Abstract
SF162 is a primary (PR), non-syncytium-inducing, macrophagetropic human immunodeficiency virus type 1 (HIV-1) clade B isolate which is resistant to antibody-mediated neutralization. Deletion of the first or second hypervariable envelope gp120 region (V1 or V2 loop, respectively) of this virus does not abrogate its ability to replicate in peripheral blood mononuclear cells and primary macrophages, nor does it alter its coreceptor usage profile. The mutant virus with the V1 loop deletion, SF162ΔV1, remains as resistant to antibody-mediated neutralization as the wild-type virus SF162. In contrast, the mutant virus with the V2 loop deletion, SF162ΔV2, exhibits enhanced susceptibility to neutralization by certain monoclonal antibodies whose epitopes are located within the CD4-binding site and conserved regions of gp120. More importantly, SF162ΔV2 is now up to 170-fold more susceptible to neutralization than SF162 by sera collected from patients infected with clade B HIV-1 isolates. In addition, it becomes susceptible to neutralization by sera collected from patients infected with clade A, C, D, E, and F HIV-1 isolates. These findings suggest that the V2, but not the V1, loop of SF162 shields an as yet unidentified region of the HIV envelope rich in neutralization epitopes and that the overall structure of this region appears to be conserved among clade B, C, D, E, and F HIV-1 PR isolates.
Several studies have reported on the differential susceptibility to antibody-mediated neutralization between primary (PR) and laboratory-adapted (LA) human immunodeficiency virus type 1 (HIV-1) isolates (1, 2, 6, 23, 24, 32, 39, 40, 45), the latter being more susceptible than the former to neutralization. To a large extent, this difference is explained by a documented differential exposure of neutralization epitopes within oligomeric envelope structures of PR and LA isolates. Such epitopes are less exposed on PR isolates than LA isolates (1, 24, 36, 40). The identification of the regions that shield neutralization epitopes and the nature of the neutralization epitopes themselves on PR isolates should shed light on the mechanism(s) by which HIV evades neutralization by serum antibodies and greatly benefit the design of effective HIV envelope-based immunogens.
It has been reported that simultaneous deletion of the V1 and V2 loops of the neutralization-sensitive LA isolate HIV-1IIIb increases even further its susceptibility to neutralization by certain monoclonal antibodies (MAbs) whose epitopes are located within the V3 loop, the CD4-binding site, and conserved gp120 regions (3). We previously reported that the individual deletion of the V1 (17 amino acids) and V2 (30 amino acids) loops of the PR, neutralization-resistant HIV-1 isolate SF162 (7, 35, 39) does not reduce the potential of the virus to replicate in peripheral blood mononuclear cells (PBMC) or alter the coreceptor function of the envelope protein (38). Immunochemical analysis of the two mutant envelopes revealed that the deleted V1 and V2 loop sequences differentially affect the exposure and conformation of specific gp120 epitopes. While deletion of the V2 loop affects the structure of certain complex epitopes which comprise elements of the CD4-binding site, deletion of the V1 loop alters the conformation and exposure of specific epitopes located in the V2 and V3 loops and the CD4-binding site. However, regardless of these specific structural changes, neither deletion alters the overall ability of the envelope to interact with CD4 or its ability to undergo conformational changes upon receptor binding (38).
In this study, we examined whether individual partial deletion of the V1 or V2 loop exposes cryptic neutralization epitopes on the surface of SF162 virions. We show that the V2, but not the V1, loop deletion renders the virus, termed SF162ΔV2, highly susceptible to serum-mediated neutralization. Most importantly, SF162ΔV2 becomes neutralizable by sera obtained from patients infected not only with clade B HIV-1 isolates but also with clade A, C, D, E, and F HIV-1 isolates. The introduced V2 loop deletion does not destabilize the gp120-gp41 subunit association, and the virus remains as resistant to antibody-mediated gp120-virion dissociation as wild-type (wt) SF162. Based on these observations, we suggest that this particular V2 loop deletion unveils an envelope region rich in neutralization epitopes whose overall structure is conserved among PR isolates. Neutralization studies conducted with various anti-gp120 MAbs failed to identify the precise epitopes forming this region, but it appears that it comprises structural elements of the CD4-binding site and conserved regions of gp120.
MATERIALS AND METHODS
MAbs, sera, and cells.
The human anti-CD4-binding site MAb 654-30D, the anti-V3 loop human MAbs 391-95D and 257D, and the human anti-V2 loop MAb 697-30D were provided by S. Zolla-Pazner. Their epitope specificity was previously reported (12, 13, 20, 34). The murine anti-V2 loop MAbs G3.4 and G3.136 were provided by Tanox, and their epitopes were determined previously (11). The human MAb IgG1b12 was obtained from D. Burton (2, 27); the human MAbs 17b and 48D were provided by J. Robinson. The human immunoglobulin G-CD4 chimera (5) was generously provided by Genentech. Clade B (GS, GSO, 5223, DT, GS25, B, LSS, and LS-BO), clade C (93BU003 and QRS 1638), clade A (93RW035 and 2743M), clade E (93TH072), and clade D (94UG117 and QRS 1523) sera were obtained from D. D. Ho, Y. Cao, and J. A. Levy and through the AIDS Research and Reference Reagent, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The HIV-1 non-clade B plasma panel was contributed by the UNAIDS Network for HIV Isolation and Characterization, courtesy of Harvey Holmes.
