Abstract
The comprehensive detection and identification of active ingredients in complex matrices is a crucial challenge. Liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS) is the most prominent analytical platform for the exploration of novel active compounds from complex matrices. However, the LC-HRMS-based analysis workflow suffers from several bottleneck issues, such as trace content of target compounds, limited acquisition for fragment information, and uncertainty in interpreting relevant MS2 spectra. Lycibarbarspermidines are vital antioxidant active ingredients in Lycii Fructus, while the reported structures are merely focused on dicaffeoylspermidines due to their low content. To comprehensively detect the new structures of lycibarbarspermidine derivatives, a “depict” strategy was developed in this study. First, potential new lycibarbarspermidine derivatives were designed according to the biosynthetic pathway, and a comprehensive database was established, which enlarged the coverage of lycibarbarspermidine derivatives. Second, the polarity-oriented sample preparation of potential new compounds increased the concentration of the target compounds. Third, the construction of the molecular network based on the fragmentation pathway of lycibarbarspermidine derivatives broadened the comprehensiveness of identification. Finally, the weak response signals were captured by data-dependent scanning (DDA) followed by parallel reaction monitoring (PRM), and the efficiency of acquiring MS2 fragment ions of target compounds was significantly improved. Based on the integrated strategy above, 210 lycibarbarspermidine derivatives were detected and identified from Lycii Fructus, and in particular, 170 potential new compounds were structurally characterized. The integrated strategy improved the sensitivity of detection and the coverage of low-response components, and it is expected to be a promising pipeline for discovering new compounds.
Keywords: Mass spectrometric detection, Weak response signals, Complex matrices, Novel lycibarbarspermidines
Graphical abstract
Highlights
-
•
The “depict” strategy was developed by sample preparation, data acquisition and data postprocessing.
-
•
The “depict” strategy was applied to in-depth characterization of Lycibarbarspermidine derivatives.
-
•
The 210 Lycibarbarspermidine derivatives were detected, 170 potential new compounds were preliminary characterized.
-
•
The integrated strategy facilitates the discovery of novel compounds in complex matrices.
1. Introduction
Owing to their wide range of physiological activities, natural products in traditional Chinese medicine (TCM) play a highly significant role in the drug discovery and development process. As most target compounds are generally present in extremely complex matrices at low levels, rapid and targeted discovery of new low-abundance active compounds has become a challenging issue.
Liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS) has become an indispensable tool for the detection and identification of secondary metabolites in plants [[1], [2], [3]]. The natural products in plants are usually complicated and have a wide content span, while chromatographic columns and mass spectrometry detectors are sophisticated and sensitive. Therefore, rigorous sample preparation is required for the detection of new compounds in complex matrices, particularly for low-abundance components. According to previous reports, solid phase extraction (SPE) reduced the matrix effect of liquid chromatography coupled with mass spectrometry (LC-MS) analysis and showed high selectivity, excellent reproducibility and superior recovery [[4], [5], [6]]. Through the purification and enrichment of the sample, the intensity of the low-abundance components was improved, and sufficient fragment information was obtained. This makes it possible to discover new compounds at low levels.
Structure identification based on LC-HRMS relies on existing databases, such as mzClound and ChemSpider. However, the low-content components are difficult to separate, and thus, the existing database is incomplete. This leads directly to inadequate identification of compounds. As natural products commonly exist in clusters, searching for their structural analogs has become an approach to discover potential new compounds. Molecular design according to the known biosynthetic reactions enables the construction of virtual compound databases, which cover as many analogs of known compounds as possible. The construction of a comprehensive database is a key driver in the discovery of new compounds.
Acquiring sufficient tandem Mass Spectrometry (MS2) information of low-abundance ingredients is the critical step for new compound discovery based on LC-HRMS. The data-dependent scanning (DDA) mode is usually applied to qualitative research. During DDA data acquisition, MS survey scans are first launched, and then the most abundant precursor ions are selected for subsequent MS2 experiments. In this setting, structural information of unknown compounds in the samples can be acquired, allowing for identification. Since the selection of precursor ions for MS2 fragmentation is intensity dependent, a potential limitation of DDA mode is that the further MS2 scan of the features with low abundance might never be triggered [7,8]. To overcome the shortcomings of DDA in acquiring MS2 spectra, the implementation of parallel reaction monitoring (PRM) could theoretically generate MS2 spectra for all precursor ions [[9], [10], [11], [12]]. Therefore, more compounds at lower concentrations could be detected and identified. In the PRM-based workflow, preselected precursor ions are selected in the quadrupole and fragmented in a higher-energy collisional dissociation (HCD) cell. Subsequently, the high-resolution mass spectrometer monitors all generated MS2 fragment ions. PRM performs MS analysis at high resolution and shows significant advantages. First, ions can be enriched in the C-trap, revealing less-abundant fragment ions in the MS2 spectrum. Second, ample MS2 information contributes to the reliable identification of compounds. Finally, a high-resolution/high mass accuracy mass spectrum provides improved specificity. As predefined precursor ions are necessary for data acquisition, the coverage of compounds in PRM is limited. It is expected to comprehensively acquire the fragment information of target analytes in complex samples through DDA coupled with PRM mode.
The raw data acquired from LC-HRMS contained a huge amount of information, making data analysis and the extraction of useful information extremely difficult. The molecular network in the Global Natural Products Social Molecular Network (GNPS, https://gnps.uscd.edu/) platform is an important tool for mining complex mass spectral information [13,14]. It is a visual network map based on the similarity of MS2 spectrometry. Structurally similar compounds tend to produce similar MS2 spectra, and their nodes cluster. After identifying the known components by comparison with the spectral library, the unknown components in the same cluster are analyzed based on the identified compounds. Molecular networks enable natural products to be rapidly identified in complex mixtures and contribute to discovering new natural products.
