Abstract
The increasing reliance on online communities for healthcare information by patients and caregivers has led to the increase in the spread of misinformation, or subjective, anecdotal and inaccurate or non-specific recommendations, which, if acted on, could cause serious harm to the patients. Hence, there is an urgent need to connect users with accurate and tailored health information in a timely manner to prevent such harm. This article proposes an innovative approach to suggesting reliable information to participants in online communities as they move through different stages in their disease or treatment. We hypothesize that patients with similar histories of disease progression or course of treatment would have similar information needs at comparable stages. Specifically, we pose the problem of predicting topic tags or keywords that describe the future information needs of users based on their profiles, traces of their online interactions within the community (past posts, replies) and the profiles and traces of online interactions of other users with similar profiles and similar traces of past interaction with the target users. The result is a variant of the collaborative information filtering or recommendation system tailored to the needs of users of online health communities. We report results of our experiments on two unique datasets from two different social media platforms which demonstrates the superiority of the proposed approach over the state of the art baselines with respect to accurate and timely prediction of topic tags (and hence information sources of interest).
Index Terms—: Topic tag prediction, healthcare information, time sensitive data, interest Forecasting, e-healthcare, health informatics
I. Introduction
PERSONAL healthcare management increasingly requires accommodating a higher degree of involvement of patients and informal caregivers. Consequently, patients and their caregivers expect healthcare providers to provide more detailed information about the disease, prognosis, and treatment. However, there is often a disconnect between healthcare providers and patients (and their caregivers) in terms of language used, and the information that is shared with the patients. The situation is not much better with information available through electronic health records (EHR) [6], [14], [19].
This situation is exacerbated by the fact that only 12% of U.S. adults have proficient health literacy [25], [55]. Health literacy is especially low among older adults, minorities, groups with low income and socio-economic status, leading to increased healthcare usage and cost and worse health outcomes [38]. Given the difficulty of comprehending health information available from healthcare providers or personal EHR, patients and caregivers increasingly turn to the internet and online communities. In such communities, patients and their caregivers interact with and seek information, e.g., regarding treatment options, side-effects of medications, etc., from their peers, in simple, lay terms.
While online communities can be useful sources of information and emotional support for patients, they also increase the risk of misinformation [35], or subjective [56], anecdotal and inaccurate or non-specific recommendations, which, if acted on by patients, could cause them serious harm. Hence, there is an urgent need to connect users with accurate and tailored health information in a timely manner to prevent such harm.
Typically, in an online community, a user seeking information posts a natural language query; Peers, or in some cases, subject matter experts (e.g., clinicians or nurses) respond to the query or point to useful sources of information [50]. Somewhat more savvy users may be able to hone in on appropriate keywords or search tokens (tags) to use in order to seek relevant information on the web (Google, Bing, etc.), medical information boards (WebMD, Mayo, etc.) or specialized platforms (HealthUnlocked, PatientsLikeMe, etc.). The keywords used in such queries are suggestive of the respective user’s information needs. However, due to low health literacy, users may not necessarily be able to identify the right keywords to use; and it can be tedious and often frustrating for patients to sift through the results retrieved in response to their queries. Against this background, we explore a solution that aims to lower the barriers for patients to obtain accurate and understandable sources, e.g., articles written for laypersons that meet their health information needs. In this study, we use data from an online community HealthUnlocked,1 i.e., a social network designed to offer health-related information and emotional support for patients and their caregivers. A longitudinal study of HealthUnlocked has shown that it has positively impacted many patients [2] in terms of health outcomes and engagement in care. An example of the data extracted from HealthUnlocked is shown in Fig. 1. Though Fig. 1 shows only one post, a typical user interacts a lot more with multiple posts and provides their corresponding keywords (tags). We observe that in such a setting, information needs of a user may involve medications, side effects, treatment options and other time critical concerns. These needs depend on not only the patient demographics and health condition, but also, where the patient is with regard to progression of the disease (e.g., stage of cancer in the case of cancer patients), or treatment being given (e.g. chemotherapy), or response to treatment (e.g., side effects, cancer remission).
Fig. 1.
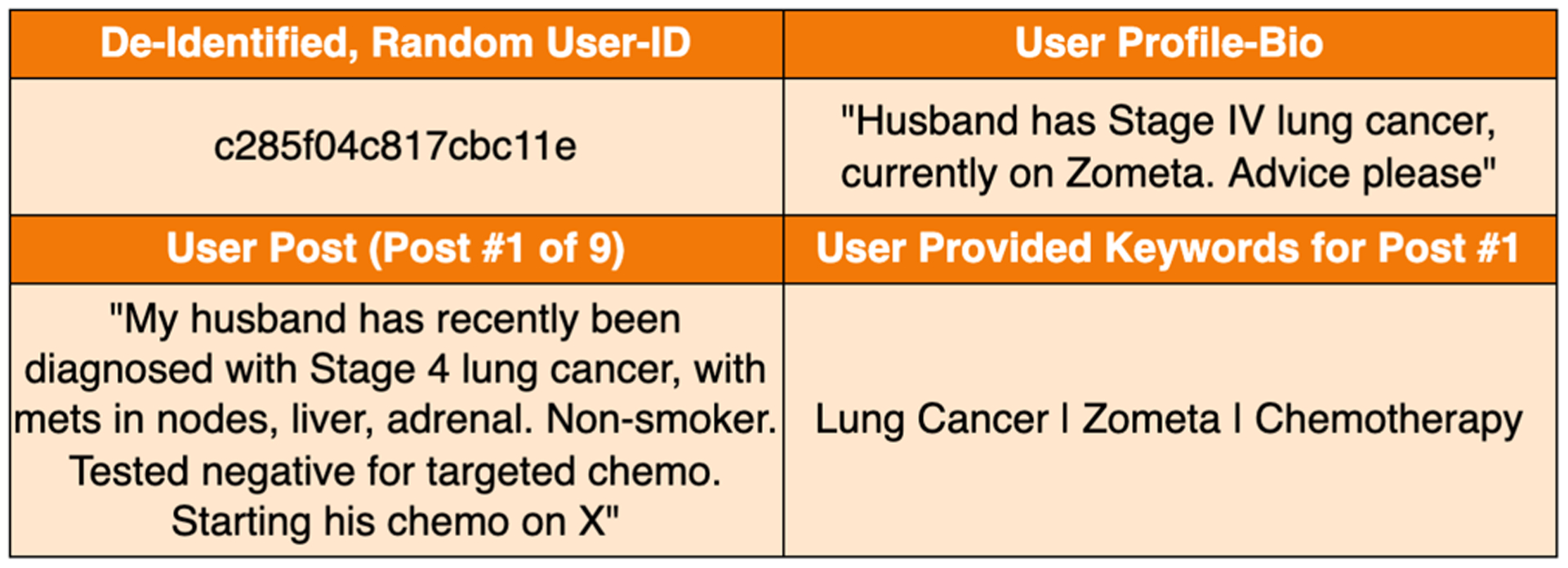
Example input snippet from HealthUnlocked; The data is de-identified by HealthUnlocked where dates and names within a post are removed. The date of the post, however, is made available.
A. Research Question
This study is designed to answer the following research question: Can the user profiles together with the traces of interactions of the user with the community (e.g., keywords used in past queries) and information from similar users be used to effectively predict contextually relevant topic tags associated with the future information needs of users?
We hypothesize that patients with similar histories of disease progression or with similar courses of treatment would have similar information needs at comparable stages. In treating many diseases, e.g., cancer physicians follow a particular course of treatment that is customized based on the patient’s profile and how they are responding to treatment. For example, the National Comprehensive Cancer Network (NCCN) has laid out guidelines for the treatment of non-small cell lung cancer [10]. If two users, say A and B, have similar histories of disease progression and treatment, we can expect that their information needs at comparable stages are likely to be similar. In Fig. 2, we illustrate an example with two users who have similar histories of disease progression and treatment. In this case, the topic recommendations for user B can be improved by utilizing user A’s past keywords. In this case, user B can be recommended articles related to “concurrent chemotherapy and radiation”.
Fig. 2.
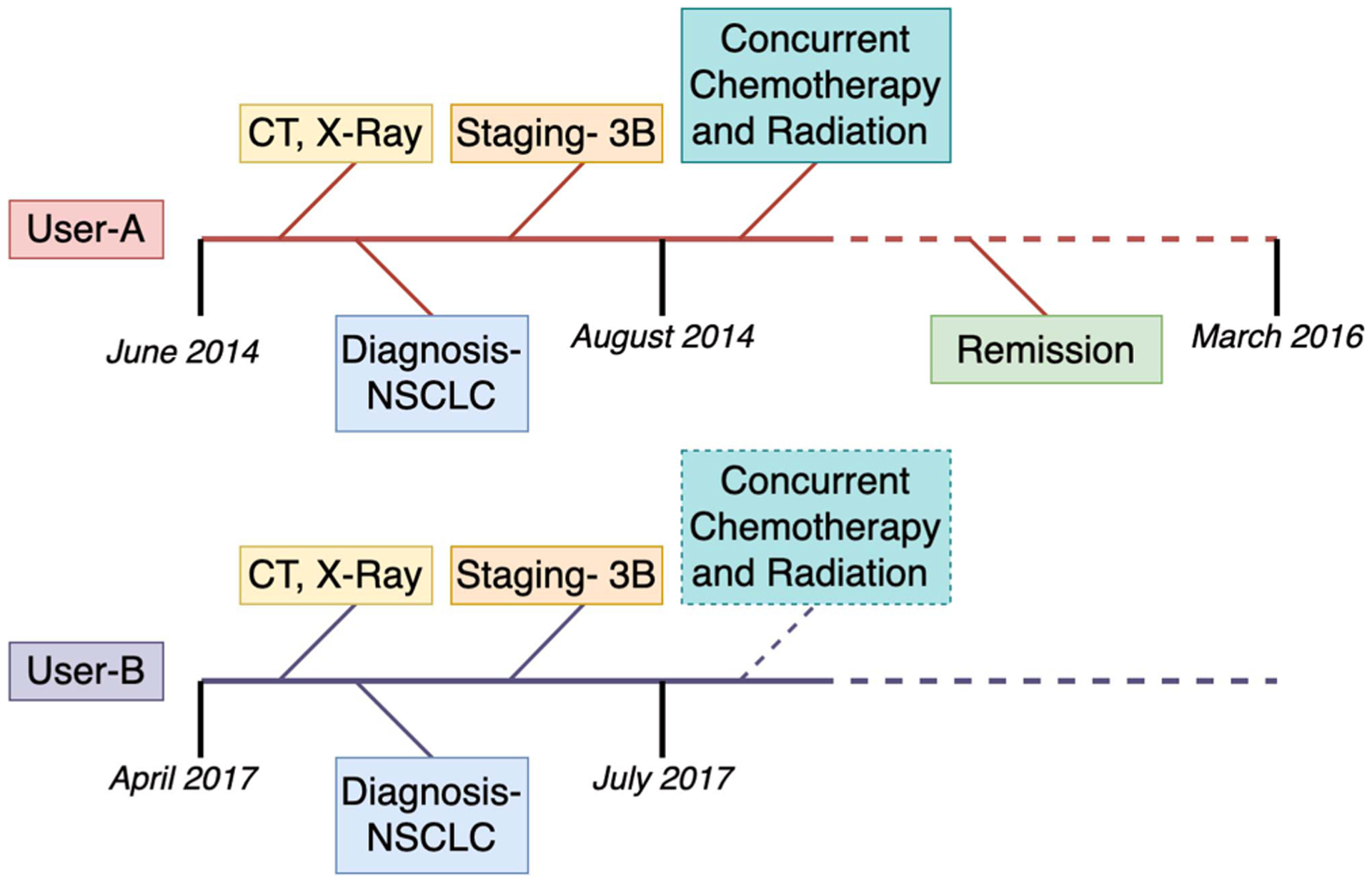
Similarity in relative disease timelines can help cluster similar users.
