Abstract
Substance use disorders (SUDs) are phenotypically and genetically correlated with each other and with other psychological traits characterized by behavioural under-control, termed externalizing phenotypes. In this study, we used genomic structural equation modelling to explore the shared genetic architecture among six externalizing phenotypes and four SUDs used in two previous multivariate genome-wide association studies of an externalizing and an addiction risk factor, respectively. We first evaluated five confirmatory factor analytic models, including a common factor model, alternative parameterizations of two-factor structures and a bifactor model. We next explored the genetic correlations between factors identified in these models and other relevant psychological traits. Finally, we quantified the degree of polygenic overlap between externalizing and addiction risk using MiXeR. We found that the common and two-factor structures provided the best fit to the data, evidenced by high factor loadings, good factor reliability and no evidence of concerning model characteristics. The two-factor models yielded high genetic correlations between factors (rgs ≥ 0.87), and between the effect sizes of genetic correlations with external traits (rg ≥ 0.95). Nevertheless, 21 of the 84 correlations with external criteria showed small, significant differences between externalizing and addiction risk factors. MiXer results showed that approximately 81% of influential externalizing variants were shared with addiction risk, whereas addiction risk shared 56% of its influential variants with externalizing. These results suggest that externalizing and addiction genetic risk are largely shared, though both constructs also retain meaningful unshared genetic variance. These results can inform future efforts to identify specific genetic influences on externalizing and SUDs.
Keywords: externalizing, genomic structural equation modelling, substance use disorders
1 |. INTRODUCTION
Substance use disorders (SUDs) are associated with substantial cost to affected individuals, their families, and society at large.1,2 Twin and family studies estimate the heritability of individual SUDs, including alcohol,3 cannabis,4 and opioid use disorders,5 to be around 50%, with a large portion of the heritability for each disorder shared across different substances.5 SUDs co-occur with other forms of psychopathology, personality and behavioural traits,6 most notably with disorders and traits characterized by under-controlled or impulsive action, often termed externalizing phenotypes.7,8 These phenotypes include psychiatric disorders, such as attention-deficit hyperactivity disorder (ADHD), conduct disorder, oppositional defiant disorder (ODD) and antisocial personality disorder,9,10 as well as personality and behavioural traits like risk taking,11 aggression,12 lack of constraint8 and antagonism.13 Mirroring phenotypic associations, SUDs are also genetically correlated with externalizing phenotypes.14–17
Evidence of strong genetic correlations among SUDs and externalizing phenotypes suggests that further investigation into their shared genetic architecture is warranted. Recent advances in statistical genetics methods allow us to leverage summary statistics from well-powered genome-wide association studies (GWAS) to model the degree to which genetic influences on individual, related phenotypes operate through broad latent factors that represent the variance shared among these phenotypes. Specifically, genomic structural equation modelling (genomic SEM)18 provides a flexible framework to apply SEM techniques to genetic covariance matrices derived from linkage disequilibrium score regression.19 This allows researchers to formally test theoretical models of the shared genetic architecture of genetically correlated traits.
Two recent large-scale studies used genomic SEM to examine the genetic architecture of externalizing phenotypes (N = 1 373 240)11 and SUDs (N = 1 019 521),20 respectively. Both identified a single, underlying latent factor that captured the majority of the variance in their indicators. A GWAS of liability to externalizing, operationalized as a latent factor indicated by ADHD, problematic alcohol use, cannabis initiation, smoking initiation, age at first sexual intercourse, number of sexual partners and general risk tolerance, identified 579 conditionally independent loci that largely operated through the latent factor.11 A GWAS of liability to addiction, indexed by problematic alcohol use, problematic tobacco use, cannabis use disorder and opioid use disorder, identified 19 independent loci.21
Results from these studies support the use of broad latent factors to interrogate the shared genetic aetiology of externalizing phenotypes and SUDs. However, the question remains as to whether SUDs and externalizing are influenced by distinct, but related, dimensions of risk, or whether they reflect the same underlying continuum of risk. In other words, is there specific genetic risk shared across SUDs that is not also shared with other externalizing outcomes? To address this question, we applied genomic SEM to six externalizing phenotypes and four SUDs previously included in the factor models of externalizing11 and addiction liability.20,21 We tested a series of a priori specified models, guided by results from the phenotypic factor analysis literature, which have produced inconsistent results with respect to the placement of SUDs in the overall structure of psychopathology. Many studies include SUDs as part of a broad externalizing factor,22–25 whereas others model it as a separate, but correlated, factor.26–29 We tested five alternative confirmatory factor analytic models, including a common factor model and alternative parameterizations of a two-factor structure to capture the genetic covariance among externalizing phenotypes and SUDs (Figure 1A–E). We followed up these analyses with exploratory factor analysis to evaluate alternative, data-driven clusters of phenotypes. Finally, to further characterize the differentiability of the factors identified in our models, we estimated their genetic correlations with other relevant traits and quantified the degree of polygenic overlap between externalizing and SUDs.
FIGURE 1.

Path diagrams and standardized factor loadings of all confirmatory models tested (A) common factor, (B) EXT-ARF model, (C) BD-SUB model, (D) EXT-resARF model, and (E) bifactor BD-SUB model.
2 |. METHODS
2.1 |. Samples
Following our preregistered analysis plan (https://osf.io/v8q2y/), we used summary statistics from well-powered GWAS previously included in investigations of externalizing11 and the addiction risk factor.20 We included the following 10 phenotypes: general risk tolerance,30 number of sexual partners,30 reverse-coded age at first sexual intercourse,30 ADHD,31 smoking initiation,32 cannabis initiation,33 problematic tobacco use,20 problematic alcohol use,34 opioid use disorder35 and cannabis use disorder.17 See Table S1 for more information about each phenotype. As previous genomic SEM studies of externalizing and addiction liability both included similar indicators for problematic alcohol use (N ~150 K in Karlsson Linnér et al.11; N ~430 K in Hatoum et al.20), we only retained the larger meta-analysis of problematic alcohol use.34 Following previous investigations using these GWAS summary statistics, we retained variants with minor allele frequency >0.01 and INFO score >0.70 for opioid use disorder and >0.90 for all other phenotypes.
2.2 |. Statistical analyses
We used genomic SEM18 to conduct all analyses. Genomic SEM is a recent statistical method that provides a flexible framework for applying SEM techniques to GWAS summary statistics, which allows for more accurate modelling of multivariate genetic covariance matrices.