PBMC were isolated from blood by Ficoll-Hypaque gradient centrifugation (6, 7) and were stimulated for 3 days with phytohemagglutinin (PHA; 3 μg/ml) prior to infection. Monocyte-derived macrophages were prepared by the plastic adherence method as previously described (8).
Generation of HIV-1SF162 mutant gp120 envelope molecules and viruses.
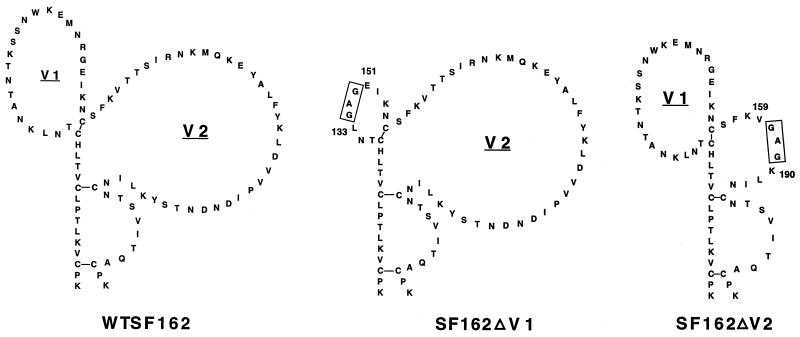
The isolation and phenotypic characterization of the infectious molecular clone of SF162 was reported previously (7). Generation of the SF162ΔV1 mutant envelope, containing a 17-amino-acid deletion within the V1 loop (comprising amino acids K134 to K150), and of the SF162ΔV2 mutant envelope, containing a 30-amino-acid deletion within the V2 loop (from T160 to Y189), and replacement of the deleted sequences by the Gly-Ala-Gly tripeptide (Fig. 1) are described elsewhere (38).
FIG. 1.
Amino acid sequences of the V1 and V2 loops of the wt SF162 and two mutant virus envelopes. Seventeen amino acids (134K to 150K) were removed in the case of SF162ΔV1, and 30 amino acids (160T to 189Y) were removed in the case of SF162ΔV2. Deleted sequences were replaced by the GAG tripeptide (boxed).
Infectious viruses were generated as described previously (7). Briefly equal amounts of the 5′ and 3′ portions of the viral genome were transfected in human 293-T cells by the calcium phosphate method. The supernatants were collected 72 h later and used to inoculate PHA-stimulated PBMC. Viral production was monitored by quantitating the p24 antigen concentration in the cell supernatant. At the peak of virus production, the supernatants were collected, aliquoted, and stored at −80°C. The 50% tissue culture infective dose (TCID50) of each stock was determined in parallel for all three viruses in PBMC.
Viral replication potential and tropism.
To assess virus replication, 10 TCID50 of each virus was added to 2 × 106 PBMC or 1.5 × 105 macrophages for 3 h at 37°C and then removed by aspiration and cell washing. Viral replication was monitored every 3 to 4 days by quantitating the p24 antigen concentration in the culture supernatant.
Neutralization studies. (i) Serum neutralization.
Neutralization experiments were performed in 96-well plates as previously described (6, 39). Sera from HIV-infected individuals were first heat inactivated (56°C, 30 min). Serially diluted sera (50 μl) were incubated in triplicate wells with 100 TCID50 of each isolate (50 μl) for 1 h at 37°C and then added to 100 μl of 4 × 106 PBMC/ml. Following an 18-h incubation at 37°C, the cells were washed three times with medium to remove residual serum anti-p24 antibodies and p24 antigens (4, 22, 43).
(ii) MAb neutralization.
The experiments were performed as described above except that the MAbs were present throughout the entire period of virus-cell incubation. The MAb concentrations indicated in the text refer to those present during the virus-MAb preincubation period.
In both serum and MAb neutralization experiments, the concentration of p24 antigen in the cell supernatant from each well was determined at the peak of viral replication of the control (monitored in parallel for each virus). The percent neutralization for a particular serum or MAb dilution was determined as (control − experimental/control) × 100, where control stands for the p24 antigen concentration in the absence of MAb or serum (0% neutralization) and experimental is the p24 concentration in wells where virus and cells were incubated in the presence of MAbs or serum. A neutralization curve is generated by plotting the percent neutralization versus the MAb concentration or serum dilution. The MAb concentrations and serum dilutions that result in 90 and 50% neutralization of infection are determined from this curve.
Antibody-mediated gp120-virion dissociation.
