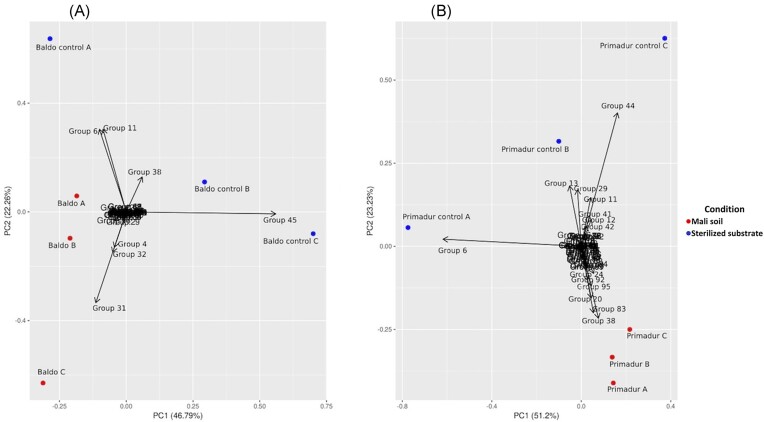
The richness (number of haplotypes obtained) ranged from 5 (Baldo, control replica C) to 21 (Baldo replica A). Baldo reference collections obtained from plants grown in sterilized sand-perlite substrate showed lower values of diversity (Shannon) as compared to collections deriving from the trapping of plants grown in Mali soil (p < 0.05, one-way ANOVA, Tukey pairwise post hoc test). Primadur collections did not shown statistically significant differences in diversity indices. The differences in the representation of the 97 RAPD haplotypes were then evaluated. Fig. 1 shows the results from a Principal Component Analysis. The endophytic collections trapped by both Baldo and Primadur plants grown on sterilized substrate were strongly separated from the ones trapped by plants grown on Mali soil. Moreover, while trapped collections were quite similar among replicates, a higher heterogeneity can be observed for replicates of controls.
Figure 1.
Principal Component Analysis from RAPD haplotype composition. (A), Baldo; (B), Primadur. The biplots, performed using the function ‘prcomp()’ in R and visualized using the package ‘ggplot2’, report the vectors related to the weight of single RAPD haplotypes (groups). Percentages of total variance explained by the first two components are indicated.
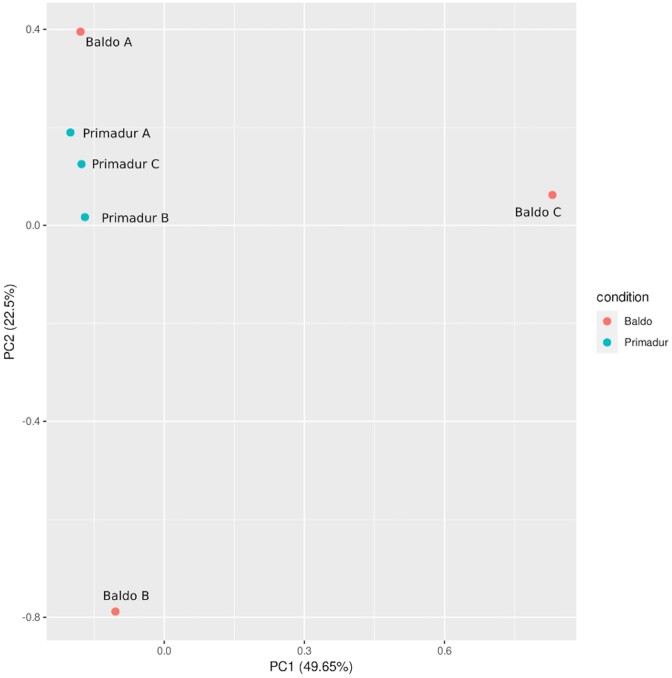
Endophytic collections recruited by rice (Baldo) and wheat (Primadur) plants were, as expected, clearly distinguished (Fig. 2), supporting the consistency of the method to effectively trap specific endophytic communities.
Figure 2.
Principal Component Analysis from RAPD haplotypes of Baldo and Primadur collections trapped from Mali soil. The number of haplotypes obtained for each biological replicates and highlighted with the letter A, B and C was as indicated in Table 1. Percentages of total variance explained by the first two components are indicated.
To further inspect the consistency of the method and the effect of crop species, the contribution of each RAPD haplotype to the separation between trapped Mali soil collection and isolates from reference plants were evaluated. The SIMPER analysis was applied to measure the amount of variance from the occurrence of each of the 97 RAPD groups in the collection.
Results of SIMPER analysis (Tables 2 and 3) indicated that for both plants the first 10 groups contributed to more than 90% of the total variance, separating Mali soil from controls, thus indicating a sharp distinction between trapped Mali soil microbiota and reference plants endophytes. Although the RAPD groups of Baldo and Primadur plants were partially different, as expected from the effect of the host plant species (Bulgarelli et al. 2015), they showed 4 groups in common (groups 6, 11, 38, 44), indicating that the two cereal crops also select a core endophytic microbiome.
Table 2.
Results from SIMPER analysis for Baldo. The first ten RAPD groups are indicated. The percentage of contribution to variance is reported.
| Haplotype | Contribution % |
|---|---|
| Group 45 | 57.28 |
| Group 11 | 10.94 |
| Group 12 | 10.34 |
| Group 6 | 8.681 |
| Group 38 | 8.043 |
| Group 44 | 2.043 |
| Group 62 | 0.1277 |
| Group 58 | 0.1277 |
| Group 59 | 0.1277 |
| Group 57 | 0.1277 |
Table 3.