In the present study, we first estimated the genetic correlations among the 10 indicators in our proposed model. Next, we tested a series of a priori specified models to investigate the genetic architecture of externalizing phenotypes and SUDs. We chose these models to reflect alternative conceptualizations of the relationships between externalizing and SUDs in the existing literature. Namely, that SUDs are a subset of externalizing36 (common factor model); that externalizing and SUDs are separable, but correlated, constructs26–28 (EXT-ARF model); that all forms of substance involvement are distinct from, but correlated with, externalizing traits reflecting behavioural disinhibition37 (BD-SUB model); that SUDs are a subset of externalizing with meaningful residual variance (EXT-resARF model); and that behavioural disinhibition and substance use are specific representations of a broad liability to externalizing36 (bifactor model). As such, we tested the following models:
Common factor model: an externalizing factor onto which all externalizing phenotypes and SUDs load (Figure 1A)
EXT-ARF model: a two-correlated factors model in which externalizing phenotypes load onto an externalizing factor (EXT) and SUDs load on an addiction risk factor (ARF; Figure 1B)
BD-SUB model: a two-correlated factors model in which all substance use phenotypes load onto a substance use factor (SUB)37 and remaining phenotypes comprise a behavioural disinhibition factor (BD; Figure 1C)
EXT-resARF model: a model in which all phenotypes load on an externalizing factor (EXT) and SUDs load onto a residual ARF (Figure 1D)
Bifactor model: a model in which liability to externalizing is captured by a general factor onto which all phenotypes load and residual liability to behavioural disinhibition (BD) and substance use (SUB) is captured by two specific factors (Figure 1E)
We assessed the performance of each alternative model using a combination of the following metrics:
Goodness-of-fit statistics, including the comparative fit index (CFI), the Akaike information criterion (AIC) and the standardized root mean square residual (SRMR), which provide absolute and relative indices of model fit. CFI and SRMR are absolute fit indices, with values greater than 0.95 and below 0.05, respectively, indicating excellent fit.38
Magnitude, median and standard deviation (SD) of the indicators’ loadings on their respective factors in each alternative model.39 If factors are representative of their indicators, indicators’ loadings will be substantial as indicated by their individual and average magnitude. If factors represent unidimensional constructs, they should be similarly defined by their constituent indicators.9,40 This is particularly important as our first aim was to determine the degree of dimensionality in the previously identified externalizing phenotypes and SUDs.
Structural validity was assessed using construct replicability (H)41 and latent variable reliability (omega).42 H conceptually reflects the extent to which a latent variable is represented by its indicators (items). It ranges from 0 to 1 and increases as a function of the magnitude of factors loadings and number of indicators on each item. Values greater than 0.70 indicate adequate replicability.41 For latent variable reliability in the common factor model, we used omega total (ωt), which reflects the percentage of total variance accounted for by the single latent construct. For the correlated factors models, we used omega subscale (ωs), which reflects the percentage of variance the latent variable accounts for in its indicators. Values greater than 0.75 indicate adequate reliability.42
Model characteristics, such as standardized factor loadings that are out of bounds (i.e., >|1|), not in the predicted direction, not significantly different from zero, or have large standard errors, were used to judge whether models are tenable.39
Sensitivity of the factors to their indicators, which was assessed by dropping one indicator in the model at a time and judging the extent to which that changed the factor loadings and their standard errors of other indicators as well as the factor correlations in the EXT-ARF and BD-SUB Models.
To explore a wider range of potential factor solutions, we next conducted exploratory factor analyses (EFA) using the stats R package.43 We tested two to four factor solutions using oblique (i.e., correlated) and orthogonal (i.e., uncorrelated) rotations. We tested solutions with a maximum of four factors to ensure factors would have more than two indicators. We conducted two sets of EFAs and follow-up confirmatory factor analyses (CFAs), which replicated results from the exploratory models, one in which we used the genetic covariance matrix of odd chromosomes to test exploratory models and the genetic covariance matrix of even chromosomes to test follow-up models, and one in which we flipped the use of odd and even chromosomes. We ran our models in this way to account for the possibility that causal variants were enriched on certain chromosomes thereby impacting the results of the exploratory models.
We next conducted genetic association analyses, estimating the zero-order genetic correlations between factors identified in the best-performing models and a wide range of preregistered phenotypes (https://osf.io/v8q2y/) from the domains of personality, risk taking, physical health, psychiatric traits and disorders, anthropometric traits, cognitive traits, socioeconomic status and reproductive health. These genetic association analyses served two purposes: (1) to explore the genetic relationships between our latent construct(s) and other relevant traits and (2) to compare patterns of genetic correlations in the best-performing two-factor models. This latter purpose allowed us to quantify the degree to which these factors provided meaningfully distinct information about genetic risk for externalizing phenotypes and SUDs. As a further test of this question, we fit models in which the correlations between the two factors and an individual external criterion variable were constrained to be equal and observed the resulting change in χ2. We note, however, that χ2 difference tests are very sensitive to small changes, especially in the presence of a large sample size, and interpret these results with caution. We used a Bonferroni corrected p value <0.05 to judge statistical significance.
Finally, to further quantify the polygenic overlap between SUDs and externalizing, we used MiXeR,44 which applies bivariate Gaussian mixture models to estimate the proportion of shared influential genetic variance between two traits. For these analyses, we used the summary statistics from the addiction risk factor21 and externalizing11 without the problematic alcohol use indicator, to reflect the two factors in the EXT-ARF model.
3 |. RESULTS
3.1 |. Model fitting
We first estimated the genetic correlations among our 10 proposed indicators. The correlations between SUDs and externalizing phenotypes varied substantially between SUDs (Figure 2). Of the four SUDs, cannabis use disorder was most strongly correlated with the externalizing phenotypes (rgs ranged from 0.45 [risk tolerance] to 0.72 [smoking initiation]) whereas problematic tobacco use was least strongly correlated with externalizing phenotypes (rgs ranged from −0.06 [cannabis initiation] to 0.45 [ADHD]).
FIGURE 2.
