Abstract
Studies on the development and function of CD4+ TH1 and TH2 cells during the progression to AIDS may increase the understanding of AIDS pathogenesis. The preferential replication of human immunodeficiency virus (HIV) in either TH1 or TH2 cells could alter the delicate balance of the immune response. TH1 (gamma interferon [IFN-γ] positive, interleukin-4 [IL-4] and IL-5 negative) and TH2 (IFN-γ negative, IL-4 and IL-5 positive) clones, developed from several healthy donors, pedigreed by reverse transcriptase PCR (RT-PCR) and enzyme linked immunosorbent assay have similar levels of cell surface expression of CD4 and several chemokine receptor cofactors necessary for viral entry. After activation by specific antigens and infection with T-cell-tropic strains of HIV type 1 (HIV-1), TH1 and TH2 clones showed similar levels of viral entry and reverse transcription. At days 3 through 14 postinfection, HIV replicated to similar levels in several TH1 and TH2 clones as measured by release of HIV p24 and total number of copies of gag RNA/total cell RNA as measured by RT-PCR. When values were normalized for viable cell number in three clones of each type, there was up to twofold more HIV RNA in TH1 than TH2 cells. In addition, several primary monocytotropic HIV-1 strains were able to replicate to similar levels in TH1 and TH2 cells. These studies suggest that the importance of TH1 and TH2 subsets in AIDS pathogenesis transcends clonal differences in their ability to support HIV replication.
During AIDS progression, CD4+ T cells are severely reduced first in biological responsiveness and then in cell numbers, leading to a degeneration of the patient’s ability to generate an effective immune response (22, 40). The propagation of human immunodeficiency virus (HIV) in vivo is not prevented by a strong cellular and humoral response against HIV type 1 (HIV-1). Antibody production and cell-mediated immunity are often reciprocal immune responses associated with distinct patterns of cytokine production by two subsets of CD4+ T-helper (TH) cells. Cells of the TH1 subset secrete interleukin-2 (IL-2) and gamma interferon (IFN-γ) but not IL-4 or IL-5 and are associated with cell-mediated responses such as delayed-type hypersensitivity; TH2 cells secrete IL-4 and IL-5 but not IFN-γ and are associated with antibody and allergic responses (36, 50, 51, 55, 57). These cytokines are also secreted by other cell types, contributing to overlapping patterns of cytokine expression which may complicate our understanding of mechanistic issues involved in the immune response.
During microbial infections, particularly chronic persistent infections, there can be a preferential development of one of the TH lineages. Simply, infections by viruses and intracellular pathogens are often better controlled by cellular (TH1 and cytotoxic T-cell) responses, whereas infections by parasites and bacteria may be controlled more effectively by antibody-TH2 responses (14, 15, 51, 57). However, while the development of the correct immune response is critical in host resistance to microbes, some infectious agents can stimulate inappropriate cytokine responses, contributing to increased disease pathology (1). As CD4+ T cells are the preferential targets of HIV, much interest and controversy have developed regarding a role for the TH1 and TH2 cells and cytokines during HIV infection and their relationship to HIV pathogenesis (3, 9–12, 31, 43–46, 49, 56).
Studies by Maggi et al. (43) suggest that HIV replicates preferentially in TH2 and TH0 rather than TH1 clones in vitro. This concept has been incorporated in recent models of HIV pathogenesis (11, 49, 56). Since the complex nature of virus-cell interactions as well as the extracellular environment can often affect the kinetics and magnitude of viral replication, HIV replication and cell survival were examined in a panel of human antigen-specific CD4+ TH1 and TH2 clones. After activation by specific antigens and infection with HIV-1, TH1 and TH2 clones, developed from healthy donors, showed similar levels of strong-stop and full-length viral DNA. Regardless of the tropism of virus used, HIV replicated to similar levels in several TH1 and TH2 clones. When values were normalized for viable cell number, there was up to twofold more HIV-1 RNA in TH1 than TH2 cells, indicating that there is little difference in the ability of TH1 and TH2 subsets to support HIV replication in vitro.
MATERIALS AND METHODS
Derivation and maintenance of antigen-specific human CD4+ T-cell clones.
Purified protein derivative (PPD)-specific, tetanus toxoid (TTx)-specific, keyhole limpet hemocyanin (KLH)- and Dermatophagoides pteronyssinus antigen (DP)-specific, and staphylococcal enterotoxin B (SEB)-reactive T-cell clones were generated as previously described (25, 28). PPD and TTx were purchased from Connaught, Inc. (Swiftwater, Pa.), SEB was purchased from the Sigma Chemical Company (St. Louis, Mo.), and DP and KLH were purchased from Miles, Inc. (Spokane, Wash.). Briefly, peripheral blood mononuclear cells (PBMCs) at a concentration of 5 × 105 cells/ml in clone medium (EHAA [Click’s] medium supplemented with l-glutamine, 2-mercaptoethanol, 2% human AB serum, 10% fetal calf serum, penicillin-streptomycin, nonessential amino acids, and sodium pyruvate; Life Technologies, Gaithersburg, Md.) were stimulated with either PPD (1 μg/ml), TTx (10 μg/ml), DP (10 IU/ml), or SEB (0.1 μg/ml) in 24-well flat-bottom plates for 7 days. Many but not all of the TH1 and TH2 clones used in these studies were derived in cultures supplemented with the TH cell selective cytokines, IL-4 and IL-12. For the generation of TH1 clones, these cultures were supplemented with recombinant human IFN-γ (rhIFN-γ; 10 U/ml; Peprotech, Rocky Hill, N.J.), rhIL-12 (50 pg/ml; Roche, Nutley, N.J.), and anti-IL-4 monoclonal antibody (MAb; 10 μg/ml; R&D Systems, Minneapolis, Minn.) over the culture period. For TH2 clones, bulk cultures were supplemented with rhIL-4 (200 U/ml; Peprotech) and anti-IFN-γ MAb (10 μg/ml; R&D Systems). Forty-eight hours after the initiation of these cultures, rhIL-2 (10 U/ml) was added to each of the wells. After 7 days of incubation, the cultures were harvested, extensively washed, replated in fresh clone medium supplemented with additional IL-2 (10 U/ml; Cellular Products, Buffalo, N.Y.), and incubated for an additional 7 to 10 days. Viable T cells were then plated in limiting-dilution cultures (0.5 cells/well) in 16 flat-bottom 96-well plates containing 2 × 105 irradiated (1,200 rads) syngeneic PBMC feeder cells, specific antigen, and IL-2 (10 U/ml) in a final volume of 200 μl. The cultures were examined daily and supplemented with the TH1- and TH2-selecting cytokines (as described above) at 10-day intervals with feeder cells and IL-2. Individual clones were isolated and then characterized for lymphokine production by enzyme-linked immunosorbent assay (ELISA) and PCR analysis and for the ability to respond to specific antigen in combination with syngeneic irradiated (1,200 rads) feeder cells.
To maintain TH clones, cells were restimulated every 14 to 21 days with specific antigen in the presence of autologous PMBCs treated with mitomycin C at 25 μg/ml to prevent outgrowth of feeder cells and IL-2 (20 U/ml). After 96 h of antigenic stimulation, cells were subjected to two successive Ficoll-Hypaque centrifugations to remove dead cells. All clones were tested for their cytokine profiles by ELISA after stimulation with a combination phorbol myristate acetate (PMA) and monoclonal anti-CD3 as well as with antigen and antigen-presenting cells to determine the phenotype of each clone.
Chemokine binding assays.