Results from SIMPER analysis for Primadur. The first ten RAPD groups are indicated. The percentage of contribution to variance is reported.
| Haplotype | Contribution % |
|---|---|
| Group 6 | 41.62 |
| Group 44 | 13.93 |
| Group 38 | 7.005 |
| Group 29 | 6.758 |
| Group 83 | 4.368 |
| Group 11 | 4.066 |
| Group 13 | 3.874 |
| Group 20 | 2.885 |
| Group 95 | 2.802 |
| Group 92 | 2.143 |
Identification of endophytic bacteria
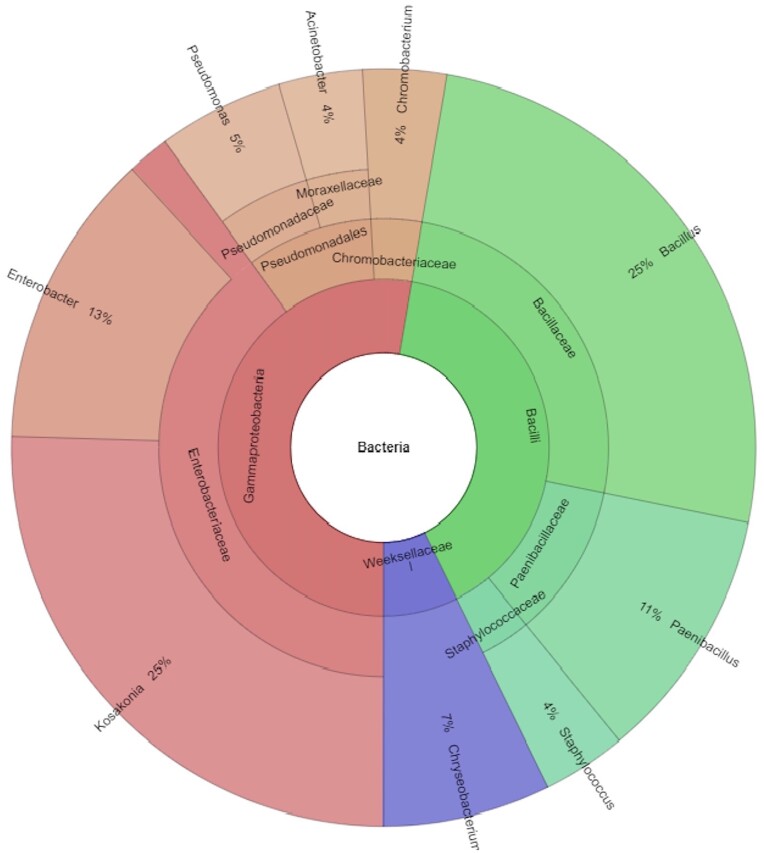
The 97 different haplotypic groups obtained in the RAPD screening were further analysed to select only the isolates coming either from Baldo or from Primadur plants. From the exclusion of the isolates captured by control plants grown in the sand-perlite substrate and those captured by both Baldo and Primadur plants, 57 isolates were obtained and selected for taxonomic identification through the partial sequencing of 16S rRNA gene (Table 4 and Table S2). Most isolates belonged to Bacillales (genus Bacillus) and to Enterobacteriales (genera Enterobacter and Kosakonia) (Table 4, Fig. 3; Table S2).
Table 4.
Genera and plant growth promoting (PGP) traits of the selected endophytes.
| Straina | Genus | IAA Production | Salt tolerance (7% NaCl) | Phosphate solubilization | nifH gene | Nitrogenase activity (nmol C2H4 plants−1 min−1) |
|---|---|---|---|---|---|---|
| BA1 | Bacillus | - | + | - | - | - |
| BA2 | Paenibacillus | - | - | - | - | - |
| BA3 | Bacillus | + | ++ | - | + | + |
| BA5 | Bacillus | + | + | - | - | - |
| BA7 | Bacillus | - | + | - | - | - |
| BA8 | Paenibacillus | - | - | - | - | - |
| BA11 | Bacillus | + | + | - | + | + |
| BA14 | Bacillus | - | + | - | - | - |
| BA18 | Bacillus | - | ++ | + | - | - |
| BA20 | Pseudomonas | - | - | - | - | - |
| BA21 | Paenibacillus | - | - | + | - | - |
| BA22 | Paenibacillus | - | - | ++ | - | - |
| BA26 | Staphylococcus | + | - | ++ | - | - |
| BA27 | Bacillus | - | + | - | - | - |
| BA29 | Bacillus | + | - | - | - | - |
| BA30 | Bacillus | - | ++ | - | - | - |
| BA38 | Bacillus | - | + | - | - | - |
| BB1 | Chryseobacterium | - | - | - | - | - |
| BB4 | Acinetobacter | - | - | - | - | - |
| BB11 | Chryseobacterium | + | - | - | - | - |
| BB14 | Chryseobacterium | - | - | - | - | - |
| BB17 | Paenibacillus | + | - | + | - | - |
| BB18 | Chryseobacterium | - | - | - | - | - |
| BB20 | Acinetobacter | - | - | - | - | - |
| BB22 | Bacillus | - | + | - | - | - |
| BB24 | Bacillus | - | + | - | - | - |
| BB27 | Kosakonia | - | - | - | + | + |
| BB30 | Bacillus | - | - | - | - | - |
| BB35 | Kosakonia | - | - | - | + | + |
| BB39 | Kosakonia | - | - | - | + | + |
| BB40 | Phytobacter | + | - | - | - | - |
| BC1 | Enterobacter | + | - | - | - | - |
| BC2 | Enterobacter | + | - | - | - | - |
| BC3 | Enterobacter | + | - | - | - | - |
| BC5 | Enterobacter | + | - | - | - | - |
| BC6 | Enterobacter | + | - | - | - | - |
| BC14 | Enterobacter | + | + | - | - | - |
| BC19 | Enterobacter | + | - | - | - | - |
| BC20 | Enterobacter | + | - | - | - | - |
| PA5 | Kosakonia | - | - | + | + | + |
| PA6 | Kosakonia | - | - | - | + | + |
| PA12 | Kosakonia | - | - | - | + | + |
| PA13 | Kosakonia | - | - | - | + | + |
| PA15 | Kosakonia | - | - | - | + | + |
| PA18 | Chromobacterium | + | - | - | - | - |
| PA20A | Paenibacillus | + | - | + | + | + |
| PA20B | Staphylococcus | + | - | + | + | + |
| PA21 | Kosakonia | - | - | - | + | + |
| PA26 | Kosakonia | - | - | - | + | + |
| PA29 | Kosakonia | - | - | - | + | + |
| PA35 | Kosakonia | - | - | - | + | + |
| PA38 | Chromobacterium | + | - | - | - | - |
| PA40 | Kosakonia | - | - | - | + | + |
| PB8 | Kosakonia | - | - | - | + | + |
| PB24 | Kosakonia | + | - | - | + | + |
| PC19 | Kosakonia | - | - | - | + | + |
| PC20 | Pseudomonas | - | + | - | - | - |
+, positive response; -, negative response.