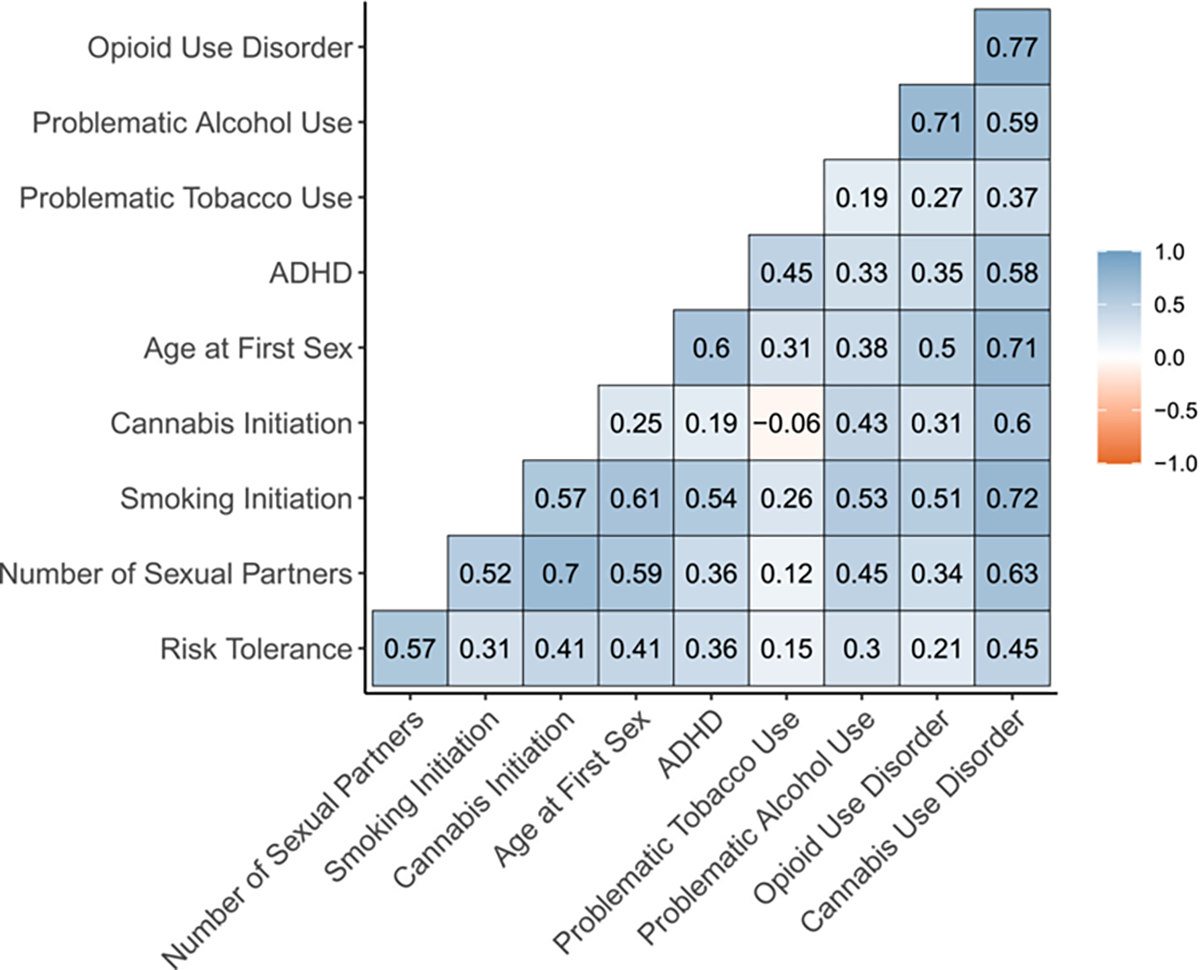
Genetic correlations among externalizing phenotypes and SUDs.
Given that SUDs were at least moderately correlated with most externalizing phenotypes, we proceeded with our prespecified set of confirmatory factor analytic models. See Table 1 for model fit and Table 2 for factor loadings and reliability indices. In addition to the CFAs presented below, we ran exploratory factor analytic (EFA) models with 2–4 factors. No clear pattern emerged from the EFAs, the results of which are reported in Tables S2–S3 and S8–S9.
TABLE 1.
Confirmatory factor analysis model fit statistics.
| Model | χ2 (df) | AIC | CFI | SRMR |
|---|---|---|---|---|
| Common factor model (Model 1) | 1708 (35)*** | 1748 | 0.86 | 0.10 |
| EXT-SUD (Model 2) | 1632 (34)*** | 1674 | 0.87 | 0.10 |
| BD-SUB (Model 3) | 1390 (34)*** | 1432 | 0.89 | 0.10 |
| EXT-residual SUD (Model 4) | 1684 (31)*** | 1732 | 0.86 | 0.09 |
| BD-SUB bifactor model (Model 5) | 183 (24)*** | 245 | 0.99 | 0.06 |
Abbreviations: AIC, Akaike information criterion; CFI, comparative fit index; SRMR, standardized root mean square residual.
p < 0.05.
TABLE 2.
Standardized factor loadings and factor reliability indices for confirmatory factor analysis models.
| Common factor | EXT-ARF | BD-SUB | EXT-resARF | Bifactor model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EXT | EXT | SUD | BD | SUB | EXT | SUD | EXT | BD | SUB | |
| Indicator | ƛ (SE) | ƛ (SE) | ƛ (SE) | ƛ (SE) | ƛ (SE) | ƛ (SE) | ƛ (SE) | ƛ (SE) | ƛ (SE) | ƛ (SE) |
| RISK | 0.52 (0.03) | 0.52 (0.03) | – | 0.55 (0.03) | – | 0.52 (0.03) | – | 0.26 (0.05) | 0.52 (0.04) | – |
| NSEX | 0.73 (0.02) | 0.74 (0.02) | – | 0.77 (0.02) | – | 0.74 (0.02) | – | 0.23 (0.08) | 1.00 (0.04) | – |
| SMOK | 0.79 (0.02) | 0.79 (0.02) | – | – | 0.82 (0.02) | 0.79 (0.02) | – | 0.58 (0.05) | – | 0.56 (0.06) |
| CANN | 0.59 (0.03) | 0.60 (0.03) | – | – | 0.61 (0.03) | 0.60 (0.03) | – | −0.04 (0.09) | – | 1.01 (0.05) |
| FSEX | 0.74 (0.02) | 0.74 (.02) | – | 0.78 (0.02) | – | 0.74 (0.02) | – | 0.69 (0.04) | 0.43 (0.07) | – |
| ADHD | 0.64 (0.03) | 0.65 (0.03) | – | 0.67 (0.04) | – | 0.65 (0.03) | – | 0.75 (0.04) | 0.22 (0.08) | – |
| PTU | 0.32 (0.03) | – | 0.34 (0.03) | – | 0.32 (0.03) | 0.31 (0.03) | 0.08 (0.05) | 0.52 (0.03) | – | −0.03 (0.05) |
| PAU | 0.62 (0.02) | – | 0.68 (0.02) | – | 0.64 (0.02) | 0.60 (0.02) | 0.28 (0.11) | 0.40 (0.05) | – | 0.50 (0.05) |
| OUD | 0.66 (0.04) | – | 0.72 (0.05) | – | 0.68 (0.04) | 0.57 (0.04) | 1.33 (0.49) | 0.57 (0.06) | – | 0.38 (0.08) |
| CUD | 0.94 (0.03) | – | 1.02 (0.04) | – | 0.96 (0.03) | 0.92 (0.03) | 0.19 (0.08) | 0.71 (0.07) | – | 0.63 (0.08) |
| Factor correlation | – | 0.88 (0.02) | 0.87 (0.02) | – | – | |||||
| Median | 0.65 | 0.69 | 0.70 | 0.72 | 0.66 | 0.62 | 0.23 | 0.55 | 0.48 | 0.53 |
| SD | 0.10 | 0.10 | 0.27 | 0.11 | 0.21 | 0.17 | 0.58 | 0.25 | 0.33 | 0.34 |
| H | 0.97 | 0.85 | 1.00 | 0.81 | 0.94 | 0.92 | 1.80 | 0.83 | 0.99 | 1.01 |
| Omega | 0.89 | 0.84 | 0.80 | 0.79 | 0.84 | 0.92 | 0.95 | 0.55 | 0.88 | 0.49 |
Abbreviations: ADHD, attention-deficit hyperactivity disorder; CANN, cannabis initiation; CUD, cannabis use disorder; FSEX, reverse coded age at first sexual intercourse; NSEX, number of sexual partners; OUD, opioid use disorder; PAU, problematic alcohol use; PTU, problematic tobacco use; RISK, general risk tolerance; SMOK, smoking initiation.