Lycibarbarspermidines are vital antioxidant active ingredients in Lycii Fructus and are considered potential anti-Alzheimer's disease drug candidates. As rare plant secondary metabolites, only 15 lycibarbarspermidines have been isolated and identified. These compounds were dicaffeoylspermidines O-glycosylated by one or two β-d-glucopyranose units [[15], [16], [17], [18]]. In addition, several investigations reported novel lycibarbarspermidines containing three or four β-d-glucopyranose units by LC-HRMS [19,20]. However, the reported structures of lycibarbarspermidines are merely concentrated on dicaffeoylspermidines and their glycosides, mainly due to their low contents in Lycii Fructus. Hydroxycinnamic acids (HCAs), such as ferulic acid, caffeic acid, sinapic acid, and p-coumaric acid, are important constituent parts of lycibarbarspermidines. Considering that HCAs are a group of compounds highly abundant in plants, we assume that new lycibarbarspermidine derivatives from Lycii Fructus could be explored using DDA coupled with PRM. However, polysaccharides, polyphenols, and carotenoids are the major constituents in Lycii Fructus and greatly interfere with the detection of lycibarbarspermidine derivatives using DDA coupled with PRM mode. Moreover, lycibarbarspermidine derivatives in Lycii Fructus are present at low levels, resulting in insufficient acquisition of fragment information. In addition, component identification based on LC-HRMS relies heavily on currently available databases, while currently available databases of lycibarbarspermidine derivatives are limited.
Herein, a “depict” strategy based on structure design, sample preparation, molecular network identification, and the capture of low response signals was developed for in-depth characterization of lycibarbarspermidine derivatives. We envision that the combinatorial approach will facilitate structural annotation of lycibarbarspermidine derivatives and is able to improve analysis coverage and the detection sensitivity of low-abundance active components. More importantly, the integrated strategy facilitates the discovery of novel components in complex matrices.
2. Experimental
2.1. Reagents and materials
Lycii Fructus, a dried and ripe fruit of the plant Lycium barbarum, was provided by Sinopharm Group Dezhong Pharmaceutical Co., Ltd. (Foshan, China). Analytical grade ethanol was purchased from Tianjin Four Fine Chemicals Co., Ltd. (Tianjin, China). Ultrapure water was prepared with a Milli-Q water purification system (Millipore, Bedford, MA, USA). LC-MS grade acetonitrile and formic acid were obtained from Thermo Fisher Scientific Inc. (Waltham, MA, USA). Oasis HLB solid phase extraction (SPE) columns (3 mL/60 mg) and Oasis MCX SPE columns (3 mL/60 mg) were purchased from Waters (Milford, MA, USA).
2.2. Sample preparation
Three grams of Lycii Fructus was accurately weighed and extracted with 50 mL of anhydrous ethanol at room temperature for 50 min by ultrasonic extraction. The extracts were evaporated to dryness under reduced pressure and then redissolved in 10.00 mL of water. The solution was extracted using Oasis HLB or Oasis MCX adsorbents.
2.2.1. Oasis HLB-SPE pretreatment
Oasis HLB-SPE columns were first conditioned with 2.0 mL of methanol followed by 2.0 mL of water. The solvents used for column conditioning were discarded. Then, the cartridge was loaded with 1.0 mL of the redissolved Lycii Fructus extract twice. The extract was allowed to pass completely through the sorbent material and was followed with 1.0 mL of water. The column eluent was discarded. To elute the analytes, 1.0 mL of methanol was added to the sorbent, and the eluent was collected. The eluate was dried with nitrogen and then redissolved in 200 μL of methanol.
2.2.2. Oasis MCX-SPE pretreatment
The Oasis MCX columns conditioned with 2 mL of methanol and equilibrated with 2 mL of water. The solvents were discarded. The cartridge was loaded with 1.0 mL of the redissolved Lycii Fructus extract twice. The column was washed with 1 mL 2% (V/V) formic acid in water to retain the alkaline and remove impurities. Then, the column was eluted with 1 mL of methanol to remove the neutral constituents. The column eluent was discarded. To elute the analytes, 1.0 mL of 5% (V/V) ammoniated methanol was added to the sorbent, and the eluent was collected. The eluate was dried with nitrogen and then redissolved in 200 μL of methanol.
All samples were centrifuged at 11,410 g for 15 min before LC-HRMS analysis.
2.3. Construction of the lycibarbarspermidine derivatives database
The biochemical reactions of HCAs and amine groups in plants were used to establish the database for lycibarbarspermidine derivatives. The structures of lycibarbarspermidine derivatives were drawn in ChemDraw to obtain molecular structures and formulas. Then, the exact mass for [M+H]+ in positive ion mode of lycibarbarspermidine derivatives was calculated by Qual Browser in Xcalibur™ 4.1 software (Thermo Fisher Scientific) and recorded in Excel.
2.4. LC-MS/MS analysis
The LC-HRMS data were obtained on a Vanquish UPLC instrument coupled with a Q Exactive Orbitrap high-resolution mass spectrometer (Waltham, MA, USA). The chromatographic analysis was carried out on a Waters ACQUITY™ BEH C18 column (2.1 mm × 100 mm, 1.8 μm) under a 300 μL/min flow rate. Mobile phases A and B were H2O with 0.1% (V/V) formic acid and acetonitrile (ACN), respectively. A 22-min gradient was established as follows: 0−2 min, 2%–10% B; 2−3 min, 10% B; 3−9 min, 10%–12% B; 9–12 min, 12%–15% B; 12−15 min, 15%–65% B; 15−20 min, 65%–100% B; and 20−22 min, 100% B. The column temperature was maintained at 38 °C. The samples were maintained at 10 °C, and the injection volume was 2 μL.