Based on the preceding discussion, we arrive at the following problem specification. For any given user U in the community, with a user profile BU, temporally ordered sequence of n prior posts , encoded by the corresponding keyword sets , and indexed by their position in the sequence as well as the past information traces of all the similar users in the network, the task is to predict the topic tags associated with a hypothetical future post that describes the future information needs of the user U.
Disambiguation:
“Tags” and “Keywords” can be used interchangeably in our work as they both describe the abstract token used to represent a larger body of text. However, we try to differentiate by using “Keywords” for tokens that have occurred prior to prediction and “Tags” as tokens that are yet to be predicted (future).
B. Contributions
We propose an innovative approach for predicting future information interests of participants in online communities as they move through different stages in their disease or treatment. We report results of our experiments on an expert curated data set from which we demonstrate the superiority of the proposed approach over the state of the art baselines with respect to accurate and timely prediction of topic tags (and hence information sources of interest).
The key contributions of this work include:
A novel approach to predicting topic keywords associated with the future information needs of users (patients, caregivers) in an online health community. We step away from traditional bag-of-words classification and towards a new sentence generation based topic prediction.
A novel methodology for collaborative-hybrid filtering based topic recommendation which leverages similarity of user disease progression timelines.
A semantic evaluation of predicted tags along with the traditional metrics such as precision and recall.
C. Related Work
The vast amounts of healthcare information available on the internet has led to the requirement of recommendation systems that help users navigate their care by enabling them with accurate information [1], [24]. Additionally, Deng and Liu [8] discuss the role of such systems in reducing risks like misinformation and time delays in access to accurate medical information. Yu et al. [53] review the recommendation models including the usage of collaborative filtering like [23], [28] and [41] and finds its relevance in healthcare topic recommendation. Unlike other user-centric recommendation systems, healthcare based models are a lot harder to be subjected to collaborative filtering as there is high degree of individuality in the information sought by the users [7]. Sahoo et al. [39] however, propose a CNN based feature aggregation approach to perform user-content based recommendation and presents improvements over personalization metrics. Similarly, Jiang et al. [18] create a similarity matching model based on heterogeneous networks to perform collaborative filtering for healthcare topic recommendation.
Extracting features from text documents is at the centre of our proposed topic recommendation system. Text documents are in essence a sequence of vectors placed together with context and grammar. Early approaches to model text included the usage of recurrent neural networks (RNNs) like in [15] and [36]. It became evident that the order (positions) of these vectors in relation to their value was vital in text based tasks. Bidirectional long-short term memory models (LSTMs) including [20], [33] and specifically healthcare applications like [17] emphasized the importance of positional embeddings of the word vectors and improved performance on tasks like translation and sequence-to-sequence text generation. Transformers, a recent advancement by [46] along with their attention modules have revolutionised feature extraction like in [49]. Bidirectional Encoder Representations from Transformers (BERTs) an advancement of transformers have additionally aided in text based tasks like classification [13], [30], summarization [29], [34] and text generation [5], [27]. They are ideal for sentence/token generation conditioned upon prior context [12].
Table I compares our work in view of current literature. The position of current healthcare recommendation literature along with advances in natural language processing technology pose the ideal setting for a novel medical information recommendation system that predicts the future interests of users through topic tags.
TABLE I.
Differentiating Existing Models and Our Approach
| # | Traditional Methods | Our Method | |
|---|---|---|---|
| 1 | Tag Prediction | Picking Tags from collection of tags [48] | Generating new words from vocabulary |
| 2 | Tag Prediction Training | Increase probability of picked tags [52] | Synthesize auxiliary sentences |
| 3 | Objective | Prediction of tags only for input document [40] [47] | Prediction of tags for unseen future document |
| 4 | User Clustering | Network interaction based [23] | Based on disease progression similarity |
| 5 | Vulnerability | Vulnerable to skewed tag distribution [45] | Robust to tag distribution |
| 6 | Evaluation Metrics | Precision@K (P@K), Recall@K (R@K) [18] | Cosine similarity in feature space along with P@K, R@K |
II. Data
We obtain our primary training data from HealthUnlocked.com (HU), which is one of the largest online health communities. The website has several sub-communities organized around specific health conditions or diseases. In our work, we focus on three such communities designated for lung cancer patients and their caregivers: Lung Cancer Support, The Roy Castle Lung Cancer Foundation and British Lung Foundation. Our choice of the communities was motivated by three considerations. First, managing chronic conditions and treatment regimes associated with lung cancer presents information needs that can be met through sustained interactions with the community. Second, chronic conditions impact the information needs of users over time, which are not adequately addressed by existing recommendation systems. Lastly, these communities represent the largest fraction of the users of the online community.
Each user has an opportunity to create a profile which includes demographic data, e.g., age, gender, etc. as well as the health condition. Once registered, a user then may pick one or more communities to join. Once in a community, users can post a “Main-Post” or post a “Comment” on an existing “Main-Post”. Additionally, each “Main-Post” can have associated “keywords” at the time of posting. These user supplied keywords can be considered to be indicative of the information needs of the user at the time and act as ground truth for training and evaluating the proposed tag prediction system.
We acquired de-identified data from the lung cancer communities for the period between 2011 and 2020. From the three lung cancer communities, we have a total of 1711 unique users. Note that ethical (or IRB) approval was not needed for our study because the data were de-identified at source (at HealthUnlocked), there is neither direct interaction nor intervention with human subjects, and there is no means to link the de-identified data to specific subjects. For this study, we focus on the subset of anonymized users with at least one post and only valid posts with a minimum of three words or more are considered. The resulting data now filters down to 1032 unique users. Each user has an anonymized hashed User-ID that links their profile-bio, posts, and keywords to the user. Among the 1032 unique users, the average posts per user is about 2.4 (minimum is 1 and maximum is 68). Each post has on average 3.2 keywords (minimum is 2 and maximum is 10). About 55% (574) of the users have a profile-bio. An example of a user’s information is provided in Fig. 1.
Due to the unstructured nature of the text in our data set, there is a need to create temporal traces for training which can be used by our model. For this purpose, we pose the user data as described below.
A. Structuring the Information Traces of Users
We organize the information associated with each user, including the profile bio and the traces of the user’s interaction with the community (information seeking posts or comments) as shown in Fig. 3. Here we assume that a user U creates a profile-bio B before they post their first post P0. At time T−1, we use the bio to predict tags where ∧ indicates the prediction. This prediction is made before the first post has been created. As time progresses, the user posts P0 at time T0 with keywords G0, We then accumulate the profile bio B, the post P0 and keywords G0 to predict the tags of a future unseen post P1. We continue to do this with a temporal window of Pn posts in the past along with the current post to predict tags of a future post. At time Tn, though we have n posts from the past, using all the posts and their keywords could add too much noise into the topic tag prediction system. We empirically question the influence of history (see Section IV-A2) and determine the ideal Pn. We then use only the last Pn posts, their associated keywords, and the profile bio which remains unchanged throughout time T0 to Tn, as well as the current post and corresponding keywords to predict a future topic tag .
Fig. 3.
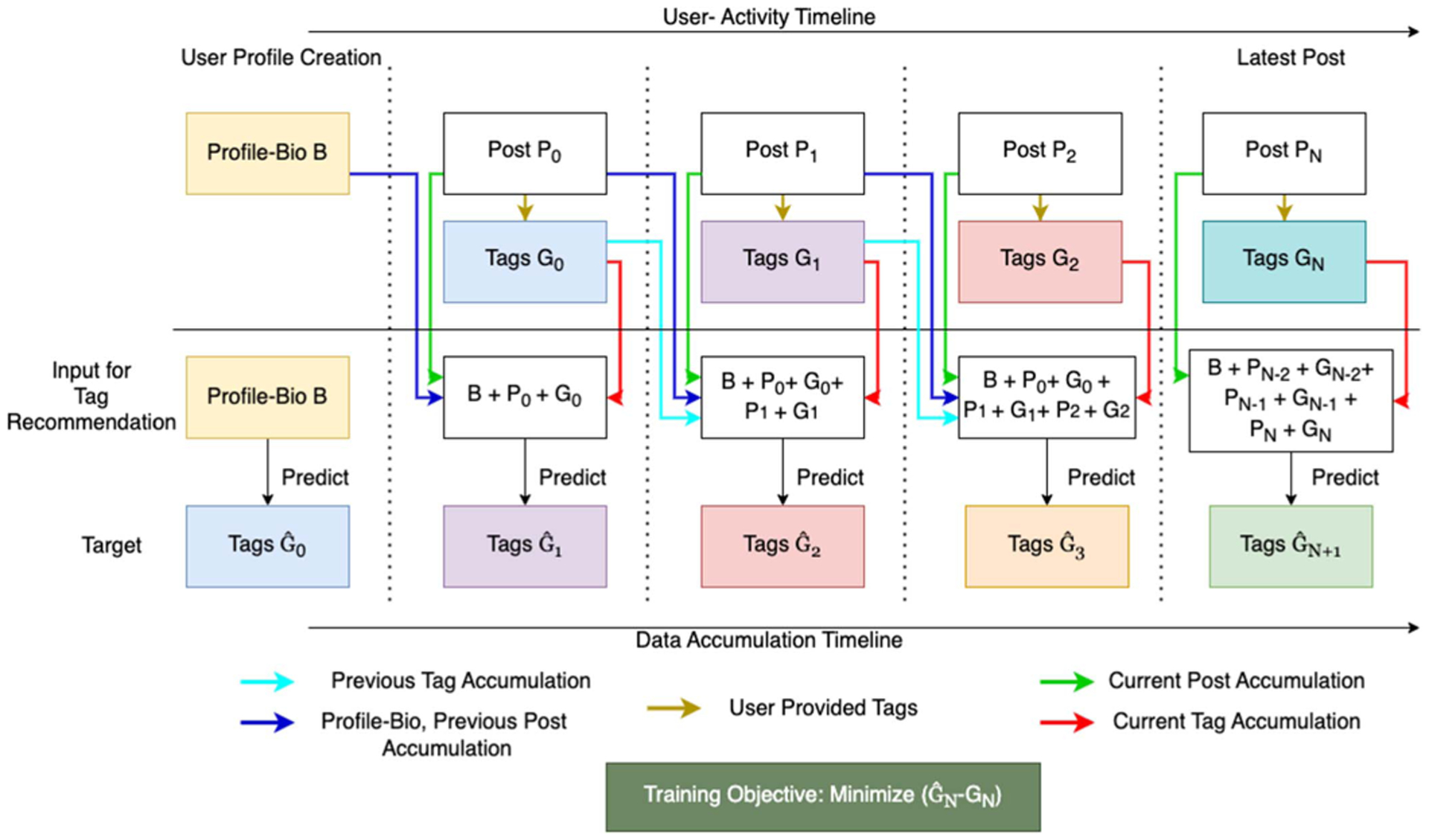
User Timeline and Data accumulation for temporal Tag Prediction.