The first letters in the name of the strains indicate the plant of origin, i.e. “B” means strain isolated from ‘Baldo’ and “P” means strain isolated from ‘Primadur’, while the second letter refers to the biological replicates (A, B, and C).
Figure 3.
Taxonomic composition of the 57 strains representing the RAPD groups of isolates trapped from Mali soil and unshared between Baldo and Primadur. The bacterial taxonomy of each isolate was obtained using SILVAngs Version: 1.9.10 / 1.4.9 run on SILVA database r138.1 (https://ngs.arb-silva.de/).
Some isolates presented the same 16S rRNA gene partial sequence (PC19, PA29, PA40, PB8 and BA7, BA18, BA27, BA30) suggesting the presence of strains of the same species but with different genomic features in wheat and rice trapped collections, respectively. When the endophytic bacterial communities of the two plant species were compared, it was found that, at the genus level, Bacillus and Enterobacter were the most abundant in the ‘Baldo’ plants, representing 32% and 19% of the 57 isolates, respectively, followed by Paenibacillus (12%). The relative abundance of other genera ranged between 2% and 7%. On the other hand, in ‘Primadur’ plants Kosakonia was the predominant genus, comprising 72% of total sequences. Chronobacterium (11.2%), Paenibacillus (5.6%), Staphylococcus (5.6%) and Pseudomonas (5.6%) were the other genera detected in these plants.
PGP traits of bacterial endophytes
All isolated endophytes were examined for IAA production, salt stress resistance, phosphate solubilization, and nitrogen fixation. The results reported in Table 4 and Table 5 showed that: i) 21 endophytes were able to produce IAA; ii) 9 endophytes were capable of solubilizing calcium phosphate and 3 of them (BA22, BA26, and BB19) were the best performers. Among these P-solubilizers, six were recruited by ‘Baldo’ plants and the other three were caught by ‘Primadur’. When the isolated endophytes were screened for salt tolerance, we found that 15 strains showed a great level of tolerance at 7% NaCl as compared to the untreated control (Table 4). Among them, three endophytes recruited by ‘Baldo’ plants (BA3, BA18, and BA30) showed the best growth in the presence of 7% NaCl. The data reported in Table 4 also showed that most of halotolerant strains have been recruited by ‘Baldo’ plants.
Table 5.
Nitrogenase activity of Oryza sativa cv. Baldo plants inoculated with the selected N-fixing endophytes.
| Strain | Nitrogenase activitya (nmol ethylene plant−1 min−1) |
|---|---|
| BA3 | 0.40 ± 0.007 |
| BA11 | 0.76 ± 0.14 |
| BB27 | 0.48 ± 0.05 |
| BB35 | 0.14 ± 0.01 |
| BB39 | 0.23 ± 0.05 |
| PA5 | 0.07 ± 0.01 |
| PA6 | 0.30 ± 0.12 |
| PA12 | 0.27 ± 0.06 |
| PA13 | 0.26 ± 0.06 |
| PA15 | 0.39 ± 0.05 |
| PA20A | 0.45 ± 0.09 |
| PA20B | 1.18 ± 0.33 |
| PA21 | 0.30 ± 0.13 |
| PA26 | 0.17 ± 0.07 |
| PA29 | 0.39 ± 0.09 |
| PA35 | 0.31 ± 0.09 |
| PA40 | 0.57 ± 0.23 |
| PB8 | 0.53 ± 0.19 |
| PB24 | 0.60 ± 0.10 |
| PC19 | 0.20 ± 0.04 |
The PCR analysis of nifH gene, which constitute an ideal biomarker to determine the potential nitrogen fixing ability in a microbial community, showed that the expected 388-bp amplicon was observed for 21 endophytic strains (35%). The results reported in Table 4 highlighted that the majority of diazotrophic strains (21 isolates) were recruited by Primadur plants and only few of them (5 isolates) were captured by ‘Baldo’ plants.