The binding of serum antibodies to virions and antibody-mediated gp120-virion dissociation were determined as previously reported, using enzyme-linked immunosorbent assays (ELISA) (36, 39). Briefly, to remove virions present in these sera which may lead to erroneous results in our assays, sera are first subjected to centrifugation under conditions that result in virion pelleting (12,500 × g, 2 h at 4°C). The supernatant from this centrifugation is then serially diluted and incubated with sucrose-purified SF162 and SF162ΔV2 virions (36, 44) for 3 h at 37°C. The virions, and the antibodies bound to them, are pelleted by centrifugation as above. Non-virion-associated gp120-antibody complexes that may have dissociated from the virion surface remain in the supernatant. The virion-associated gp120 molecules (present in the viral pellet) are captured onto wells (96-well plates) precoated with sheep polyclonal sera (D6205; International Enzymes) directed against the carboxy-terminal 15 amino acids of gp120. The relative quantity of serum antibodies bound to these gp120 molecules is determined following the addition of goat anti-human antibodies coupled to alkaline phosphatase (Zymed), by recording the optical density at 490 nm (OD490) (36, 37, 39). In parallel, the supernatant of the viral pellet, which may contain gp120 molecules that became dissociated from the virion surface upon antibody binding, are added to separate D6205-coated wells. The amount of serum antibodies bound to such gp120 molecules is determined as described above. The sum of the OD490 signals recorded from the pellet and the supernatant represents the total amount of serum antibodies bound to virions during the 3-h incubation period of viruses with anti-HIV sera. By dividing the OD490 signal recorded from the supernatant by the sum of the OD490 signals of the pellet and supernatant, we determine the percentage of gp120 molecules that became dissociated from the virion surface upon antibody binding. Spontaneous gp120-virion dissociation, i.e., in the absence of antibody-virion binding, is also quantified by incubating virions in the absence of anti-HIV sera for 3 h at 37°C and quantitating (as described above) the proportion of gp120 molecules that become dissociated from the viral surface during this period.
The sera used in our studies were collected from HIV-infected individuals and thus may already have contained soluble gp120-antibody complexes. These complexes can also be captured to D6205-coated wells and thus contribute to the OD490 signal recorded in our ELISA. To control for such an eventuality, serially diluted sera were directly added to D6205-coated wells. For each serum dilution, the OD490 signal recorded in this way was subtracted from the OD490 signal obtained from wells containing the supernatant of the virion pellet.
RESULTS
Deletion of the V1 or V2 loop of the SF162 envelope does not affect envelope function of the mutant viruses.
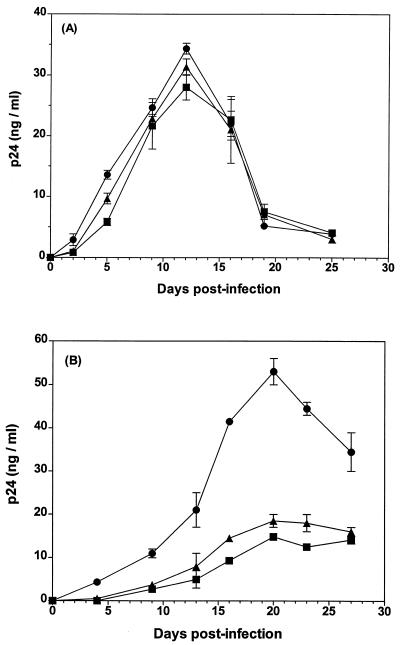
The amino acid sequence of the V1-V2 region of each of the three isolates is shown in Fig. 1. The functionality of the mutated envelopes was evaluated by comparing the replicative potential of mutants SF162ΔV1 and SF162ΔV2 to that of the wt virus, SF162. Both mutant viruses replicate to similar high titers and with similar replication kinetics as SF162 in PBMC (38) (Fig. 2A), indicating that the introduced deletions do not affect the potential of the envelope to mediate virus-PBMC entry. The two deletion mutants also replicated in macrophages, but more slowly and to lower titers than SF162 (Fig. 2B).
FIG. 2.
Replication in PBMC and macrophages. PHA-stimulated PBMC (A) and macrophages (B) were inoculated with 10 TCID50 of SF162 (•), SF162ΔV1 (▴), or SF162ΔV2 (■). The inoculum was removed 3 h later. Viral replication was followed over time by measuring the p24 concentration in the culture medium at the indicated days postinfection. Data represent the mean and standard deviation from three infections.
Deletion of the V2, but not the V1, loop renders the virus more susceptible to neutralization by MAbs directed against conserved CD4-binding site epitopes.