Binding conditions for CC chemokines MIP-1α, MIP-1β, RANTES, MCP-1 and MCP-3 and the CXC chemokine IL-8 were as previously described (58, 61). Briefly, 2 × 106 cells were incubated in duplicate or triplicate (depending on the availability of the clones) with increasing concentrations of 125I-labeled chemokines in a modified binding medium (RPMI 1640 with 1 mg of bovine serum albumin per ml, 25 mM HEPES, and 0.05% sodium azide [pH 7.4]) in a total volume of 200 μl. The residual nonspecific binding was determined by parallel incubation of 125I-labeled chemokine in the presence of a 100-fold excess of unlabeled chemokine. After incubation at 4°C or room temperature for 90 min, the cells were pelleted through a 10% sucrose–phosphate-buffered saline (PBS) cushion. The tips of the tubes containing cells were cut, and radioactivity was quantitated in a gamma counter. The residual nonspecific bound radioactivity associated with cells in the presence of unlabeled chemokine was subtracted from the total bound radioactivity to yield specific binding. The data were analyzed with the Biosoft RADLIG program.
Chemokine receptor flow cytometric analysis.
Phycoerythrin and fluorescein isothiocyanate-labeled rabbit antibodies specific for human CXCR4, CCR5, CXCR1, CXCR2, and CD4 were obtained from R&D Systems. Polyclonal rabbit anti-CCR1 antibody was generously provided by Richard Horuk (Berlex Biosciences, Richmond, Calif.). Flow cytometric staining and analysis were performed as previously described (52, 61). The data are expressed as percent positive (± standard deviation) and/or as mean channel fluorescence.
HIV infection of T-cell clones.
Antigen-stimulated clones (2 × 106 cells/0.5 ml), 4 to 7 days postactivation (>95% viable by trypan blue exclusion), were inoculated with cell-free viral isolates with a total of 100 pg of HIV p24 and allowed to adsorb for 90 min at 37°C in a shaking water bath before complete aspiration of medium, washing with PBS, and addition of fresh growth medium containing IL-2 (20 U/ml). Cells were aliquoted at 106 cells/ml in 24-well plates. Three laboratory T-cell-tropic (syncytium-inducing [SI]) strains of HIV used were BP1 (48), IIIB, and MN (purified 1,000-fold; kindly provided by the AIDS vaccine program, Frederick, Md.). These viral stocks were grown in H9 cells. The primary monocytotropic (non-SI [NSI]) viruses, SF162, US657, US714, and US727, were obtained from the AIDS Reference and Reagent Program. These stocks were grown in primary PBMCs.
Quantitation of cytokine and p24 production by T-cell clones.
Quantitative determinations for lymphokines (human IL-2, IL-4, IL-5, IL-10, and IFN-γ) from the 48-h supernatants of antigen-stimulated T-cell clones were done by ELISA (Quantikine; R&D Systems) by following the manufacturers’ instructions. The results are expressed in either nanograms/milliliter or units/milliliter based on a standard curve determined by using recombinant cytokine within the ELISAs. Cytokine analyses after HIV infection were performed with cell-free supernatants and were quantitated by ELISA for IL-4, IL-5 (R&D Systems), and IFN-γ (Medigenix) with sensitivities of 3 pg/ml, 1 pg/ml, and 1 IU/ml, respectively. Viral p24 antigen was determined by ELISA (Cellular Products) with a sensitivity of 10 pg/ml. To determine the portion of the cells productively infected, flow cytometry for intracellular expression of HIV-1 p24 was performed with rhodamine-conjugated anti-p24 antibody.
Detection of HIV-1 DNA in T-cell clones.
For the detection of viral DNA, cell lysates were made by incubating 106 cells in 100 μl of lysis buffer (10 mM Tris HCl [pH 8], 1 mM EDTA, 0.001 mM Triton X-100–sodium dodecyl sulfate, 1 mg of proteinase K per ml) at 60°C for 1 h followed by 99°C for 10 min to inactivate the proteinase K. Quantitative PCR amplification was performed with one oligonucleotide of each pair end labeled with 33P, and 25 ng was used in each reaction (5 × 105 to 5 × 106 cpm). The samples were denatured for 5 min at 94°C followed by 25 cycles of denaturation for 1 min at 91°C and annealing-extension for 2 min at 65°C (63). Primers for minus strong-stop HIV-1 R and U5 (140 bp), sense (5′-GGCTAACTAGGGAACCCACGT-3′) and antisense (5′-CTGCTAGAGATTTTCCACACTGAC-3′), and for HIV-1 long terminal repeat (LTR) and gag (200 bp), sense (5′-CTGCTAACTAGGGAACCCACGT-3′) and antisense (5′-CCTGCGTCGAGAGAGCTCCTCTGG-3′), were previously described by Zack et al. (63). The primers LA1 and LA2 (63) for human β-globin were included as a control for amplification. Products were separated by electrophoresis on an 8% nondenaturing acrylamide gel. Dried gels were analyzed on a PhosphorImager (Molecular Dynamics, San Diego, Calif.), quantitated with ImageQuant and Microsoft Excel software, and exposed to Kodak XAR-5 film at −70°C overnight. HIV-1 copy numbers per 50,000 cells/lane were estimated by comparing graded doses of ACH-2 DNA lysate; this cell line contains 1 proviral copy/cell (26).
RT-PCR analysis of cytokine and HIV-1 RNA.
RNA was prepared from 106 cells by using RNA Stat 60 (Tel Test Inc., Friendswood, Tex.). Reverse transcription of 5 μg of RNA was performed with Superscript II reverse transcriptase (RT) (Life Technologies). One to two microliters of the reaction mixture was used in each amplification. Primer pairs for cytokine detection were purchased as RT-PCR Amplimer sets (Clontech, Palo Alto, Calif.). Primers SK38 and SK39 for HIV gag (positions 1543 to 1570 and 1630 to 1657) (6) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (371 to 388, 546 to 565) (29) were previously described. Amplification was carried out in the presence of [32P]dCTP as previously described (17). Products were separated by electrophoresis on an 8% nondenaturing acrylamide gel. Dried gels were analyzed on a PhosphorImager (Molecular Dynamics), quantitated with ImageQuant and Microsoft Excel software, and exposed to Kodak XAR-5 film at −70°C for 1 to 4 h. GAPDH was always used as a control for amplification.
In vitro synthesis of HIV-1 RNA.
HIV-1 RNA synthesized from a DNA template produced by the amplification of HIV-1 proviral DNA by using modified HIV-1 gag region-specific primers SK38 and SK39 (64) was kindly provided by B. Poiesz (State University of New York, Syracuse). Briefly, primer SK38 was altered by the addition of the T3 RNA polymerase promoter sequence (5′-CCCTATAGTGAGTCGTATTA-3′), in inverse complementary orientation, to the 5′ end of the original primer sequence. An additional five bases (GGTCG) upstream of the promoter site were included to ensure more efficient binding of the RNA polymerase. The modified primer, RPSK38, was used with primer SK39 to amplify 1 μg of HUT 78/HIVAAV DNA by PCR. The resulting product was separated by electrophoresis on and excised from a native 10% polyacrylamide gel and eluted into 50 μl of diethyl pyrocarbonate-treated H2O. Ten microliters of this HIV-1 gag DNA was then mixed with 40 μl of RNA synthesis cocktail containing 10 μl of 5× reaction buffer (Life Technologies), 400 U of RNasin (Promega, Madison, Wis.), 100 U of T7 RNA polymerase (Stratagene, La Jolla, Calif.), 2.5 μl of 100 mM dithiothreitol, and 2 μl each of 10 mM deoxynucleoside triphosphates in diethyl pyrocarbonate-treated H2O. After incubation at 37°C for 1 h, 4 U of RQ1 DNase was added to digest the template DNA. The resulting HIV-1 single-stranded gag RNA was separated on and eluted from a native 10% polyacrylamide gel and serially used as an RNA standard in RT-PCR assays. For quantitation, RNA was diluted until its signal was in the linear range of the standard curve, using 1,000 to 2,000 RNA molecules, and dried gels were analyzed on a PhosphorImager (Molecular Dynamics), quantitated with ImageQuant and Microsoft Excel software, and exposed to Kodak XAR-5 film at −70°C for 1 to 4 h.
Statistics.
Means and standard error of the mean (SEM) were calculated for the results of HIV p24 antigen determination.
RESULTS
Generation and characterization of antigen-specific TH clones.