The mass spectrometer was equipped with an electrospray ionization source, and mass detection was operated in positive mode. The parameters were set as follows: spray voltage, 3.5 kV (+); capillary temperature, 320 °C; auxiliary gas heater temperature, 350 °C; sheath gas velocity, 35 arb; auxiliary gas velocity, 10 arb; sweep gas pressure, 0 arb; and S-lens RF level, 50 V. Full Scan/dd-MS2 was used to acquire untargeted data. The resolutions of MS and MS2 acquisition were set as 70,000 and 17,500, respectively. The scan range was from m/z 150 to 2,000. The normalized collision energies (NCEs) were 20%, 40%, and 60%. The top 10 most intense ions were selected to perform MS2 acquisition. PRM was used to acquire the targeted data.
2.5. Data analysis
The LC-HRMS raw data was processed using Xcalibur™ 4.1 software (Thermo Fisher Scientific Inc.). The full scan/ddMS2 raw data were analyzed by the GNPS platform to display the structural relationship of higher content constituents, and then Cytoscape 3.9.1 was used for network visualization. In the GNPS workflow, the maximum mass tolerance of the parent ions and fragment ions was set to 0.02 Da. The minimum cosine score was set to 0.6. The minimum matched fragment ions required for node-to-node connection was set as 6.
Compound Discoverer 3.2 (Fisher Scientific Inc.) was used to screen the potential lycibarbarspermidine derivatives in the samples. The database containing 10,560 compounds was imported into Compound Discoverer 3.2, and then the precursor ions with intensities greater than 100,000 in raw data were searched against the database with a mass tolerance of 5 ppm. The potential lycibarbarspermidine derivatives were exported to Excel for further PRM experiments. Then, exact mass and MS2 fragment ions were applied to identify the lycibarbarspermidine derivatives in the PRM data.
3. Results
3.1. Construction of lycibarbarspermidine derivative database
3.1.1. The selection of building blocks for lycibarbarspermidine derivatives based on the biosynthetic pathway
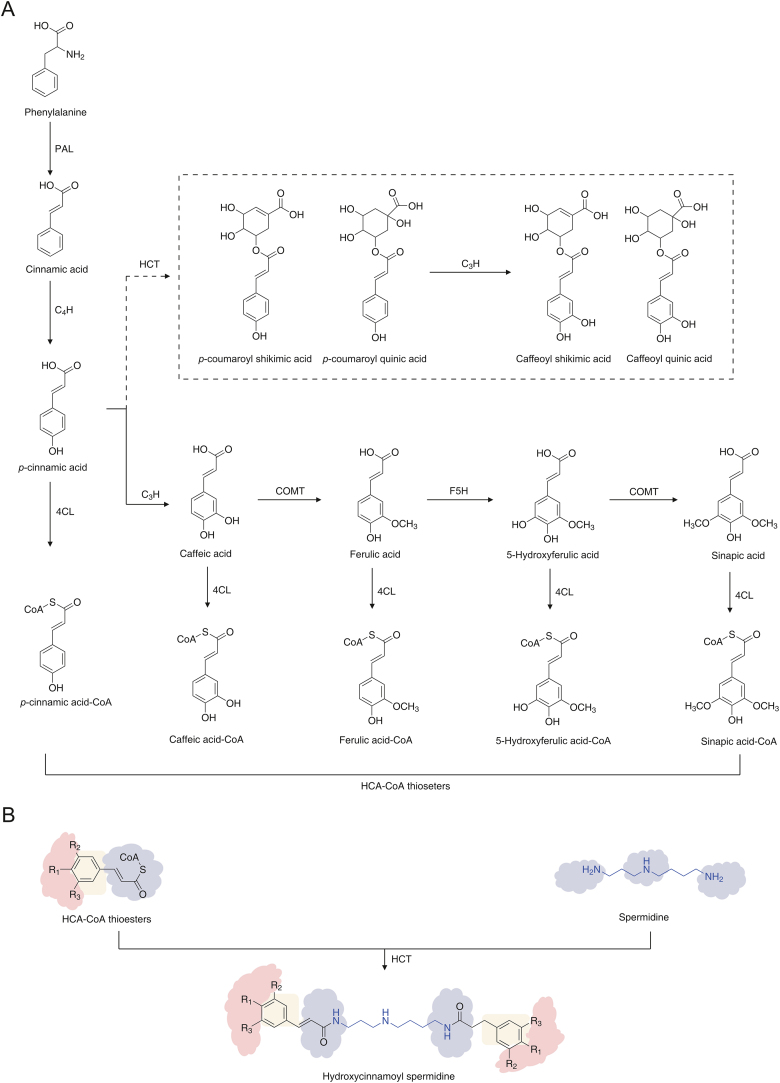
For the exploration of more lycibarbarspermidine analogs and the extension of analytical coverage, a comprehensive database based on the biosynthetic pathway of HCAs was constructed. The biosynthesis of lycibarbarspermidine derivatives is based on the phenylalanine pathway [[21], [22], [23], [24]]. In plants, phenylalanine ammonia lyase (PAL) is responsible for the first step in the phenylalanine pathway and transforms phenylalanine to trans-cinnamate. Then, trans-cinnamate is catalyzed to form ρ-coumaric acid by 4-hydroxylase (C4H) and converted to other hydroxycinnamates, such as caffeic acid, ferulic acid and sinapic acid. These HCAs are esterified into hydroxycinnamoyl-CoA esters by 4-cinnamate, CoA ligase (4CL), and serve as substrates for entry into various branch pathways (Fig. 1A). For hydroxycinnamoyl spermidine biosynthesis, the ultimate step is the condensation of hydroxycinnamoyl-CoA thioesters with spermidine catalyzed by hydroxycinnamoyl transferases (HCTs) (Fig. 1B). In addition, ρ-coumaryl quinic acid/ρ-coumaryl shikimic acid and cafeoyl quinic acid/cafeoyl shikimic acid might be produced under the catalysis of HCT (Fig. 1). In conclusion, three types of building blocks were screened out. Hydroxycinnamic acids, hydrocinnamoyl-quininic acids and hydrocinnamoyl-shikimic acids all have the potential to bind to spermidine (Fig. S1).