It has been reported that the simultaneous deletion of the V1 and V2 loops of the neutralization-sensitive LA isolate HIV-1 IIIb increases its susceptibility to neutralization by anti-V3 loop and anti-CD4-binding-site MAbs (3). It was of interest, therefore, to determine whether deletion of the V1 or V2 loop of a PR, neutralization-resistant HIV-1 isolate also alters its susceptibility to neutralization by similar MAbs. We observed that SF162ΔV1 displayed the same pattern of MAb-mediated neutralization susceptibility as SF162 (Table 1). That is, it was highly susceptible to neutralization by MAbs IgGCD4 and IgG1b12 and resistant to neutralization by the anti-V3 loop MAbs 391-95D and 257D, the anti-V2 loop MAbs G3.4, G3.136, and 697-30D, the CD4-induced MAbs 17b and 48d, and the anti-CD4-binding-site MAb 654-30D. In contrast, the SF162ΔV2 isolate was highly susceptible to neutralization by MAb 654-30D (90% neutralization at 5 μg/ml) and to a lesser extent, susceptible to neutralization by MAbs 17b and 48d (50% neutralization at 7 and 10 μg/ml, respectively). At the same time, SF162ΔV2 became less susceptible than SF162 to neutralization by MAb IgG1b12, most likely because this MAb recognizes a complex epitope comprising structural elements of the CD4-binding site and of the V2 loop (9, 21), and deletion of the V2 loop decreases the binding of this MAb to the virion surface (38). SF162ΔV2 remained resistant to neutralization by both anti-V3 loop MAbs tested. All three isolates were similarly susceptible to neutralization by several anti-gp41 MAbs tested (data not shown).
TABLE 1.
Neutralization by anti-gp120 MAbs
| MAb (highest concn [μg/ml] tested) | Epitope | MAb concn (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|---|
| SF162
|
SF162ΔV1
|
SF162ΔV2
|
|||||
| 90% | 50% | 90% | 50% | 90% | 50% | ||
| 391-95D (10) | V3 loop | — | — | — | — | — | — |
| 257D (10) | V3 loop | — | — | — | — | — | — |
| G3.4 (20) | V2 loop | — | — | — | — | — | — |
| G3.136 (20) | V2 loop | — | — | — | — | — | — |
| 697-30D (20) | V2 loop | — | — | — | — | — | — |
| IgGCD4 (20) | CD4-binding site | 5 (2) | 0.5 (0.3) | 5 (2) | 0.25 (0.2) | 4 (2) | 0.1 (0.05) |
| 654.30D (10) | CD4-binding site | — | — | — | — | 5 (1) | 1.5 (1) |
| IgG1b12 (20) | Complex | 1 (0.7) | 0.1 (0.05) | 2 (1) | 0.1 (0.05) | — | 10 (5) |
| 17b (20) | Conserved | — | — | — | — | — | 7 (4) |
| 48d (20) | Conserved | — | — | — | — | — | 10 (3) |
Mean concentration required to inhibit by 90 or 50% the infection of PBMC by the three viruses. Values in parentheses represent the standard deviation from three to five independent neutralization experiments. —, neutralization not observed with highest MAb concentration used.
SF162ΔV2 is highly susceptible to neutralization by polyclonal clade B antisera.
Since a modest increase in neutralization susceptibility by anti-CD4-binding-site and anti-CD4-induced MAbs is observed upon V2 loop deletion, we next examined whether the SF162ΔV2 virus is also more susceptible to neutralization by polyclonal anti-HIV sera. Sera obtained from seven individuals infected with clade B HIV-1 isolates were tested. Sera LSS and LS-BO were obtained from patients classified as long-term nonprogressors, and the remaining sera were from patients at various stages of infection (4, 6). Sera GJ, GSO, DT, 5223, and GS25 were collected early during the AIDS epidemic (in the mid-1980s), while LSS and LS-BO were collected later (in the early 1990s). Regardless of the time of collection and the state of the disease of the patient, we found that SF162ΔV2 was significantly (up to 170-fold) more susceptible to neutralization than SF162 by all seven sera tested (Table 2). Similar results were obtained when neutralization experiments were performed with 300 instead of 100 TCID50 of SF162ΔV2 (data not shown). Furthermore, the difference in neutralization susceptibility between SF162 and SF162ΔV2 was the same when determined at 7, 10, or 15 days postinfection. In contrast, SF162ΔV1 was moderately more susceptible to neutralization than SF162 by sera GS25 and GJ but was comparable to SF162 in neutralization sensitivity by the remaining sera.
TABLE 2.
Neutralization by clade B HIV-positive sera
| Serum | Titera
|
Fold increaseb | ||
|---|---|---|---|---|
| SF162 | SF162ΔV1 | SF162ΔV2 | ||
| GJ | 100 | 700 | 5,000 | 50 |
| DT | 100 | 100 | 1,000 | 10 |
| 5223 | 200 | 216 | 1,222 | 6 |
| GS0 | 50 | 75 | 4,750 | 95 |
| GS25 | 233 | 666 | 2,000 | 8.5 |
| LSS | 50 | NDc | 2,222 | 45 |
| LS-BO | 100 | ND | 17,000 | 170 |
Reciprocal of serum dilution that resulted in 90% inhibition of PBMC infection. Neutralization experiments were performed as described in Materials and Methods. The lowest serum dilution tested was 1:50, and the highest was 1:40,000.
Increase in neutralization susceptibility by the partial deletion of the V2 loop at 90% neutralization.
ND, not determined.
SF162ΔV2 is also highly susceptible to cross-clade neutralization.