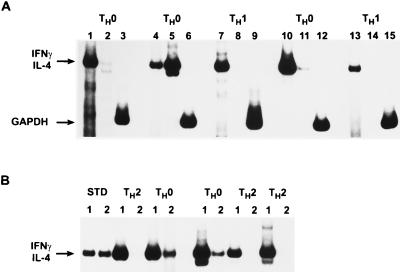
TH cell clones were derived from three different non-HLA-matched healthy PBMC donors as described in Materials and Methods. In contrast to mice, human TH1 and TH2 phenotypes are less restricted in cytokine expression in that both subsets produce IL-2 and IL-10 (14, 55, 56). Therefore, we used RT-PCR to in addition to ELISAs to pedigree each of these antigen-specific T-cell clones. Using these criteria, we found numerous TH0 clones regardless of whether TH1- or TH2-promoting media were used (Fig. 1). The TH1 clones used in these studies were specific for either TTx, DP, SEB, or PPD, while TH2 clones were specific for DP or TTx. All of the T-cell clones used expressed similar cell surface phenotypes (CD4+, CD8−, CD3+, CD19−, CD56−, CD16−, and CD14−) as determined by flow cytometric analysis and were found to proliferate in response to specific but not irrelevant antigens (data not shown). All TH1 clones used were negative for IL-4 and IL-5 and positive for IFN-γ by ELISA (Tables 1 and 2) and IL-4 negative and IFN-γ positive by RT-PCR (representative clones are shown in Fig. 1A). Similarly, all of the TH2 clones used were negative for IFN-γ and positive for IL-4 and IL-5 by ELISA (Tables 1 and 2) and IFN-γ negative and IL-4 positive by RT-PCR (representative clones are shown in Fig. 1B). All TH1 and four of seven TH2 clones also produced IL-2. All clones producing a combination of TH1 and TH2 lymphokine mRNAs were designated TH0 (Fig. 1; Table 2) and not used for HIV infection.
FIG. 1.
Characterization of human TH clones by RT-PCR. Single-cell cloning of human peripheral blood was performed as described in Materials and Methods. Presence of mRNA for IFN-γ and IL-4 in these clones was measured 7 days after antigen activation, using RT-PCR as described in Materials and Methods. (A) Clones isolated under TH1 conditions. Lane 1, IFN-γ; lane 2, IL-4; lane 3, GAPDH. Arrows indicate positions of cytokine standards. (B) Clones isolated under TH2 conditions. Lane 1, standard (STD) for IL-4 (427 bp); lane 2, standard for IFN-γ (459 bp) (Clontech). GAPDH (not shown) was used as a loading control.
TABLE 1.
Characterization of cytokine production in human TH1 and TH2 clones in vitro
| Clonea | Type | Antigen specificity | Concn (pg/ml)b
|
||
|---|---|---|---|---|---|
| IL-4 | IFN-γ | IL-5 | |||
| A57.D4 | TH1 | SEB | 0 | 900 | 0 |
| C01.D6 | TH1 | SEB | 0 | 875 | 0 |
| B95.E1 | TH2 | SEB | 128 | 0 | 65 |
| A057 | TH1 | DP | 0 | 2,625 | 0 |
| D.D6 | TH1 | PPD | 0 | 1,625 | 0 |
| A57.G1 | TH2 | DP | 54 | 0 | 125 |
| 57.D10 | TH2 | DP | 125 | 0 | 46 |
| H1.15 | TH1 | TTx | 0 | 400 | 0 |
| H1.12 | TH1 | TTx | 0 | 110 | 0 |
| H1.18 | TH1 | TTx | 0 | 110 | 0 |
| H2.25 | TH2 | TTx | 302 | 0 | 104 |
| H2.29 | TH2 | TTx | 401 | 0 | 126 |
| H2.33 | TH2 | TTx | 720 | 0 | 225 |
| H2.18 | TH2 | TTx | 104 | 0 | 132 |
T-cell clones with the indicated antigenic specificities were generated from PBMCs of three HIV-seronegative donors (SEB represents one donor, DP and PPD represent a second donor, and TTx represents a third donor) maintained and activated as described in Materials and Methods.
Cytokine production was determined by ELISA at 7 days after antigen activation.
TABLE 2.
Phenotypic summary of TH cell clones derived from two different human donorsa
| Clone | Type | Lymphokineb
|
Chemokine receptorsc
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IFN-γ | IL-4 | IL-5 | IL-13 | CXCR1 | CXCR2 | CXCR4 | CCR1 | CCR5 | ||
| H1.1 | TH1 | + | − | − | − | − | + | + | + | + |
| H1.2 | TH1 | + | − | − | − | − | + | + | + | + |
| H1.3 | TH1 | + | − | − | − | − | + | + | − | + |
| H1.5 | TH1 | + | − | − | − | ±e | + | + | + | + |
| H1.11 | TH1 | + | − | − | − | − | − | NDd | ND | ND |
| H1.12 | TH1 | + | − | − | − | − | + | + | + | + |
| H1.15 | TH1 | + | − | − | − | − | − | + | + | + |
| H1.18 | TH1 | + | − | − | − | ND | + | ND | ND | ND |
| H2.1 | TH2 | − | + | + | + | − | + | + | + | + |
| H2.2 | TH2 | − | + | + | + | − | + | + | − | + |
| H2.5 | TH2 | − | + | + | + | − | + | ND | ND | ND |
| H2.8 | TH2 | − | + | + | + | − | + | ND | ND | ND |
| H2.10 | TH2 | − | + | + | + | − | + | + | + | + |
| H2.11 | TH2 | − | + | + | + | − | + | + | + | + |
| H2.15 | TH2 | − | + | + | + | ND | ND | ND | ND | ND |
| H2.25 | TH2 | − | + | + | + | − | + | + | + | + |
| H2.29 | TH2 | − | + | + | + | − | + | + | − | + |
| 500.F7 | TH0 | + | + | + | + | − | + | + | + | + |
| 500.G3 | TH0 | + | + | + | + | − | + | + | + | + |
| 500.E10 | TH0 | + | + | + | + | − | + | ND | ND | ND |
| 100.F10 | TH0 | + | + | + | + | − | + | + | + | + |
Human T-cell clones were obtained through limiting-dilution analysis as described in Materials and Methods. All of the H-series clones are TTx specific, while the remaining clones are KLH specific.
Supernatants obtained from activated T-cell clones were tested for the production of various cytokines by ELISA, and cytokine mRNA expression was tested by RT-PCR analysis as described in Materials and Methods.
Both quiescent and activated T-cell clones were examined by flow cytometric analysis for the presence of various chemokine receptors on their cell surface. In addition, many of these clones were also examined by radiolabeled binding assays for chemokine binding sites and affinity as described in Materials and Methods.
ND, not determined.
±, not consistently positive.
Similar levels of HIV receptors on the cell surface of antigen-activated TH cell clones.
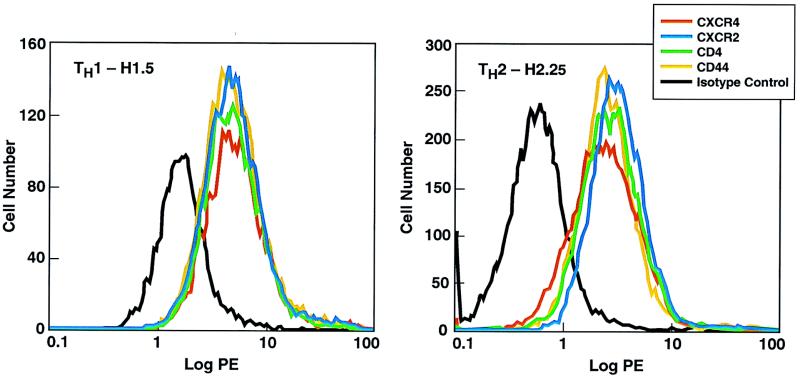
The recent demonstration that CC chemokines can inhibit HIV-1 infection (13) followed by the finding that selected CC and CXC chemokine receptors (8, 16, 19, 20, 23, 38) act as cofactors with CD4+ to mediate viral entry into cells represents a significant advance in our understanding of HIV infectivity. Different strains of HIV-1 use different chemokine receptors for cell entry: macrophage-tropic HIV-1 strains mainly use CCR5 (8, 16, 20) and to a lesser extent CCR3 and CCR2b (19), while T-cell-tropic strains use CXCR4 (23, 38). Therefore, we analyzed the cell surface expression of HIV-1 receptors on TH1, TH2, and TH0 clones 4 to 5 days after antigen activation. Regardless of the TH subset of the clones, they expressed detectable levels of CXCR4 and CCR5 by flow cytometry analysis whereas expression of CXCR1 and CCR1 was variable (Table 2). The densities (the amount of antigen as measured by the mean channel fluorescence) of these molecules as well as CD4 and CD44 expression on the cell surface were also similar on TH1 and TH2 clones (Fig. 2; Table 3). The density of CXCR4 was consistently higher than the density of CCR5 on these activated TH cells regardless of subset. Scatchard analysis showed that the number and affinity (Kd) of binding sites for CC and CXC chemokines were similar on TH1 and TH2 clones (Table 4).
