Abstract
Background
The relevance of iron status biomarkers for coronary artery disease (CAD), heart failure (HF), ischemic stroke (IS), and type 2 diabetes (T2D) is uncertain. We compared the observational and Mendelian randomization (MR) analyses of iron status biomarkers and hemoglobin with these diseases.
Methods and Results
Observational analyses of hemoglobin were compared with genetically predicted hemoglobin with cardiovascular diseases and diabetes in the UK Biobank. Iron biomarkers included transferrin saturation, serum iron, ferritin, and total iron binding capacity. MR analyses assessed associations with CAD (CARDIOGRAMplusC4D [Coronary Artery Disease Genome Wide Replication and Meta‐Analysis Plus The Coronary Artery Disease Genetics], n=181 522 cases), HF (HERMES [Heart Failure Molecular Epidemiology for Therapeutic Targets), n=115 150 cases), IS (GIGASTROKE, n=62 100 cases), and T2D (DIAMANTE [Diabetes Meta‐Analysis of Trans‐Ethnic Association Studies], n=80 154 cases) genome‐wide consortia. Observational analyses demonstrated J‐shaped associations of hemoglobin with CAD, HF, IS, and T2D. In contrast, MR analyses demonstrated linear positive associations of higher genetically predicted hemoglobin levels with 8% higher risk per 1 SD higher hemoglobin for CAD, 10% to 13% for diabetes, but not with IS or HF in UK Biobank. Bidirectional MR analyses confirmed the causal relevance of iron biomarkers for hemoglobin. Further MR analyses in global consortia demonstrated modest protective effects of iron biomarkers for CAD (7%–14% lower risk for 1 SD higher levels of iron biomarkers), adverse effects for T2D, but no associations with IS or HF.
Conclusions
Higher levels of iron biomarkers were protective for CAD, had adverse effects on T2D, but had no effects on IS or HF. Randomized trials are now required to assess effects of iron supplements on risk of CAD in high‐risk older people.
Keywords: coronary artery disease, heart failure, hemoglobin, iron status, ischemic stroke, type 2 diabetes
Subject Categories: Diet and Nutrition, Epidemiology, Cardiovascular Disease, Primary Prevention
Nonstandard Abbreviations and Acronyms
- CMD
cardiometabolic diseases
- GRS
genetic risk score
- IS
ischemic stroke
- LAGE
localized average causal effects
- MR
Mendelian randomization
- PheWAS
phenome‐wide associations
- T2D
type 2 diabetes
- TIBC
total iron binding capacity
- TSAT
transferrin saturation
- UKB
UK Biobank
Clinical Perspective.
What Is New?
Higher levels of genetically predicted iron status biomarkers were inversely associated with coronary artery disease (7%–14% lower risk for 1 SD higher iron status biomarkers), positively associated with diabetes, but there was no evidence of any associations with ischemic stroke or heart failure.
What Are the Clinical Implications?
Mendelian randomization analyses demonstrated modest protective effects of higher iron status biomarkers for coronary artery disease, adverse effects for type 2 diabetes, and no support from Mendelian randomization for heart failure or ischemic stroke.
The results could guide the design of large‐scale randomized trials of iron supplements in high‐risk older people for prevention of coronary artery disease.
Iron deficiency is a common nutritional deficiency worldwide that preferentially affects women, older people, and low‐income populations. 1 Iron overload is a less common problem that chiefly affects men and people of Celtic ancestry in which excess iron is deposited in multiple organs, such as the heart, liver, or pancreas. 2 , 3 Previous studies reported higher risks of cardiometabolic diseases (CMD) associated with iron status outside the normal range. 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 Moreover, iron deficiency is a therapeutic target for the treatment of patients with heart failure with reduced ejection fraction. 20 , 21 , 22 , 23 , 24 Given the fact that there is no consensus on the optimum biomarkers for assessing iron status and its effects on health outcomes, 25 , 26 it is important to assess the levels of iron status by multiple biomarkers 27 , 28 and explore their associations with CMD and other selected diseases.
Most of the body stores of iron are located in hemoglobin within circulating erythrocytes, where their primary role is to transport oxygen to body tissues. 1 Previous observational studies have examined associations of hemoglobin levels with CMD and other diseases, 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 but their results have been inconsistent, ranging from null to positive, linear, and nonlinear (U‐shaped or J‐shaped) relationships, in addition to differences by sex. Moreover, few studies have explored the strength of associations of hemoglobin levels and iron biomarkers with CMD, 29 in addition to their causal relevance using linear and nonlinear Mendelian randomization (MR) analyses in the same study. 30 , 31
In contrast to other well‐established iron status biomarkers such as transferrin saturation (TSAT), serum iron, ferritin, and total iron binding capacity (TIBC), hemoglobin has been more extensively examined in large‐scale cohort studies. The UK Biobank (UKB), with detailed data collected by questionnaire and health outcomes in relation to hemoglobin levels and genetic data, 32 affords a unique opportunity to conduct observational and genetic analyses of iron status markers and hemoglobin levels with disease outcomes. Assessing the causal relevance of these associations could guide the design of large‐scale randomized trials to assess the effects of iron supplements on the risk of coronary artery disease (CAD), heart failure (HF), ischemic stroke (IS), and type 2 diabetes (T2D).
The aims of this study were to (1) examine the observational associations of hemoglobin with CMD in UKB; (2) assess the shape and strength of associations of hemoglobin with CMD using linear and nonlinear 1‐sample MR analyses in UKB; (3) compare the genetic associations of iron status markers (including TSAT, serum iron, ferritin, and TIBC) with hemoglobin; and (4) investigate the associations of iron status markers with CAD, HF, IS, and T2D through 2‐sample MR analyses using summary data from published genome‐wide consortia. Additional analyses included phenome‐wide associations (PheWAS) of genetically predicted differences in hemoglobin with a wide range of disease outcomes in UKB and sex‐specific analyses of observational and genetic associations of hemoglobin with CMD in UKB.
METHODS
Study Population
The study design and methods used in UKB have been previously reported. 32 , 33 , 34 In brief, UKB is a prospective study of over 500 000 participants, aged 37 to 73 years when recruited from 22 assessment centers in the United Kingdom between 2006 and 2010 (Table 1; Figure S1; Table S1). Participants provided information on their health and lifestyle using questionnaires supplemented by interviews and had physical measurements recorded and blood samples collected for biochemical and genetic measurements. 32 , 33 , 34 Among the 502 507 participants in the UKB, we excluded individuals who reported current use of iron supplements (n=115 035), or who withdrew from the study (n=99), or had missing data on hemoglobin (n=24 331), leaving 368 406 participants for the present analyses (Table 1). Ethics approval for UKB was provided by the National Information Governance Board for Health and Social Care and the National Health Service North West Multicenter Research Ethics Committee (REC reference: 11/NW/03820). All participants provided written informed consent.
Table 1.