B. Example Generation
To create a training example, we need an input sequence (input text document) containing profile-bio, posts and keywords and a target sequence (Tags of future post). This is commonly known as sequence-to-sequence training. In our setting, we increase our training example pairs by extracting all possible pairs of training data as shown in Fig. 4. In order to maintain chronological patterns in the data, we pick pairs that have information from consecutive time steps. For example, if Post-1 and Keywords-1 are from the latest time step, the target can only be Keywords-2 and cannot be used to predict Keywords-3. Using such a rule, a single user’s timeline is converted into several chronological training pairs. Following this, we obtain a total of 10,791 training pairs from the 1032 unique users. From this dataset, we use 8415 random pairs (78%) for training and the remaining 2376 (22%) pairs for testing.
Fig. 4.
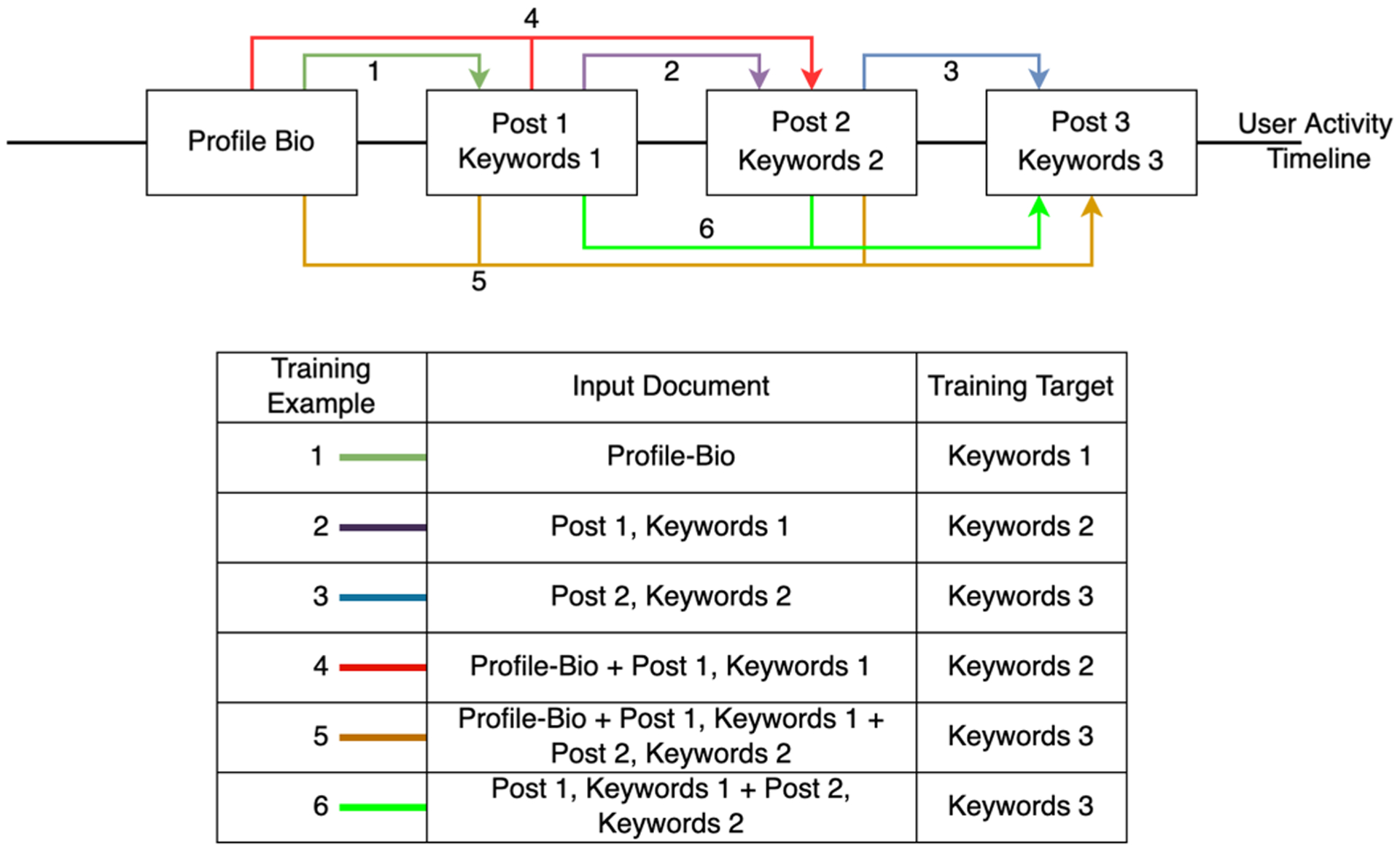
Timeline from a single user with a profile-bio and 3 posts is converted into 6 training examples.
C. Tag Statistics
Our training set has 472 unique tags while the test set has 375 tags. Among the 375 tags, 18 of them are not in the training set and are completely new words. However, since these words occur in the posts of the users, they are a part of the vocabulary that the models can observe and thus are not deleted from the test set. Fig. 5 shows the distribution of the top 20 tags in the test set. It is evident that ‘Chemotherapy’, ‘Cancer and Tumors’ occur several times more frequently than other tags. Tag prediction models designed around classification could incorrectly pick frequently occurring tags leading to poorer performance. Additionally, their metrics tend to be sensitive to the tag frequency in the training data. We address this issue of skewness and the resulting bias in Section IV-C.
Fig. 5.
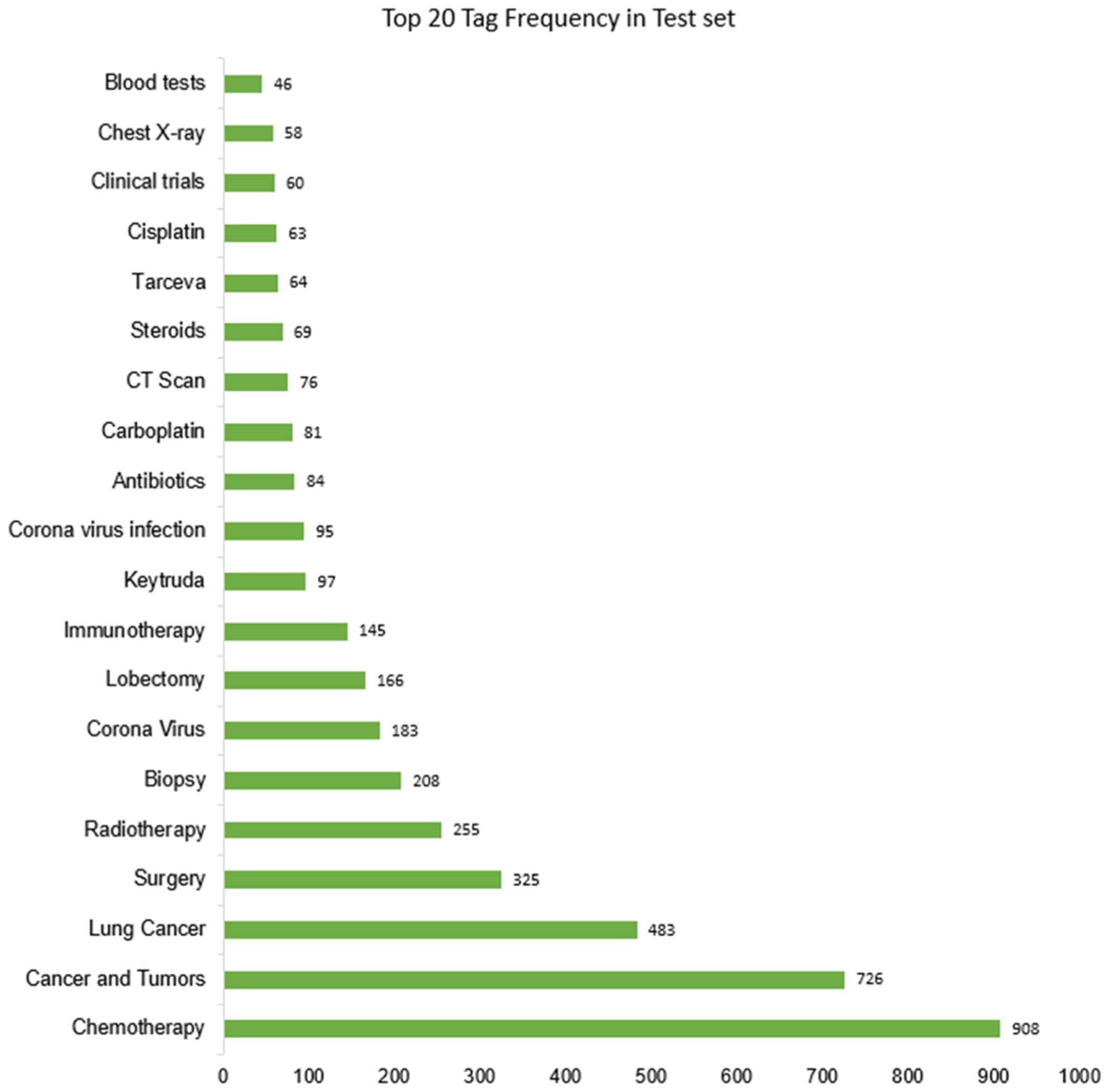
Plot of 20 most frequently occurring tags in test set.
D. Validation on Facebook Data
In order to validate generalizability of our approach to data from other social media platforms, upon training our proposed model, we ran validation on data extracted from a Facebook group named Chronic Pain Support Group.2 The data was extracted through web-crawling of posts between 2015 to 2021. All identifiable information like user-names, mentions and locations were removed. From the complete list of 11219 users who had posts, we selected those who have at least 4 consecutive posts. This resulted in a total of 1253 users. It should be noted that the training is only performed on the HU data and the Facebook data is reserved for testing. Unlike the HU dataset, the Facebook data does not contain ground truth tags provided by users. Here, we instead use a term frequency-inverse document frequency (tf-idf) method to extract 5 tags from each post and use them as the ground truth reference. Similar to the HU dataset, we first structure the information traces and generate examples for the purpose of validation.
III. Methodology
Recent advancements in the field of natural language processing (NLP) have opened a realm of possibilities due to a better understanding of context from text data. Bidirectional Encoder Representations from Transformers (BERT) [9] have further allowed text representations to be learnt from vast sources like the Book Corpus and the English Wikipedia with more than 3300 million words. Their utility has been recognized in the bio-informatics community through the proposal of Bio-BERT [26], an extension of the BERT, trained over the PubMed and PMC medical word corpora. The extension of the pre-trained models helps in inferring context, more effectively than the traditional frequency based models, from text containing medically relevant words including those from online health communities. We leverage the Bio-BERT to extract illness-relevant information and predict future topics of interest based upon the users’ needs.