Fig. 1.
The biosynthesis of lycibarbarspermidine derivatives. (A) The transformation of hydroxycinnamic acids (HCAs). (B) The biosynthetic pathway of hydroxycinnamoyl spermidine. PAL: phenylalanine ammonia-lyase; C4H: cinnamate 4-hydroxylase; C3H: p-coumarate 3-hydroxylase; 4CL: 4-cinnamate:CoA ligase; HCT: hydroxycinnamoyl transferase; COMT: caffeic acid O-methyltransferase; F5H: ferulate 5-hydroxylase; CoA: coenzymeA.
3.1.2. Structural design of lycibarbarspermidine derivatives
The structures of several glycosylated dicaffeoylspermidines obtained from Lycii Fructus have been identified, and their structures are shown in Fig. S2. In this study, all potential lycibarbarspermidine derivatives were predicted based on the glycosylation reaction of HCAs and the conjugation reaction between HCA/HCA-O glycosides and spermidine. On the one hand, the hydrogen atom on the benzene ring of the hydroxycinnamoyl group might be replaced by various numbers of OH and OCH3, and the C–C double bond of cinnamic acid derivatives might be reduced, leading to a diversity of substituents. On the other hand, 1 to 3 primary and imine groups in spermidine can interact with various HCAs to form amide derivatives. The accurate mass of [M+H]+ in positive ion mode of lycibarbarspermidine derivatives was recorded in the Supplementary Data. A total of 10,560 lycibarbarspermidine derivatives were generated.
3.2. Enrichment of lycibarbarspermidine derivatives by solid phase extraction
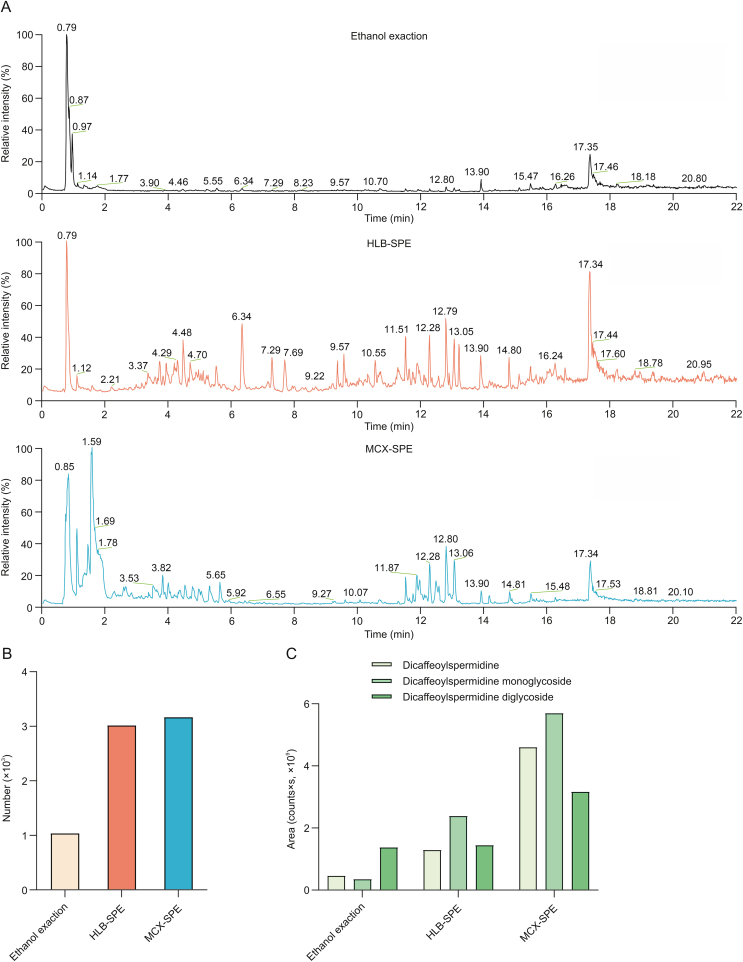
Based on previous studies, we found that lycibarbarspermidines eluted slightly later than impurities in reversed-phase columns, suggesting that the polarity of lycibarbarspermidines is different from that of impurities. Targeted components and impurities could be separated based on their polarity, which is called the polarity-oriented analysis method. To remove the impurities and enhance the detection sensitivity of mass spectrometry, we chose SPE technique. By investigating the literatures, we summarized various SPE sorbent materials, such as reversed-phase sorbent materials, strong cation exchange, strong anion exchange, weak cation exchange, weak anion exchange, and magnetic beads [[25], [26], [27], [28]]. Considering the polarity and pH of lycibarbarspermidine derivatives, special attention was concentrated on the reversed-phase hydrophilic-lipophilic balance sorbent (Oasis HLB) and the mixed-mode strong cation exchange/reversed-phase sorbent (Oasis MCX). Oasis HLB is a universal sorbent and has been widely used for the determination of compounds with different polarities in complex substrates. Oasis MCX could establish reversed-phase and ion-exchange interactions with the analytes. Their performance was evaluated by comparing the peak numbers and peak areas of feature peaks in each chromatogram processed by different SPE columns. The results showed that 1,036 features were detected in the crude extract of Lycii Fructus, and 3,011 and 3,162 features were observed after treatment by Oasis HLB-SPE columns and Oasis MCX-SPE columns, respectively. Furthermore, the total peak area of representative lycibarbarspermidines was the largest after Oasis MCX-SPE extraction (Fig. 2). Oasis MCX is a better option and thus was selected for further study.