Baseline Characteristics of Participants in the Observational Analysis in UK Biobank, by Sex
| Men | Women | All | |
|---|---|---|---|
| Number of participants | 177 535 (48.2) | 190 871 (51.8) | 368 406 |
| Demographic factors | |||
| Age, y | 56.8 (8.2) | 56.5 (8.0) | 56.6 (8.1) |
| Race, White, % | 167 472 (94.3) | 181 554 (95.1) | 349 026 (94.7) |
| Socioeconomic deprivation tertiles | |||
| 1 (least deprived) | 59 105 (33.3) | 63 690 (33.4) | 122 795 (33.3) |
| 2 | 58 231 (32.8) | 64 573 (33.8) | 122 804 (33.3) |
| 3 (most deprived) | 60 199 (33.9) | 62 608 (32.8) | 122 807 (33.3) |
| Education, university/college, % | 59 133 (33.3) | 57 582 (30.2) | 116 715 (31.7) |
| Lifestyle factors | |||
| Current smoker, % | 22 444 (12.6) | 17 366 (9.1) | 39 810 (10.8) |
| Current drinker, % | 166 031 (93.5) | 172 515 (90.4) | 338 546 (91.9) |
| Postmenopausal (Yes in women), % | … | 116 375 (61.0) | 116 375 (31.6) |
| Physical activity, metabolic equivalent of task, h/week | 29.8 (13.2, 61.5) | 28.0 (12.9, 55.6) | 29.0 (13.0, 58.8) |
| Family history of diseases, N (%) | |||
| Family history of cardiovascular disease | 124 389 (70.1) | 145 489 (76.2) | 269 878 (73.3) |
| Family history of diabetes | 36 514 (20.6) | 42 879 (22.5) | 79 393 (21.6) |
| Medical history, N (%) | |||
| Use of antihypertensive drugs | 44 920 (25.3) | 35 045 (18.4) | 79 965 (21.7) |
| Use of cholesterol‐lowering drugs | 41 536 (23.4) | 25 334 (13.3) | 66 870 (18.2) |
| Baseline coronary artery disease | 15 160 (8.5) | 6399 (3.4) | 21 559 (5.9) |
| Baseline ischemic stroke | 702 (0.4) | 306 (0.2) | 1008 (0.3) |
| Baseline heart failure | 1454 (0.8) | 465 (0.2) | 1919 (0.5) |
| Baseline type 2 diabetes | 11 665 (6.6) | 6351 (3.3) | 18 016 (4.9) |
| Clinical measurements | |||
| Body mass index, kg/m2 | 27.9 (4.2) | 27.2 (5.2) | 27.5 (4.8) |
| Waist circumference, cm | 97.1 (11.3) | 84.9 (12.6) | 90.8 (13.5) |
| Waist‐to‐hip ratio | 0.93 (0.89, 0.98) | 0.81 (0.77, 0.86) | 0.88 (0.81, 0.94) |
| Systolic blood pressure, mm Hg | 141.1 (17.5) | 135.7 (19.2) | 138.3 (18.6) |
| Diastolic blood pressure, mm Hg | 84.2 (10.0) | 80.8 (10.0) | 82.4 (10.1) |
| Random glucose, mmol/L | 4.95 (4.60, 5.36) | 4.91 (4.60, 5.28) | 4.93 (4.60, 5.32) |
| Hemoglobin A1c, % | 5.38 (5.15, 5.65) | 5.37 (5.15, 5.61) | 5.38 (5.15, 5.63) |
| Hemoglobin, mg/dL | 15.0 (14.4, 15.7) | 13.5 (12.9, 14.1) | 14.2 (13.4, 15.1) |
| High‐density lipoprotein cholesterol, mmol/L | 1.28 (0.31) | 1.59 (0.38) | 1.44 (0.38) |
| Low‐density lipoprotein cholesterol, mmol/L | 3.47 (0.87) | 3.63 (0.87) | 3.56 (0.87) |
| Triglycerides, mmol/L | 1.69 (1.18, 2.44) | 1.34 (0.97, 1.90) | 1.49 (1.05, 2.16) |
| Total cholesterol, mmol/L | 5.48 (1.14) | 5.88 (1.13) | 5.69 (1.15) |
| C‐reactive protein, mg/L | 1.29 (0.67, 2.56) | 1.40 (0.66, 3.01) | 1.34 (0.66, 2.78) |
Values are shown as numbers (percentages) for categorical variables and means (SD) or median (interquartile range) for continuous variables.
Ascertainment of Exposures and Outcomes
Among samples collected at recruitment, hemoglobin concentrations (Field ID 30020, mg/dL) were measured in EDTA‐vacutainers using the Sysmex XN‐1000 hematology analyzer at the laboratory of the UK Biocenter (https://biobank.ctsu.ox.ac.uk/crystal/crystal/docs/haematology.pdf). 35 , 36 The primary outcomes included incident cases of CAD, HF, IS, and T2D, which were identified through linkage to Hospital Episodes Statistics and mortality data, with diagnostic criteria defined by the International Classification of Diseases, Ninth Revision (ICD‐9) and Tenth Revision (ICD‐10) codes (Table S2). Data from UKB are available to bona fide researchers on application to https://www.ukbiobank.ac.uk. Data from other genetic consortia are also available on application to the relevant consortia.
Statistical Analysis
Observational Analyses of Hemoglobin With CMD
Logistic regression was used to estimate the odds ratios (OR) and their 95% CIs for CAD, HF, IS, and T2D in individuals who were free of cardiovascular disease (CVD) or diabetes at baseline. All analyses were adjusted for age, sex, ethnicity, education, socioeconomic status, family history of CVD/diabetes, smoking status, alcohol intake, physical activity, body mass index, baseline diabetes, medication use, systolic blood pressure, and triglycerides where appropriate. Townsend Deprivation Index was used as an area‐based proxy measure for socioeconomic status, with lower levels indicating higher socioeconomic status. 37 Body mass index was calculated by dividing body weight in kilograms by the square of height in meters (kg/m2).
Cubic spline curves 38 were used to assess the shape of the associations of hemoglobin levels with incident disease outcomes. Nonlinearity was evaluated using a Wald test. 38 All analyses were stratified by sex and any heterogeneity by sex was assessed using a Cochran's Q test. 39
Genetic Analyses
Genetic analyses in UKB were restricted to individuals of White‐British descent, and individuals were excluded if they were related to each other (third‐degree relatives or closer), had mismatched information between self‐reported and genetic sex, had missing values and outliers for hemoglobin levels, or had missing data for any of the covariates, resulting in 331 964 participants for the present analysis (Figure S1; Table S1).
MR Analyses of Hemoglobin With CMD
In linear MR analysis, we generated a weighted genetic risk score (GRS) consisting of 532 single‐nucleotide variant (SNVs, instrument strength: F‐statistics=22335.7, R 2=13%), derived from a recent genome‐wide association study (GWAS) of 563 085 European ancestry participants, 40 as an instrumental variable for blood hemoglobin concentrations (Tables S3 and S4). A 2‐stage least squares analysis was performed to assess the associations of genetically predicted hemoglobin levels with prevalent and incident CAD, HF, IS, and T2D. Both stages of the least square regression were adjusted for age, sex, assessment centers, top 10 principal components, and genotyping array. To assess the consistency of results, an independent set of instrumental variables (140 SNVs reported by Astle et al. 41 F‐statistics=12934.7, R 2=7%) and 2‐sample MR were conducted using summary data from the most recent publicly available genome‐wide consortia: CARDIOGRAMplusC4D (Coronary Artery Disease Genome Wide Replication and Meta‐Analysis Plus the Coronary Artery Disease Genetics), HERMES (Heart Failure Molecular Epidemiology for Therapeutic Targets), GIGASTROKE, and DIAMANTE (Diabetes Meta‐Analysis of Trans‐Ethnic Association Studies).
We used nonlinear MR 30 , 42 with fractional polynomials to assess the nonlinear associations of genetically determined hemoglobin levels with CMD. First, the study participants were categorized into 4 strata according to the residual levels of hemoglobin after regressing on the GRS of hemoglobin. Second, we calculated the localized average causal effects within each stratum, which are the ratio of coefficients of the GRS‐hemoglobin–outcome association to those of the GRS‐hemoglobin–hemoglobin association. Third, these the localized average causal effects estimates were meta‐regressed against the mean levels of hemoglobin in each stratum by fitting fractional polynomial exposure–outcome models. Maximum likelihood estimates of model parameters were obtained among all possible fractional polynomial models using 1 or 2 degrees of freedom and the likelihood ratio test statistic was used to select the best‐fitting models. P values from the quadratic test and Cochran's Q test were reported for nonlinearity.
For 1‐sample and nonlinear MR analyses, we performed sex‐stratified analyses in UKB to explore whether the effect of hemoglobin on CMD differed by sex using Cochran's Q test for heterogeneity. All statistical analyses were performed in R (version 3.6.3), in addition to Plink (version 2.0) software for the GRS calculation and the “nlmr” package for nonlinear MR analysis.