Our pipeline is intended to extract context from the unstructured data and recommend accurate and time-relevant tags for each user in the online health community database. We make several modifications from works in the literature to accommodate our data and increase the overall accuracy of tag prediction. Among these, is our usage of Bio-BERT model as a sentence generator instead of a simple bag-of-words classifier. Additionally, we show the need for collaborative filtering but move away from traditional K-means method to a disease timeline based clustering method. In this section, we will describe our proposed tag prediction procedure, the model architecture, the objective function for performing the training and the metrics used to compare the models in our experiments.
A. Tag Prediction
Our primary objective in this work is to predict future topic tags for users in order to forecast their medical interests. Unlike existing tag prediction algorithms, we do not conform to a classification approach where a model picks the most probable tag from a set of tags. Instead, we perform an auxiliary sentence generation (ASG) task. The ASG task is similar to sentence completion where the last word is masked out using the [MASK] token. In our set-up, the sentence generated is a set of comma separated future tags with no grammar or punctuation. We fine-tune a pretrained Bio-BERT model to read a given input document and pick a word from its entire vocabulary that fits the context and position of the mask. A similar training approach has been employed by [5] for generating questions based on a paragraph of context and an answer sentence. Since we are using a Bio-BERT model Tag generation, we refer to our model as BBERTTg. An example of the training process has been shown in Fig. 6. First, we have a [CLS] token to indicate the start of the input document. The text in Orange is the profile-bio followed by a [SEP] separation token. Similarly, in Blue, we have the past and current posts, the Green text corresponds to tags from previous and current posts, the tags from neighboring users or shared tags (see Section III-D) are in Red and the predicted future tags are in Purple. We experimented with the positioning of these individual elements in the input document. However, they did not lead to a significant difference in the results. Having accumulated all the parts of the input document, we first clean the text through removal of stop words and lemmatization. Following this, the words are converted to vectors using a BERT word-embedding and tokenization module which converts the sentences into tokens based on the position of the words in the sentence and the position of the sentence in the input document. This encoded data is then fed to the Bio-BERT model along with the BERT-tokens like [CLS], [SEP], [MASK] etc.
Fig. 6.

Example of a sequential training routine of BBERTTg with 2 tag generations. The input document has multiple sentences and each sentence is separated by [SEP] token. Each iteration has a specific target. The target token at each iteration is appended at the position of the [MASK] token in the next iteration. A [SEP] token is added after each tag is predicted.
The pre-trained Bio-BERT model uses the originally prescribed BERTbase model with 12 layers, 768 hidden dimensions and 12 attention heads. We followed the training procedure set by [5] with the usage of an Adamax optimizer set with an initial learning rate of 5e-5. The training is performed over 15 epochs with 8415 examples while 2376 examples are used for testing. We use softmax as the final layer of the model where the Bio-BERT computes the probabilities of 200K+ words. Following literature, we adapt the Beam search [11] method to pick the top words and decode the predicted Bio-BERT output into regular words (tags).
Modification for Bio-BERT with long text Documents
The traditional transformer model is meant for short sentences as computing attention over large input documents is computationally expensive and requires lengthy training. Our input documents range from about 14 words to 550+ words when multiple data sources like profile-bio, posts, keywords etc. are combined. We modify our fine-tuned model based on the recommendations provided by [3] where a “longformer” is proposed. This reduces the attention computation from quadratic to linear space. Gated Recurrent Units (GRUs) and Long-Short Term Memory (LSTM) models, though not entirely bidirectional in attention, are however capable of handling such large documents without any modifications.
B. Objective Function
In our sequence-to-sequence based learning approach the input sequence is the input document containing profile bio and posts and the output sequence is the target tags set. Both the sequences are assumed to come from a single long sentence where the target sequence is masked. We first tokenize the input sequence x into n tokens, {xi}, i = 1, 2, …n, and predict the sentence fragment xp:q where p and q are positions such that p < q < n using the specially masked input which can be denoted as x/p:q. Unlike traditional tasks like translation, x does not have a paired y but we instead have to rely on the conditional probability of a token appearing next in the sequence given the past tokens (until the r-th position, p ≤ r ≤ q) of the sequence. Let us denote the domain from which x is obtained as χ, the log likelihood objective to learn the model parameters θ then becomes:
| (1) |
Expanding the conditional probability as a product of probabilities at each position between p and q, we obtain (2). Here, r is the position at which the [MASK] token is placed.
| (2) |
The position, or r, can be controlled to dictate the amount of prior information in the input document. In our experiments, we always have a non-empty set of tokens for prior data coming from either Profile Bios or Posts. However, (2) can be used with null prior data where it will generalize into the OpenAI GPT model for generating text [42].
C. Metrics
To evaluate the tag generation model, we can view it as a recommendation system which recommends topic tags. This is similar to medication recommendation evaluation by [31]. We can generate any number of tags once the model is trained as this would only involve generating longer sentences. Traditionally, if K is the number of tags recommended, Recall@K and Precision@K are used to evaluate the success of the recommendation. In order to enable fair comparison using both Recall and Precision, we use the popular F1@K metric where K is the number of predictions made and F1 is calculated with equal weights to both recall and precision.
We go beyond the traditional evaluation and calculate cosine similarity in the feature space between the recommended tags and the ground truth because the tag generation network is not picking words from a list but instead generating new words with a certain degree of variety. To calculate the cosine similarity, we first extract the word embeddings (features) of the K predicted tags and individually check their similarity to the word embeddings of the ground truth tags. If the ground truth tag is ‘radiotherapy’ and the predicted tag is ‘radiation therapy’ the penalty will not be too large.
D. Model Improvement Through Collaborative Filtering
Our approach for model improvement comes from the intuition that “similar users seek similar information”. Collaborative filtering is an approach to view a user as a parametric function of interests defined by other similar users in the community. Multiple recommendation systems including [21] and [44] utilize collaborative filtering to extract a deeper understanding of user needs and provide recommendations based on other users in the group.
Identifying similar users is a crucial step in performing collaborative filtering. In our setting, with healthcare data, we identify similar users by matching relative disease timelines. Users with similar illness progression tend to search for similar information. Information search history of users who have already traversed the timeline can be utilized to recommend topics to users trailing along similar paths. We hypothesize that the similarity in treatment profiles for chronic conditions like non-small cell lung cancer (NSCLC), can be leveraged to recommend information at different time intervals based on the disease progression.
Neighbor Tags:
For a query user U with a post PU and keywords GU posted on date T, we identify nearest neighbors h1, h2, ..hn using the novel disease timeline matching method described below. For each neighbor hi, we check for posts that have occurred prior to T. The keywords associated with such a post will be then referred to as Neighbor Tags or ST and are appended to the input document after the posts and their corresponding keywords (see Fig. 6). The entire input document is then used for future topic tag prediction.
Disease Timeline Matching for Identifying Neighbors:
We describe the procedure for our novel approach using the following steps:
The unstructured profile-bios are converted into a structured 13-column disease timeline using a Named Entity Recognition (NER) Bio-BERT model.
A Bio-BERT model was also used to extract features from the disease timelines and were clustered in the feature space using t-Stochastic Neighborhood Embedding (t-SNE).
The h nearest neighbors are identified using Euclidean distance in the two dimensional t-SNE feature space.
Keywords of neighbors which have occurred prior to query post are used as neighbor tags (ST).
Neighbor tags are then appended to the input document before being provided to the BBERTTg model.
Conversion of Profile-Bios:
In order to provide structure to the profile bios, we contextually sorted the text into various classes. The classes created are shown in Fig. 7. Where “Other” class corresponds to words that do not belong to any of the other 12 classes. “Current” refers to the time when the bio was written, “Past” and “Future” are with respect to the “current” time. As it can be observed, the data is forced to take a temporal shape in order to extract user-disease timeline. This annotation was done by two subject experts who manually read each profile bio and filled in the corresponding columns. We compared the similarity of the annotations and obtained a cosine similarity of 0.811 and a Cohen’s kappa of 0.784, which is only 0.16 short of “perfect agreement” [32]. An intersection of the labels between the annotators is used as ground truth for the NER text classification task.
Fig. 7.
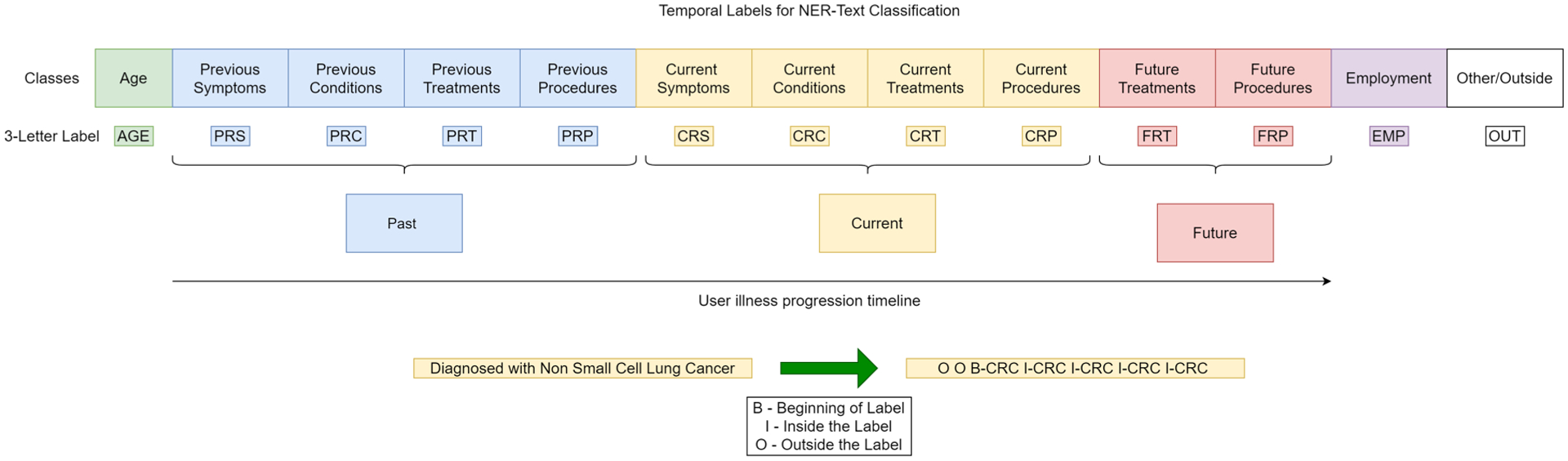
Temporal Named Entity Recognition task Labels based on illness progression.