To confirm the diazotrophic nature of the nifH-positive endophytes, an ARA test was carried out on 10-day-old O. sativa L. cv. Baldo plants inoculated with the strains that were positive in PCR analysis of nifH gene. For all the tested strains, a nitrogenase activity was observed for the inoculated plants when the uninoculated ones were used as reference (Table 5). The highest activity was measured for the strains BA11, PA20b, PA40, PB8, and PB24. Among these 5 strains, two (BA11 and PA20B) showed three of the four PGP traits analyzed in our study.
The data obtained in this study and reported in Table 4 and Table 5 demonstrated that: (i) 71% of the diazotrophic strains were recruited by wheat plants, while the remaining 29% were trapped by rice plants; (ii) 83% of endophytes isolated from wheat plants were diazotrophs; (iii) 93% of halotolerant strains were captured by rice plants; (iv) about 40% of all isolates produced IAA; (v) the majority of phosphate-solubilizing isolates were recruited by rice plants.
Discussion
The supply of food for the increasing human population is one of the greatest challenges for the agrarian community. The main strategy for the improvement of plant production is the use of agrochemicals, which however cause deteriorating effects on soil health and fertility, and in turn negatively influence the productivity and sustainability of crops (Le Mouël and Forslund 2017). In this context, the use of naturally abundant microorganisms as biofertilizers to supply nutrients to plants takes on particular significance both in terms of costs and the environment (Ke et al. 2021).
Despite the numerous studies carried out in this field, the role of microbial communities in promoting crops productivity and environmental sustainability is not yet fully explored. Indeed, most of the studies are related to model plant species, while non-model crops, landraces and local varieties have received little attention (Vaccaro et al. 2022). Moreover, beneficial microbial systems involved in plant growth promotion are frequently not only crop-specific but also strongly influenced by agronomic practices and ecological factors, whose effects are magnified in the current global climate changes (Sessitsch et al. 2019). To address such problems, plant species or varieties adapted to growth in extreme environments could be considered a hot spot of helpful biodiversity to be used as source of beneficial microorganisms capable of promoting adaptation of crop plants to harsh environmental conditions (Massa et al. 2022, Fagorzi et al. 2023)
However, the selection of beneficial microorganisms from plants adapted to different extreme environments and their use in promoting the growth of the crops of interest is a complex procedure facing the need to isolate a native microbial community directly from plants collected in remote areas. Since plants shape their microbiome (Bulgarelli et al. 2015, Xiong et al. 2021), we can hypothesize that plants (e.g. local landraces) grown in harsh environments could harbor a microbiome enriched in relevant functions, which could help crops cope with different environmental conditions.
Bianco et al. (2021) demonstrated that some culturable endophytic bacteria isolated from O. glaberrima plants grown in Mali were able to colonize quite efficiently the domesticated Asian rice Oryza sativa cv. Baldo, selected to grow in the typical cold and wet environmental conditions of Northern Italy. They found that out of 70 strains isolated from O. glaberrima plants, 36 (51.4%) belonged to the Enterobacterales order and 5 (7.1%) to Bacillales. They also demonstrated that inoculated Baldo plants showed high activity of enzymes involved in salt stress response (Bianco et al. 2021). These results suggested that plants adapted to extreme ecosystems could harbor a microbiome enriched in strains with functions relevant to other host plants, such as rice (O. sativa cv. Baldo) and wheat (Triticum durum Primadur).
In the present study, the soil surrounding O. glaberrima roots grown in Mali (West Africa) was transferred to laboratory conditions and used to identify culturable endophytes potentially useful for promoting the growth of Italian commercial rice and wheat varieties.
We demonstrated that the ex-situ procedure used was robust in capturing the microbial diversity from soil and in minimizing the effect of “environmental” contamination from other endophytic bacteria. Indeed, the alpha diversity of the isolates obtained from plants grown in Mali soil was higher than the reference plants grown in sterilized perlite-sand mix. In addition, the beta diversity indicated a high consistency of replicates of the endophytic collections from Mali soil, while those from references were quite dissimilar. The high consistency of trapping replicates and the strong separation from reference plants suggested that this method was robust.
Considering that the substrate used for the growth of control plants was sterilized, this result could derive either from vertical transmission of few endophytes present in the seeds of the Baldo and Primadur plants or from stochastic contamination by environmental bacteria during plant growth in the green chamber.
We focused our attention on the haplotypes distinctive of the two host plants, selecting 57 haplotypes out of the 97. The trapped haplotypes showed a differential occurrence in the two host plants, allowing to define two different groups for the two host plants, respectively. Interestingly 3 groups where present in both plants, suggesting that they may belong to a core endophytic microbiota able to colonize both rice and wheat (Escudero-Martinez and Bulgarelli 2019, Bianco et al. 2021, Riva et al. 2022).
The taxonomic identification of the isolated haplotypes highlighted greater taxonomic diversity in the collection obtained from ‘Baldo’ plants compared to ‘Primadur’ ones, probably due to the closer phylogenetic affiliation of O. sativa to O. glaberrima. Indeed, Bacillus (32%), Enterobacter (19%), and Paenibacillus (12%) were the main genera detected in ‘Baldo’ plants. On the other hand, in ‘Primadur’ plants Kosakonia was the predominant genus, comprising 72% of total sequences.
The differences in taxonomic composition between endophytes collected in-situ from O. glaberrima and the endophytes trapped ex-situ from O. sativa could be attributed to many factors, including plant species, plant growth stage, in vitro cultivation, and changes in soil microbiota during its transfer from the field to the laboratory. However, the aim of the applied ex-situ plant trapping was to isolate useful microbial diversity and produce information on the effects of plant species, and not to infer ecological patterns of native soil or plant tissues.