3.1.1 |. Model 1: Common factor model
Fit statistics for the common factor model, in which all externalizing phenotypes and SUDs loaded onto a single factor, did not meet our prespecified threshold for good model fit. Nevertheless, the standardized factor loadings of the indicators were high (median ƛ = 0.65) and uniform (SD of ƛ = 0.10), indicating that the common factor was well and similarly defined by its indicators. In further support of this, H and ωt were high (0.97 and 0.89, respectively).
3.1.2 |. Model 2: Two-correlated factors model with externalizing and addiction risk factors (EXT-ARF)
We next fit the EXT-ARF model, in which externalizing phenotypes loaded onto an externalizing (EXT) factor and SUDs loaded on an addiction risk factor (ARF). Fit statistics for this model also did not meet thresholds for good fit, although fit was somewhat improved relative to the common factor model. The median standardized factor loadings were high (ƛ = 0.66), although the SD of the loadings on the ARF factor were somewhat higher (SD of ƛ = 0.27), indicating greater variability in the degree to which these factors were represented by their indicators. The EXT and ARF factors were very strongly genetically correlated (rgs = .88), suggesting they reflect largely overlapping dimensions of genetic risk. H and ωs were high for both factors, although it is important to note that the H for the ARF factor is uninterpretable given that cannabis use disorder’s loading exceeded 1.
3.1.3 |. Model 3: Two-correlated factors model with substance use and behavioural disinhibition factors (BD-SUB)
Fit statistics for the BD-SUB model, in which all substance use phenotypes loaded onto a substance use (SUB) dimension and remaining phenotypes loaded onto a behavioural disinhibition factor (BD) were improved relative to the common factor and EXT-ARF models, but still did not meet threshold for good fit. The median factor loadings were high (ƛ = 0.72), although the variability of loadings on the SUB factor was somewhat high (SD of ƛ = 0.21). The BD and SUB factors were strongly correlated (rgs = 0.87), again suggesting that they represent similar dimensions of risk. H and ωs were high for both factors.
3.1.4 |. Model 4: Externalizing factor with residual addiction risk factor (EXT-resARF)
The EXT-resARF model, in which all phenotypes loaded onto an externalizing (EXT) factor and SUDs loaded additionally onto a residual addiction risk factor (resARF), did not meet the threshold for good fit. The median loading on the residual ARF factor was low (median ƛ = 0.23) and the loadings were variable (SD of ƛ = 0.58). In addition, the residual variance of the opioid use disorder (OUD) indicator was negative.
3.1.5 |. Model 5: Bifactor model with externalizing general factor and specific behavioural disinhibition and substance use factors (Bifactor BD-SUB)
Finally, we fit the BD-SUB bifactor model, in which all phenotypes loaded on a general externalizing factor and residual variation among the phenotypes was captured by behavioural disinhibition and substance use specific factors. This model fit the data well based on fit statistics, but several indicators had negative residual variances or nonsignificant or negative loadings on the general and specific factors, likely reflecting overfitting. In addition, the median standardized factors loadings of the general and specific factors were low, and the SDs were high. H was high for most factors, although this is strongly influenced by the loadings greater than 1.0. ω was below the threshold of good reliability for the general factor and substance use specific factor (0.55 and 0.49, respectively).
3.1.6 |. Summary of model fitting
Across all models, problematic tobacco use had the lowest loadings on its respective factors (ƛ ranged from 0.32 to 0.34 in the common factor, EXT-ARF and BD-SUB models) and cannabis use disorder had the highest loadings (ƛ ranged from 0.94 to 1.02 in the common factor, EXT-ARF and BD-SUB models). We tested a series of models dropping the problematic tobacco use indicators to test the impact this indicator had on model fit and found that model fit was not substantially impacted (see Table S4).
No models met our preregistered criteria for good fit based on model fit indices. Nevertheless, we evaluated each model against the other evaluation criteria outlined in the methods (i.e., magnitude, median and SD of the indicators’ loadings, structural validity and model characteristics) to decide which models to carry forward into subsequent analyses. We determined that the EXT-resARF and Bifactor BD-SUB models were untenable given evidence of low factor loadings, high standard errors, poor reliability and negative residual variances. The three remaining models, the common factor, EXT-ARF and BD-SUB models, all had high loadings, good reliability, and no evidence of concerning model characteristics (i.e., high standard errors or negative residual variances). The factors in both the EXT-ARF and BD-SUB models were very highly genetically correlated, suggesting that the two dimensions may not be meaningfully distinct. We carried forward all three remaining models into the sensitivity and genetic correlation analyses to further evaluate this question.
3.2 |. Sensitivity analyses
3.2.1 |. Post hoc models with residual correlations
Given that none of our models reached our thresholds of good fit, we ran the common factor, EXT-ARF and BD-SUB models and included data-driven residual correlations among indicators. The primary purpose of these analyses was to determine the extent to which the parameter estimates in our primary models were influenced by inadequate fit. Factor loadings and correlations were stable across models with and without residual correlations (Figure S1, Table S5). The factor correlations between the externalizing and addiction risk factors (EXT-ARF model) were 0.90 and 0.88 in models with and without residual correlations, respectively. Factor correlations between the behavioural disinhibition and substance use factors (BD-SUB model) were 0.87 and 0.86 in models with and without residual correlations, respectively.