In order to automate this conversion between unstructured profile bios to structured timeline data, we trained a NER model to perform a 13-class classification of profile-bio text. We perform classification only to obtain the temporal disease timeline and not the topic tags. We trained two models namely (i) Bio-BERT [26] and (ii) LUKE-Deep Contextualized Entity Representations with Entity-aware Self-attention [51]. The Bio-BERT is only fine-tuned on our dataset while LUKE was trained entirely from scratch on our dataset. This was done to evaluate the value of information provided by the Bio-BERT pre-training. For both models, we used the B,I,O labelling procedure (Beginning of a label, Inside a label and Outside a label) to maintain standardized practices followed in the NLP community and to increase reproducibility. In the training set, we used 414 profile bios with 44,320 total words. In the non overlapping test set, there were 19,014 words from 160 bios. The models were trained for 25 epochs on a Nvidia DGX Tesla V-100 GPU for approximately 3.5 hours for fine-tuning Bio-BERT and 18 hours for training the LUKE. The Bio-BERT model outperformed the LUKE classifier in the following metrics- (a) The overall accuracy of the Bio-BERT model was 80.59% while LUKE achieved 73.63%. (b) The average recall percentage for Bio-BERT was 74.98% while only 66.29% for LUKE. The imbalance in the dataset skews the predictions which lead to a high false positive rate (see Fig. 8).The precision for each of the classes is low due to the large number of words in the “Other (OUT)” category. The confusion matrix for Bio-BERT along with its class-wise precision and recall is shown as well. For all the downstream tasks that follow, we use the Bio-BERT model fine tuned on our dataset as the primary feature extractor. In cases when the user has not provided an input for any of the above mentioned classes, or if the model fails to pick the provided input, the model predicts an empty string for that particular class and we use an ‘NA’ token instead and proceed with feature extraction.
Fig. 8.
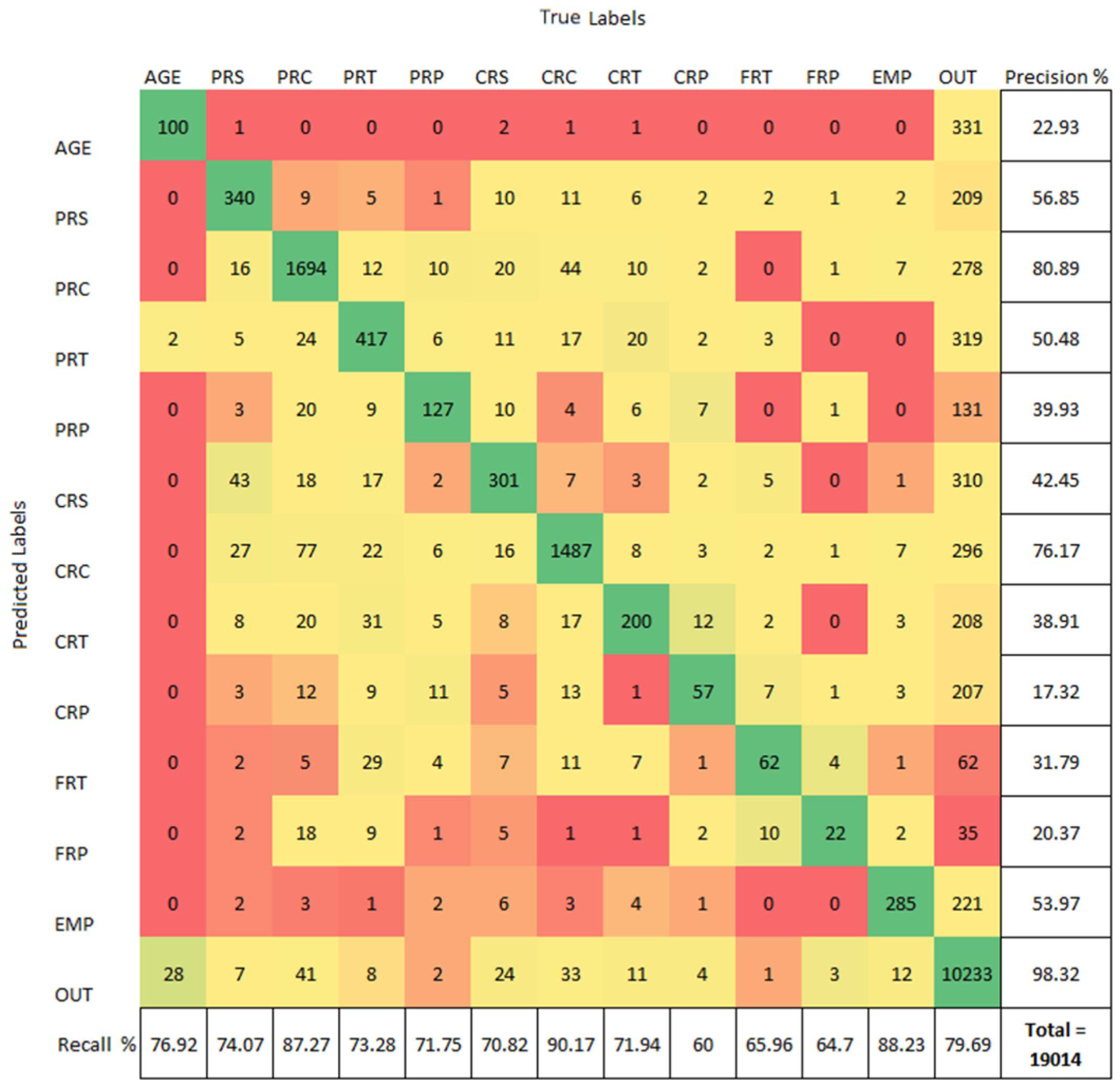
Confusion Matrix from Bio-BERT NER classification of profilebio text in test set.
Feature Extraction and t-SNE:
The next step is a clustering process to perform unsupervised clustering based on similarities in the column values. The 13 columns can be treated as a sentence vector for feature extraction. We employed the previously used Bio-BERT fine tuned on profile-bios to extract the features of the structured text data and map it to a two dimensional t-distributed stochastic neighbor embedding (t-SNE) space. Unlike K-means [54], with t-SNE, we do not have fixed clusters. Areas in the 2D plot (see Fig. 9) can be loosely defined as topics and users can belong to more than one particular topic similar to soft-clustering [22]. Features extraction can be performed from our filtered temporal disease timelines as well as directly from the profile bios. The comparison of the two feature extractions is visualized as shown in the t-SNE plots acquired after 750 iterations in Fig. 9. As it can be seen, features extracted from temporal timelines following NER form natural clusters compared to features directly extracted from profile bios. This is primarily due to the noise removal inherently performed by the NER task. For illustration, we pick a random user (#248) and check the nearest 3 users’ profile-bios. Inferred from Fig. 9(b), the clustering is around COPD and Emphysema. For the purpose of clustering, we find the nearest h users to the primary user using Euclidean distances in the t-SNE feature space. We accumulate all keywords of the neighbourhood users (ST), occurring prior to time T of current post and append it to the input document of the primary user. At the end of clustering, a particular user has the following items in their input document: (a) The profile-bio BU, (b) The previous posts and their corresponding keywords (c) The current post and its keywords and (d) Neighbor tags from similar users (ST). This input document is used to predict future tags .
Fig. 9.
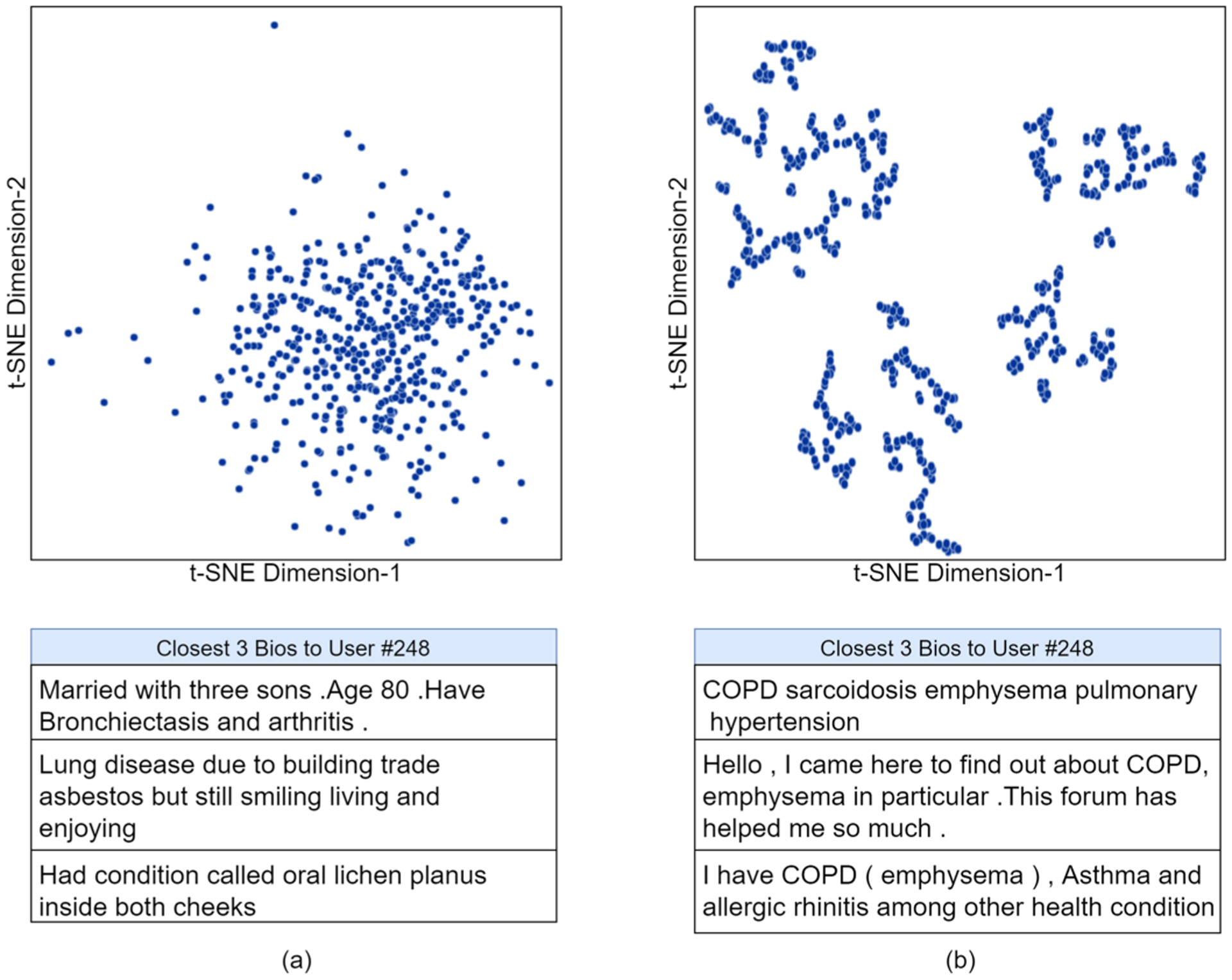
(a) t-SNE plot of features extracted directly from profile-bios of 414 users, Closest 3 bios (shortened for image) to random user #248; (b) t-SNE plot of features extracted from NER temporal-classified user timeline of 414 users, Closest 3 bios (shortened for image) to random user #248.
IV. Experiments
A. Model Ablation
We first determine the best settings for our proposed BBERTTg for the obtaining the highest tag prediction F1 score.
We experiment with 4 settings namely
Input Document Accumulation: Accumulation of different elements of the input document- (1) Profile Bios only (BU), (2) Profile Bios & Posts (BU+P U) (3) Profile Bios, Posts & Corresponding keywords (BU+PU+GU).
Influence of History: The number of past posts () to use for predicting future tags.
Different types of user clustering.
Ideal number of neighbors for collaborative-filtering.
1). Input Document Accumulation: Significance:
To understand the importance of different components of the input document.