Concerning the identified bacterial genera, the Bacillus one is among the most abundant in the rhizosphere of extreme environments (Deng et al. 2023). Moreover, Bacillus species are the most widespread and abundant metabolite-producing bacterial endophytes (Tsotetsi et al. 2022), representing an important member of the “core” cultivated plant-associated bacterial taxa (Radhakrishnan et al. 2017). Their plant growth promoting activities have been associated with the production of secondary metabolites, production of plant hormone (e.g. IAA), nitrogen fixation, solubilization of zinc, potassium and phosphorous, up-regulation of stress regulating genes, and synthesis of bio-control agents (Radhakrishnan et al. 2017). The first commercial bacterial fertilizer, Alinit, was developed from Bacillus spp. and resulted in a 40% increase in crop yield (Romano et al. 2020). Results reported in this study showed that the endophytes identified as IAA producers, halotolerant strains, and P-solubilizers were mostly Bacilli. These data suggest that the isolated Bacillus endophytes could be used to improve the response of host plants (rice and wheat) to abiotic stresses.
As far as Kosakonia is concerned, in the last few years, several members of this genus have been identified as endophyte of different crops (e.g. wheat, maize, tomato, pea, and cruciferous vegetable) and their growth-promoting effects and plant yield improvement have been reported (Hoang et al. 2016, Kämpfer et al. 2016). Furthermore, in recent years, genome sequences for several members of this genus have been released (Mosquito et al. 2020). However, since this genus is relatively young, many of its features have yet to be studied. Results here reported clearly show that about 85% of isolated N-fixers endophytes were Kosakonia strains, thus indicating that nitrogen fixation could be considered as one of the characteristic traits of this genus. These data confirmed our previous results regarding O. glaberrima endophytes (Bianco et al. 2021) and the results obtained with other cereal crops (Bloch et al. 2020, Chen et al. 2020). Besides confirming that different host plants select different endophytic microbiomes in terms of taxa, our study also showed that they differentiated in term of functions: ‘Baldo’ plants mainly recruited halotolerant and P-solubilizer strains, whereas ‘Primadur’ plants mainly engaged N-fixing strains and IAA-producers. These results could arise from different nutritional requirements or from a different sensitivity to environmental stimuli, which could mean that in nature these plants select the microbial partners populating soil according to their needs (Olanrewaju et al. 2019; Rolfe et al. 2019, Rolli et al. 2021). Indeed, rice is very sensitive to salinity stress and is currently listed as the most salt sensitive cereal crop with a threshold of 3 dSm−1 for most cultivated varieties (Wang et al. 2021), whereas a soil is generally considered as salt-affected if it has an electrical conductivity of its saturation extract above 4 dSm−1 (Hoang et al. 2016). Rice loses 10% of its yield even when the electrical conductivity is 3.5 dSm−1. On the other side, mature wheat grain has a higher protein content (accounting for 10–18% of endosperm dry weight) compared with other major cereals, such as rice (Oryza sativa), maize (Zea mays), rye (Secale cereale), and millet (Pennisetum glaucum) (Wang et al. 2021). N plays the most important role in determining protein content, dough quality, and processing characteristic of wheat. Furthermore, wheat is the major crop with the lowest nitrogen use efficiency (NUE), and only 30–35% of applied N fertilizer could be absorbed (Coskun et al. 2017).
We suggest that the selective action on the soil microbiome, which favored the recruitment of specific taxa having particular functions, could result from: i) ‘Baldo’ and ‘Primadur’ plants producing different input resources, such as the exudates (Chai and Schachtman 2022); ii) ‘Baldo’ and ‘Primadur’ plants having different immune system (Oukala et al. 2021), which may have favored the colonization and multiplication of endophytes able to adapt or avoid the defense machinery of the host plants (Rolfe et al. 2019, Rolli et al. 2021). However, since the differential taxonomy (i.e. Bacillus vs. Kosakonia) of the collections co-occur with different PGP functions, we cannot define if the two host plants select endophytes based on the taxonomy or the PGP functions. In the present study, we demonstrate that the applied ex-situ trapping method can effectively enrich a culture collection with nitrogen-fixing bacteria, as well as P-solubilizers, features for which no efficient plate enrichment methods yet exist. Our data also strengthened the concept that IAA-producers are widespread among plant-associated bacteria (Spaepen and Vanderleyden 2011).
Further studies will allow us to understand the differential enrichments observed for ‘Baldo’ and ‘Primadur’ plants and provide a mechanistic interpretation for these differences.
Data reported in this manuscript highlight the following concepts: i) endophytes associated with plants grown in harsh conditions hold adaptive features, which could be exploited to improve the growth of cultivated crops under stressful environmental conditions; ii) the ex-situ plant trapping is an efficient system for the isolation of wide range of genetic diversity of endophytic bacteria and to overcome the difficulties of carrying out experiments in remote areas.
Supplementary Material
Acknowledgement
We thank Marco Petruzziello and Francesca Segreti for technical assistance. We are grateful to Carlotta Volterrani and Luciano Spaccino (Research Institute on Terrestrial Ecosystems, National Research Council, Porano, Italy) for the quantitative determination of C and N levels in soil.
Contributor Information
Maria Laura Amenta, National Research Council, Institute of Biosciences and BioResources, via P. Castellino 111, 80131 Naples, Italy.
Francesca Vaccaro, Department of Biology, University of Florence, 50019 Sesto Fiorentino, Italy.
Stefano Varriale, National Research Council, Institute of Biosciences and BioResources, via P. Castellino 111, 80131 Naples, Italy.