The original externalizing model11 included residual correlations between smoking initiation and problematic alcohol use and cannabis initiation and age at first sex. Although we dropped these correlations in the current analyses, as they would have necessitated cross-factor residual correlations, we tested an additional common factor model including these original residual correlations. The addition of these parameters resulted in substantial improvement in model fit (χ2 = 907, p < 0.001, AIC = 951, CFI = 0.93, SRMR = 0.10).
3.2.2 |. Variability of factor loadings
We first compared the loadings of each indicator across the common factor, EXT-ARF and BD-SUB models. The loadings were relatively stable (Figure 3A), indicating that each indicator had a similar loading on its respective factor in each model. We next ran a series of analyses to test the sensitivity of factors to their indicators by dropping one indicator from the model at a time. Figure 3B,C shows the range of factor loadings and standard errors of an individual indicator when each other indicator was dropped from the model one at a time. Overall, factors were not very sensitive to their indicators, as evidenced by the low variability of loadings (Figure 3B) and their standard errors (Figure 3C). We observed the most variability for the standard errors of SUDs in the EXT-ARF model. Factor correlations in the EXT-ARF and BD-SUB models were also stable, ranging from 0.78 to 0.94 and 0.84 to 0.90, respectively.
FIGURE 3.
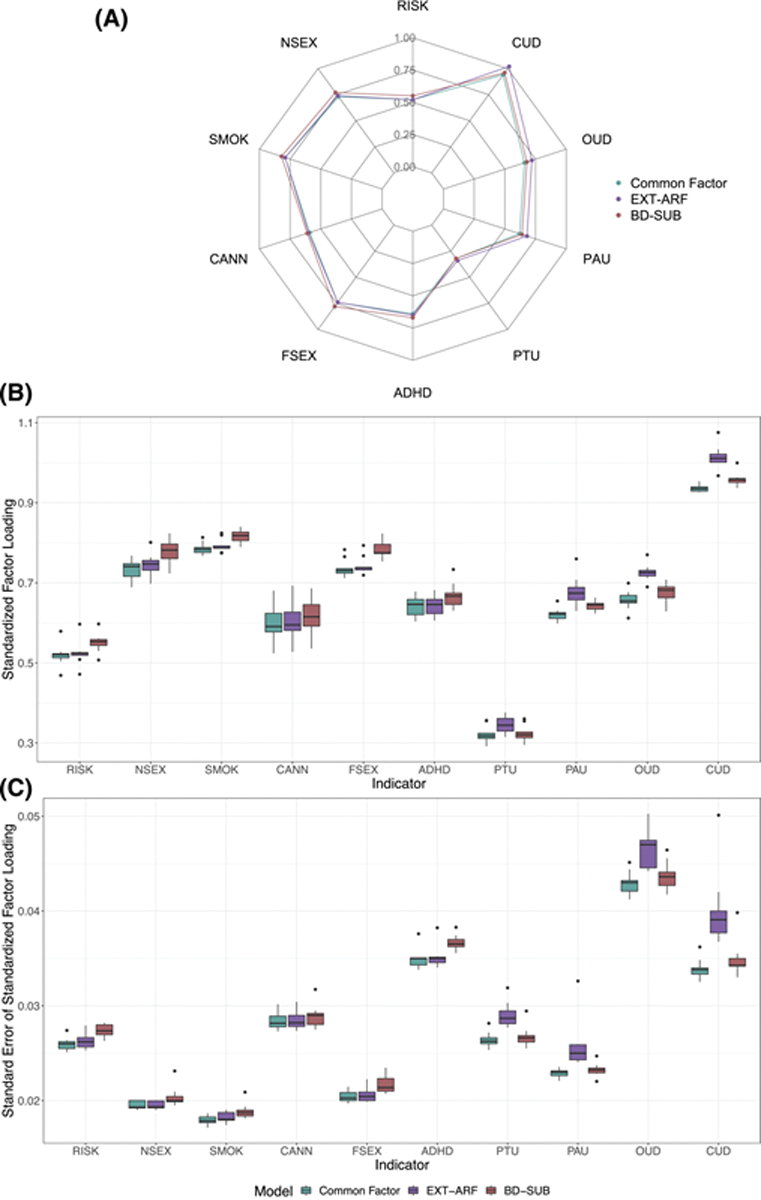
(A) Indicators’ loadings in the common factor, EXT-ARF and BD-SUB models. Variability of (B) factor loadings and (C) standard errors of the factor loadings of indicators when one indicator was dropped from the model at a time.
3.3 |. Genetic correlations
We estimated the genetic correlations between the factors in the common factor, EXT-ARF and BD-SUB models and 84 preregistered phenotypes (Figures 4 and 5 and Tables S6 and S7).
FIGURE 4.

Comparison of genetic correlations of factors in the EXT-ARF and BD-SUB models with psychological, personality and substance-use traits. * = Statistically significant decrement in model fit when the genetic correlations were constrained to be equal.
FIGURE 5.

Genetic correlations between externalizing factor in the common factor model and psychological, personality and substance-use traits.
3.3.1 |. Are externalizing and SUDs differentially genetically correlated with other phenotypes?
The magnitude of the correlations of the EXT and ARF factors with external criteria were generally similar and their confidence intervals were often overlapping (Figure 4A). Further, the correlation between the effect sizes for the two factors was 0.96 and the median absolute difference in correlations was 0.06 (Figure 4B). The χ2 difference tests indicated that externalizing and the addiction risk factor were statistically significantly differentially correlated with 21 of 84 external criteria (see Figure 4A and Table S6). However, many of these differences were small in magnitude, reflecting the sensitivity of the test to large sample size. The greatest differences EXT and ARF were observed for tobacco-related phenotypes (age of initiation [rgARF = −0.57, rgEXT = −0.69, |Δrg| = 0.12] and cigarettes per day [rgARF = 0.49, rgEXT = 0.34, |Δrg| = 0.15]); and certain personality traits (neuroticism [rgARF = 0.37, rgEXT = 0.12, |Δrg| = 0.25] and extraversion [rgARF = 0.11, rgEXT = 0.34, |Δrg| = 0.23]); and other forms of psychopathology (major depressive disorder [rgARF = 0.56, rgEXT = 0.45, |Δrg| = 0.12 and schizophrenia [rgARF = 0.43, rgEXT = 0.27, |Δrg| = 0.16).