We conducted three experiments with increasing amount of information presented in the input document. We start with the profile bio if available and then add posts and their corresponding keywords in the subsequent trials. For this experiment, we use 2 past posts along with the current post (Pn = 3). The average results with 4 runs each are shown in Table II.
TABLE II.
Influence of Data Accumulation in Input Document, Pn = 3, Averaged Over 4 Individual Runs
| K= 1 | K=3 | K=5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Input Document | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 |
| BU (Only profile bio) | 0.205 | 0.487 | 0.491 | 0.288 | 0.430 | 0.296 | 0.476 | 0.350 | 0.483 | 0.187 | 0.428 | 0.269 |
| B U + P U | 0.274 | 0.791 | 0.866 | 0.407 | 0.517 | 0.442 | 0.830 | 0.476 | 0.594 | 0.363 | 0.826 | 0.450 |
| BU+PU+GU (Full mode) | 0.296 | 0.874 | 0.909 | 0.442 | 0.590 | 0.584 | 0.891 | 0.586 | 0.681 | 0.402 | 0.887 | 0.505 |
2). Influence of History: Significance:
To understand the translation of user interests over time through posts.
Our dataset is primarily based on chronic health conditions, therefore, utilizing the past information through past posts is vital for predicting future topics of interest. In this experiment, we determine the ideal number of previous posts () required. corresponds to using only the current post while accumulates the last 3 posts along with the current post. For this experiment, we use the BU + PU + GU, referred to as full mode of data accumulation. Table III details the average results for 4 runs of this experiment.
TABLE III.
Influence of Past Posts in Input Document With Full Mode Data Accumulation, Averaged Over 4 Individual Runs
| Number of past posts in | K= 1 | K=3 | K= 5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Input Document | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 |
| Pn=l (Only current post) | 0.271 | 0.791 | 0.866 | 0.403 | 0.536 | 0.453 | 0.856 | 0.491 | 0.597 | 0.381 | 0.832 | 0.465 |
| Pn=2 (One previous post) | 0.277 | 0.820 | 0.909 | 0.414 | 0.545 | 0.567 | 0.879 | 0.555 | 0.657 | 0.395 | 0.876 | 0.493 |
| Pn=3 (Two previous posts) | 0.296 | 0.874 | 0.909 | 0.442 | 0.590 | 0.584 | 0.891 | 0.586 | 0.681 | 0.402 | 0.887 | 0.505 |
| Pn=4 (Three previous posts) | 0.286 | 0.856 | 0.887 | 0.428 | 0.593 | 0.577 | 0.887 | 0.584 | 0.671 | 0.381 | 0.852 | 0.486 |
| Pn=5 (Four previous posts) | 0.284 | 0.823 | 0.871 | 0.422 | 0.555 | 0.502 | 0.874 | 0.527 | 0.649 | 0.377 | 0.834 | 0.476 |
3). Type of Clustering: Significance:
To identify the best performing user clustering method. We experiment with three forms of clustering namely:
Grouping of user profile-bios using K-means clustering.
Using profile-bio text features extracted from Bio-BERT model for feature space clustering (t-SNE).
Novel disease timeline based similarity matching of users, which is our proposed and adopted approach.
For K-means clustering, we vectorized the words using scikit-learn [37] TfidfVectorizer package and used a gap statistic based elbow approach [43] to identify the ideal number of clusters. We experimented with 2 to 80 clusters and found the characteristic elbow at 34 clusters. Following this, we fit the profile bios into 34 multi-user clusters. The average number of users in each of these clusters were approximately 11. We chose clusters with at least 4 users (1 query user and 3 neighbors).
In the case of profile-bio based feature extraction, we replaced the TfidfVectorizer with a Bio-BERT model to improve the semantic representation of profile-bios in the feature space. We reduced the Bio-BERT feature vectors to a two dimensional t-SNE space and found the 3 (h = 3) nearest neighbors using simple euclidean distances.
The final method compared is our novel temporal disease timeline matching approach described in Section III-D. The comparison is performed through improvement in topic tag prediction F1 score as detailed in Table IV. Please note the improvement in F1 score at K=5 level with 5 tag predictions.
TABLE IV.
Comparison of User Grouping Approaches With h = 3 Neighbors, Pn = 3 and Full Mode Data Accumulation, Averaged Over 4 Individual Runs; No Neighbors and Random Neighbors Are Included as Baselines
| K= 1 | K=3 | K=5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clustering Method | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 |
| No Clustering, No Neighbors | 0.296 | 0.874 | 0.909 | 0.442 | 0.590 | 0.584 | 0.891 | 0.586 | 0.681 | 0.402 | 0.887 | 0.505 |
| No Clustering, 3 Random Neighbors | 0.277 | 0.813 | 0.763 | 0.413 | 0.510 | 0.526 | 0.730 | 0.517 | 0.597 | 0.383 | 0.700 | 0.466 |
| K-Means of Profile-Bios [54] | 0.290 | 0.865 | 0.812 | 0.434 | 0.572 | 0.545 | 0.742 | 0.558 | 0.610 | 0.384 | 0.765 | 0.471 |
| BBERT Features of Profile-Bios | 0.301 | 0.880 | 0.911 | 0.448 | 0.629 | 0.612 | 0.892 | 0.620 | 0.708 | 0.491 | 0.887 | 0.579 |
| BBERT Features of NER Disease Timelines | 0.317 | 0.952 | 0.916 | 0.475 | 0.661 | 0.649 | 0.892 | 0.654 | 0.739 | 0.524 | 0.890 | 0.613 |
4). Ideal Number of Neighbors: Significance:
To identify the ideal number of neighbors to be used for tag sharing.
Having established the ideal clustering model, we experiment with number of neighbors to determine influence of similar users in the community. Table V presents the results with 5 levels of neighbors.
TABLE V.
Influence of Neighbourhood Users in Input Document With Pn = 3 and Full Mode Data Accumulation, Averaged Over 4 Individual Runs
| K=1 | K=3 | K=5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neighbourhood Users | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 |
| h=0 (Full Mode) | 0.296 | 0.874 | 0.909 | 0.442 | 0.590 | 0.584 | 0.891 | 0.586 | 0.681 | 0.402 | 0.887 | 0.505 |
| h= 1 | 0.302 | 0.898 | 0.911 | 0.451 | 0.601 | 0.597 | 0.891 | 0.598 | 0.712 | 0.437 | 0.889 | 0.541 |
| h=2 | 0.319 | 0.957 | 0.916 | 0.478 | 0.648 | 0.636 | 0.891 | 0.641 | 0.730 | 0.495 | 0.889 | 0.589 |
| h=3 | 0.317 | 0.952 | 0.916 | 0.475 | 0.661 | 0.649 | 0.892 | 0.654 | 0.739 | 0.524 | 0.890 | 0.613 |
| h=A | 0.316 | 0.948 | 0.916 | 0.474 | 0.655 | 0.640 | 0.890 | 0.647 | 0.733 | 0.501 | 0.877 | 0.595 |
| h=5 | 0.309 | 0.912 | 0.908 | 0.461 | 0.652 | 0.638 | 0.888 | 0.644 | 0.725 | 0.460 | 0.871 | 0.562 |
B. Model Comparison
1). Comparison With HealthUnlocked Dataset:
In the second set of experiments, we compare our best model (Full-Mode with shared tags, Pn = 3, h = 3) with the current state-of-the-art (SOTA) topic tag recommendation techniques using the same metrics. We also establish a baseline using Latent Dirichlet allocation (LDA) method of tag prediction with an implementation similar to [4], [16]. We identified two SOTA models based on architectural and feature extraction differences. The first is HashRec [48] and the second is Attention-based Multimodal Neural Network Model for Hashtag Recommendation (AMNN) [52]. HashRec is a hashtag recommendation model for social media text, specifically, conversational text on Twitter and Weibo. The model uses two encoders to encode the user posts and the conversation individually, followed by a bi-attention module to capture their interactions. The extracted features are further merged and fed into the hashtag decoder consisting of sequential Gated Recurrent Units (GRUs). We provide the first encoder with Profile Bios and the second encoder with a combination of prior posts, their keywords and shared tags from the neighborhood. We train the model to produce future tags sequentially. The default (Twitter) hyper-parameters and settings were used. AMNN is an attention based model for predicting tags from multi-modal data. The model uses two individual encoders, one for text and one for images consisting of bi-directional Long- Short Term Memory (LSTM) modules followed by an attention unit. The features are concatenated and fed to a sequential GRU model to predict tags. We modified the network to have two text encoders by removing the image encoder and the remainder of the network is unchanged. Similar to the previous model, the profile bios are provided to one of the encoders and the remaining input document is given to the next text encoder. We used the default hyper-parameters and settings to obtain results. The SOTA models are trained and tested on the same training set and test set used for our Bio-BERT (BBERTTg) model. Table VI reports the results of this experiment. The best performance is in bold while the second best is underlined.
TABLE VI.
Model Comparison With State of the Art Works and BbertCl
| K= 1 | K=3 | K=5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 | Recall | Precision | Cosine Similarity | F1 |
| LDA | 0.137 | 0.419 | 0.642 | 0.206 | 0.320 | 0.445 | 0.619 | 0.372 | 0.480 | 0.203 | 0.606 | 0.285 |
| AMNN | 0.264 | 0.794 | 0.847 | 0.396 | 0.585 | 0.529 | 0.795 | 0.555 | 0.627 | 0.497 | 0.715 | 0.554 |
| HashRec | 0.278 | 0.836 | 0.771 | 0.417 | 0.602 | 0.563 | 0.736 | 0.581 | 0.681 | 0.503 | 0.773 | 0.578 |
| BBERTCl (Ours) | 0.266 | 0.797 | 0.834 | 0.398 | 0.536 | 0.499 | 0.718 | 0.516 | 0.593 | 0.438 | 0.695 | 0.503 |
| BBERTTg (Ours) | 0.317 | 0.952 | 0.916 | 0.475 | 0.661 | 0.649 | 0.892 | 0.654 | 0.739 | 0.524 | 0.890 | 0.613 |
2). Validation on Facebook Data:
In this experiment, we use the previously trained models to predict topics of interest for users of the Facebook group Chronic Pain Support Group (data described in Section II-D). Each model is provided with 2 previous posts and 1 current post (3 posts in total) and asked to predict the topics of the unseen 4th post. A total of 1310 examples from 1253 users were used for the validation. This out of distribution prediction is a challenging task as the models are only trained on lung cancer data while the validation is entirely on the chronic pain dataset. We compare our top model (BBERTTg) against other models with 5 (K = 5) tag predictions. The average results from 4 runs of this experiment are presented in Table VII.
TABLE VII.
Validation of Models With Facebook “Chronic Pain” Dataset, K = 5
| K=5 | ||||
|---|---|---|---|---|
| Model | Recall | Precision | Cosine Similarity | F1 |
| LDA | 0.197 | 0.184 | 0.208 | 0.190 |
| AMNN | 0.350 | 0.223 | 0.346 | 0.272 |
| HashRec | 0.398 | 0.275 | 0.341 | 0.325 |
| BBERTTg | 0.644 | 0.519 | 0.726 | 0.574 |
C. Data Bias Experiments
Classification based models even with Bio-BERT embeddings do not perform as accurately as text generation models due to the imbalance in the prediction classes (tags). We show this by comparing a Bio-BERT classification (BBERTCl) model against our proposed tag generation model (BBERTTg). This is included in Table VI. Very frequently occurring tags can skew results as they make it easier for models to guess the most frequent tag multiple times. We checked for the robustness of LDA and the BBERTTg models by deleting the top two tags (Chemotherapy and Cancer and Tumors) from the train and test sets. This experiment is to measure the impact of tag frequency on the models. As each example in the test set had 5 or more tag targets, dropping two tags did not require omission of test examples. Table VIII shows the F1 scores when (K = 3) tags are predicted with the two modified models.