FIG. 2.
Flow cytometric analysis of a human TH1 clone and a human TH2 clone for cell surface expression of chemokine receptors. A total of 106 cells of each of the T-cell clones H1.5 and H2.25 were suspended in PBS containing 1% heat-inactivated human AB serum and 0.5% sodium azide and stained with phycoerythrin (PE)-labeled anti-CXCR2, -CXCR4, -CD4, or -CD44 or an isotype control labeled immunoglobulin G antibody. After staining, the cells were extensively washed and then fixed with 1% paraformaldehyde. The clones were analyzed on a FACStar Plus flow cytometer.
TABLE 3.
Chemokine receptor expression by human TTx-specific TH1 and TH2 clonesa
| Clone | % Positive (mean channel fluorescence)b
|
|||||
|---|---|---|---|---|---|---|
| Control | CXCR1 | CXCR2 | CXCR4 | CCR1 | CCR5 | |
| H1.1 | 4.2 (3.2) | 3.5 (3.4) | 88.9 (42.2) | 98.7 (106.7) | 43.5 (38.6) | 55.3 (96.2) |
| H1.3 | 2.1 (4.3) | 2.7 (4.4) | 87.4 (37.8) | 96.5 (118.7) | 3.3 (5.6) | 43.2 (94.1) |
| H1.15 | 5.6 (3.5) | NDc | 6.7 (4.2) | 97.4 (123.5) | 49.6 (46.2) | 64.7 (98.2) |
| H1.5 | 2.1 (3.2) | 11.4 (3.6) | 67.4 (54.2) | 99.2 (114.2) | 52.3 (37.2) | 49.6 (76.4) |
| H2.5 | 1.4 (2.4) | 2.2 (2.4) | 78.9 (32.1) | 97.6 (122.3) | ND | 69.5 (96.2) |
| H2.25 | 3.2 (1.6) | 3.2 (2.1) | 84.3 (42.3) | 98.9 (118.7) | 39.8 (34.5) | 54.3 (78.6) |
| H2.29 | 2.4 (2.6) | 2.6 (2.2) | 89.5 (45.7) | 99.2 (123.4) | 5.8 (3.6) | 88.4 (97.2) |
A total of 106 cells were suspended in PBS and stained with fluorescein isothiocyanate-labeled MAb to CXCR1, CXCR2, CXCR4, CCR1, or CCR5. After staining, the cells were washed and then fixed with 1% paraformaldehyde.
All were analyzed on a FACStar Plus flow cytometer.
ND, not determined.
TABLE 4.
Similar numbers of cell surface chemokine binding sites on human TH1 and TH2 clonesa
| Chemokine | H1.5
|
H2.25
|
||
|---|---|---|---|---|
| Receptors/cell | Kd (nM) | Receptors/cell | Kd (nM) | |
| MIP-1α | 1,525 | 0.8 | 2,143 | 1.1 |
| MIP-1β | 890 | 0.8 | 1,334 | 1.0 |
| RANTES | 2,156 | 0.6 | 3,420 | 0.7 |
| MCP-1 | 1,325 | 0.8 | 2,132 | 0.9 |
| MCP-3 | 3,467 | 1.2 | 4,786 | 0.9 |
| IL-8 | 3,216 | 1.2 | 4,556 | 1.4 |
Binding conditions for the CC chemokines MIP-1α, MIP-1β, RANTES, MCP-1, and MCP-3 and the CXC chemokine IL-8 were as described in Materials and Methods. The data were analyzed with the Biosoft RADLIG program.
Similar levels of HIV-1 entry in antigen-activated TH cell clones.
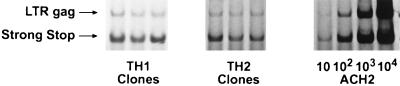
Three different TH1 and TH2 clones were exposed to a filtered RNase-free DNase I-treated preparation of HIV-1IIIB for 90 min at 37°C, washed, and recultured. At various time points postinfection (p.i.), aliquots of cells (105) were removed and lysates were prepared for PCR analysis of HIV-1 DNA. In the PCR method previously described by Zack et al. (63), primer pairs were designed to detect certain steps of the reverse transcription process by using the accepted model for reverse transcription of retroviral RNA. Amplification using the R-U5 primer pair detects a DNA region representing initial reverse transcription, and nearly complete reverse transcription is detected with the LTR-gag primer pair. The sensitivity of the amplification was similar in TH1 and TH2 clones, equaling 5 to 10 copies of HIV DNA. As there is evidence that HIV-1 virions can contain short transcripts and DNA (42, 60), we used a heat-inactivated virus preparation as a control in all infections. No signal was detected either in this control or in aliquots taken 90 min after HIV exposure (data not shown). By 24 h p.i., 2,000 to 5,000 copies of R-U5 and 100 to 500 copies of LTR-gag DNA (Fig. 3) were seen. No significant differences in copy number of R-U5 or LTR-gag DNA were observed between TH1 and TH2 clones at 24 h (Fig. 3), indicating similar levels of viral entry and reverse transcription in these antigen-specific TH1 and TH2 clones.
FIG. 3.
HIV-1 DNA detection in TH1 and TH2 clones. Lysates were prepared 24 h p.i. A primer pair was used to detect the earliest region of DNA formed by reverse transcription (141 bp of the minus strong-stop strand). Another primer pair was used to detect full-length HIV-1 DNA (200 bp of LTR-gag). The sense primers of each pair were radiolabeled. Three different TH1 (H1.12, H1.15, and H1.18) and TH2 (H2.25, H2.29, and H2.33) clones shown for each infection are representative of three separate experiments. Tenfold serial dilutions of the ACH-2 cell line, containing one integrated copy of HIV-1 DNA per cell, were made in a background of uninfected T cells and shown for estimation of copy number.
Similar levels of replication by SI HIV strains in antigen-activated TH cell clones.
Next, the ability of these clones to support HIV replication in vitro was evaluated. Clones (>95% viable following Ficoll-Hypaque separation) were infected between 4 and 7 days after antigen stimulation, using the specific antigen and mitomycin C-treated autologous PBMC feeder cells. Using two T-cell-tropic laboratory viral strains, BP1 (48) and IIIB, all seven TH1 and seven TH2 clones released substantial amounts of extracellular p24 at days 7 and 14 p.i. (Table 5). At day 7, for example, infected TH1 and TH2 clones released similar levels of virus, with ranges of p24 being 22,500 to 47,500 pg/ml for TH1 clones and 23,000 to 57,000 pg/ml for TH2 clones (Table 5). Furthermore, results obtained with clones infected with the BP1 strain (Table 5, clones 1 to 7) were indistinguishable from those obtained with clones infected with the IIIB strain (Table 5, clones 8 to 14). Infected TH1 cells (day 10 p.i.) maintained a TH1 phenotype in that the infected cultures were positive for IFN-γ mRNA but not for IL-4 mRNA; infected TH2 cells remained IFN-γ negative and IL-4 mRNA positive (data not shown). By fluorescence-activated cell sorting analysis, for HIV-1 p24, the number of positive cells at day 7 ranged from 47 to 72%, with no differences seen between infected TH1 and TH2 clones (data not shown).