PheWAS of Hemoglobin With Multiple Diseases
Among the 331 964 participants who passed quality control, we conducted a PheWAS 43 to examine the associations of GRS‐hemoglobin with a range of diseases using PHESANT. 44 All disease outcomes were obtained from the first occurrence of each disease (Category 1712) in UKB that were derived from hospital inpatient records, death registers, disease registers, and self‐reported health conditions. We restricted analyses to diseases involving more than 1000 cases (377 diseases) and used multivariable logistic regression to assess associations with diseases after adjusting for age, sex, assessment center, top 10 principal components of ancestry, and genotyping array for study participants. Power calculations for PheWAS analysis 45 suggested that a sample size of 1000 cases had 80% power to detect an OR of 1.25 or 0.76 for any disease outcome. We also performed sex‐specific PheWAS analyses using a threshold of 500 cases, leaving 319 and 376 diseases in men and women, respectively. All statistical tests were 2‐sided and corrected for multiple testing using a 5% false discovery rate. 46
MR Analyses of Iron Status Biomarkers With Hemoglobin and CMD
We selected the SNVs independently contributing to 4 biomarkers of iron status: TSAT (12 SNVs, N=131 471, F‐statistics [R 2]=8157.8 [6%]), serum iron (16 SNVs, N=163 511, F‐statistics [R 2]=6412.2 [4%]), ferritin (42 SNVs, N=246 139, F‐statistics [R 2]=5981.0 [2%]), and TIBC (16 SNVs, N=135 430, F‐statistics [R 2]=12989.5 [9%]), based on recent GWAS 47 (Tables S4 and S5). For SNVs associated with iron status biomarkers, we retrieved GWAS summary statistics from outcome data sets obtained from publicly available genetic databases, for CAD using CARDIOGRAMplusC4D (N=181 522 cases), 48 HF using HERMES (N=115 150 cases), 49 IS using GIGASTROKE (N=62 100 cases), 50 and T2D using DIAMANTE (N=80 154 cases), 51 and for cardiovascular risk factors using other publicly genetic databases. Additional details of data sets included in the analysis are provided in Table S6.
Before implementing the conventional MR analyses, we estimated genetic correlation (R g) for iron markers with hemoglobin, CMD, and cardiometabolic risk factors (Figures S2 and S3), to evaluate the shared genetic background among these phenotypes. R g was estimated by using a range of publicly available GWAS summary statistics through the ‐rg option of linkage disequilibrium score regression 52 , 53 , 54 (https://github.com/bulik/ldsc), with the values > or <0 indicating positive or negative correlations, respectively.
The inverse‐variance weighted method was used to estimate the associations of iron markers with hemoglobin and CMD, respectively, which were accessed by regression of the SNV‐outcome (ie, hemoglobin, CAD, and HF) associations on the SNV‐exposure (4 iron markers) associations. Sensitivity analyses investigated the possible relevance of directional (unbalanced horizontal) pleiotropic effects: (1) MR‐Egger provides a statistical test for the presence of pleiotropic effects due to aggregation of invalid genetic instruments, assuming absence of dose–response confounding of SNVs through pleiotropic pathways 55 ; (2) weighted median MR should provide a valid causal effect estimate if more than 50% of the information were derived from the genetic instrumental variables 56 ; and (3) the model‐based estimate is consistent when the largest subset of individual‐instruments which identify the causal effect are valid instruments. 57
For iron markers–hemoglobin association, bidirectional MR analyses were conducted to check whether genetically predicted differences in iron status biomarkers were associated with hemoglobin concentrations and vice versa. To identify the role of established risk factors that potentially mediate the associations of iron status biomarkers with CMD, we further explored whether genetic predisposition for higher iron status was associated with higher levels of cardiometabolic risk factors (Table S6), including anthropometric measurements (ie, body mass index, waist circumference, waist‐to‐hip ratio, birth weight, and height), glycemic traits (ie, fasting glucose, fasting insulin, and hemoglobin A1c), blood lipids (ie, low‐density lipoprotein cholesterol, high‐density lipoprotein cholesterol, triglycerides, and total cholesterol), blood pressure traits (ie, systolic blood pressure, diastolic blood pressure, and pulse pressure), imaging phenotypes (ie, liver fat, liver iron measurement, and pancreas iron measurement), and other risk factors (ie, smoking status, educational attainment, and C‐reactive protein). All statistical analyses were performed in R (version 3.6.3) using the “TwoSampleMR” package. 58
RESULTS
The Central Illustration outlines the study design and key results of the 4 components of this study: (1) observational analyses of hemoglobin with CMD in UKB; (2) MR analyses of genetically predicted hemoglobin with disease outcomes in UKB; (3) MR analyses of genetically predicted iron with hemoglobin; and (4) MR analyses of genetically predicted iron with CAD, HF, IS, and T2D in worldwide GWAS consortia.
Among the 368 406 participants included in the observational analyses in UKB (Table 1), the mean (SD) age of participants was 56.6 (8.1) years, and 51.8% were women. About 10.8% were current smokers, and 91.9% were alcohol drinkers, respectively. The median (interquartile range) hemoglobin concentration was 15.0 (14.4, 15.7) mg/dL in men and 13.5 (12.9, 14.1) mg/dL in women. The characteristics of individuals in the genetic analyses (Table S1) were comparable with those in the observational analyses in UKB.
Observational and Genetic Associations of Hemoglobin With CMD
The observational analyses in Figure 1 demonstrated U‐shaped associations of hemoglobin levels with CAD, wherein both lower and higher levels of hemoglobin were each associated with higher risks of CAD (reference level 14 mg/dL). However, there was no evidence of nonlinearity in the MR analyses (Cochran Q P=0.853, quadratic test P=0.703, Table S7). For the associations of hemoglobin with HF and IS, the associations were L‐shaped (leveling off at about 14 mg/dL and increasing slightly after 16 mg/dL) and U‐shaped in the observational analyses, but no significant association, in addition to nonlinear associations in the genetic analyses. For hemoglobin–T2D relationship, both conventional and genetic analyses demonstrated positive associations whereby higher levels of hemoglobin were associated with T2D at hemoglobin levels ≥14 mg/dL. No significant linear association was observed with hemoglobin levels <14 mg/dL in the observational analyses, but a modest protective effect was found in the genetic analyses (Cochran Q P=0.065, quadratic test P=0.177 in MR analysis, Table S7). Overall, there was no evidence of nonlinear associations of hemoglobin with either CAD, HF, IS, or T2D in the overall population, with concordant associations obtained from linear 2‐sample MR analysis (Table S8).
Figure 1. Associations of hemoglobin with coronary artery disease, heart failure, ischemic stroke, and type 2 diabetes in UK Biobank.
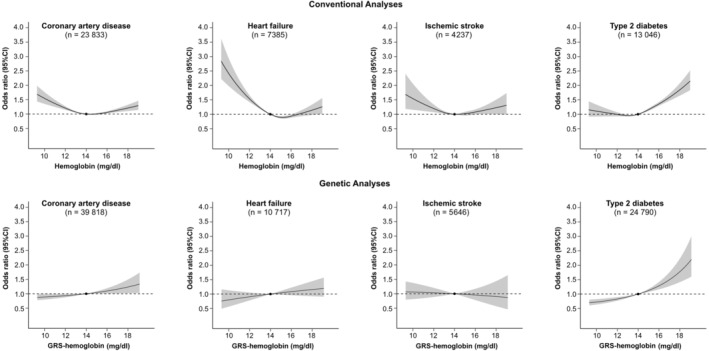
Associations of hemoglobin with coronary artery disease, heart failure, ischemic stroke, and type 2 diabetes were obtained from conventional analyses and nonlinear Mendelian randomization analyses in UK Biobank. GRS indicates genetic risk score.
The results from sex‐stratified analyses were comparable with those obtained for the overall population (Figures S4 and S5; Tables S9 and S10), with the exception of the stronger associations of hemoglobin with CAD in men and the potential nonlinear association of hemoglobin‐T2D in women in the MR analyses (Cochran Q P=0.030, quadratic test P=0.027).