Fig. 2.
Comparison of sample preparation methods. (A) Total ion chromatogram (TIC) of samples with different sample preparation methods. (B) The number of features detected under different sample preparation conditions. (C) The peak area of representative compounds under different sample preparation conditions. HLB: hydrophilic-lipophilic balance; SPE: solid phase extraction; MCX: mixed-mode strong cation exchange/reversed-phase.
3.3. Summary of fragmentation patterns based on the literature
In this section, the fragmentation patterns of known lycibarbarspermidines were integrated, and the characteristic fragment ions were selected for MS parameter optimization. In addition, the characteristic fragment ions of potential lycibarbarspermidine derivatives were predicted based on the fragmentation patterns. This enabled rapid and comprehensive identification.
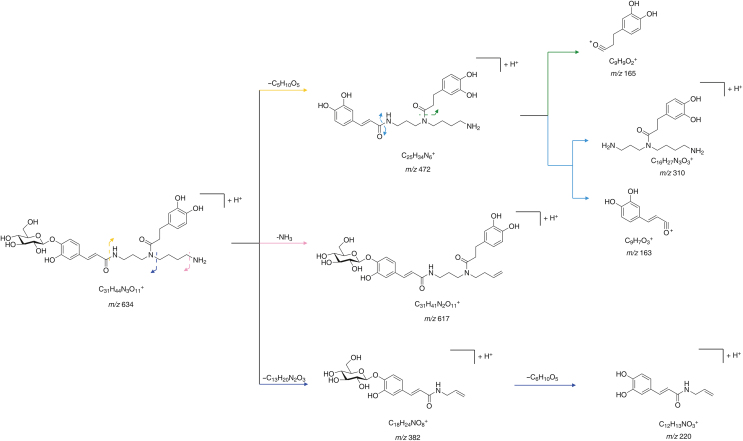
We summarized the fragmentation patterns of lycibarbarspermidines based on the previous literature (Fig. 3) [19,20,29]. Under HCD, the glucoside bonds of lycibarbarspermidines tend to break and lose glucose units. Then, the amide bonds break, producing various fragment ions. In addition, the cleavage of the central C–N bond and the neutral loss of terminal amines are also classical fragmentation pathways. For instance, N1-caffeoyl, N5-dihydrocaffeoyl spermidine monoglycoside produced the parent ion at m/z 634 (C25H33N3O6, calculated m/z 634.29703). After HCD, the ion m/z 472 [M + H–C6H10O5]+ was produced by loss of the glucose unit. Subsequently, m/z 310 [M + H–C6H10O5–C9H7O3]+ and m/z 163 [Caffeoyl]+ were produced by the cleavage of the N1-amide bond. By cleavage of the N5-amide bond, m/z 165 [dihydrocaffeoyl]+ was produced. In addition, m/z 617 [M + H–NH3]+ was produced by the neutral loss of the terminal amine. Finally, m/z 382 [M + H–C13H20N2O3]+ was yielded by breaking the C4–N5 bond, and then m/z 220 [M + H–C13H20N2O3–C6H10O5]+ was yielded by the neutral loss of the glucose unit. The ions at m/z 163, 220, 293, 310, 382, and 472 could be regarded as the key MS2 spectral information to identify N1-caffeoyl, N5-dihydrocaffeoyl spermidine monoglycoside.
Fig. 3.
The fragmentation patterns of N1-caffeoyl,N5-dihydrocaffeoyl spermidine monoglycoside. Reprinted from Refs. [19,20,29] with permission.
3.4. Characterization of potential new compounds using DDA followed by PRM
3.4.1. Characterization by molecular networks
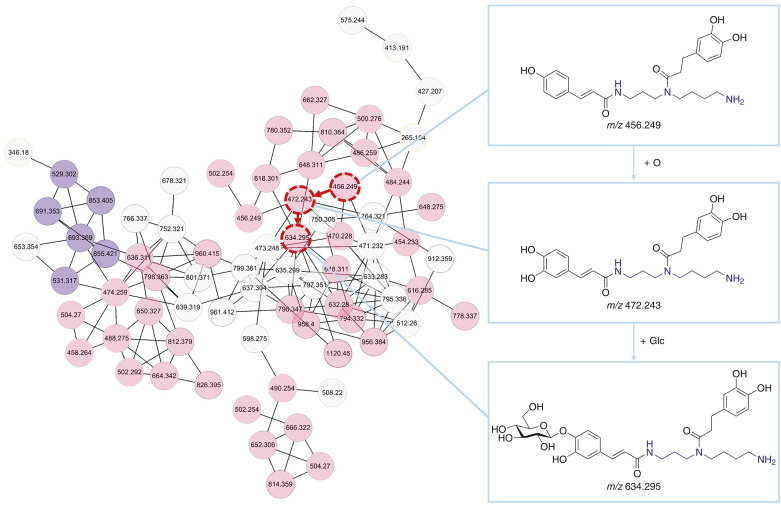
To improve the efficiency of unknown compound identification, molecular networks were introduced to process the raw data in DDA mode (Fig. 4). As MS2 spectra are closely related to chemical structures, compounds with similar structures are associated. The datasets were clustered into visual networks by MS2 spectral similarity scoring. The known lycibarbarspermidines were identified through the spectral library and published literature, which were used to successively elucidate the potential new lycibarbarspermidine derivatives on adjacent nodes. Compounds PB1-PB100 and PB103-PB155 were identified by the above method (Figs. S3−S5).