3.3.2 |. Are behavioural disinhibition and substance use phenotypes differentially genetically correlated with other phenotypes?
A similar pattern emerged for the behavioural disinhibition and substance use factors in the BD-SUB model such that point estimates were similar and confidence intervals were often overlapping. χ2 difference tests indicated that 21 of 84 criteria were differentially associated with the behavioural disinhibition and substance use factors (see Figure 4C and Table S6). The correlation between the effect sizes for the two factors was 0.95 (0.01), and the median difference in correlations was 0.06 (Figure 4D). The greatest differences were observed for drinks per week (rgBD = 0.36, rgSUB = 0.57, |Δrg| = 0.20), maternal smoking around birth (rgBD = 0.65, rgSUB = 0.83, |Δrg| = 0.18), antisocial behaviour (rgBD = 0.74, rgSUB = 0.59, |Δrg| = 0.15).
3.3.3 |. With what is externalizing genetically correlated?
In Figure 5 and Table S5, we report the correlations of the expanded externalizing factor that emerged from the common factor model with external criteria. Expanded externalizing was significantly genetically correlated with other psychiatric disorders, personality traits, socioeconomic and substance use phenotypes. The strongest correlations were observed with drug exposure (rg = 0.91 [0.06]), prenatal tobacco exposure (rg = 0.77 [0.02]), the Townsend Index reflecting material deprivation (rg = 0.74 [0.04]), suicide attempt (rg = 0.73 [0.05]), antisocial behaviour (rg = 0.69 [0.07]), age of smoking initiation (rg = −0.68 [0.03]) and agreeableness (rg = −0.62 [0.14]).
3.3.4 |. Summary of genetic correlations
In each of the two-factor models (EXT-ARF and BD-SUB), the factors were similarly genetically correlated with external criteria such that the correlations of effect sizes were ≥0.95 and statistically significant differences were, for the most part, small in magnitude. Nevertheless, there were potentially meaningful differences in the associations of externalizing and SUDs with substance use outcomes and other forms of psychopathology. For example, addiction risk was significantly more strongly associated with cigarettes per day, major depressive disorder and schizophrenia. The expanded externalizing factor in the common factor model was strongly genetically correlated with a variety of relevant psychological, substance use and personality traits.
3.4 |. Polygenic overlap between externalizing and SUDs
Results from MiXeR44 indicated that addiction risk was influenced by 16 202 (SE = 1833) SNPs and externalizing was influenced by 11 050 (SE = 159) SNPs. Externalizing shared approximately 81% of its influential variants with addiction risk whereas addiction risk shared 56% of its influential variants with externalizing (Figure 6). Within this shared component, the variants that influence both addiction risk and externalizing had high concordance of direction of effects (95%, SE = 0.05). The estimated genetic correlation between the two traits was 0.66 (SE = 0.01). Finally, model fit statistics supported a model in which addiction risk and externalizing modeled as distinct traits that share substantial polygenic overlap, rather than models in which there was complete or no polygenic overlap (Table S10).
FIGURE 6.

Polygenic overlap between externalizing and addiction risk (A) Venn diagram depicting the estimated number of influencing variants in thousands shared (grey) between and unique to addiction risk (blue) and externalizing (orange). Subparts (B) and (C) show conditional quantile–quantile (Q–Q) plots of observed versus expected −log10 p values in addiction risk and externalizing as a function of the significance of association with the other trait. (D) Log likelihood curves showing goodness of model fit as a function of the number of shared influential variants.
4 |. DISCUSSION
4.1 |. Interpretation of findings
In this study, we investigated the structure of the shared genetic architecture among externalizing phenotypes and SUDs. To accomplish this, we tested a series of factor analytic models using genomic SEM.18 We further explored the potential differences in genetic architecture between externalizing and SUDs by estimating their genetic correlations with other relevant variables and quantifying the degree of polygenic overlap between externalizing and addiction risk. No models met our criteria for good fit; however, we identified three models with homogeneous, high and stable factor loadings, good reliability and no evidence of concerning model characteristics. We note, however, that the factors in the two-factor models were very highly correlated, suggesting they may not represent meaningfully distinct constructs.
In genetic correlation analyses, the correlations of addiction risk and externalizing with external traits were largely similar in magnitude, although χ2 difference tests indicate that there were statistically significant differences in magnitude in 21 out of 84 traits tested. The greatest differences in the associations of externalizing and addiction risk were observed for tobacco-related phenotypes and other forms of psychopathology. For example, age of smoking initiation was more strongly correlated with externalizing than with the addiction risk factor, which is consistent with evidence from twin studies suggesting that genetic variance for general externalizing, rather than variance specific to substance use, predicts age of initiation.45 The finding that the addiction risk factor is more strongly associated with neuroticism, major depression and schizophrenia and less strongly associated with extraversion, suggests that what distinguishes SUDs from externalizing is not necessarily variance specific to risk for addiction, but rather risk shared with other forms of psychopathological distress. This may also reflect differences in ascertainment such that participants in the SUD GWAS were drawn from clinical populations and therefore likely have a higher rate of psychopathology, whereas many externalizing traits were drawn from GWAS of the general population. Similar results emerged for the genetic correlations of the factors representing behavioural disinhibition and substance use.
Finally, we used MiXeR44 to quantify the degree of polygenic overlap between externalizing and addiction risk. These results suggested that the two constructs share a substantial proportion of their influential genetic variants, with significant unique variants remaining for each trait. We believe that these findings, along with evidence that the two latent traits are differentially genetically correlated with certain psychiatric traits, suggest that, despite a high degree of overlap, addiction risk and externalizing retain meaningful unique genetic variance.
We note that the genetic correlation estimated by MiXeR was more modest than that estimated in the latent variable model. This difference may be explained by differences in methods; whereas Genomic SEM estimates genetic correlations between latent variables in a measurement model and thus only includes genetic effects that act homogeneously through the latent factor, MiXeR uses summary statistics, which include genetic effects that act through the latent factor as well as those that influence individual indicators (i.e., Q-SNPs). Use of summary statistics may therefore result in downward bias of genetic correlations. This may also suggest that the influential variants for addiction risk that are unshared with externalizing impact one or a few SUDs, rather than the broad addiction risk factor.