TABLE VIII.
Impact of Frequent Tags in Tag Prediction Model With K = 3 (Brackets Indicate the Change in Absolute Value)
| K=3 | ||||
|---|---|---|---|---|
| Model | Recall | Precision | Cosine Similarity | F1 |
| LDA | 0.320 | 0.445 | 0.619 | 0.372 |
| BBERTTg | 0.661 | 0.649 | 0.892 | 0.654 |
| LDA −2Tg | 0.257 (−0.063) | 0.372 (−0.073) | 0.429 (−0.190) | 0.303 (−0.069) |
| BBERT−2Tg | 0.658 (−0.003) | 0.643 (−0.006) | 0.878 (−0.014) | 0.650 (−0.004) |
V. Interpretation of Results
A:
Modeling information seeking behavior in a healthcare setting requires identifying key variables like current stage of disease, disease similarity and history of interests. We designed our ablation studies to identify these factors and leverage the information to develop an accurate temporal tag prediction model.
A1:
From Table II we observe that the amount of information presented to the BBERTTg model largely affects the model’s ability to understand topics of interest. While the profile bios are instrumental in capturing overarching context and history, they poorly correlate with the users’ dynamic interests when considered individually, yielding the lowest F1 score. Thus utilizing multiple data sources like the posts and their corresponding keywords help in improving predictions.
A2:
As anticipated, the number of previous posts in the input document causes variations in the tag prediction metrics (see Table III). The posts in our dataset are asynchronous which means the time gap between them can be as small as 5 minutes or as large as multiple years. As the disease progresses, users have varying interests and if the time gap is larger, two or more consecutive posts will have completely different topics discussed. This confuses the model to pay attention to incorrect topic-keywords which are no longer of interest. We notice that the F1 score is the highest when 2 previous posts along with the current post which is the ideal trade-off point.
A3-A4:
Improvements to the model come through collaborative filtering, an additional source of information regarding similar users in the database. In Table IV, experimenting with random neighbors yielded lower F1 scores at all levels indicating that including search tags from random users only caused the model to pay attention to noisy inputs. While on the other hand, Bio-BERT features extracted from cleaned NER disease timelines yielded the cleanest set of neighbors who positively contributed to the tag prediction metrics. The shared tags (St) from neighbouring users helped the Bio-BERT model predict better as it is likely that they have queried about this topic in the past. An interesting observation from Table V is that the number of nearest users did not affect the cosine similarity significantly. We believe the reason for this is that since cosine similarity is calculated in the feature space, the user clusters are also defined in a similar feature space. Due to this, adding more users does not negatively affect cosine similarity. It did however point out that using more than 3 (h > 3) neighbors could induce noise in the model due to uniqueness in the timelines. Hence we use h = 3 or three neighbors for our final model.
B1:
In Table VI, we illustrate comparison of our proposed BBERTTg model against the state of the art networks. Tag prediction models tend to perform well when the tags to be predicted depend only on the input document provided. However, understanding context and predicting the a future tag is a non-trivial task that leads to low F1 score with the HashRec and AMNN models. To overcome this, we pose the problem as a sentence generation task instead of the traditional classification task and obtain benchmark performance. We even use the Bio-BERT embeddings in a classification task (BBERTCl) to show that sentence generation performs better due to robustness to skewed tag distribution which is usually the case with social media based text data. To show that our model is truly understanding the context, we show an example of generated tags for a future post by providing the previous posts, tags, profile-bio etc to all the models in comparison. The future post not provided to the model was: “My partner was diagnosed with stage 4 lung cancer (nsclc) in June 2009. He has had Chemotherapy, Radiotherapy and Tarceva. He started taking the new unlicensed drug Afatinib yesterday. So far no side effects! He has been told that the side effects can be more severe than Tarceva. I really hope this drug works for him as we are fast running out of options.” Our BBERTTg model was the only one to predict the tag Afatinib and also understand that the patient is undergoing a drug trial.
B2:
In this validation experiment using Facebook data, Table VII demonstrates the superiority of BBERTTg across all metrics even without collaborative filtering as it extracts and understands context and relies on the vocabulary to generate tags unlike other models which are restricted to their trained keywords. Please note that due to the unavailability of Profile-Bios in the Facebook dataset, we do not perform collaborative filtering and thus we only use the BBERTTG(full-mode) for experiments. The model’s ability is exemplified in the qualitative comparison presented in the second part of Table IX. The Facebook post from the Chronic Pain dataset that was not provided to the model was: “Amitriptyline was most effective but once the disc tore it stopped working, and the dosage was raised but only caused fatigue and weight gain, and withdrawal caused the itching to start that has not gone away completely.” In the validation trial, BBERTTg was able to predict tags unrelated to lung cancer and closer to chronic pain while all other models had some cancer related tags.
TABLE IX.
Qualitative Comparison of Tag Prediction
| Ground Truth Ibgs for HealthUnlocked Dataset (Future Post): Chemotherapy / Radiolherapy / Tarceva / Afatinib / Cancer and Tumors | |
|---|---|
| Model | Predicted Tags |
| LDA | Chemotherapy / Cancer / Lung / Treatment / Medication |
| HashRec | Chemotherapy / Cough / Cancer / Infection / Medication |
| AMNN | Chemotherapy / Tumor / Medication / Respiratory / Cough |
| BBERTCl | Chemotherapy / Infection / Cancer / Tumors / Cough |
| BBERTTg (Full Mode) (No-Neighbors) | Chemotherapy / Infection / Tumors / Radiotherapy / Cancer |
| BBERTTg (Ours) (3 Neighbors) | Chemotherapy / Radiotherapy / Afatinib / Cancer and Tumors / Drug Trial |
| Ground Truth Tbgs for Face book Chronic Pain Dataset (Future Post): Fatigue / Lumbar / Disc Tear / Weight Gain / Withdrawal | |
| Model | Predicted Tags |
| LDA | |
| HashRec | Back Pain / Cancer / Side Effects / Headache / Medication |
| AMNN | Sitting / Tumor / Back Pain / Metastasis / Surgery |
| BBERTTg (Full Mode) (No-Neighbors) | Tiredness / Side Effects / Weight Gain / Lower Back / Tylenol |
C:
Finally, we emphasize that our BBERTTg model is robust to tag occurrence frequency and show that even when the 2 most frequent tags are removed, the performance does not change by much when compared to a traditional LDA model as shown in Table VIII.
VI. Summary AND Discussion
In summary, our topic recommendation system, a hybrid system combining collaborative-filtering and content based ideas is designed for online health community users and utilises patient timeline similarity to group similar users. We use a pretrained Bio-BERT to perform the tasks of classification and sequential sentence generation. Our predictions of future topics yield accurate and contextual tags. We compare our model against models proposed for similar tasks like HashRec and AMNN and observe a superior performance.
We have empirically presented the quantitative advantage of our methodology over existing models. This improvement in precision and recall metrics across two unique datasets is attributed to two specific contributions- (1) An auxiliary sentence generation task for tag generation instead of tag classification and (2) Usage of “disease timelines” to match similar users and perform collaborative filtering. Additionally, we also performed ablation studies to understand the importance of several factors like history and influence of similar users in the network. As a part of future work, we will focus on two specific aspects. First, we intend to improve the matching of similar users by leveraging interactions and graphs. Second, we plan to utilize the predicted tags to retrieve tailored medical articles from trusted sources like WebMD and Mayo clinic.
Acknowledgment
We also would like to thank HealthUnlocked.com for providing the data used in this work.
This work was supported in part by the NSF Center for Health Organization and Transformation (CHOT), in part by the Penn State Institute of Computational and Data Sciences (ICDS), and in part by the Federal Ministry of Education and Research (BMBF), Germanyunder the project LeibnizKILabor under Grant 01DD20003.
Footnotes
[Online]. Available: https://healthunlocked.com/
Contributor Information
Amogh Subbakrishna Adishesha, College of Information Sciences and Technology, Pennsylvania State University, State College, PA 16801 USA.
Lily Jakielaszek, College of Information Sciences and Technology, Pennsylvania State University, State College, PA 16801 USA.
Fariha Azhar, College of Engineering, Pennsylvania State University, State College, PA 16801 USA.
Peixuan Zhang, Department of Industrial and Manufacturing Engineering, Pennsylvania State University, State College, PA 16801 USA.
Vasant Honavar, College of Information Sciences and Technology, Pennsylvania State University, State College, PA 16801 USA.
Fenglong Ma, College of Information Sciences and Technology, Pennsylvania State University, State College, PA 16801 USA.
Chandra Belani, Department of Medicine, Penn State College of Medicine, Hershey, PA 17033 USA.
Prasenjit Mitra, College of Information Sciences and Technology, Pennsylvania State University, State College, PA 16801 USA.
Sharon Xiaolei Huang, College of Information Sciences and Technology, Pennsylvania State University, State College, PA 16801 USA.