Jean Rodrigue Sangaré, Institut d'Economie Rurale (IER), Centre Régional de Recherche Agronomique (CRRA) de Sikasso, B.P: 16, Mali.
Roberto Defez, National Research Council, Institute of Biosciences and BioResources, via P. Castellino 111, 80131 Naples, Italy.
Alessio Mengoni, Department of Biology, University of Florence, 50019 Sesto Fiorentino, Italy.
Carmen Bianco, National Research Council, Institute of Biosciences and BioResources, via P. Castellino 111, 80131 Naples, Italy.
Funding
This work was supported by the grant “MICRO4Legumes” (Il microbioma vegetale simbionte come strumento per il miglioramento delle leguminose foraggere), D.M.n.89267 (Italian Ministry of Agriculture). This work was also supported by the project PON ARS01_00783 ALI-FUN- “Development of functional foods for the traditional Italian food products innovation”. This study was also carried out within the Agritech National Research Center and received funding from the European Union Next-GenerationEU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR)—MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4–D.D. 1032 17/06/2022, CN00000022). This manuscript reflects only the authors' views and opinions, neither the European Union nor the European Commission can be considered responsible for them. F.V. is supported by a PhD fellowship from D.M. n. 351/2022 (Italian Ministry of University and Research).
Author contributions
Maria Laura Amenta (Investigation, Methodology), Francesca Vaccaro (Data curation, Investigation, Methodology), Stefano Varriale (Data curation, Investigation, Methodology), Jean Rodrigue Sangaré (Resources), Roberto Defez (Writing – review & editing), Alessio Mengoni (Funding acquisition, Writing – review & editing), and Carmen Bianco (Conceptualization, Funding acquisition, Supervision, Writing – original draft)
Conflict of interest statement. The authors have no conflict of interest to declare.
Data availability statement
Sequence data are available in the GenBank database under the accessions OR186984-OR187002 and OR187006-OR187042.
References
- Afzal I, Shinwari ZK, Sikandar S et al. Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol Res. 2019;221:36–49. [DOI] [PubMed] [Google Scholar]
- Aguilar-Paredes A, Valdes G, Araneda N et al. Microbial community in the composting process and its positive impact on the soil biota in sustainable agriculture. Agronomy. 2023;13:542. [Google Scholar]
- Alsharif W, Saad MM, Hirt H. Desert microbiome for boosting sustainable agriculture in extreme environments. Front Microbiol. 2020;11:1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreozzi A, Prieto P, Mercado-Blanco et al. Efficient colonization of the endophytes herbaspirillum huttiense RCA24 and Enterobacter cloacae RCA25 influences the physiological parameters of Oryza sativa L. cv. Baldo rice. Environ Microbiol. 2019;21:3489–504. [DOI] [PubMed] [Google Scholar]
- Bianco C, Andreozzi A, Romano S et al. Endophytes from African Rice (Oryza glaberrima L.) efficiently colonize Asian rice (Oryza sativa L.) stimulating the activity of its antioxidant enzymes and increasing the content of nitrogen, carbon, and chlorophyll. Microorganisms. 2021;9:1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch SE, Clark R, Gottlieb SS et al. Biological nitrogen fixation in maize: optimizing nitrogenase expression in a root-associated diazotroph. J Exp Bot. 2020;71:4591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouri M, Mehnaz S, Şahin F. Extreme environments as potential sources for PGPR. In: Secondary Metabolites and Volatiles of PGPR in Plant-Growth Promotion. Springer; Cham: 2022; 10.1007/978-3-031-07559-9_12. [DOI] [Google Scholar]
- Bulgarelli D, Garrido-Oter R, Münch PC et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangioli L, Mancini M, Napoli M et al. Differential response of wheat rhizosphere bacterial community to plant variety and fertilization. Int J Mol Sci. 2022;23:3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai YN, Schachtman DP. Root exudates impact plant performance under abiotic stress. Trends Plant Sci. 2022;27:80–91. [DOI] [PubMed] [Google Scholar]
- Chen Y, Huang Z, Li J et al. Complete genome sequence of Kosakonia radicincitans GXGL-4A, a nitrogen-fixing bacterium with capability to degrade TEX. Curr Microbiol. 2020;77:1848–57. [DOI] [PubMed] [Google Scholar]
- Chepsergon J, Moleleki LN. Rhizosphere bacterial interactions and impact on plant health. Current Opinion in Microbiology. 2023;73:102297. 10.1016/j.mib.2023.102297 [DOI] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. 10.1016/j.mib.2023.102297. [DOI] [Google Scholar]
- Compant S, Cambon MC, Vacher C et al. The plant endosphere world—bacterial life within plants. Env Microbiol. 2021;23:1812–29. [DOI] [PubMed] [Google Scholar]
- Compant S, Samad A, Faist H et al. A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J Adv Res. 2019;19:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun D, Britto DT, Shi W et al. How plant root exudates shape the nitrogen cycle. Trends Plant Sci. 2017;22:8. [DOI] [PubMed] [Google Scholar]
- Defez R, Andreozzi A, Bianco C. The overproduction of indole-3-acetic acid (IAA) in endophytes up-regulates nitrogen-fixation in both bacterial cultures and inoculated rice plants. Microb Ecol. 2017;74:441–52. [DOI] [PubMed] [Google Scholar]
- Delgado-Baquerizo M. Simplifying the complexity of the soil microbiome to guide the development of next-generation SynComs. Journal of Sustainable Agriculture and Environment. 2022;1:9–15. [Google Scholar]