4.2 |. Implications
The finding that SUDs share a large proportion of variance with externalizing phenotypes is consistent with previous phenotypic SEM and twin literature. Many structural models of psychopathology include SUDs in the externalizing dimension,22,23 and twin studies have found that genetic and environmental influences on SUDs largely operate through a broad externalizing factor.8,46 Many structural models, however, place SUDs within specific subdimensions of externalizing, thus acknowledging variance that is unique to SUDs and not shared with all other forms of externalizing. For example, in the externalizing spectrum model,10 substance use related traits are a subdimension of general externalizing and, in the HiTOP model,36 SUDs are part of an externalizing subdimension representing disinhibited forms of externalizing, which are distinct from antagonistic forms. The number of well-powered GWAS of externalizing phenotypes may limit our ability to detect dimensionality in the current analyses and, as more GWAS of relevant traits become available, a more complex, hierarchical genetic architecture may be uncovered.
Results from this paper suggest that the degree of genetic overlap between externalizing and SUDs warrants exploration of genetic effects that jointly and uniquely influence these phenotypes. This is particularly important as gene identification efforts for SUDs have lagged behind consumption- and initiation-related traits and other forms of psychopathology, largely due to issues of power.47 Simultaneous analysis of SUDs and genetically correlated traits, such as the externalizing phenotypes included in the current study, can boost power to detect associations for SUDs.18 The use of genomic SEM, in particular, also allows for the study of residual phenotypes (e.g., variance specific to cannabis use disorder unique from what it shares with other externalizing traits), which provides insights into genetic architecture that is specific to a given trait.48–50 Pursuing these dual goals will allow us to overcome barriers to gene identification in SUDs while still investigating any potential disorder-specific effects.
4.3 |. Limitations and future directions
This project is marked by four notable limitations. First, due to limited availability of GWAS summary statistics and technical issues involved with including multiple ancestries in a single genomic SEM model, our analyses are limited to individuals of European ancestries. Second, we note that none of our models met our preregistered criteria for good fit. One explanation for this is the use of residual correlations in the original externalizing model, which were excluded from the main analyses here, but in sensitivity analyses were shown to improve model fit substantially. Third, our model does not account for other sources of genetic variance (e.g., internalizing and thought disorders) shared among externalizing and SUD phenotypes and which may contribute to the increased overlap among these phenotypes. These sources of variance may be especially important, given evidence that the addiction risk and externalizing factors are differentially associated with internalizing and thought disorder psychopathology. Finally, the definitions of cases in the GWAS used in this study include any person who meets criteria for the relevant diagnosis without regard to comorbid conditions for which they may also meet criteria. High rates of phenotypic correlations and comorbidity among psychiatric disorders suggest a high likelihood that many individuals designated as cases in, for example, the AUD GWAS, also had other psychiatric diagnoses. This means that univariate GWAS necessarily tag genetic variance related to other, correlated traits that is not explicitly modelled by genomic SEM.
5 |. CONCLUSION
In this study, we show that externalizing and addiction risk share a substantial proportion of their genetic influences in common, but that each construct retains meaningful unique genetic variance. These results can be carried forward into future studies, in which we can leverage the genetic correlations among these phenotypes to boost power to detect associations for SUDs, the gene-identification efforts of which have lagged behind other psychiatric traits. This will also facilitate a more fine-grained exploration of the genetic influences unique to each externalizing addiction risk phenotype, thereby allowing exploration of both broad and specific dimensions of genetic risk.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the National Institute on Drug Abuse (R01DA050721). Additional funding for investigator effort was provided by the National Institute on Alcohol Abuse and Alcoholism (grants T32AA028254 [H. E. P.], K01AA030083 [A. S. H.], and P50AA022537 [D. M. D.]) and the National Institute on Drug Abuse (grant P50DA037844 [A. A. P.]).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study were publicly available genome-wide association study summary statistics, which are available from the authors or online repositories of the respective publications.
REFERENCES
- 1.Florence CS, Zhou C, Luo F, Xu L. The economic burden of prescription opioid overdose, abuse, and dependence in the United States, 2013. Medicine. 2016;54(10):901–906. doi: 10.1097/MLR.0000000000000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD. 2010 national and state costs of excessive alcohol consumption. Am J Prev Med. 2015;49(5):e73–e79. doi: 10.1016/j.amepre.2015.05.031 [DOI] [PubMed] [Google Scholar]
- 3.Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med. 2015;45(5):1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verweij KJ, Zietsch BP, Lynskey MT, et al. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010;105(3):417–430. doi: 10.1111/j.1360-0443.2009.02831.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160(4):687–695. doi: 10.1176/appi.ajp.160.4.687 [DOI] [PubMed] [Google Scholar]
- 6.Kendler KS, Myers J. The boundaries of the internalizing and externalizing genetic spectra in men and women. Psychol Med. 2014;44(3):647–655. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr PB, Dick DM. The genetics of externalizing problems. Curr Top Behav Neurosci. 2020;47:93–112. doi: 10.1007/7854_2019_120 [DOI] [PubMed] [Google Scholar]
- 8.Krueger H, Patrick C, Iacono MG. Etiological connections among substance dependence, antisocial behavior, and personality: modeling the externalizing Spectrum. J Abnorm Psychol. 2002;111(3):411–424. doi: 10.1037/0021-843X.111.3.411 [DOI] [PubMed] [Google Scholar]
- 9.Watts AL, Poore HE, Waldman ID. Riskier tests of the validity of the bifactor model of psychopathology. Clin Psychol Sci. 2019;7(6):1285–1303. doi: 10.1177/2167702619855035 [DOI] [Google Scholar]
- 10.Krueger Markon, Patrick Benning, Kramer. Linking antisocial behavior, substance use, and personality: an integrative quantitative model of the adult externalizing spectrum. J Abnorm Psychol. 2007;116(4):645–666. doi: 10.1037/0021-843X.116.4.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlsson Linnér R, Mallard TT, Barr PB, et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci. 2021;24(10):1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Card NA, Little TD. Proactive and reactive aggression in childhood and adolescence: a meta-analysis of differential relations with psychosocial adjustment. Int J Behav Dev. 2006;30(5):466–480. doi: 10.1177/0165025406071904 [DOI] [Google Scholar]
- 13.Watts AL, Poore HE, Lilienfeld SO, Waldman ID. Clarifying the associations between big five personality domains and higher-order psychopathology dimensions in youth. J Res Pers. 2019;82:103844. doi: 10.1016/j.jrp.2019.07.002 [DOI] [Google Scholar]