To assess the breadth of neutralization sensitivity of SF162ΔV2, we compared its susceptibility to neutralization by sera obtained from individuals infected with clade A, C, D, E, and F HIV-1 isolates with that of SF162 (Table 3). As expected, the wt virus SF162 is resistant to neutralization by these sera (90% neutralization was not achieved at the lowest serum dilution, 1:50, tested). In contrast, the introduced V2 loop deletion renders the virus highly susceptible to neutralization by all non-clade B sera tested. Thus, 90% neutralization can now be achieved at serum dilutions of between 1:100 and 1:1,000.
TABLE 3.
Neutralization by non-clade B HIV-positive sera
| Serum (clade) | Titera
|
||
|---|---|---|---|
| SF162, 50% | SF162ΔV2
|
||
| 90% | 50% | ||
| 93RW035 (A) | 100 | 100 | 333 |
| 2743M (A) | 66 | 1,000 | 5,000 |
| 91BU003 (C) | 50 | 100 | 1,000 |
| QRS1638 (C) | —b | 100 | 500 |
| 94UG117 (D) | — | 100 | 1,500 |
| QRS1523 (D) | 50 | 100 | 222 |
| 93TH072 (E) | 100 | 100 | 500 |
| 93BR029 (F) | 142 | 500 | 5,000 |
Reciprocal of serum dilution that resulted in 90 and 50% inhibition of PBMC infection. Neutralization experiments were performed as described in Materials and Methods. The lowest serum dilution tested was 1:50, and the highest was 1:40,000. For SF162, 90% neutralization was not observed at the lowest dilution used for all sera.
—, neutralization not achieved at the lowest serum dilution used.
The increase in neutralization susceptibility of SF162ΔV2 is not due to a destabilization of the gp120-gp41 association.
A major mechanism for neutralization of LA strains is the induction of gp120 shedding as a result of antibody binding (26). Structural perturbation of the envelope by the V2 loop introduced might affect the stability of the gp120-gp41 association. To determine whether this mechanism is responsible for the increase in sensitivity to serum neutralization of SF162ΔV2, we compared the degree to which gp120 molecules become dissociated from the surface of SF162 and SF162ΔV2 virions upon virus-serum incubation. For these experiments we used two sera, GJ and LS-BO, which neutralize SF162ΔV2 very efficiently (Table 2). We found that in the absence of anti-HIV sera, only a small fraction (approximately 5%) of virion-associated gp120 molecules become spontaneously dissociated from the virion surface of SF162 and SF162ΔV2 (Table 4). Incubation of these two viruses with anti-HIV sera results in a modest increase in gp120-virion dissociation. No difference, however, in the degree of antibody-mediated gp120-virion dissociation was observed between SF162 and SF162ΔV2.
TABLE 4.
Stability of the gp120-gp41 association
| Serum dilution | % Dissociationa
|
|||
|---|---|---|---|---|
| GJ
|
LS-BO
|
|||
| SF162 | SF162ΔV2 | SF162 | SF162ΔV2 | |
| 500 | 10 (5) | 8 (2) | 13 (3) | 10 (3) |
| 1,000 | 4 (2) | 5 (3) | 7 (3) | 5 (3) |
| 5,000 | 4 (3) | 4 (3) | 5 (2) | 4 (3) |
| 10,000 | 5 (3) | 3 (3) | 5 (3) | 4 (3) |
| 50,000 | 4 (3) | 3 (3) | 3 (3) | 2 (2) |
| No serum | 5 (3)b | 4 (2)b | ||
Percentage of gp120 that became dissociated from the virion surface upon incubation of virions with serially diluted sera for 3 h at 37°C, as described in Materials and Methods. Values in parentheses represent the standard deviation from three experiments.
Spontaneous gp120-virion dissociation, i.e., in the absence of anti-HIV serum, during the 3-h incubation of virions at 37°C.
DISCUSSION
HIV-1 PR isolates resist, while LA isolates are susceptible to, neutralization by serum antibodies and MAbs (6, 10, 15, 23, 24, 32, 39, 40, 45). This difference in neutralization susceptibility between PR and LA isolates is due largely to a more efficient concealment of neutralization epitopes on the surface of PR isolates than on LA isolates and the instability of the gp120-gp41 association of LA envelope proteins (1, 24, 26, 40). Identification of the envelope region(s) that shield neutralization epitopes on PR isolates and protect them from in vitro and in vivo antibody-mediated neutralization, as well as the identification of the concealed epitopes themselves, will provide important information regarding the mechanisms by which PR HIV-1 isolates escape neutralization by serum antibodies and may assist in the development of more effective envelope-based anti-HIV strategies.
Previous studies conducted with the LA isolate HIV-1IIIb have shown that deletion of the variable loops from gp120 results in exposure of previously hidden epitopes (46, 47). Some of these epitopes are neutralization epitopes, since the simultaneous deletion of the V1 and V2 loops increases three- to fivefold the susceptibility of this virus to neutralization by certain MAbs that bind to the V3 loop, the CD4-binding site, and conserved gp120 regions (3). In this study, we examined whether neutralization epitopes can also be exposed by introducing individual partial deletions in the V1 and V2 loops of SF162, a neutralization-resistant, PR clade B HIV-1 isolate (6).