Genetic Associations of Hemoglobin With Multiple Diseases
In PheWAS analyses (Figure 2), we investigated the associations of GRS‐hemoglobin with 377 diseases in UKB after correction for multiple testing. Strong associations of genetically predicted higher hemoglobin levels were associated with iron deficiency anemia, other causes of anemia, and disorders of mineral metabolism, with ORs of 0.90 (95% CI, 0.89–0.92), 0.88 (95% CI, 0.86–0.89), and 1.23 (95% CI, 1.18–1.27), respectively. Moreover, 1 SD higher levels of hemoglobin were associated with 3% to 12% higher risks of acute myocardial infarction, hypertension, diabetes, disorders of porphyrin and bilirubin metabolism, fibrosis and cirrhosis of liver, duodenal ulcer, and psoriasis. Sex‐specific PheWAS results are shown separately in Figures S6 and S7 for men and women, respectively (>500 cases), with 11 out of 319 diseases and 8 out of 376 diseases exceeding thresholds for multiple‐testing correction.
Figure 2. Associations of a genetic risk score for hemoglobin with different disease outcomes in UK Biobank.
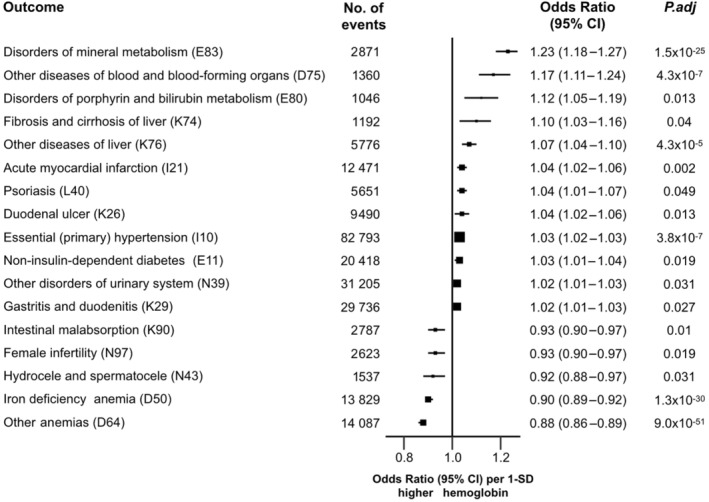
Odds ratios obtained from PheWAS represent the estimates for the effect of 1‐unit higher genetic risk scores for hemoglobin (GRS‐hemoglobin), after adjusting for age, sex, center, genotyping array, and genetic principal components (the first 10 principal components). Analyses were restricted to outcomes with greater than 1000 cases (377 diseases) and showed the associations that survived from the multiple testing. International Classification of Diseases, Tenth Revision codes were also shown in the parentheses for each outcome. Error bars represent 95% CIs. All statistical tests were 2‐sided, following a correction for multiple testing using a strategy controlling for the false discovery rate. GRS indicates genetic risk score; and PheWAS, phenome‐wide associations.
MR Analyses of Iron Status Biomarkers With Hemoglobin
Table 2 shows that using all iron biomarkers, genetically predicted higher iron status was associated with higher levels of hemoglobin, with an increase of 0.37 (95% CI, 0.20–0.54) mg/dL for TSAT, 0.49 (95% CI, 0.30–0.67) mg/dL for serum iron, 0.51 (95% CI, 0.26–0.76) mg/dL for ferritin, and −0.16 (95% CI, −0.34 to 0.02) mg/dL for TIBC (reflecting lower systemic iron) obtained from an inverse‐variance weighted MR method. Table S11 presents the results of additional bi‐directional MR analyses for genetically predicted hemoglobin levels with each iron biomarker.
Table 2.
Causal Effects of a 1 SD Higher Plasma Level of Iron Biomarkers on Hemoglobin Concentrations
| MR method | Transferrin saturation (n=131 471) | Serum iron (n=163 511) | Serum ferritin (n=246 139) | Total iron binding capacity (n=135 430) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Beta (95% CI) | P | SNP | Beta (95% CI) | P | SNP | Beta (95% CI) | P | SNP | Beta (95% CI) | P | |
| Inverse variance weighted | 10 | 0.37 (0.20 to 0.54) | 1.5×10−5 | 14 | 0.49 (0.30 to 0.67) | 2.1×10−7 | 37 | 0.51 (0.26 to 0.76) | 7.9×10−5 | 15 | −0.16 (−0.34 to 0.02) | 0.077 |
| Weighted median | 10 | 0.41 (0.36 to 0.45) | 1.2×10−73 | 14 | 0.60 (0.56 to 0.64) | 2.7×10−200 | 37 | 0.16 (0.09 to 0.24) | 9.0×10−6 | 15 | −0.08 (−0.14 to −0.02) | 0.007 |
| Weighted mode | 10 | 0.44 (0.41 to 0.47) | 3.0×10−10 | 14 | 0.59 (0.56 to 0.63) | 8.4×10−14 | 37 | 0.13 (0.06 to 0.20) | 8.3×10−4 | 15 | −0.07 (−0.10 to −0.03) | 2.6×10−3 |
| MR Egger | 10 | 0.48 (0.25 to 0.71) | 3.5×10−3 | 14 | 0.66 (0.41 to 0.90) | 2.0×10−4 | 37 | 0.72 (0.26 to 1.18) | 4.4×10−3 | 15 | −0.16 (−0.40 to 0.08) | 0.214 |
| Egger intercept | −0.02 (−0.05 to 0.01) | 0.223 | −0.02 (−0.04 to 0.00) | 0.088 | −0.01 (−0.03 to 0.01) | 0.294 | −0.00 (−0.04 to 0.03) | 0.948 | ||||
Estimates represent the effect of a 1‐SD increase in levels of transferrin saturation, serum iron, log‐transformed serum ferritin, and total iron binding capacity on hemoglobin. MR was implemented through the “TwoSampleMR” package in R‐version 3.6.3. The F‐statistics (R2) of iron markers were 8157.8 (0.06) for transferrin saturation (12 SNVs), 6412.2 (0.04) for serum iron (16 SNVs), 5981.0 (0.02) for serum ferritin (42 SNVs), and 12989.5 (0.09) for total iron binding capacity (16 SNVs), respectively. MR indicates Mendelian randomization; and SNV single‐nucleotide variant.
MR Analyses of Iron Status Biomarkers With CMD
Figure 3 shows the results of MR analysis for iron markers with CMD, which are reported as ORs per 1 SD higher level of each iron status biomarker. There were modest inverse associations of genetically predicted higher iron status biomarkers with CAD, with ORs of 0.93 (95% CI, 0.88–0.98) for TSAT, 0.91 (95% CI, 0.83–0.99) for serum iron, 0.86 (95% CI, 0.77–0.96) for (log‐transformed) ferritin, and 1.04 (95% CI, 0.96–1.12) for TIBC (reflecting lower systemic iron). In contrast, we found adverse effects of higher levels of iron status with 7% higher risks of T2D per 1 SD higher level of TSAT but not for other iron status biomarkers. There was no evidence of associations of iron status markers with HF or IS or with other CVD outcomes (Table S12). Sensitivity analyses using different analytical methods or different sets of SNVs demonstrated consistent results with the overall analyses (Tables S13 through S15).
Figure 3. Associations of genetically determined iron markers with coronary artery disease, heart failure, ischemic stroke, and type 2 diabetes.