- Deng A, Wang T, Wang J et al. Adaptive mechanisms of Bacillus to near space extreme environments. Sci Tot Environ. 2023;886:163952. 0048-9697. [DOI] [PubMed] [Google Scholar]
- Escudero-Martinez C, Bulgarelli D. Tracing the evolutionary routes of plant–microbiota interactions. Curr Opin Microbiol. 2019;49:34–40. [DOI] [PubMed] [Google Scholar]
- Fagorzi C, Passeri I, Cangioli L et al. When biodiversity preservation meets biotechnology: the challenge of developing synthetic microbiota for resilient sustainable crop production. J Sust Agric Environ. 2023;2:5–15. [Google Scholar]
- Favero VO, de Carvalho RH, Leite ABC et al. Characterization and nodulation capacity of native bacteria isolated from mung bean nodules used as a trap plant in Brazilian tropical soils. Appl Soil Ecol. 2021;167:104041. [Google Scholar]
- Fu X, Huang Y, Fu Q et al. Critical transition of soil microbial diversity and composition triggered by plant rhizosphere effects. Front Plant Sci. 2023;14:: 1252821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves OS, Creevey CJ, Santana MF. Designing a synthetic microbial community through genome metabolic modeling to enhance plant–microbe interaction. Environmental Microbiome. 2023;18:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarãesa AA, Jaramillob PMA, Nóbregaa RSA et al. Genetic and symbiotic diversity of nitrogen-fixing bacteria isolated from agricultural soils in the Western Amazon by using cowpea as the trap plant. Appl Environ Microb. 2012;78:6726–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizani A, Askri H, Amenta ML et al. Drought responsiveness in six wheat genotypes: identification of stress resistance indicators. 2023;14:1–17. 10.3389/fpls.2023.1232583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T, Biosciences I, Carlsbad C et al. BioEdit: an important software for molecular biology. GERF Bulletin of Biosciences. 2011; 2:60–1. [Google Scholar]
- Hoang TML, Tran TN, Nguyen TKT et al. Improvement of salinity stress tolerance in rice: challenges and opportunities. Agronomy. 2016; 6:54. [Google Scholar]
- Kämpfer P, McInroy JA, Doijad S et al. Kosakonia pseudosacchari sp. nov., an endophyte of Zea mays. Syst Appl Microbiol. 2016;39:1–7. [DOI] [PubMed] [Google Scholar]
- Kandasamy GD, Kathirvel P. Insights into bacterial endophytic diversity and isolation with a focus on their potential applications—a review. Microbiol Res. 2023;266:127256. 10.1016/j.micres.2022.127256. [DOI] [PubMed] [Google Scholar]
- Ke J, Wang B, Yoshikuni Y. Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2021;39:244–61. [DOI] [PubMed] [Google Scholar]
- Le Mouël C, Forslund A. How can we feed the world in 2050? A review of the responses from global scenario studies. Europ Rev Agricult Econom. 2017;44:541–91. [Google Scholar]
- Mapelli F, Mengoni A, Riva V et al. Bacterial culturing is crucial to boost sustainable agriculture. Trends Microbiol. 2022;31:1–4. [Google Scholar]
- Marasco R, Fusi M, Ramond J-B et al. The plant rhizosheath-root niche is an edaphic “mini-oasis” in hyper arid deserts with enhanced microbial competition. ISME Communications. 2022;2:47. 10.1038/s43705-022-00130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco R, Ramond J-B, Van Goethem MW et al. Diamonds in the rough: dryland microorganisms are ecological engineers to restore degraded land and mitigate desertification. Microb Biotechnol. 2023;16:1603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa F, Defez R, Bianco C. Exploitation of plant growth promoting bacteria for sustainable agriculture: hierarchical approach to link laboratory and field experiments. Microorganisms. 2022;10:865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengoni A, Maida I, Chiellini C et al. Antibiotic resistance differentiates Echinacea purpurea endophytic bacterial communities with respect to plant organs. Res Microbiol. 2014;165:686–94. [DOI] [PubMed] [Google Scholar]
- Mohanram S, Kumar P. Rhizosphere microbiome: revisiting the synergy of plant-microbe interactions. Ann Microbiol. 2019;69:307–20. [Google Scholar]
- Mosquito S, Bertani I, Licastro D et al. In planta colonization and role of T6SS in two rice Kosakonia endophytes. MPMI. 2020;33:349–63. [DOI] [PubMed] [Google Scholar]
- Mushtaq S, Shafiq M, Tariq MR et al. Interaction between bacterial endophytes and host plants. Front Plant Sci. 2023;13. 10.3389/fpls.2022.1092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170:265–70. [DOI] [PubMed] [Google Scholar]
- Olanrewaju OS, Ayangbenro AS, Glick BR et al. Plant health:feedback effect of root exudates-rhizobiome interactions. Appl Microbiol Bitechnol. 2019;103:1155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oukala N, Aissat K, Pastor V. Bacterial endophytes: the hidden actor in plant immune responses against biotic stress. Plants. 2021;10:1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øyvind H, Harper DAT, Paul D et al. PAST: paleontological statistics software package for education and data analysis. Palaeont Electr. 2001;4.1:9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. Accessed 10 June 2023. [Google Scholar]
- Pathan SI, Ceccherini MT, Sunseri F et al. Rhizosphere as Hotspot for Plant-soil-microbe interaction. In: Datta R, Meena R, Pathan S, Ceccherini M. (eds) Carbon and Nitrogen Cycling in Soil. Springer, Singapore: 2020. 10.1007/978-981-13-7264-3_2. [DOI] [Google Scholar]