References
- [1].Aceto G, Persico V, and Pescapé A, “The role of information and communication technologies in healthcare: Taxonomies, perspectives, and challenges,” J. Netw. Comput. Appl, vol. 107, pp. 125–154, 2018. [Google Scholar]
- [2].Barrett M, Oborn E, and Orlikowski W, “Creating value in online communities: The sociomaterial configuring of strategy, platform, and stakeholder engagement,” Inf. Syst. Res, vol. 27, no. 4, pp. 704–723, 2016. [Google Scholar]
- [3].Beltagy I, Peters ME, and Cohan A, “Longformer: The long-document transformer,” 2020, arXiv:2004.05150. [Google Scholar]
- [4].Blei DM, Ng AY, and Jordan MI, “Latent dirichlet allocation,” J. Mach. Learn. Res, vol. 3, pp. 993–1022, 2003. [Google Scholar]
- [5].Chan Y-H and Fan Y-C, “A recurrent Bert-based model for question generation,” in Proc. 2nd Workshop Mach. Reading Question Answering, 2019, pp. 154–162. [Google Scholar]
- [6].Day FC et al. , “Feasibility study of an EHR-integrated mobile shared decision making application,” Int. J. Med. Inform, vol. 124, pp. 24–30, 2019. [DOI] [PubMed] [Google Scholar]
- [7].Deng X and Huangfu F, “Collaborative variational deep learning for healthcare recommendation,” IEEE Access, vol. 7, pp. 55679–55688, 2019. [Google Scholar]
- [8].Deng Z and Liu S, “Understanding consumer health information-seeking behavior from the perspective of the risk perception attitude framework and social support in mobile social media websites,” Int. J. Med. Inform, vol. 105, pp. 98–109, 2017. [DOI] [PubMed] [Google Scholar]
- [9].Devlin J, Chang M-W, Lee K, and Toutanova K, “BERT: Pre-training of deep bidirectional transformers for language understanding,” 2018, arXiv:1810.04805. [Google Scholar]
- [10].Ettinger DS et al. , “NCCN guidelines insights: Non–small cell lung cancer, version 1.2020: Featured updates to the NCCN guidelines,” J. Nat. Comprehensive Cancer Netw, vol. 17, no. 12, pp. 1464–1472, 2019. [DOI] [PubMed] [Google Scholar]
- [11].Freitag M and Al-Onaizan Y, “Beam search strategies for neural machine translation,” 2017, arXiv:1702.01806. [Google Scholar]
- [12].Gao S et al. , “Limitations of transformers on clinical text classification,” IEEE J. Biomed. Health Inform, vol. 25, no. 9, pp. 3596–3607, Sep. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Garg S and Ramakrishnan G, “BAE: Bert-based adversarial examples for text classification,” in Proc. Conf. Empirical Methods Natural Lang. Process, 2020, pp. 6174–6181. [Google Scholar]
- [14].Genes N, Violante S, Cetrangol C, Rogers L, Schadt EE, and Chan Y-FY, “From smartphone to EHR: A case report on integrating patient-generated health data,” NPJ Digit. Med, vol. 1, no. 1, pp. 1–6, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guo L, Zhang D, Wang L, Wang H, and Cui B, “CRAN: A hybrid CNN-RNN attention-based model for text classification,” in Proc. Int. Conf. Conceptual Model, 2018, pp. 571–585. [Google Scholar]
- [16].Hassan HAM, Sansonetti G, Gasparetti F, and Micarelli A, “Semantic-based tag recommendation in scientific bookmarking systems,” in Proc. 12th ACM Conf. Recommender Syst, 2018, pp. 465–469. [Google Scholar]
- [17].Jelodar H, Wang Y, Orji R, and Huang S, “Deep sentiment classification and topic discovery on novel Coronavirus or COVID-19 online discussions: NLP using LSTM recurrent neural network approach,” IEEE J. Biomed. Health Inform, vol. 24, no. 10, pp. 2733–2742, Oct. 2020. [DOI] [PubMed] [Google Scholar]
- [18].Jiang L and Yang CC, “User recommendation in healthcare social media by assessing user similarity in heterogeneous network,” Artif. Intell. Med, vol. 81, pp. 63–77, 2017. [DOI] [PubMed] [Google Scholar]
- [19].Joukes E, de Keizer NF, de Bruijne MC, Abu-Hanna A, and Cornet R, “Impact of electronic versus paper-based recording before EHR implementation on health care professionals’ perceptions of EHR use, data quality, and data reuse,” Appl. Clin. Inform, vol. 10, no. 2, pp. 199–209, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kågebäck M and Salomonsson H, “Word sense disambiguation using a bidirectional LSTM,” 2016, arXiv:1606.03568. [Google Scholar]
- [21].Kaur H, Kumar N, and Batra S, “An efficient multi-party scheme for privacy preserving collaborative filtering for healthcare recommender system,” Future Gener. Comput. Syst, vol. 86, pp. 297–307, 2018. [Google Scholar]
- [22].Kim J, Yoon J, Park E, and Choi S, “Patent document clustering with deep embeddings,” Scientometrics, vol. 123, pp. 563–577, 2020. [Google Scholar]
- [23].Klašnja-Milićević A, Ivanović M, Vesin B, and Budimac Z, “Enhancing e-learning systems with personalized recommendation based on collaborative tagging techniques,” Appl. Intell, vol. 48, no. 6, pp. 1519–1535, 2018. [Google Scholar]
- [24].Kruse CS and Beane A, “Health information technology continues to show positive effect on medical outcomes: Systematic review,” J. Med. Internet Res, vol. 20, no. 2, 2018, Art. no. e8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kutner M, Greenburg E, Jin Y, and Paulsen C, “The health literacy of America’s adults: Results from the 2003 national assessment of adult literacy. NCES 2006–483,” Nat. Center Educ. Statist, Washington, DC, USA, NCES 2006–483, 2006. [Google Scholar]
- [26].Lee J et al. , “BioBERT: A pre-trained biomedical language representation model for biomedical text mining,” Bioinformatics, vol. 36, pp. 1234–1240, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li J, Tang T, Zhao WX, and Wen J-R, “Pretrained language models for text generation: A survey,” 2021, arXiv:2105.10311. [Google Scholar]
- [28].Liao Y-S, Lu J-Y, and Liu D-R, “News recommendation based on collaborative semantic topic models and recommendation adjustment,” in Proc. IEEE Int. Conf. Mach. Learn. Cybern, 2019, pp. 1–6. [Google Scholar]
- [29].Liu Y and Lapata M, “Text summarization with pretrained encoders,” in Proc. Conf. Empirical Methods Natural Lang. Process. 9th Int. Joint Conf. Natural Lang. Process, 2019, pp. 3730–3740. [Google Scholar]
- [30].Luo X, Gandhi P, Storey S, and Huang K, “A deep language model for symptom extraction from clinical text and its application to extract COVID-19 symptoms from social media,” IEEE J. Biomed. Health Inform, vol. 26, no. 4, pp. 1737–1748, Apr. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mahyari A, Pirolli P, and Leblanc JA, “A deep recurrent neural network with user-profile attention-based real-time physical exercises recommendation system in mhealth,” IEEE J. Biomed. Health Inform, vol. 26, no. 8, pp. 4281–4290, Aug. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McHugh ML, “Interrater reliability: The kappa statistic,” Biochemia Medica, vol. 22, no. 3, pp. 276–282, 2012. [PMC free article] [PubMed] [Google Scholar]
- [33].Melamud O, Goldberger J, and Dagan I, “Context2vec: Learning generic context embedding with bidirectional LSTM,” in Proc. 20th SIGNLL Conf. Comput. Natural Lang. Learn, 2016, pp. 51–61. [Google Scholar]
- [34].Miller D, “Leveraging Bert for extractive text summarization on lectures,” 2019, arXiv:1906.04165. [Google Scholar]
- [35].Morahan-Martin J and Anderson CD, “Information and misinformation online: Recommendations for facilitating accurate mental health information retrieval and evaluation,” CyberPsychol. Behav, vol. 3, no. 5, pp. 731–746, 2000. [Google Scholar]
- [36].Nallapati R, Xiang B, and Zhou B, “Sequence-to-sequence RNNs for text summarization,” in Proc. Int. Conf. Learn. Representations Workshop, 2016. [Google Scholar]
- [37].Pedregosa F et al. , “Scikit-learn: Machine learning in Python,” J. Mach. Learn. Res, vol. 12, pp. 2825–2830, 2011. [Google Scholar]
- [38].Rasu RS, Bawa WA, Suminski R, Snella K, and Warady B, “Health literacy impact on national healthcare utilization and expenditure,” Int. J. Health Policy Manage, vol. 4, no. 11, 2015, Art. no. 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sahoo AK, Pradhan C, Barik RK, and Dubey H, “DeepReco: Deep learning based health recommender system using collaborative filtering,” Computation, vol. 7, no. 2, 2019, Art. no. 25. [Google Scholar]
- [40].Shu J, Shen X, Liu H, Yi B, and Zhang Z, “A content-based recommendation algorithm for learning resources,” Multimedia Syst, vol. 24, no. 2, pp. 163–173, 2018. [Google Scholar]
- [41].Singh A, Nagwani N, and Pandey S, “TAGme: A topical folksonomy based collaborative filtering for tag recommendation in community sites,” in Proc. 4th Multidisciplinary Int. Social Netw. Conf, 2017, pp. 1–7. [Google Scholar]
- [42].Song K, Tan X, Qin T, Lu J, and Liu T-Y, “MASS: Masked sequence to sequence pre-training for language generation,” in Proc. Int. Conf. Mach. Learn, 2019, pp. 5926–5936. [Google Scholar]
- [43].Tibshirani R, Walther G, and Hastie T, “Estimating the number of clusters in a data set via the gap statistic,” J. Roy. Stat. Soc.: Ser. B (Stat. Methodol.), vol. 63, no. 2, pp. 411–423, 2001. [Google Scholar]
- [44].Tran VC, Hwang D, and Nguyen NT, “Hashtag recommendation approach based on content and user characteristics,” Cybern. Syst, vol. 49, no. 5/6, pp. 368–383, 2018. [Google Scholar]
- [45].Tuarob S, Pouchard LC, and Giles CL, “Automatic tag recommendation for metadata annotation using probabilistic topic modeling,” in Proc. IEEE/ACM-CS 13th Joint Conf. Digit. Libraries, 2013, pp. 239–248. [Google Scholar]
- [46].Vaswani A et al. , “Attention is all you need,” in Proc. Adv. Neural Inf. Process. Syst, 2017, pp. 5998–6008. [Google Scholar]
- [47].Wang H, Zhang F, Xie X, and Guo M, “DKN: Deep knowledge-aware network for news recommendation,” in Proc. World Wide Web Conf, 2018, pp. 1835–1844. [Google Scholar]
- [48].Wang Y, Li J, King I, Lyu MR, and Shi S, “Microblog hashtag generation via encoding conversation contexts,” in Proc. Conf. North Amer. Chapter Assoc. Comput. Linguistics: Hum. Lang. Technol, 2019, pp. 1624–1633. [Google Scholar]
- [49].Wang Z, Yin Z, and Argyris YA, “Detecting medical misinformation on social media using multimodal deep learning,” IEEE J. Biomed. Health Inform, vol. 25, no. 6, pp. 2193–2203, Jun. 2021. [DOI] [PubMed] [Google Scholar]
- [50].White RW and Horvitz E, “From health search to healthcare: Explorations of intention and utilization via query logs and user surveys,” J. Amer. Med. Inform. Assoc, vol. 21, no. 1, pp. 49–55, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yamada I, Asai A, Shindo H, Takeda H, and Matsumoto Y, “LUKE: Deep contextualized entity representations with entity-aware self-attention,” 2020, arXiv:2010.01057. [Google Scholar]
- [52].Yuan J, Jin Y, Liu W, and Wang X, “Attention-based neural tag recommendation,” in Proc. Int. Conf. Database Syst. Adv. Appl, 2019, pp. 350–365. [Google Scholar]
- [53].Yue W, Wang Z, Zhang J, and Liu X, “An overview of recommendation techniques and their applications in healthcare,” IEEE/CAA J. Automatica Sinica, vol. 8, no. 4, pp. 701–717, Apr. 2021. [Google Scholar]
- [54].Zarzour H, Al-Sharif Z, Al-Ayyoub M, and Jararweh Y, “A new collaborative filtering recommendation algorithm based on dimensionality reduction and clustering techniques,” in Proc. IEEE 9th Int. Conf. Inf. Commun. Syst, 2018, pp. 102–106. [Google Scholar]
- [55].Zheng M et al. , “The relationship between health literacy and quality of life: A systematic review and meta-analysis,” Health Qual. Life Outcomes, vol. 16, no. 1, pp. 1–10, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhou J and Wang C, “Improving cancer survivors’e-health literacy via online health communities (OHCS): A social support perspective,” J. Cancer Survivorship, vol. 14, pp. 244–252, 2020. [DOI] [PubMed] [Google Scholar]