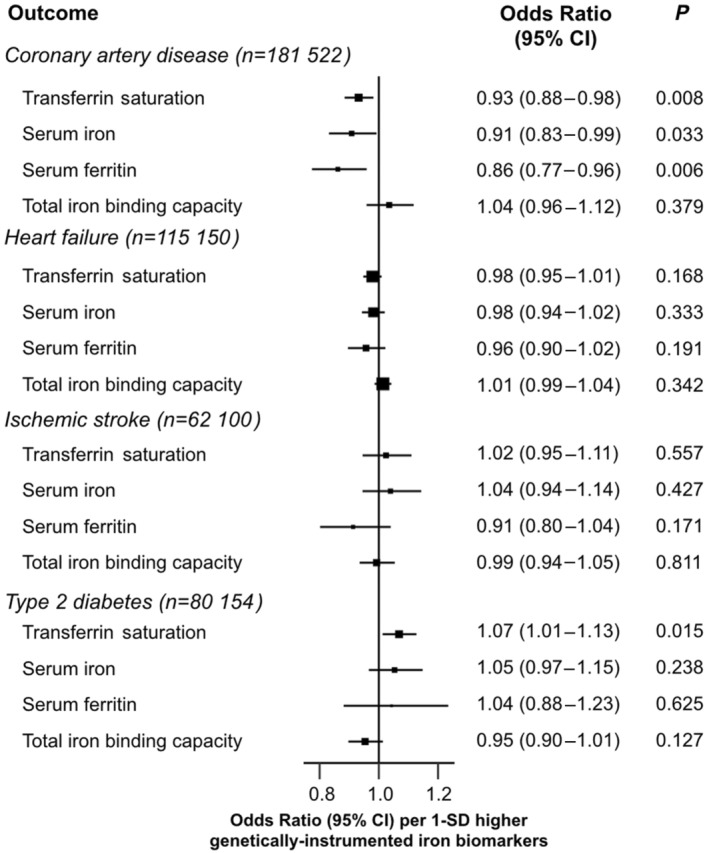
Odds ratios represent the estimates for the effect of a 1‐SD increase in levels of transferrin saturation, serum iron, log‐transformed serum ferritin, and total iron binding capacity, respectively, on disease outcomes.
For the associations of iron markers with cardiometabolic risk factors, Tables S16 through S18 show that a 1 SD higher level of iron status biomarkers was associated with higher levels of waist circumference, height, triglycerides, diastolic blood pressure, estimated glomerular filtration rate, and C‐reactive protein and lower levels of hemoglobin A1c, LDL, and total cholesterol, respectively.
Sex‐Specific Associations of Hemoglobin With CMD
Figure 4 shows that higher levels of hemoglobin were associated with lower risks of CAD in women in the observational analyses, but genetic analyses from 1‐sample MR analyses did not provide support for the causal relevance of such associations. In addition, a 1 SD higher genetically predicted hemoglobin was associated with 8% higher risks of CAD in men (OR, 1.08 [95% CI, 1.04–1.13]). There was evidence of heterogeneity by sex for hemoglobin‐CAD associations in the genetic analyses (P for heterogeneity 0.003). For the hemoglobin‐HF relationship, strong inverse associations were observed in both men and women in the observational analyses but not in the genetic analyses. The findings from both observational and genetic analyses were consistent with hemoglobin associations with T2D, showing 10% to 12% and 10% to 13% higher risks per 1 SD higher hemoglobin, respectively, in both men and women. There were no significant associations for hemoglobin with IS in the observational analyses. However, we found a potential protective effect of hemoglobin on IS in women only (OR, 0.86 [95% CI, 0.74–1.01]), with P for heterogeneity by sex of 0.039 in the genetic analyses.
Figure 4. Sex‐specific associations of a 1 SD higher level of hemoglobin on risk of cardiometabolic diseases in UK Biobank.
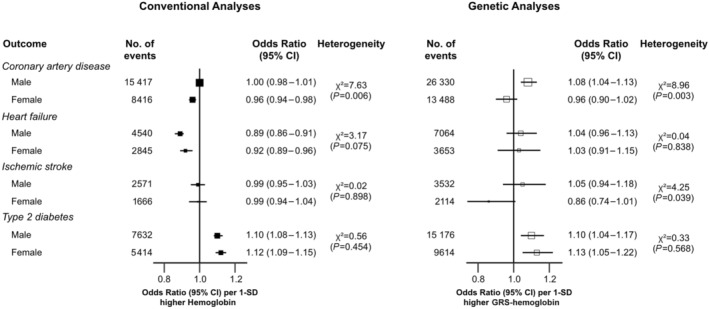
We used multivariable logistic regression in observational analysis to estimate the risk of incident coronary artery disease, ischemic stroke, heart failure, and type 2 diabetes among individuals by 1‐SD increase in levels of hemoglobin after adjusting sociodemographic factors (age, sex, ethnicity, and education), socioeconomic status (Townsend deprivation index), family history of cardiovascular diseases or diabetes, smoking status, alcohol intake, physical activity, body mass index, baseline diabetes, medication use (antihypertensive or cholesterol‐lowering drugs), systolic blood pressure, and triglycerides where appropriate. Odds ratios from Mendelian randomization represent the estimates for the effect of a 1‐unit higher genetic risk score for hemoglobin (GRS‐hemoglobin). GRS indicates genetic risk score.
DISCUSSION
In this large study of UKB and worldwide GWAS consortia, we observed linear positive associations of higher genetically predicted hemoglobin with risk for CAD and T2D but found no evidence of any associations with HF or IS. However, MR analyses of iron status biomarkers demonstrated that 1 SD higher genetically instrumented levels of iron status were associated with 10% lower risks of CAD but higher risks of T2D.
This is the first study to assess both the shape and strength of associations of hemoglobin with CMD, in addition to assessing possible heterogeneity by sex using both observational and genetic analyses. Previous observational studies of hemoglobin concentrations with disease outcomes in diverse populations have reported conflicting associations with CVD, 4 , 5 , 6 , 7 , 8 , 10 , 11 dementia, 12 and all‐cause mortality. 5 , 9 The Korean Heart Study involving 407 858 participants, 8 reported that men with lower or higher hemoglobin levels had higher risks of total CVD, coronary heart disease, and stroke, and higher risk of total CVD related to higher hemoglobin only in women. The REGARDS (Reasons for Geographical and Racial Differences in Stroke) study of 16 332 participants 4 reported higher risks of coronary heart disease in both the lowest and highest quintiles of hemoglobin without any differential effects by sex, whereas in a separate report by Panwar et al, 6 there was a U‐shaped association of hemoglobin with incident stroke in women only. Among the few studies that explored possible causality, Wang et al 29 reported that higher genetically determined hemoglobin levels were associated with higher risks of CAD, but they did not assess effects on other diseases. In the present study, higher genetically determined hemoglobin levels were associated with higher risks of CAD and T2D but not with HF and IS. Moreover, the present study reported heterogeneity in hemoglobin‐CAD associations, by sex (P interaction <0.05, male only). The genetic associations of hemoglobin with CVD outcomes were more likely to be linear than the observational analyses, perhaps reflecting incomplete control of confounding or reverse causality due to smoking, kidney function, chronic obstructive pulmonary disease, and other clinical or subclinical diseases in the observational analyses. 59 , 60 , 61 , 62 , 63 , 64 Consistent with previous studies, the present study suggested additional potential targets for higher genetically predicted hemoglobin, including hypertension 59 , 60 , 61 , 62 and liver disease, 63 , 64 but these associations will require further investigation.
In addition to hemoglobin concentrations, the present study also evaluated the causal relevance and strength of associations of iron status biomarkers with CAD, stroke, T2D, and HF. A meta‐analysis of 17 studies 19 conducted before 2014, involving 9236 cases of coronary heart disease and 156 427 participants, demonstrated an inverse association of TSAT and coronary heart disease, and no such associations were found for serum iron, ferritin, and TIBC. Moreover, previous MR studies reported that higher genetically predicted iron status, instrumented by 3 loci obtained from the Genetics of Iron Status consortium involving 48 972 participants, 65 were associated with lower risk for CAD 66 , 67 and higher risk for stroke 68 and T2D. 69 Using the latest summary results of genetic studies that yielded 62 independent variants associated with iron status biomarkers at 56 loci with sample sizes ranging from 131 471 to 246 139 participants, 47 the present study demonstrated modest protective effects of higher iron status for CAD, possible hazards for T2D, but no evidence of any associations with HF or IS.