TABLE 5.
Replication of HIV-1 in human TH1 and TH2 clones
| Clonea
|
Type | Antigen specificity | HIV p24 (pg/ml)b
|
||
|---|---|---|---|---|---|
| No. | Name | Day 7 | Day 14 | ||
| 1 | A57.D4 | TH1 | SEB | 47,500 ± 7.3 | 35,000 ± 4.2 |
| 2 | C01.D6 | TH1 | SEB | 31,500 ± 2.4 | 34,000 ± 4.3 |
| 3 | B95.E1 | TH2 | SEB | 42,000 ± 5.7 | 39,500 ± 2.7 |
| 4 | A057 | TH1 | DP | 27,000 ± 3.1 | 34,000 ± 1.9 |
| 5 | D.D6 | TH1 | PPD | 22,500 ± 1.5 | 27,000 ± 3.3 |
| 6 | A57.G1 | TH2 | DP | 57,000 ± 8.1 | 42,000 ± 5.1 |
| 7 | 57.D10 | TH2 | DP | 49,500 ± 6.3 | 37,000 ± 3.5 |
| 8 | H1.15 | TH1 | TTx | 23,000 ± 1.6 | 17,000 ± 1.5 |
| 9 | H1.12 | TH1 | TTx | 37,000 ± 2.9 | 35,000 ± 3.7 |
| 10 | H1.18 | TH1 | TTx | 43,500 ± 5.5 | 42,000 ± 5.2 |
| 11 | H2.25 | TH2 | TTx | 23,000 ± 2.1 | 29,500 ± 3.3 |
| 12 | H2.29 | TH2 | TTx | 45,000 ± 6.7 | 41,000 ± 5.2 |
| 13 | H2.33 | TH2 | TTx | 54,000 ± 7.3 | 61,000 ± 8.3 |
| 14 | H2.18 | TH2 | TTx | 29,000 ± 3.3 | 23,500 ± 1.7 |
T-cell clones were generated, maintained, and activated as described in Materials and Methods.
Clones were infected with either HIV-1BP1 (clones 1 to 7) or HIV-1IIIB (clones 8 to 14) as described in Materials and Methods. At day 7, 80% of the medium was replaced with fresh growth medium and cell-free supernatants were used for HIV p24 assays (SEM [103] for triplicates of two experiments).
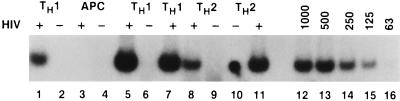
In addition, the number of HIV gag RNA molecules per microgram of total RNA was determined by using a dilution of a standard number of RNA molecules determined as previously described (64) (Fig. 4). At day 2 p.i. HIV-1-infected TH1 cells had approximately 500 gag RNA molecules/μg of RNA (Fig. 4, lane 1), while mitogen-stimulated mitomycin-treated feeder cells expressed no viral RNA (Fig. 4, lane 3). At day 7, a substantial number of gag RNA molecules were present in both TH1 and TH2 clones (Fig. 4). To obtain more precise numbers, the viral RNA was diluted to 1,000 to 2,000 molecules/reaction so that the signal obtained under the amplification conditions used was in the linear range of the standard curve. Using this approach, we determined that similar numbers of gag RNA molecules were present in different TH1 and TH2 clones throughout the duration of the infection (Table 6). Thus, although different clones varied two- to threefold in number of mRNA molecules present (8,000 to 25,000 molecules/μg of RNA), neither the kinetics nor the magnitude of HIV expression in TH2 clones was significantly higher than in TH1 clones regardless of the viral isolate used to infect the clones (Tables 5 and 6). We also investigated whether the antigen specificity of the clones made a difference by measuring RNA at day 7 p.i.; in the case of a SEB TH1 and TH2 clone, there was little difference in number of viral RNA molecules (9,500 versus 10,800).
FIG. 4.
Analysis of HIV gag mRNA in HIV-infected TH1 and TH2 clones. HIV infection of antigen-activated T-cell clones, RT-PCR analysis, and use of HIV gag RNA standards were performed as described in Materials and Methods. Cells were infected with HIVMN. Lanes 1 and 2, TH1 H1.15 with and without HIV, day 2; lanes 3 and 4, mitogen-stimulated antigen-presenting cells (APC) with and without HIV, day 2; lanes 5 and 6, TH1 H1.15 with and without HIV, day 7; lane 7, TH1 H1.12 with HIV, day 7; lanes 8 and 9, TH2 H2.29 with and without HIV, day 7; lane 10, TH2 H2.29; lane 11, TH2 H2.29 with HIV, day 14; lanes 12 to 16, RNA standard curve.
TABLE 6.
Levels of HIV gag mRNA in TH1 and TH2 clones
| Clonea | Type | Antigen specificity | HIV gag RNA (mol/μg of RNA)b
|
||
|---|---|---|---|---|---|
| Day 2 | Day 5 | Day 10 | |||
| H1.15 | TH1 | TTx | 10,600 | 25,300 | 41,000 |
| H1.12 | TH1 | TTx | 8,400 | 20,100 | 36,800 |
| H1.18 | TH1 | TTx | 9,100 | 23,700 | 39,200 |
| H2.25 | TH2 | TTx | 11,700 | 31,100 | 52,600 |
| H2.29 | TH2 | TTx | 2,000 | 10,800 | 19,100 |
| H2.33 | TH2 | TTx | 7,300 | 21,400 | 37,700 |
Derivation, antigen activation, and HIVIIIB infection of T-cell clones were performed as described in Materials and Methods.
Construction and use of an RNA standard and RT-PCR analysis are described in Materials and Methods. For quantitation of number of HIV gag mRNA molecules, the gel was scanned on a PhosphorImager (Molecular Dynamics), using ImageQuant and Microsoft Excel programs, in comparison to a standard curve in a linear range on each gel. Viral samples were diluted to fall within the standard curve.
Since accelerated cell death has been seen in HIV-infected T cells following in vitro stimulation (2, 24, 32, 39, 47, 59), it is possible that if the data were based on input cell number, the results could vary between clones. When the number of HIV gag molecules per 105 viable cells (measured by trypan blue exclusion and Ficoll-Hypaque separation) was determined, the infected TH1 cells were found to contain roughly twofold more RNA molecules than infected TH2 clones (Table 7). In addition, when a TH1 clone activated with either specific antigen or PMA–anti-CD3 was infected with HIV, similar levels of replication were observed in this TH1 clone (Table 7). Thus, even when potential differences in cell viability or polyclonal activation are taken into account, we did not observe any preferential replication of HIV-1 in TH subsets.
TABLE 7.
Levels of HIV gag mRNA/105 viable cells in TH1 and TH2 clones after HIV-1 infection
| Clonea | Type | Antigen specificity | HIV gag RNA (mol/105 viable cells)b
|
|
|---|---|---|---|---|
| Day 7 | Day 14 | |||
| H1.19 | TH1 | TTx | 7,300 | 108,000 |
| H1.19 | TH1 | PMA–anti-CD3 | 8,400 | 205,000 |
| H1.15 | TH1 | TTx | 12,700 | 231,000 |
| H1.18 | TH1 | TTx | 9,700 | 196,000 |
| H2.29 | TH2 | TTx | 4,600 | 87,000 |
| H2.33 | TH2 | TTx | 5,300 | 95,000 |
| H2.25 | TH2 | TTx | 3,800 | 79,000 |
Derivation, antigen activation, and HIVIIIB infection of T-cell clones were performed as described in Materials and Methods. The H1.19 clone was not previously described.