Our neutralization studies with anti-gp120 MAbs indicate that V2 loop deletion of SF162 resulted in a modest increase in susceptibility to neutralization by MAbs 17b and 48d of the mutant virus (Table 1). These two MAbs recognize epitopes that are CD4 induced and are located within conserved gp120 regions (42). In addition, SF162ΔV2 became susceptible to neutralization by the anti-CD4-binding site MAb 654-30D. However, contrary to what was reported for HIV-1IIIb, SF162ΔV2 remained resistant to neutralization by the anti-V3 loop MAbs tested (Table 1). Interestingly, as mentioned above, the simultaneous deletion of the V1 and V2 loops from HIV-1IIIb renders this neutralization-sensitive LA virus even more susceptible to neutralization by MAb 17b, by certain anti-CD4-binding-site MAbs, and by certain anti-V3 loop MAbs (3). These findings suggest therefore that V2 loop deletion exposes an envelope region whose structure is partially shared among PR and LA isolates.
Of more interest are the observations made with the use of polyclonal anti-HIV sera. Partial deletion of the V2, but not the V1, loop increases up to 170-fold the virus susceptibility to neutralization by polyclonal sera collected from patients infected with clade B viruses (Table 2). More importantly, while SF162 is resistant to neutralization by sera collected from patients infected with non-clade B isolates, SF162ΔV2 is highly susceptible to neutralization irrespective of the clade of the sera tested (Table 3). Several possibilities could account for this switch in the viral phenotype. We show here that this switch is not due to an increased susceptibility of the SF162ΔV2 virus to antibody-mediated gp120-virion dissociation (Table 4). It could be argued that SF162ΔV2 is more susceptible to neutralization because it replicates more slowly in susceptible cells, especially macrophages (Fig. 2). However, our assays were performed in PBMC, where the replication rates of the mutant viruses are comparable to that of wt SF162 (Fig. 2A). Furthermore if replication competence of the virus is the underlying basis for the difference in neutralization susceptibility, we would expect the SF162ΔV1 isolate to be also more susceptible to neutralization than SF162. This is clearly not the case (Tables 1 to 3). The simplest explanation for this enhanced susceptibility in neutralization of SF162ΔV2 is that the V2 loop deletion exposes a region, rich in neutralization epitopes, whose overall structure is conserved among PR isolates regardless of the clade they are clustered in. The V2 loop is exposed on the surface of both PR and LA isolates (25, 30, 31, 36). Although it appears to be a target for neutralization on LA isolates (11), we previously demonstrated (39), and confirmed here (Table 1), that binding of certain anti-V2 loop MAbs on the surface of SF162 does not block its infectivity. This observation implies that the V2 loop plays a crucial role as a shield, protecting the virus from neutralizing antibodies, while itself not being a target for antibody-mediated neutralization on PR isolates. Furthermore, the region of the envelope shielded by the V2 loop appears to be highly immunogenic, since all sera tested so far contained antibodies directed against it (Tables 2 and 3). Last, the finding that the degree to which the sera neutralized SF162ΔV2 was independent of the time of their collection during the AIDS epidemic suggests that the overall structure of the exposed region remains unaltered over time, which is indicative of protection from immune selection. Taken together, our observations underline the importance of this envelope region in the HIV life cycle, implicating it as a desirable target for envelope-based anti-HIV immunization methodologies.
If the exposed epitopes are highly conserved among PR isolates, how then would one explain the fact that some sera (like GSO, GJ, LSS, and LS BO) are so potent in neutralizing SF162ΔV2 and not SF162? One possible explanation is that antibody access to the region containing neutralization epitopes on the surface of wt SF162 virions is very efficiently blocked by the V2 loop. Therefore, even if high titers of antibodies directed against this region are present in these sera, the virus manages to evade neutralization. However, upon V2 loop deletion and exposure of this region, antibodies are capable of binding and neutralizing HIV infection very efficiently. Alternatively, in addition to exposing cryptic neutralization epitopes, deletion of the V2 loop modulates the envelope structure in such a way that fewer enhancing epitopes are exposed on the HIV envelope. The presence of enhancing antibodies in the sera of HIV-infected individuals is well documented (14, 16, 17, 19, 29). Enhancing antibodies bind to several regions of the viral envelope, including the V3 loop, the CD4-binding site, and the gp41 subunit (18, 28, 33, 39, 41). Understanding the precise mechanism(s) responsible for the observed increase in antibody-mediated susceptibility of HIV upon V2 loop deletion is important, since it will provide valuable information regarding the biology of HIV and help in the development of envelope-based immunogens.
What is the nature of this putative highly conserved neutralization region? The finding that SF162ΔV2 is still susceptible to neutralization by MAb IgG1b12, whose epitope comprises elements of the CD4-binding site and the V2 loop (21, 27), and is now susceptible to neutralization by MAbs 17b and 48d, whose epitopes become exposed upon receptor binding, and by the anti-CD4 binding site MAb 654-30D indicates that the epitopes recognized by these MAbs could be part of a much larger region which becomes exposed upon V2 loop deletion. Although the precise nature of this region is unknown, we propose, based on data presented in Table 1, that it comprises elements of the CD4-binding site and conserved gp120 sequences.