Strengths and Limitations
This MR study assessed a broad panel of laboratory biomarkers of iron status (because no single biomarker provides all relevant information 27 , 28 ) to derive optimum instrumental variables for MR analyses. Bidirectional MR analyses provided support for the causal relevance of higher levels of hemoglobin with iron status biomarkers (TSAT, serum iron, ferritin, and TIBC). The study demonstrated the protective effects of higher iron biomarkers for CAD but adverse effects of higher hemoglobin on CAD. The reasons for these discrepant results are not fully understood but may reflect the effects of iron status markers on established CVD risk factors or direct effects of iron independent of hemoglobin. The analyses used linear and nonlinear MR methods 30 to assess the shape and strength of the causal associations of hemoglobin with CAD and T2D in addition to PheWAS analyses 43 to identify other possible targets of altered hemoglobin levels. The use of 2‐sample MR analyses involving worldwide genetic consortia enhanced the statistical power of the present study when compared with previous MR studies. 70 , 71 , 72 , 73 , 74 , 75 , 76 , 77 , 78 However, the present study also had several limitations. The use of iron supplements was evaluated by self‐reported questionnaires, which may have resulted in some misclassification of iron use and possible bias in the results. Randomized trials are required to assess the effects of iron supplements on CVD in high‐risk individuals (with stratification for use of multivitamins with or without added minerals). The detailed phenotyping data available in UKB enabled the assessment of associations of genetically predicted hemoglobin with multiple diseases in PheWAS analyses, but this could have introduced some misclassification bias due to the differential case ascertainment derived from multiple sources. However, access to the large sample size in UKB, limiting PheWAS to a minimum of 200 cases, 45 and correction for multiple testing should have minimized the effects of chance findings. MR analyses could also be biased if there was pleiotropy where variants influenced disease outcomes through pathways other than those mediated by iron biomarkers. 79 However, results of sensitivity analyses using other statistical methods and different genetic instruments yielded concordant results with the main analyses. Hence, it is unlikely that pleiotropy could have influenced our results. MR studies using genetic instruments approximate the average effects over the life course, so the strength and shape of the associations may not be comparable with the short‐term effects of iron supplements on disease outcomes. 79 To minimize the risk of possible bias due to population stratification, the present genetic analyses were restricted to participants of White‐British descent. Although this limits the application of the findings for other ancestry populations, it does not affect the internal validity of the present study findings.
CONCLUSIONS
This MR study demonstrated modest protective effects of higher levels of iron status biomarkers for CAD, possible hazards for T2D, but no evidence for any associations with HF or IS. The discrepant results of protective effects of higher iron status biomarkers for CAD, but adverse effects of higher hemoglobin, are unexplained, raising questions about the likely effects of iron supplements for the prevention of CAD. Large‐scale randomized trials are required to assess the efficacy and safety of iron supplements for the prevention of CAD in high‐risk older adults, but trials of treatments with support from MR studies are much more likely to have a successful outcome.
Sources of Funding
R.C. and D.A.B. were supported by the British Heart Foundation and the Medical Research Council (United Kingdom), and W.G. was supported by Novo Nordisk.
Disclosures
None.
Supporting information
Tables S1–S8
Figures S1–S7
This article was sent to Jacquelyn Y. Taylor, PhD, PNP‐BC, RN, FAHA, FAAN, Guest Editor, for review by expert referees, editorial decision, and final disposition.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.123.031732
For Sources of Funding and Disclosures, see page 11.
References
- 1. Pasricha SR, Tye‐Din J, Muckenthaler MU, Swinkels DW. Iron deficiency. Lancet. 2021;397:233–248. doi: 10.1016/S0140-6736(20)32594-0 [DOI] [PubMed] [Google Scholar]
- 2. Bulaj ZJ, Ajioka RS, Phillips JD, LaSalle BA, Jorde LB, Griffen LM, Edwards CQ, Kushner JP. Disease‐related conditions in relatives of patients with hemochromatosis. N Engl J Med. 2000;343:1529–1535. doi: 10.1056/NEJM200011233432104 [DOI] [PubMed] [Google Scholar]
- 3. Fleming RE, Ponka P. Iron overload in human disease. N Engl J Med. 2012;366:348–359. doi: 10.1056/NEJMra1004967 [DOI] [PubMed] [Google Scholar]
- 4. Houghton DE, Koh I, Ellis A, Key NS, Douce DR, Howard G, Cushman M, Safford M, Zakai NA. Hemoglobin levels and coronary heart disease risk by age, race, and sex in the reasons for geographic and racial differences in stroke study (REGARDS). Am J Hematol. 2020;95:258–266. doi: 10.1002/ajh.25703 [DOI] [PubMed] [Google Scholar]
- 5. Lee G, Choi S, Kim K, Yun JM, Son JS, Jeong SM, Kim SM, Park SM. Association of hemoglobin concentration and its change with cardiovascular and all‐cause mortality. J Am Heart Assoc. 2018;7:007723. doi: 10.1161/JAHA.117.007723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Panwar B, Judd SE, Warnock DG, McClellan WM, Booth JN III, Muntner P, Gutierrez OM. Hemoglobin concentration and risk of incident stroke in community‐living adults. Stroke. 2016;47:2017–2024. doi: 10.1161/STROKEAHA.116.013077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gotoh S, Hata J, Ninomiya T, Hirakawa Y, Nagata M, Mukai N, Fukuhara M, Ikeda F, Ago T, Kitazono T, et al. Hematocrit and the risk of cardiovascular disease in a Japanese community: the Hisayama study. Atherosclerosis. 2015;242:199–204. doi: 10.1016/j.atherosclerosis.2015.07.014 [DOI] [PubMed] [Google Scholar]
- 8. Kim MY, Jee SH, Yun JE, Baek SJ, Lee DC. Hemoglobin concentration and risk of cardiovascular disease in Korean men and women ‐ the Korean heart study. J Korean Med Sci. 2013;28:1316–1322. doi: 10.3346/jkms.2013.28.9.1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zakai NA, Katz R, Hirsch C, Shlipak MG, Chaves PH, Newman AB, Cushman M. A prospective study of anemia status, hemoglobin concentration, and mortality in an elderly cohort: the cardiovascular health study. Arch Intern Med. 2005;165:2214–2220. doi: 10.1001/archinte.165.19.2214 [DOI] [PubMed] [Google Scholar]
- 10. Sarnak MJ, Tighiouart H, Manjunath G, MacLeod B, Griffith J, Salem D, Levey AS. Anemia as a risk factor for cardiovascular disease in the atherosclerosis risk in communities (ARIC) study. J Am Coll Cardiol. 2002;40:27–33. doi: 10.1016/S0735-1097(02)01938-1 [DOI] [PubMed] [Google Scholar]
- 11. Gagnon DR, Zhang TJ, Brand FN, Kannel WB. Hematocrit and the risk of cardiovascular disease the Framingham study: a 34‐year follow‐up. Am Heart J. 1994;127:674–682. doi: 10.1016/0002-8703(94)90679-3 [DOI] [PubMed] [Google Scholar]
- 12. Wolters FJ, Zonneveld HI, Licher S, Cremers LGM; Heart Brain Connection Collaborative Research G , Ikram MK, Koudstaal PJ, Vernooij MW, Ikram MA. Hemoglobin and anemia in relation to dementia risk and accompanying changes on brain MRI. Neurology. 2019;93:e917–e926. doi: 10.1212/WNL.0000000000008003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu J, Li Q, Yang Y, Ma L. Iron metabolism and type 2 diabetes mellitus: a meta‐analysis and systematic review. J Diabetes Investig. 2020;11:946–955. doi: 10.1111/jdi.13216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diaz‐Lopez A, Iglesias‐Vazquez L, Palleja‐Millan M, Rey Renones C, Flores Mateo G, Arija V. Association between iron status and incident type 2 diabetes: a population‐based cohort study. Nutrients. 2020;12:3249. doi: 10.3390/nu12113249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang L, Wang K, Lo K, Zhong Y, Yang A, Fang X, Akezhuoli H, Song Z, Chen L, An P, et al. Sex‐specific association of circulating ferritin level and risk of type 2 diabetes: a dose‐response meta‐analysis of prospective studies. J Clin Endocrinol Metab. 2019;104:4539–4551. doi: 10.1210/jc.2019-00495 [DOI] [PubMed] [Google Scholar]