- 14.Walters RK, Polimanti R, Johnson EC, et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21(12):1656–1669. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waldman ID, Poore HE, Luningham JM, Yang J. Testing structural models of psychopathology at the genomic level. World Psychiatry. 2020;19(3):350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Roige S, Palmer AA, Fontanillas P, et al. Genome-wide association study meta-analysis of the alcohol use disorders identification test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2019;176(2):107–118. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson EC, Demontis D, Thorgeirsson TE, et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry. 2020;7(12):1032–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grotzinger AD, Rhemtulla M, de Vlaming R, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3(5):513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatoum AS, Johnson EC, Colbert SMC, et al. The addiction risk factor: a unitary genetic vulnerability characterizes substance use disorders and their associations with common correlates. Neuropsychopharmacology. 2022;47:1739–1745. doi: 10.1038/s41386-021-01209-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatoum AS, Colbert SMC, Johnson EC, et al. Multivariate genome-wide association meta-analysis of over 1 million subjects identifies loci underlying multiple substance use disorders. Nature Mental Health. 2023;1(3):210–223. doi: 10.1038/s44220-023-00034-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes MK, Greene AL, Levin-Aspenson HF, et al. Three recommendations based on a comparison of the reliability and validity of the predominant models used in research on the empirical structure of psychopathology. J Abnorm Psychol. 2021;130(3):297–317. doi: 10.1037/abn0000533 [DOI] [PubMed] [Google Scholar]
- 23.Lahey BB, Krueger RF, Rathouz PJ, Waldman ID, Zald DH. A hierarchical causal taxonomy of psychopathology across the life span. Psychol Bull. 2017;143(2):142–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene AL, Eaton NR. The temporal stability of the bifactor model of comorbidity: an examination of moderated continuity pathways. Compr Psychiatry. 2017;72:74–82. doi: 10.1016/j.comppsych.2016.09.010 [DOI] [PubMed] [Google Scholar]
- 25.Keyes KM, Eaton NR, Krueger RF, et al. Thought disorder in the meta-structure of psychopathology. Psychol Med. 2013;43(8):1673–1683. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellanos-Ryan N, Briere FN, O’Leary-Barrett M, et al. The structure of psychopathology in adolescence and its common personality and cognitive correlates. J Abnorm Psychol. 2016;125(8):1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verona E, Javdani S, Sprague J. Comparing factor structures of adolescent psychopathology. Psychol Assess. 2011;23(2):545–551. doi: 10.1037/a0022055 [DOI] [PubMed] [Google Scholar]
- 28.Vollebergh WA, Iedema J, Bijl RV, de Graaf R, Smit F, Ormel J. The structure and stability of common mental disorders: the NEMESIS study. Arch Gen Psychiatry. 2001;58(6):597–603. doi: 10.1001/archpsyc.58.6.597 [DOI] [PubMed] [Google Scholar]
- 29.Blanco C, Wall MM, He J-P, et al. The space of common psychiatric disorders in adolescents: comorbidity structure and individual latent liabilities. J am Acad Child Adolesc Psychiatry. 2015;54(1):45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlsson Linnér R, Biroli P, Kong E, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51(2):245–257. doi: 10.1038/s41588-018-0309-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demontis D, Walters RK, Martin J, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M, Jiang Y, Wedow R, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51(2):237–244. doi: 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasman JA, Verweij KJH, Gerring Z, et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal effect of schizophrenia liability. Nat Neurosci. 2018;21(9):1161–1170. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H, Sealock JM, Sanchez-Roige S, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23(7):809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H, Rentsch CT, Cheng Z, et al. Association of OPRM1 functional coding variant with opioid use disorder: a genome-wide association study. JAMA Psychiatry. 2020;77(10):1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krueger RF, Hobbs KA, Conway CC, et al. Validity and utility of hierarchical taxonomy of psychopathology (HiTOP): II. Externalizing super-spectrum. World Psychiatry. 2021;20(2):171–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoeler T, Baldwin J, Allegrini A, et al. Novel biological insights into the common heritable liability to substance involvement: a multivariate genome-wide association study. Biol Psychiatry. 2022;93(6):524–535. doi: 10.1016/j.biopsych.2022.07.027 [DOI] [Google Scholar]
- 38.Hu L, Bentler PM. Cutoff criteria for fit indexes in covariance structure analysis: conventional criteria versus new alternatives. Struct Equ Modeling. 1999;6(1):1–55. doi: 10.1080/10705519909540118 [DOI] [Google Scholar]
- 39.Waldman ID, King CD, Poore HE, et al. Recommendations for adjudicating among alternative structural models of psychopathology. Clin Psychol Sci. 2023;11(4):616–640. doi: 10.1177/21677026221144256 [DOI] [Google Scholar]
- 40.Graham JM. Congeneric and (essentially) tau-equivalent estimates of score reliability: what they are and how to use them. Educ Psychol Meas. 2006;66(6):930–944. doi: 10.1177/0013164406288165 [DOI] [Google Scholar]
- 41.Hancock GR, Mueller RO. Rethinking construct reliability within latent variable systems. In: Cudeck R, du Toit S, Sorbom D, eds. Structural equation modeling: present and future—a festschrift in honor of Karl Jöreskog. Scientific Software International; 2001:195–216. [Google Scholar]
- 42.Revelle W, Zinbarg RE. Coefficients alpha, Beta, omega, and the glb: comments on Sijtsma. Psychometrika. 2009;74(1):145–154. doi: 10.1007/s11336-008-9102-z [DOI] [Google Scholar]
- 43.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria; 2022. [Google Scholar]
- 44.Frei O, Holland D, Smeland OB, et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat Commun. 2019;10(1):2417. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dick DM, Adkins AE, Kuo SIC. Genetic influences on adolescent behavior. Neurosci Biobehav Rev. 2016;70:198–205. doi: 10.1016/j.neubiorev.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young Stallings, Corley Krauter, Hewitt. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet. 2000;96(5):684–695. doi: 10.1002/1096-8628(20001009)96:53.0.CO;2-G [DOI] [PubMed] [Google Scholar]
- 47.Deak JD, Johnson EC. Genetics of substance use disorders: a review. Psychol Med. 2021;51(13):2189–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demange PA, Malanchini M, Mallard TT, et al. Investigating the genetic architecture of noncognitive skills using GWAS-by-subtraction. Nat Genet. 2021;53(1):35–44. doi: 10.1038/s41588-020-00754-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barr PB, Mallard TT, Sanchez-Roige S, et al. Parsing genetically influenced risk pathways: genetic loci impact problematic alcohol use via externalizing and specific risk. Transl Psychiatry. 2022;12:420. doi: 10.1038/s41398-022-02171-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallard TT, Savage JE, Johnson EC, et al. Item-level genome-wide association study of the alcohol use disorders identification test in three population-based cohorts. Am J Psychiatry. 2022;179(1):58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study were publicly available genome-wide association study summary statistics, which are available from the authors or online repositories of the respective publications.