In summary, our studies indicate that major deletions within the first and second hypervariable regions of gp120 from macrophagetropic PR HIV-1 isolates will not abrogate the potential of these viruses to replicate into PBMC and macrophages. Deletion of the V2 loop, but not of the V1 loop, exposes highly conserved neutralization epitopes located within the core of the envelope protein and results in a dramatic increase in the susceptibility of the virus to neutralization by antibodies present in sera collected from patients infected with pan-clade HIV isolates. The envelope of SF162ΔV2 could be used as an immunogen to generate antibodies against the exposed region. We believe that such antibodies would have a more potent cross-clade neutralizing potential than antibodies generated against the envelope of SF162.
ACKNOWLEDGMENTS
L.S. is an AmFAR Scholar (award 70479-19-RF, made in memory of Bernard C. Hirsh). This work was supported by grant CA 72822 (C.C.-M.) and by PAF grant PG-50617 (L.S.).
We thank J. A. Levy, D. D. Ho, C. Y. Cao, D. Burton, S. Z. Pazner, and J. Robinson for their generous gifts of sera and MAbs.
REFERENCES
- 1.Bou-Habib D C, Roderiquez G, Oravesz T, Berman P W, Lusso P, Norcross M A. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton D R, Pyati J, Koduri R, Sharp S J, Thorton G B, Parren P H I, Sawyer L S W, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F I. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 3.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Y, Qin L, Zhang L, Safrit J, Ho D D. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. J Exp Med. 1995;332:201–208. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 5.Capon D J, Chamow S M, Mordenti J, Marsters S A, Gregory T, Mitsuya H, Byrn R A, Lucas C, Wurm F M, Groopman J E, Broder S, Smith D H. Designing CD4 immunoadhesins for AIDS therapy. Nature (London) 1989;337:525–531. doi: 10.1038/337525a0. [DOI] [PubMed] [Google Scholar]
- 6.Cheng-Mayer C, Homsy J, Evans L A, Levy J A. Identification of human immunodeficiency virus subtypes with distinct patterns of sensitivity to serum neutralization. Proc Natl Acad Sci USA. 1988;85:2815–2819. doi: 10.1073/pnas.85.8.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng-Mayer C, Weiss C, Seto D, Levy J A. Isolates of human immunodeficiency virus type 1 from brain may constitute a special group of the AIDS virus. Proc Natl Acad Sci USA. 1989;86:8575–8579. doi: 10.1073/pnas.86.21.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ditzel H J, Binley J M, Moore J P, Sodroski J, Sullivan N, Sawyer L S W, Hendry R M, Yang W-P, Barbas III C F, Burton D R. Neutralizing recombinant human antibodies to a conformational V2- and CD4-binding site-sensitive epitope of HIV-1 gp120 isolated by using an epitope-masking procedure. J Immunol. 1995;154:893–906. [PubMed] [Google Scholar]
- 10.Fouts T R, Binley J M, Trkola A, Robinson J E, Moore J P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fung M S C, Sun C R Y, Gordon W L, Liou R-S, Chang T W, Sun W N C, Daar E S, Ho D D. Identification and characterization of a neutralization site within the second variable region of human immunodeficiency virus type 1. J Virol. 1992;66:848–856. doi: 10.1128/jvi.66.2.848-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorny M K, Moore J P, Conley A J, Karwowska S, Sodroski J, Williams C, Burda S, Boots L J, Zolla-Pazner S. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J Virol. 1994;68:8312–8320. doi: 10.1128/jvi.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorny M K, Xu J-Y, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- 14.Gras G, Strub T, Dormont D. Antibody-dependent enhancement of HIV infection. Lancet. 1988;i:1285. [PubMed] [Google Scholar]
- 15.Hanson C V. Measuring vaccine-induced HIV neutralization: report of a workshop. AIDS Res Hum Retroviruses. 1994;10:645–648. doi: 10.1089/aid.1994.10.645. [DOI] [PubMed] [Google Scholar]
- 16.Homsy J, Meyer M, Tateno S, Clarkson S, Levy J A. The Fc and not CD4 receptor mediates antibody enhancement of HIV infection in human cells. Science. 1989;244:1357–1360. doi: 10.1126/science.2786647. [DOI] [PubMed] [Google Scholar]
- 17.Homsy J, Tataeno M, Levy J A. Antibody-dependent enhancement of HIV infection. Lancet. 1988;i:1285–1286. [PubMed] [Google Scholar]
- 18.Kliks S C, Shioda T, Haigwood N L, Levy J A. V3 variability can influence the ability of an antibody to neutralize or enhance infection by diverse strains of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1993;90:11518–11522. doi: 10.1073/pnas.90.24.11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostrikis L G, Cao Y, Ngai H, Moore J P, Ho D D. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J Virol. 1996;70:445–458. doi: 10.1128/jvi.70.1.445-458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laal S, Burda S, Gorny M K, Karwowska S, Buchbinder A, Zolla-Pazner S. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J Virol. 1994;68:4001–4008. doi: 10.1128/jvi.68.6.4001-4008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo H, Stamatatos L, Ip J E, Barbas C F, Parren P W H I, Burton D R, Moore J P, Ho D D. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. J Virol. 1997;71:6869–6874. doi: 10.1128/jvi.71.9.6869-6874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore J P, Cao Y, Leu J, Qin L, Korber B, Ho D D. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 serotypes. J Virol. 1996;70:427–444. doi: 10.1128/jvi.70.1.427-444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore J P, Ho D D. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS. 1995;9:117–136. [PubMed] [Google Scholar]