- 16. Silvestre OM, Goncalves A, Nadruz W Jr, Claggett B, Couper D, Eckfeldt JH, Pankow JS, Anker SD, Solomon SD. Ferritin levels and risk of heart failure‐the atherosclerosis risk in communities study. Eur J Heart Fail. 2017;19:340–347. doi: 10.1002/ejhf.701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klip IT, Voors AA, Swinkels DW, Bakker SJ, Kootstra‐Ros JE, Lam CS, van der Harst P, van Veldhuisen DJ, van der Meer P. Serum ferritin and risk for new‐onset heart failure and cardiovascular events in the community. Eur J Heart Fail. 2017;19:348–356. doi: 10.1002/ejhf.622 [DOI] [PubMed] [Google Scholar]
- 18. Podmore C, Meidtner K, Schulze MB, Scott RA, Ramond A, Butterworth AS, Di Angelantonio E, Danesh J, Arriola L, Barricarte A, et al. Association of multiple biomarkers of iron metabolism and type 2 diabetes: the EPIC‐InterAct study. Diabetes Care. 2016;39:572–581. doi: 10.2337/dc15-0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Das De S, Krishna S, Jethwa A. Iron status and its association with coronary heart disease: systematic review and meta‐analysis of prospective studies. Atherosclerosis. 2015;238:296–303. doi: 10.1016/j.atherosclerosis.2014.12.018 [DOI] [PubMed] [Google Scholar]
- 20. Savarese G, von Haehling S, Butler J, Cleland JGF, Ponikowski P, Anker SD. Iron deficiency and cardiovascular disease. Eur Heart J. 2023;44:14–27. doi: 10.1093/eurheartj/ehac569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalra PR, Cleland JGF, Petrie MC, Thomson EA, Kalra PA, Squire IB, Ahmed FZ, Al‐Mohammad A, Cowburn PJ, Foley PWX, et al. Intravenous ferric derisomaltose in patients with heart failure and iron deficiency in the UK (IRONMAN): an investigator‐initiated, prospective, randomised, open‐label, blinded‐endpoint trial. Lancet. 2022;400:2199–2209. doi: 10.1016/S0140-6736(22)02083-9 [DOI] [PubMed] [Google Scholar]
- 22. Docherty KF, Welsh P, Verma S, De Boer RA, O'Meara E, Bengtsson O, Kober L, Kosiborod MN, Hammarstedt A, Langkilde AM, et al. Investigators D‐H and committees. Iron deficiency in heart failure and effect of dapagliflozin: findings from DAPA‐HF. Circulation. 2022;146:980–994. doi: 10.1161/CIRCULATIONAHA.122.060511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Butler J, Khan MS, Friede T, Jankowska EA, Fabien V, Goehring UM, Dorigotti F, Metra M, Pina IL, Coats AJS, et al. Health status improvement with ferric carboxymaltose in heart failure with reduced ejection fraction and iron deficiency. Eur J Heart Fail. 2022;24:821–832. doi: 10.1002/ejhf.2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anker SD, Ponikowski P, Khan MS, Friede T, Jankowska EA, Fabien V, Goehring UM, Metra M, Pina IL, Coats AJS, et al. Responder analysis for improvement in 6‐min walk test with ferric carboxymaltose in patients with heart failure with reduced ejection fraction and iron deficiency. Eur J Heart Fail. 2022;24:833–842. doi: 10.1002/ejhf.2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Camaschella C. Iron‐deficiency anemia. N Engl J Med. 2015;372:1832–1843. doi: 10.1056/NEJMra1401038 [DOI] [PubMed] [Google Scholar]
- 26. Lopez A, Cacoub P, Macdougall IC, Peyrin‐Biroulet L. Iron deficiency anaemia. Lancet. 2016;387:907–916. doi: 10.1016/S0140-6736(15)60865-0 [DOI] [PubMed] [Google Scholar]
- 27. Restrepo‐Gallego M, Diaz LE, Rondo PHC. Classic and emergent indicators for the assessment of human iron status. Crit Rev Food Sci Nutr. 2021;61:2827–2840. doi: 10.1080/10408398.2020.1787326 [DOI] [PubMed] [Google Scholar]
- 28. Wish JB. Assessing iron status: beyond serum ferritin and transferrin saturation. Clin J Am Soc Nephrol. 2006;1(suppl 1):S4–S8. doi: 10.2215/CJN.01490506 [DOI] [PubMed] [Google Scholar]
- 29. Wang K, Shi X, Zhu Z, Hao X, Chen L, Cheng S, Foo RSY, Wang C. Mendelian randomization analysis of 37 clinical factors and coronary artery disease in east Asian and European populations. Genome Med. 2022;14:63. doi: 10.1186/s13073-022-01067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Staley JR, Burgess S. Semiparametric methods for estimation of a nonlinear exposure‐outcome relationship using instrumental variables with application to Mendelian randomization. Genet Epidemiol. 2017;41:341–352. doi: 10.1002/gepi.22041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burgess S, Butterworth A, Malarstig A, Thompson SG. Use of Mendelian randomisation to assess potential benefit of clinical intervention. BMJ. 2012;345:e7325. doi: 10.1136/bmj.e7325 [DOI] [PubMed] [Google Scholar]
- 32. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J, et al. The UK biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Littlejohns TJ, Sudlow C, Allen NE, Collins R. UK biobank: opportunities for cardiovascular research. Eur Heart J. 2019;40:1158–1166. doi: 10.1093/eurheartj/ehx254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oskarsson GR, Oddsson A, Magnusson MK, Kristjansson RP, Halldorsson GH, Ferkingstad E, Zink F, Helgadottir A, Ivarsdottir EV, Arnadottir GA, et al. Predicted loss and gain of function mutations in ACO1 are associated with erythropoiesis. Commun Biol. 2020;3:189. doi: 10.1038/s42003-020-0921-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takeuchi H, Kawashima R. A prospective study on the relationship between iron supplement intake, hemoglobin concentration, and risk of parkinsonism. Nutrients. 2022;14:4671–4684. doi: 10.3390/nu14214671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tyrrell J, Jones SE, Beaumont R, Astley CM, Lovell R, Yaghootkar H, Tuke M, Ruth KS, Freathy RM, Hirschhorn JN, et al. Height, body mass index, and socioeconomic status: mendelian randomisation study in UK biobank. BMJ. 2016;352:i582. doi: 10.1136/bmj.i582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Desquilbet L, Mariotti F. Dose‐response analyses using restricted cubic spline functions in public health research. Stat Med. 2010;29:1037–1057. doi: 10.1002/sim.3841 [DOI] [PubMed] [Google Scholar]
- 39. Gan W, Bragg F, Walters RG, Millwood IY, Lin K, Chen Y, Guo Y, Vaucher J, Bian Z, Bennett D, et al. Genetic predisposition to type 2 diabetes and risk of subclinical atherosclerosis and cardiovascular diseases among 160,000 Chinese adults. Diabetes. 2019;68:2155–2164. doi: 10.2337/db19-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vuckovic D, Bao EL, Akbari P, Lareau CA, Mousas A, Jiang T, Chen MH, Raffield LM, Tardaguila M, Huffman JE, et al. The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182(1214–1231):e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros‐Mckay F, Kostadima MA, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burgess S, Davies NM, Thompson SG and Consortium EP‐I . Instrumental variable analysis with a nonlinear exposure‐outcome relationship. Epidemiology. 2014;25:877–885. doi: 10.1097/EDE.0000000000000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown‐Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome‐wide scan to discover gene‐disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Millard LAC, Davies NM, Gaunt TR, Davey Smith G, Tilling K. Software application profile: PHESANT: a tool for performing automated phenome scans in UK biobank. Int J Epidemiol. 2018;47:29–35. doi: 10.1093/ije/dyx204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Verma A, Bradford Y, Dudek S, Lucas AM, Verma SS, Pendergrass SA, Ritchie MD. A simulation study investigating power estimates in phenome‐wide association studies. BMC Bioinformatics. 2018;19:120. doi: 10.1186/s12859-018-2135-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benjamini Y, Hochberg Y. Controlling the false discovery rate ‐ a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 47. Bell S, Rigas AS, Magnusson MK, Ferkingstad E, Allara E, Bjornsdottir G, Ramond A, Sorensen E, Halldorsson GH, Paul DS, et al. A genome‐wide meta‐analysis yields 46 new loci associating with biomarkers of iron homeostasis. Commun Biol. 2021;4:156. doi: 10.1038/s42003-020-01575-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913 [DOI] [PubMed] [Google Scholar]