Construction and use of an RNA standard and RT-PCR analysis are described in Materials and Methods. For quantitation of number of HIV gag mRNA molecules, the gel was scanned on a PhosphorImager (Molecular Dynamics), using ImageQuant and Microsoft Excel programs, in comparison to a standard curve on each gel. Viable cells were separated by Ficoll-Hypaque density centrifugation.
Infection of TH1 and TH2 clones by primary NSI HIV-1 strains.
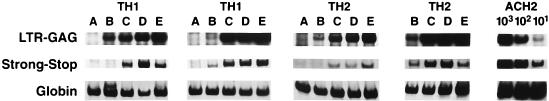
The different tropism (primary macrophages and T cells but not T-cell lines) of primary NSI strains from SI strains suggested a possible difference in infecting different T-cell clones. To investigate this, we infected three TH1 and three TH2 clones highly susceptible to infection by laboratory SI strains with four primary well-characterized NSI isolates. As discussed earlier, amplification using the R-U5 primer pair detects a DNA region representing initial reverse transcription and nearly complete reverse transcription is detected with the LTR-gag primer pair (Fig. 5). At 12 h p.i., the SF162 strain did not show detectable viral entry in two of three clones of each type, while the other three NSI strains showed equivalent strong-stop DNA in each clone, with little variation between TH1 and TH2 clones. At 7 days p.i., the amount of full-length viral DNA present in the cells (Fig. 5) and the amount of HIV-1 p24 in the supernatant (Table 8) show clear clonal variations in ability to support viral infection. The TH1 clones H1.20 and H1.25 and the TH2 clones H2.10 and H2.5 supported vigorous replication with three of four NSI viral isolates, while the TH1 clone H1.15 and the TH2 clone H2.33 poorly supported NSI viral replication (Table 8). There was also variation between viral isolates, with SF162 being poorly replicative in the four clones in which the three other NSI viruses replicated well.
FIG. 5.
HIV-1 DNA detection in TH1 and TH2 clones. Lysates were prepared at 10 and 24 h p.i. A primer pair was used to detect the earliest region of DNA formed by reverse transcription (141 bp of the minus strong-stop strand). Another primer pair was used to detect full-length HIV-1 DNA (200 bp of LTR-gag) and human β-globin. The sense primers of each pair were radiolabeled. Two different TH1 (H1.20 and H1.25) and TH2 (H2.10 and H2.25) clones are shown. Lanes: A, control; B, SF162 infection; C, US657; D, US714; E, US727. Tenfold serial dilutions of the ACH-2 cell line, containing one integrated copy of HIV-1 DNA per cell, were made in a background of uninfected T cells and shown for estimation of copy number.
TABLE 8.
Replication of NSI HIV-1 strains in human TH1 and TH2 clones
| Clonea
|
HIV p24 (pg/ml)b
|
||||
|---|---|---|---|---|---|
| No. | Name | SF162 | US657 | US714 | US727 |
| 1 | H1.20 | 300 ± 0.3 | 12,500 ± 1.3 | 15,200 ± 1.5 | 22,000 ± 2.3 |
| 2 | H1.25 | 0 | 21,500 ± 1.7 | 29,700 ± 2.3 | 27,000 ± 3.3 |
| 3 | H1.15 | 550 ± 0.5 | 420 ± 0.5 | 1,300 ± 1.0 | 600 ± 0.5 |
| 4 | H2.10 | 2,100 ± 1.3 | 18,500 ± 1.7 | 21,000 ± 3.0 | 23,000 ± 2.7 |
| 5 | H2.33 | 0 | 1,700 ± 0.7 | 2,200 ± 1.5 | 950 ± 0.3 |
| 6 | H2.5 | 2,500 ± 1.3 | 23,000 ± 2.7 | 17,000 ± 1.7 | 34,500 ± 3.3 |
T-cell clones were generated, maintained, and activated as described in Materials and Methods.
Clones were infected with the indicated viral isolates as described in Materials and Methods. At day 7, cell-free supernatants were used for HIV p24 assays (SEM [103] for triplicates of two experiments).
DISCUSSION
CD4+ T cells, the preferential targets of HIV-1, can be divided into functional TH1 and TH2 subsets which are responsible for initiating the immune response against different classes of foreign invaders (14, 15, 50, 51, 57). Since alterations in the TH1 and TH2 responses can increase microbial pathogenesis, much effort has gone into determining the role of TH1 and TH2 cells and cytokines during HIV infection and their relationship to HIV pathogenesis. Whether there is an alteration in TH1 and TH2 responses during AIDS progression remains controversial (3, 9–12, 31, 43–46, 49, 56).
One study by Maggi et al. (43) has suggested that HIV replicates preferentially in TH2 and TH0 clones rather than TH1 clones in vitro. This concept could have major implications in AIDS pathogenesis and has been incorporated in recent models of HIV pathogenesis (11, 49, 56). As the kinetics and magnitude of a viral infection can often be affected by the nature of virus-cell interactions as well as extracellular environment, we examined HIV infectivity in a panel of defined human TH cell clones derived from different donors. Since cytokine secretion by TH cells is a continuum and TH2 and TH1 subsets represent the polar ends of the TH cell spectrum (36, 51, 55, 57), we used well-defined TH1 clones (IFN-γ+ IL-4− IL-5−) and TH2 clones (IFN-γ− IL-4+ IL-5+) in this study. Neither mitogen (PMA–anti-CD3) treatment nor HIV infection altered the cytokine patterns produced by TH1 and TH2 clones.
We observed no significant differences between TH1 and TH2 clones with respect to (i) the cell surface expression of CD4 and the chemokine receptor cofactors (both CXC and CC classes), (ii) viral entry and reverse transcription (measured by strong-stop and full-length HIV-1 DNA), and (iii) HIV replication (measured by release of HIV p24 and total number of copies of gag RNA per total cell RNA). The results were similar whether the virus used was a laboratory SI strain or a primary NSI strain. In the case where a virus replicated poorly, it did so in both types of TH cells. Even though the amount of gag mRNA molecules could be an overestimate due to the presence of some viral genomic RNA, there is unlikely to be significantly more genomic RNA in one subset than another. Thus, we did not observe any preferential HIV infection of activated TH2 over TH1 clones in vitro. In the production of viral p24, additional parameters, such as the use of different T-cell donors for cell cloning and the use of different antigenic specificities, had no effect on the results. The presence of substantial amounts of CD4, CD44 (21), and CCR5 (8, 16, 19, 20) on the cell surface of TH1 and TH2 clones used supports the similar production seen with monocytotropic HIV-1 strains and suggests these T-cell clones are more like primary T cells (30, 63) than T-cell lines.
In using NSI strains of HIV to infect these clones, we found, as previously reported, clonal variation in that some clones would not support replication of these viruses (27, 44) very well as well as variation in the ability of these viruses to replicate in permissive clones. However, these differences were not restricted to either TH1 or TH2 clones. Thus, there are not likely to be any intrinsic differences in the ability of different types of T-cell clones to support HIV-1 replication. However, there could be many environmental reasons for differences in HIV replication, such the amount of chemokines released (13, 22), the absence of coreceptors on certain types of clones (41), and the ability of immune stimulation to preferentially downregulate the CCR5 coreceptor (4, 7).
In the study by Maggi et al. (43) and other studies (34, 54, 57), TH clones were activated by mitogens (such as PMA–anti-CD3), as it was believed that cells express a more heterogeneous cytokine pattern after mitogen activation than after antigen activation. Therefore, mitogen stimulation would result in uncovering more TH0 clones. However, we did not find any difference between antigen and mitogen activation on viral replication or cytokine production in TH1 clones. Indeed, our results are similar to those of other studies which, when measuring cytokine production at the single-cell level, found no qualitative differences in cytokine profiles between antigen and mitogen stimulation (36, 54). In addition, HIV-infected TH1 and TH2 clones still maintained the same phenotype as measured by cytokine profile 10 days after infection. Recent evidence that memory and naive CD4+ T cells (precursors of both TH1 and TH2 cells) from HIV-infected individuals have similar rates of decline during AIDS progression (45, 46), and the ability to obtain high percentages of both TH1 and TH2 clones from PBMCs of late-stage AIDS patients (43, 47, 56) despite the daily loss and replacement of CD4+ T cells during HIV infection (33, 62) makes it unlikely that there is preferential infection of CD4+ TH cell subsets in vivo during AIDS progression.