- 24.Moore J P, McKeating J A, Huang Y, Askenazi A, Ho D D. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore J P, Sattentau Q J, Yoshiyama H, Thali M, Charles M, Sullivan N, Poon S-W, Fung M S, Traincard F, Pinkus M, Robey G, Robinson J E, Ho D D, Sodroski J. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J Virol. 1993;67:6136–6151. doi: 10.1128/jvi.67.10.6136-6151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poignard P, Fouts T, Naniche D, Moore J P, Sattentau Q J. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J Exp Med. 1995;183:473–484. doi: 10.1084/jem.183.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roben P, Moore J P, Thali M, Sodroski J, Barbas C F I, Burton D R. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson J R, E. W, Gorny M K, Xu J-Y, Mitchell W M, Zolla-Pazner S. Two immunodominant domains of gp41 bind antibodies which enhance human immunodeficiency virus type 1 infection in vitro. J Virol. 1991;65:4169–4176. doi: 10.1128/jvi.65.8.4169-4176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson W E, JR, Montefiori D C, Mitchell W M. Antibody-dependent enhancement of human immunodeficiency virus type 1 infection. Lancet. 1988;i:790–794. doi: 10.1016/s0140-6736(88)91657-1. [DOI] [PubMed] [Google Scholar]
- 30.Sattentau Q J, Moore J P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomers. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawyer L S, Wrin M T, Crawford-Miksza L, Potts B, Wu Y, Weber P A, Alfonso R D, Hanson C V. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J Virol. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schutten M, Andeweg A C, Bosch M L, Osterhaus A D M E. Enhancement of infectivity of a non-syncytium inducing HIV-1 by sCD4 and by human antibodies that neutralize syncytium inducing HIV-1. Scand J Immunol. 1995;41:18–22. doi: 10.1111/j.1365-3083.1995.tb03528.x. [DOI] [PubMed] [Google Scholar]
- 34.Seligman S J, Binley J M, Gorny M K, Burton D R, Zolla-Pazner S, Sokolowski K A. Characterization by serial competition ELISAs of HIV-1 V3 loop epitopes recognized by monoclonal antibodies. Mol Immunol. 1996;33:737–745. doi: 10.1016/0161-5890(96)00044-2. [DOI] [PubMed] [Google Scholar]
- 35.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature (London) 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 36.Stamatatos L, Cheng-Mayer C. Structural modulations of the envelope gp120 glycoprotein of human immunodeficiency virus type 1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J Virol. 1995;69:6191–6198. doi: 10.1128/jvi.69.10.6191-6198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatatos L, Werner A, Cheng-Mayer C. Differential regulation of cellular tropism and sensitivity to soluble CD4 neutralization by the envelope gp120 of human immunodeficiency virus type 1. J Virol. 1994;68:4973–4979. doi: 10.1128/jvi.68.8.4973-4979.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatatos L, Wiskerchen M, Cheng-Mayer C. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV-1 isolate on viral envelope structure, cell-entry and replication. AIDS Res Hum Retroviruses. 1998;14:1129–1139. doi: 10.1089/aid.1998.14.1129. [DOI] [PubMed] [Google Scholar]
- 39.Stamatatos L, Zolla-Pazner S, Gorny M, Cheng-Mayer C. Binding of antibodies to virion-associated gp120 molecules of primary-like human immunodeficiency virus type 1 (HIV-1) isolates: effect on HIV-1 infection of macrophages and peripheral blood mononuclear cells. Virology. 1997;229:360–369. doi: 10.1006/viro.1997.8443. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda A, Tuazon C U, Ennis F A. Antibody-enhanced infection by HIV-1 via Fc receptor-mediated entry. Science. 1988;242:580–583. doi: 10.1126/science.2972065. [DOI] [PubMed] [Google Scholar]
- 42.Thali M, Moore J P, Furma C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber J, Fenyo E-M, Beddows S, Kaleebu P, Bjorndal A the WHO Network for HIV Isolation and Characterization. Neutralizing serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. J Virol. 1996;70:7827–7832. doi: 10.1128/jvi.70.11.7827-7832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werner A, Levy J A. Human immunodeficiency virus type 1 envelope gp120 is cleaved after incubation with recombinant soluble CD4. J Virol. 1993;67:2566–2574. doi: 10.1128/jvi.67.5.2566-2574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wrin T, Loh T P, Vennari J C, Schuitemaker H, Nunberg J H. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol. 1995;69:39–48. doi: 10.1128/jvi.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]