Fig. 4.
Molecular network of lycibarbarspermidine derivatives. The pink nodes represent lycibarbarspermidine derivatives, and the purple nodes represent lycibarbarspermidines.
3.4.2. Screening of novel potential lycibarbarspermidine derivatives in DDA mode by database searching
DDA mode is usually used to investigate untargeted components in samples. The raw data of lycibarbarspermidine derivatives in the samples were completely acquired in DDA mode. Subsequently, the potential lycibarbarspermidine derivatives were screened based on the self-established database and were recorded to create a list for further PRM experiments.
The raw data were processed using Compound Discoverer 3.2 and manual methods. As a result, 267 potential lycibarbarspermidine derivatives were screened out by Compound Discoverer 3.2. Then, manual methods were used to confirm the correctness of the software screening. These results showed that software screening not only ensures accuracy but also improves the efficiency of data processing. The exact mass of the screened lycibarbarspermidine derivatives in the samples was summarized as an ion list for further PRM experiments. Most potential lycibarbarspermidine derivatives in samples might have 1 or 2 HCAs conjugated to spermidine. Only a small proportion of them belong to the products of hydroxycinnamoyl-quininic acids or hydroxycinnamoyl-shikimic acids conjugated to spermidine (Fig. 5).
Fig. 5.
Structural relationship network of potential lycibarbarspermidine derivatives. The central node represents spermidine. Cluster A represents potential monohydroxycinnamoyl spermidine. Cluster B represents potential mono-dihydro (hydroxycinnamoyl) spermidine. Cluster C represents potential dihydroxycinnamoyl spermidine. Cluster D represents potential hydroxycinnamoyl-dihydro (hydroxycinnamoyl) spermidine. Cluster E represents potential di-dihydro (hydroxycinnamoyl) spermidine. Other nodes represent the products of hydroxycinnamoyl-quininic acids or hydroxycinnamoyl-shikimic acids conjugated to spermidine.
3.4.3. Characterization using subsequent PRM data
To obtain MS2 information of potential lycibarbarspermidine derivatives adequately, the parameters of PRM acquisition were optimized. Aiming to ensure that the precursor and characteristic fragment ions have the appropriate abundance for compound identification, the spray voltage, capillary temperature and normalized collision energy (NCE) were optimized. The peak area of the parent ions generated by representative lycibarbarspermidine (tR = 5.71, C31H43N3O11) was used to optimize the spray voltage and capillary temperature, which are significant factors for ionization efficiency. NCE is a crucial parameter that affects the quality of the MS2 spectrum. The peak area of the characteristic fragment ions produced by representative lycibarbarspermidine (tR = 5.71, C31H43N3O11) was used to optimize NCE. As shown in Fig. S6, the stepped NCE at 20%, 40% and 60%, spray voltage at 3.4 kV and capillary temperature at 320 °C showed the best performance. The optimized MS2 spectrum is shown in Fig. S7. Targeted data acquisition was performed under the optimized conditions. Both the parent and daughter ions are of appropriate intensity.
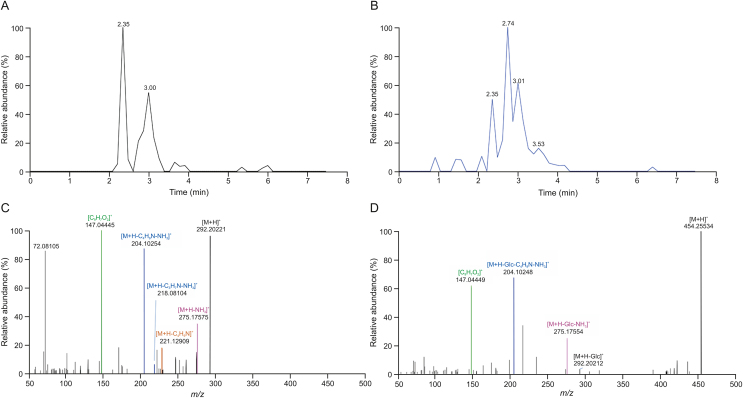
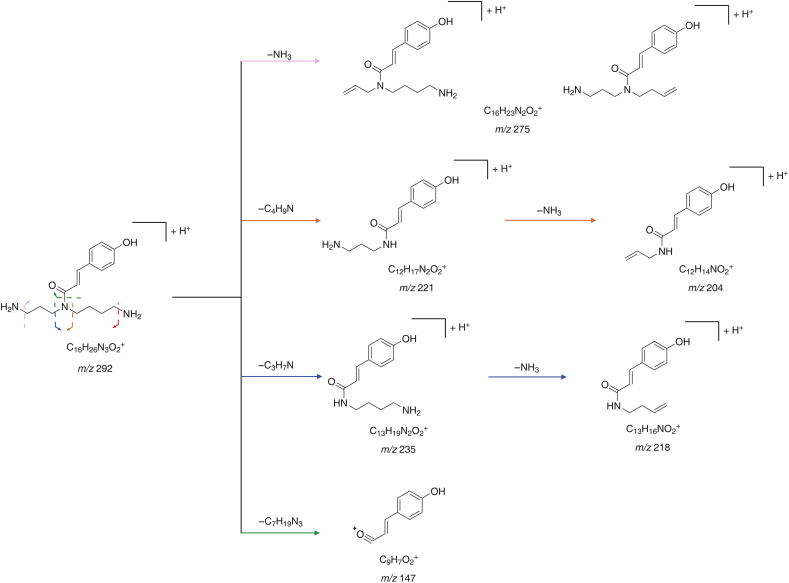
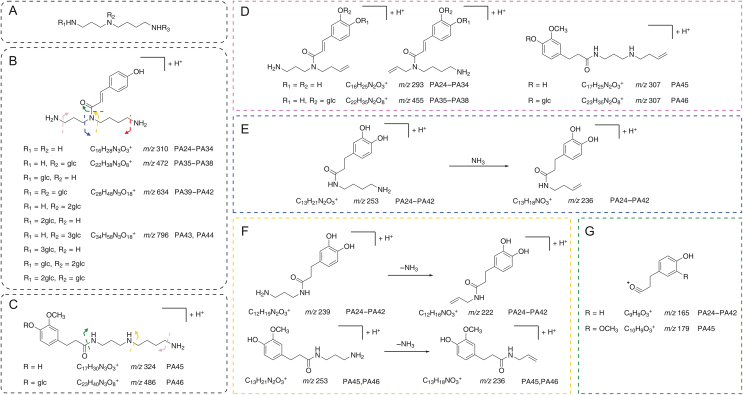