Maggi et al. (43), who found no evidence for in vitro infection of TH1 clones by HIV, examined only one time period (20 days p.i.) in their study. While the differences between the two studies could be due to several factors (differences in experimental design, differences in the panel of clones used, methods of T-cell activation, etc.), the time point used to measure HIV-1 production is close to the limit of T-cell clone survival without further stimulation with antigen and feeder cells. Since both infected and uninfected T cells from HIV-infected individuals undergo activation induced apoptosis in vitro (2, 24, 32, 39, 47, 59) probably through Fas-mediated killing (5, 18, 34, 35, 37, 53), one possible reason for the discrepancy between the two studies is differential cell death of the infected TH1 and TH2 clones. Differences in virus production in vitro at later times of infection could be due to more rapid killing of one subset. However, when values were normalized for viable cell number, more gag RNA molecules were found in TH1 cells. These studies indicate that any role of TH1 and TH2 subsets in AIDS pathogenesis transcends clonal differences in their ability to support HIV replication.
ACKNOWLEDGMENTS
We thank Bernard Poiesz for providing the HIV gag RNA standard and Cari Petrow, Jason Troxell, and Anne Meyers for excellent technical assistance. The HIV primary isolates were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
REFERENCES
- 1.Aggarwal B, Puri R, editors. Human cytokines: their role in disease and therapy. Cambridge, Mass: Blackwell Science; 1995. [Google Scholar]
- 2.Ameisen J. Programmed cell death and AIDS: from hypothesis to experiment. Immunol Today. 1992;13:388–391. doi: 10.1016/0167-5699(92)90086-M. [DOI] [PubMed] [Google Scholar]
- 3.Barcellini W, Rizzardi G, Borghi M, Fain C, Lazzarin A, Meroni P. TH1 and TH2 cytokine production by peripheral blood mononuclear cells from HIV-infected patients. AIDS. 1994;8:757–762. doi: 10.1097/00002030-199406000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Bleul C, Wu L, Hoxie J, Springer T, MacKay C. The HIV coreceptors CXCR4 and CCR5 are differently expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunner T, Mogli R, LaFace D, Yoo N, Mahboubi A, Echeverri F, Martin S, Force W, Lynch D, Ware C, Green D. Cell-autonomous Fas (CD95)/Fas-ligand interaction mediates activation-induced apoptosis in T-cell hybridomas. Nature. 1995;373:441–444. doi: 10.1038/373441a0. [DOI] [PubMed] [Google Scholar]
- 6.Byrne B, Li J, Sninsky J, Poiesz B. Detection of HIV RNA sequences by in vitro DNA amplications. Nucleic Acids Res. 1988;16:4165. doi: 10.1093/nar/16.9.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll R, Riley J, Levine B, Feng Y, Kaushal S, Ritchey D, Bernstein W, Weislow O, Brown C, Berger E, June C, St. Louis D. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science. 1997;276:273–276. doi: 10.1126/science.276.5310.273. [DOI] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 9.Clerici M, Shearer G. A TH1-TH2 switch is a critical step in the etiology of HIV infection. Immunol Today. 1993;14:107–110. doi: 10.1016/0167-5699(93)90208-3. [DOI] [PubMed] [Google Scholar]
- 10.Clerici M, Hakim F, Venzon D, Blatt S, Hendrix C, Wynn T, Shearer G. Changes in interleukin-2 and interleukin-4 production in asymptomatic, human immunodeficiency virus-seropositive individuals. J Clin Invest. 1993;91:759–765. doi: 10.1172/JCI116294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clerici M, Shearer G. The Th1-Th2 hypothesis of HIV infection: new insights. Immunol Today. 1994;15:575–581. doi: 10.1016/0167-5699(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 12.Clerici M, Wynn T, Berzofsky J, Blatt S, Hendrix C, Sher A, Coffman R, Shearer G. Role of interleukin-10 in T helper cell dysfunction in asymptomatic individuals infected with human immunodeficiency virus. J Clin Invest. 1994;93:768–775. doi: 10.1172/JCI117031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Del Prete G, De Carli M, Mastromauro C, Macchia D, Biagiotti R, Ricci M, Romagnani S. Purified protein derivative of Mycobacterium tuberculosis and excretory-secretory antigen(s) of Toxocara canis expand in vitro human T cells with stable and opposite (type 1 T helper or type 2 T helper) profile of cytokine production. J Clin Invest. 1991;88:346–352. doi: 10.1172/JCI115300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Prete G, Romagnani S. The role of TH1 and TH2 subsets in human infectious diseases. Trends Microbiol. 1994;2:4–6. doi: 10.1016/0966-842x(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio P, Marmon S, Sutton R, Hill C, Davis C, Peiper S, Schall T, Littman D, Landau N. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Derse D, Mikovits J, Polianova M, Felber B K, Ruscetti F. Virions released from cells transfected with a molecular clone of human T-cell leukemia virus type I give rise to primary and secondary infections of T cells. J Virol. 1995;69:1907–1912. doi: 10.1128/jvi.69.3.1907-1912.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhein J, Walczak H, Baumler C, Debatin K, Krammer P. Autocrine T-cell suicide mediated by APO-1/(Fas/CD95) Nature. 1995;373:438–441. doi: 10.1038/373438a0. [DOI] [PubMed] [Google Scholar]
- 19.Doranz B, Rucker J, Yi Y, Smyth R, Samson M, Peiper S, Parmentier M, Collman R, Doms R. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 20.Dragic T, Litwin V, Allaway G, Martin S, Huang Y, Nagashima K, Cayanan C, Maddon P, Koup R, Moore J, Paxton W. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 21.Dukes C, Yu Y, Rivadeneira E, Sauls D, Liao D H-X, Haynes B, Weinberg J. Cellular CD44S as a determinant of human immunodeficiency virus type 1 infection and cellular tropism. J Virol. 1995;69:4000–4005. doi: 10.1128/jvi.69.7.4000-4005.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fauci A. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 23.Feng T, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 24.Finkel T, Tudor-Williams G, Banda N, Cotton M, Curiel T, Monks C, Baba T, Ruprecht R, Kupfer A. Apoptosis occurs predominately in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat Med. 1995;1:129–134. doi: 10.1038/nm0295-129. [DOI] [PubMed] [Google Scholar]
- 25.Fitch F, Gajewski T. Production of T cell clones. In: Coligan J, Kruisbeek A, Margulies D, Shevach E, Strober W, editors. Current protocols in immunology. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 3.13.1–3.13.9. [Google Scholar]
- 26.Folks T, Powell D, Lightfoote M, Benn S, Martin M, Fauci A. Induction of HTLV-III/LAV from a non-virus producing T cell line: implications for latency. Science. 1986;231:600–602. doi: 10.1126/science.3003906. [DOI] [PubMed] [Google Scholar]
- 27.Fouchier R, Meyaard L, Brouwer M, Hovenkamp E, Schuitemaker H. Broader tropism and higher cytopasticity for CD4+ T cells of a syncytium-inducing compared to a non-syncytium-inducing HIV-1 isolate as a mechanism for accelerated CD4+ T cell decline in vivo. Virology. 1996;291:87–95. doi: 10.1006/viro.1996.0225. [DOI] [PubMed] [Google Scholar]