- 49. Shah S, Henry A, Roselli C, Lin H, Sveinbjornsson G, Fatemifar G, Hedman AK, Wilk JB, Morley MP, Chaffin MD, et al. Genome‐wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun. 2020;11:163. doi: 10.1038/s41467-019-13690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten‐Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, et al. Multiancestry genome‐wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, et al. Fine‐mapping type 2 diabetes loci to single‐variant resolution using high‐density imputation and islet‐specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bulik‐Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen C, Psychiatric Genomics C; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control C , Duncan L, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bulik‐Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J; Schizophrenia Working Group of the Psychiatric Genomics C , Patterson N, Daly MJ, Price AL, Neale BM. LD score regression distinguishes confounding from polygenicity in genome‐wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Finucane HK, Bulik‐Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, et al. Partitioning heritability by functional annotation using genome‐wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–1998. doi: 10.1093/ije/dyx102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR‐base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Timoteo VJ, Chiang KM, Pan WH. Positive or U‐shaped association of elevated hemoglobin concentration levels with metabolic syndrome and metabolic components: findings from Taiwan biobank and UK biobank. Nutrients. 2022;14:4007. doi: 10.3390/nu14194007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tapio J, Vahanikkila H, Kesaniemi YA, Ukkola O, Koivunen P. Higher hemoglobin levels are an independent risk factor for adverse metabolism and higher mortality in a 20‐year follow‐up. Sci Rep. 2021;11:19936. doi: 10.1038/s41598-021-99217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lim WH, Choi EK, Han KD, Lee SR, Cha MJ, Oh S. Impact of hemoglobin levels and their dynamic changes on the risk of atrial fibrillation: a nationwide population‐based study. Sci Rep. 2020;10:6762. doi: 10.1038/s41598-020-63878-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou J, Zhao R, Wang D, Gao Q, Zhao D, Ouyang B, Hao L, Peng X. Sex‐specific association between iron status and the predicted 10‐year risk for atherosclerotic cardiovascular disease in hypertensive patients. Biol Trace Elem Res. 2022;200:4594–4607. doi: 10.1007/s12011-021-03060-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scheiner B, Semmler G, Maurer F, Schwabl P, Bucsics TA, Paternostro R, Bauer D, Simbrunner B, Trauner M, Mandorfer M, et al. Prevalence of and risk factors for anaemia in patients with advanced chronic liver disease. Liver Int. 2020;40:194–204. doi: 10.1111/liv.14229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Singh S, Manrai M, Parvathi VS, Kumar D, Srivastava S, Pathak B. Association of liver cirrhosis severity with anemia: does it matter? Ann Gastroenterol. 2020;33:272–276. doi: 10.20524/aog.2020.0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, Gogele M, Anderson D, Broer L, Podmore C, et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2014;5:4926. doi: 10.1038/ncomms5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grote Beverborg N, Said MA, van der Wal HH, Verweij N, van der Meer P, van der Harst P. Genetically determined high levels of iron parameters are protective for coronary artery disease. Circ Genom Precis Med. 2020;13:e002544. doi: 10.1161/CIRCGEN.119.002544 [DOI] [PubMed] [Google Scholar]
- 67. Gill D, Del Greco MF, Walker AP, Srai SKS, Laffan MA, Minelli C. The effect of iron status on risk of coronary artery disease: a Mendelian randomization study‐brief report. Arterioscler Thromb Vasc Biol. 2017;37:1788–1792. doi: 10.1161/ATVBAHA.117.309757 [DOI] [PubMed] [Google Scholar]
- 68. Gill D, Monori G, Tzoulaki I, Dehghan A. Iron status and risk of stroke. Stroke. 2018;49:2815–2821. doi: 10.1161/STROKEAHA.118.022701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang X, Fang X, Zheng W, Zhou J, Song Z, Xu M, Min J, Wang F. Genetic support of a causal relationship between iron status and type 2 diabetes: a Mendelian randomization study. J Clin Endocrinol Metab. 2021;106:e4641–e4651. doi: 10.1210/clinem/dgab454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grammer TB, Kleber ME, Silbernagel G, Pilz S, Scharnagl H, Tomaschitz A, Konig W, Marz W. Hemoglobin, iron metabolism and angiographic coronary artery disease (the Ludwigshafen Risk and Cardiovascular Health study). Atherosclerosis. 2014;236:292–300. doi: 10.1016/j.atherosclerosis.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 71. Grammer TB, Scharnagl H, Dressel A, Kleber ME, Silbernagel G, Pilz S, Tomaschitz A, Koenig W, Mueller‐Myhsok B, Marz W, et al. Iron metabolism, hepcidin, and mortality (the Ludwigshafen risk and cardiovascular health study). Clin Chem. 2019;65:849–861. doi: 10.1373/clinchem.2018.297242 [DOI] [PubMed] [Google Scholar]
- 72. Liu D, Zhang Y, Wang CC, Xiao‐Hong E, Zuo H. Markers of iron metabolism and stroke risk: cross‐sectional and longitudinal findings from the China Health and Nutrition Survey (CHNS). Iran J Public Health. 2022;51:115–123. doi: 10.18502/ijph.v51i1.8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Del Greco MF, Foco L, Pichler I, Eller P, Eller K, Benyamin B, Whitfield JB; Genetics of Iron Status C, Consortium CK , Pramstaller PP, Thompson JR. Serum iron level and kidney function: a Mendelian randomization study. Nephrol Dial Transplant. 2017;32:273–278. [DOI] [PubMed] [Google Scholar]
- 74. Gill D, Benyamin B, Moore LSP, Monori G, Zhou A, Koskeridis F, Evangelou E, Laffan M, Walker AP, Tsilidis KK, et al. Associations of genetically determined iron status across the phenome: a mendelian randomization study. PLoS Med. 2019;16:e1002833. doi: 10.1371/journal.pmed.1002833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gill D, Brewer CF, Monori G, Tregouet DA, Franceschini N, Giambartolomei C, Consortium I, Tzoulaki I, Dehghan A. Effects of genetically determined iron status on risk of venous thromboembolism and carotid atherosclerotic disease: a Mendelian randomization study. J Am Heart Assoc. 2019;8:e012994. doi: 10.1161/JAHA.119.012994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang F, Bao Q, Wang Z, Ma M, Shen J, Ye F, Xie X. Sex‐specific genetically predicted iron status in relation to 12 vascular diseases: a Mendelian randomization study in the UK biobank. Biomed Res Int. 2020;2020:6246041. doi: 10.1155/2020/6246041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhou J, Liu C, Francis M, Sun Y, Ryu MS, Grider A, Ye K. The causal effects of blood iron and copper on lipid metabolism diseases: evidence from phenome‐wide Mendelian randomization study. Nutrients. 2020;12:3174. doi: 10.3390/nu12103174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang T, Cheng J, Wang Y. Genetic support of a causal relationship between iron status and atrial fibrillation: a Mendelian randomization study. Genes Nutr. 2022;17:8. doi: 10.1186/s12263-022-00708-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Davies NM, Holmes MV, Davey SG. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S8
Figures S1–S7