MS2 information of potential lycibarbarspermidine derivatives acquired by subsequent PRM experiments was used for structural identification. Mass fragmentation rules of lycibarbarspermidines were integrated to annotate the structure of the unknown compound. As an example, PA1−PA6 were identified as p-coumaroyl spermidine in Figs. 6A and B. A set of isomers, PA1 and PA2, were observed in PRM. As shown in Fig. 6C, m/z 292.20221 [M+H]+ (0.9 ppm, C16H25N3O2) was generated. Then, m/z 275.17575 was generated by the neutral loss of NH3. The ion at m/z 147.04445 indicated that the substituent was hydroxycinnamoyl. The ion at m/z 221.12909 was produced by C6–N5 dissociation, and the ion at m/z 204.10254 was produced by the subsequent neutral loss of NH3. The ion m/z 218.08104 produced by the breakage of C4–N5 followed by the loss of neutral NH3 indicated that the hydroxycinnamoyl group is at the N5 position. Since PA1 and PA2 had similar fragment ions, they were inferred to be N5-hydroxycinnamoyl spermidine. A set of isomers PA3−PA6 was observed at m/z 454.25534 [M+H]+ (1.2 ppm, C22H35N3O7). As shown in Fig. 6D, PA4 yielded ions similar to PA1 at m/z 292.20212, 275.17554, 204.10248, and 147.04449, and due to the lack of the ion at m/z 218, PA4 was identified as N10-hydroxycinnamoyl spermidine monoglycoside. Since PA3, PA5 and PA6 had similar fragment ions to PA4, they were also inferred to be N10-hydroxycinnamoyl spermidine monoglycosides. The fragmentation pathway of PA1 is shown in Fig. 7. Notably, 46 monohydroxycinnamoyl spermidines were identified for the first time in Lycii Fructus (Fig. 8, Fig. 9). Their structures consisted of a hydroxycinnamic acid attached to spermidine. In addition, products of spermidine bound to other organic acids were found, and the proposed structure and characteristic fragment ions are shown in Fig. S8.
Fig. 6.
Extracted ion chromatogram (EIC) and MS2 spectrum of representative compounds. (A) EIC of PA1–PA2; (B) EIC of PA3–PA6; (C) MS2 spectrum of PA1; (D) MS2 spectrum of PA4.
Fig. 7.
Fragmentation pathway of compound PA1.
Fig. 8.
The inferred components and the characteristic fragment ions of monohydroxycinnamoyl spermines. (A) The skeleton of spermidine. (B–D) Tentative identification of lycibarbarspermidines. (E–H) Corresponding fragment ions of the inferred components. Box color indicates cleavage site.
Fig. 9.
The inferred components and the characteristic fragment ions of mono-dihydro (hydroxycinnamoyl) spermines. (A) The skeleton of spermidine. (B,C) Tentative identification of lycibarbarspermidines. (D–G) Corresponding fragment ions of the inferred components. Box color indicates cleavage site.
A total of 210 lycibarbarspermidine derivatives were tentatively characterized, and 170 potential new compounds, including 164 hydroxycinnamoyl spermidines and 6 hydroxycinnamoyl-quinic spermidines, were identified. All compounds were tentatively identified by comparing the predicted MS2 fragment ions based on the fragmentation pathway of the known lycibarbarspermidins. The corresponding structure and characteristic fragment ions of other lycibarbarspermidine derivatives are shown in Figs. 8, 9, S3−S5 and S8.
4. Conclusion
To enhance the detection sensitivity of low-response components in complex matrices by LC-HRMS, an integrated pipeline based on sequential stages, including sample preparation, database construction, data acquisition and data postprocessing, was developed. As applicability illustration, structural identification was conducted for 210 lycibarbarspermidine derivatives in Lycii Fructus. The combination of SPE extraction, targeted database, DDA coupled with PRM and integrated mass fragmentation rule analysis significantly improved the detection sensitivity and analysis coverage and benefited the LC-HRMS-based discovery of new compounds in complicated matrices.