- Petipas RH, Geber MA, Lau JA. Microbe-mediated adaptation in plants. Ecol Lett. 2021;24:: 1302–17. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Egidi E, Liu H et al. New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol Adv. 2019;37:107371. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JM, Kiers ET. Rewilding plant microbiomes. Science. 2022;378:599–600. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, Paulitz TC, Steinberg C et al. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2009;321:341–61. [Google Scholar]
- Radhakrishnan R, Hashem A, Allah EA. Bacillus: a biological tool for crop improvement through bio-molecular changes in adverse environments. Front Physiol. 2017;8:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Lakshmanan V, Labbé JL et al. Microbe to microbiome: a paradigm shift in the application of microorganisms for sustainable agriculture. Front Plant Sci. 2020;11:622926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva V, Mapelli F, Bagnasco A et al. A meta-analysis approach to defining the culturable core of plant endophytic bacterial communities. Appl Env Microbiol. 2022; 88:02537–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JLM, Melotto M. Naturally engineered plant microbiomes in resources-limited ecosystems. Trends Microbiol. 2023;31:329–31. 10.1016/j.tim.2023.02.006. [DOI] [PubMed] [Google Scholar]
- Rolfe SA, Griffiths J, Ton J. Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr Opin Microbiol. 2019;49:73–82. 10.1016/j.mib.2019.10.003. [DOI] [PubMed] [Google Scholar]
- Rolli E, Vergani L, Ghitti E et al. ‘Cry-for-help’ in contaminated soil: a dialogue among plants and soil microbiome to survive in hostile conditions. Env Microbiol. 2021;23:5690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano I, Ventorino V, Ambrosino P et al. Development and application of low-cost and eco-sustainable bio-stimulant containing a new plant growth-promoting strain Kosakonia pseudosacchari TL13. Front Microbiol. 2020;11:2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Sandino T, Delgado-Baquerizo M, Egidi E et al. New microbial tools to boost restoration and soil organic matter. Appl Microbiol Int. 2023;16:2019–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki S, Takeda A, Shimazu T. x Assay of inorganic phosphate in the mild pH range, suitable measurement of glycogen phosphorylase activity. Anal Biochem. 1985;148:277–81. [DOI] [PubMed] [Google Scholar]
- Santoyo G, Moreno-Hagelsieb G, del Carmen Orozco-Mosqueda M et al. Plant growth-promoting bacterial endophytes. Microbiol Res. 2016;183:92–9. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Bulgarelli D. The plant microbiome at work. MPMI. 2015; 28:212–7. [DOI] [PubMed] [Google Scholar]
- Schmitz L, Yan Z, Schneijderberg M et al. Synthetic bacterial community derived from a desert rhizosphere confers salt stress resilience to tomato in the presence of a soil microbiome. ISME J. 2022;16:1907–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz VA, McGivern BB, Daly RA et al. Variation in root exudate composition influences soil microbiome membership and function. Appl Env Microbiol. 2022;88:00226–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch A, Pfaffenbichler N, Mitter B. Microbiome applications from lab to field: facing complexity. Trends Plant Sci. 2019;24:194–8. [DOI] [PubMed] [Google Scholar]
- Sie M, Sanni K, Futakuchi K et al. Towards a rational use of African rice (Oryza glaberrima Steud.) for breeding in Sub-Saharan Africa. Genes Genomes Genetics. 2012;6:1–7. https://hdl.handle.net/20.500.11766/5034. Accessed date 15 April 2023 [Google Scholar]
- Singh BK, Trivedi P, Egidi E et al. Crop microbiome and sustainable agriculture. Nat Rev Micro. 2020;18:601–2. [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harb Perspect Biol. 2011;3:a001438–8. 10.1101/cshperspect.a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Zhang F, Chen P et al. Rhizosphere bacterial communities of three minor grain crops exhibit distinct environmental adaptations and assembly processes. Eu J Soil Sci. 2022;73:e13314. [Google Scholar]
- Tsotetsi T, Nephali L, Malebe M et al. Bacillus for plant growth promotion and stress resilience: what have we learned?. Plants. 2022;11:2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro F, Cangioli L, Mengoni A et al. Synthetic plant microbiota challenges in non-model species. Trends Microbiol. 2022;30:922–4. [DOI] [PubMed] [Google Scholar]
- Van Andel T. African rice (Oryza glaberrima Steud.): lost crop of the enslaved Africans discovered in Suriname. Econ Bot. 2010;64:1–10. 10.1007/s12231-010-9111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He T, Gen S et al. Soil properties and agricultural practices shape microbial communities in flooded and rainfed croplands. Appl Soil Ecol. 2020;147:0929–1393. [Google Scholar]
- Wang Y, Wang D, Tao Z et al. Impacts of nitrogen deficiency on wheat (Triticum aestivum L.) grain during the medium filling stage: transcriptomic and metabolomic comparisons. Front Plant Sci. 2021;12:674433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. https://link.springer.com/book/10.1007/978-3-319-24277-4. Accessed 10 June 2023 [Google Scholar]
- Xiong C, Zhu Y-G, Wang J et al. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021;229:1091–104. [DOI] [PubMed] [Google Scholar]
- Yan D, Tajima H, Cline LC et al. Genetic modification of flavone biosynthesis in rice enhances biofilm formation of soil diazotrophic bacteria and biological nitrogen fixation. Plant Biotechnol J. 2022;20:2135–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are available in the GenBank database under the accessions OR186984-OR187002 and OR187006-OR187042.