- 28.Gajewski T F, Lancki D W, Stack R, Fitch F W. “Anergy” of TH0 helper T lymphocytes induces downregulation of TH1 characteristics and a transition to a TH2-like phenotype. J Exp Med. 1994;179:481–491. doi: 10.1084/jem.179.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gendelman H, Friedman R, Joe S, Baca L, Turpin J, Dveksler G, Meltzer M, Dieffenbaker C. A selective defect of interferon-alpha production in human immunodeficiency virus-infected monocytes. J Exp Med. 1990;172:1433–1440. doi: 10.1084/jem.172.5.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granelli-Piperno A, Mose B, Pope M, Chen D, Wei Y, Isdell F, O’Doherty U, Paxton W, Koup R, Mojsov S, Bhardwaj N, Clark-Lewis I, Baggiolini M, Steinman R M. Efficient interaction of HIV-1 with purified dendritic cells via multiple chemokine coreceptors. J Exp Med. 1996;184:2433–2438. doi: 10.1084/jem.184.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graziosi C, Pantaleo G, Gantt K, Fortin J, Demarest J, Cohen O, Sekaly R, Fauci A. Lack of evidence for the dichotomy of TH1 and TH2 predominance in HIV-infected individuals. Science. 1994;265:248–252. doi: 10.1126/science.8023143. [DOI] [PubMed] [Google Scholar]
- 32.Groux H, Torpier G, Monte D, Mouton Y, Capron A, Ameisen J. Activation-induced death by apoptosis in CD4+ T cells from human immunodeficiency virus-infected asymptomatic individuals. J Exp Med. 1992;175:331–340. doi: 10.1084/jem.175.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho D, Neumann A, Perelson A, Chen W, Leonard J, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 34.Ju S-T, Panka D, Cul H, Ettinger R, El-Khatib M, Sherr D, Stanger B, Marshak-Rothstein A. Fas(CD95)/FasL interactions required for programmed cell death after T-cell activation. Nature. 1995;373:444–448. doi: 10.1038/373444a0. [DOI] [PubMed] [Google Scholar]
- 35.Katsikis P D, Wunderlich E S, Smith C A, Herzenberg L A, Herzenberg L A. Fas antigen stimulation induces marked apoptosis of T lymphocytes in human immunodeficiency virus-infected individuals. J Exp Med. 1995;181:2029–2036. doi: 10.1084/jem.181.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelso A. Th1 and Th2 subsets: paradigms lost? Immunol Today. 1995;16:374–379. doi: 10.1016/0167-5699(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi N, Hamamoto Y, Yamamoto N, Ishii A, Yonehara M, Yonehara S. Anti-Fas monoclonal antibody is cytocidal to human immunodeficiency virus-infected cells without augmenting viral replication. Proc Natl Acad Sci USA. 1990;87:9620–9624. doi: 10.1073/pnas.87.24.9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 39.Laurent-Crawford A, Krust B, Muller S, Riviere Y, Cuille M, Bechet J, Montanier L, Hovanessian A. The cytopathic effect of HIV is associated with apoptosis. Virology. 1991;185:829–839. doi: 10.1016/0042-6822(91)90554-o. [DOI] [PubMed] [Google Scholar]
- 40.Levy J. HIV pathogenesis and long-term survival. AIDS. 1993;7:1401–1410. doi: 10.1097/00002030-199311000-00001. [DOI] [PubMed] [Google Scholar]
- 41.Loetscher P, Uguccioni M, Bondoli L, Baggiolini M, Moser B, Chizzolini C, Dayer J. CCR5 is characteristic of Th1 lymphocytes. Nature. 1998;391:344–345. doi: 10.1038/34814. [DOI] [PubMed] [Google Scholar]
- 42.Lori F, Veronese F, Del Vico A, Lusso P, Reitz M, Gallo R. Viral DNA carried by human immunodeficiency virus type 1 virions. J Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maggi E, Mazzetti M, Ravina A, Annunziato F, De Carli M, Piccinni M, Manetti R, Carbonari M, Pesce A, Del Prete G, Romagnani S. Ability of HIV to promote a Th1 to Th0 shift and to replicate preferentially in Th2 and Th0 cells. Science. 1994;265:244–248. doi: 10.1126/science.8023142. [DOI] [PubMed] [Google Scholar]
- 44.Meyaard L, Fouchier R, Brouwer M, Hovenkamp E, Miedema F. Syncytium-inducing HIV-1 replicate equally well in all types of T-helper cell clones. AIDS. 1996;10:1598–1600. doi: 10.1097/00002030-199611000-00023. [DOI] [PubMed] [Google Scholar]
- 45.Meyaard L, Hovenkamp E, Keet I P M, Hooibrink B, deJong I H, Otto S A, Miedema F. Single-cell analysis of IL-4 and IFN-γ production by T cells from HIV-infected individuals. J Immunol. 1996;157:2712–2718. [PubMed] [Google Scholar]
- 46.Meyaard L, Otto S, Hooibrink B, Miedema F. Quantitative analysis of CD4+ T cell function in the course of human immunodeficiency virus infection. J Clin Invest. 1994;94:1947–1952. doi: 10.1172/JCI117545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyaard L, Otto S, Jonker R, Mijnster M, Keet R, Miedema F. Programmed death of T cells in HIV-1 infection. Science. 1992;257:217–219. doi: 10.1126/science.1352911. [DOI] [PubMed] [Google Scholar]
- 48.Mikovits J, Raziuddin, Gonda M, Ruta M, Lohrey N, Kung H-F, Ruscetti F. Negative regulation of human immune deficiency virus replication in monocytes. J Exp Med. 1990;171:1705–1720. doi: 10.1084/jem.171.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mosmann T. Cytokine patterns during the progression to AIDS. Science. 1994;265:193–194. doi: 10.1126/science.8023139. [DOI] [PubMed] [Google Scholar]
- 50.Mosmann T, Cherwinski H, Bond M, Giedlin M, Coffman R. Two types of murine helper T-cell clones. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2355. [PubMed] [Google Scholar]
- 51.Mosmann T, Coffman R. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 52.Murphy W, Tian Z, Asai O, Strieter R, Kunkel S, Longo D L, Taub D. Chemokines and lymphocyte activation. II. Facilitation of human cell trafficking in severe combined immunodeficiency mice. J Immunol. 1996;156:2104–2111. [PubMed] [Google Scholar]
- 53.Nagata S, Golstein P. The Fas death factor. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 54.Openshaw P, Murphy E, Hosken N, Maino V, Davis K, Murphy K, O’Garra A. Heterogeneity of intracellular cytokine synthesis at the single cell in polarized T helper 1 and T helper 2 populations. J Exp Med. 1995;182:1357–1367. doi: 10.1084/jem.182.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romagnani S. Human TH1 and TH2: doubt no more. Immunol Today. 1991;12:256–261. doi: 10.1016/0167-5699(91)90120-I. [DOI] [PubMed] [Google Scholar]
- 56.Romagnani S, Maggi E, Del Prete G. An alternative view of the Th1/Th2 switch hypothesis in HIV infection. AIDS Res Hum Retroviruses. 1994;10:iii–ix. doi: 10.1089/aid.1994.10.iii. [DOI] [PubMed] [Google Scholar]
- 57.Swain S. Generation and in vivo persistence of polarized TH1 and TH2 memory cells. Immunity. 1994;1:543–552. doi: 10.1016/1074-7613(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 58.Taub D D, Lloyd A, Wang J M, Oppenheim J. α and β chemokines. In: Coligan J, Kruisbeek A, Margulies D, Shevach E, Strober W, editors. Current protocols in immunology. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 6.12.1–6.12.28. [Google Scholar]
- 59.Terai C, Kornbluth R, Pauza R C, Richman D, Carson D. Apoptosis as a mechanism of cell death in cultured T lymphoblasts acutely infected with HIV-1. J Clin Invest. 1991;87:1710–1715. doi: 10.1172/JCI115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J M, McVicar D, Oppenheim J, Kelvin D. Identification of RANTES receptors on human monocytic cells: competition for binding and desensitization by homologous chemotactic cytokines. J Exp Med. 1993;177:699. doi: 10.1084/jem.177.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei X, Ghosh S, Taylor M, Johnson V, Emini E, Deutsch P, Lifson J, Bonhoeffer S, Nowak M, Hahn B, Saag M, Shaw G. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 63.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 64.Zhang H, Zhang Y, Spicer T, Abbott L, Abbott M, Poiesz B. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res Hum Retroviruses. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]