CRediT author statement
Chen Han: Investigation, Writing - Original draft preparation; Zhixin Zhang: Investigation; Zhiyang Feng: Resources; Chuanjia Zhai: Investigation, Formal analysis; Xuejiao Li and Yulian Shi: Visualization; Xiang Li and Miao Li: Validation; Ying Wang and Gan Luo: Writing - Reviewing and Editing; Xiaoyan Gao: Writing - Reviewing and Editing, Conceptualization.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities in China (Grant No.: 2020-JYB-ZDGG-033).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.10.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Dong M., Tian Z., Ma Y., et al. Rapid screening and characterization of glucosinolates in 25 Brassicaceae tissues by UHPLC-Q-exactive orbitrap-MS. Food Chem. 2021;365 doi: 10.1016/j.foodchem.2021.130493. [DOI] [PubMed] [Google Scholar]
- 2.Feng J., Yu P., Zhou Q., et al. An integrated data filtering and identification strategy for rapid profiling of chemical constituents, with Arnebiae Radix as an example. J. Chromatogr. A. 2020;1629 doi: 10.1016/j.chroma.2020.461496. [DOI] [PubMed] [Google Scholar]
- 3.Fu S., Cheng R., Deng Z., et al. Qualitative analysis of chemical components in Lianhua Qingwen capsule by HPLC-Q Exactive-Orbitrap-MS coupled with GC-MS. J. Pharm. Anal. 2021;11:709–716. doi: 10.1016/j.jpha.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chi X., Liu J., Yu M., et al. Analysis of bromophenols in various aqueous samples using solid phase extraction followed by HPLC-MS/MS. Talanta. 2017;164:57–63. doi: 10.1016/j.talanta.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Klont F., Joosten M.R., Ten Hacken N.H.T., et al. Quantification of the soluble Receptor of Advanced Glycation End-Products (sRAGE) by LC-MS after enrichment by strong cation exchange (SCX) solid-phase extraction (SPE) at the protein level. Anal. Chim. Acta. 2018;1043:45–51. doi: 10.1016/j.aca.2018.09.050. [DOI] [PubMed] [Google Scholar]
- 6.Li X., Li S., Kellermann G. Simultaneous determination of three estrogens in human saliva without derivatization or liquid-liquid extraction for routine testing via miniaturized solid phase extraction with LC-MS/MS detection. Talanta. 2018;178:464–472. doi: 10.1016/j.talanta.2017.09.062. [DOI] [PubMed] [Google Scholar]
- 7.Guo J., Huan T. Comparison of full-scan, data-dependent, and data-independent acquisition modes in liquid chromatography-mass spectrometry based untargeted metabolomics. Anal. Chem. 2020;92:8072–8080. doi: 10.1021/acs.analchem.9b05135. [DOI] [PubMed] [Google Scholar]
- 8.Li C., Yang J., Tong X., et al. Precursor ion scan enhanced rapid identification of the chemical constituents of Danhong injection by liquid chromatography-tandem mass spectrometry: An integrated strategy. J. Chromatogr. A. 2019;1602:378–385. doi: 10.1016/j.chroma.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 9.Bai J., Chen W., Huang J., et al. Transformation of stilbene glucosides from Reynoutria multiflora during processing. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.757490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson A.C., Russell J.D., Bailey D.J., et al. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schilling B., MacLean B., Held J.M., et al. Multiplexed, scheduled, high-resolution parallel reaction monitoring on a full scan QqTOF instrument with integrated data-dependent and targeted mass spectrometric workflows. Anal. Chem. 2015;87:10222–10229. doi: 10.1021/acs.analchem.5b02983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J., Liu H., Liu Y., et al. Development and evaluation of a parallel reaction monitoring strategy for large-scale targeted metabolomics quantification. Anal. Chem. 2016;88:4478–4486. doi: 10.1021/acs.analchem.6b00355. [DOI] [PubMed] [Google Scholar]
- 13.Pan H., Zhou H., Miao S., et al. An integrated approach for global profiling of multi-type constituents: Comprehensive chemical characterization of Lonicerae Japonicae Flos as a case study. J. Chromatogr. A. 2020;1613 doi: 10.1016/j.chroma.2019.460674. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Wang Z., Li Y., et al. A strategy for comprehensive identification of sequential constituents using ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometer, application study on chlorogenic acids in Flos lonicerae Japonicae. Talanta. 2016;147:16–27. doi: 10.1016/j.talanta.2015.09.039. [DOI] [PubMed] [Google Scholar]
- 15.Chen D., Guo S., Zhou J., et al. Chemical constituents from Lycium barbarum (Solanaceae) and their chemophenetic significance. Biochem. Syst. Ecol. 2021;97 [Google Scholar]
- 16.Lopatriello A., Previtera R., Pace S., et al. NMR-based identification of the major bioactive molecules from an Italian cultivar of Lycium barbarum. Phytochemistry. 2017;144:52–57. doi: 10.1016/j.phytochem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Qian D., Chen J., Lai C., et al. Dicaffeoyl polyamine derivatives from bitter goji: Contribution to the bitter taste of fruit. Fitoterapia. 2020;143 doi: 10.1016/j.fitote.2020.104543. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z.-Q., Fan H.-X., He R.-R., et al. Lycibarbarspermidines A–O, new dicaffeoylspermidine derivatives from wolfberry, with activities against alzheimer’s disease and oxidation. J. Agric. Food Chem. 2016;64:2223–2237. doi: 10.1021/acs.jafc.5b05274. [DOI] [PubMed] [Google Scholar]
- 19.Ahad H., Jin H., Liu Y., et al. Chemical profiling of spermidines in goji berry by strong cation exchange solid-phase extraction (SCX-SPE) combined with ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF/MS/MS) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2020;1137 doi: 10.1016/j.jchromb.2019.121923. [DOI] [PubMed] [Google Scholar]
- 20.dos Santos G.S., de Almeida Veiga A., Carlotto J., et al. Identification and fingerprint analysis of novel multi-isomeric Lycibarbarspermidines and Lycibarbarspermines from Lycium barbarum L. by liquid chromatography with high-resolution mass spectrometry (UHPLC-Orbitrap) J. Food Compos. Anal. 2022;105 [Google Scholar]
- 21.Boerjan W., Ralph J., Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 22.Leonard W., Zhang P., Ying D., et al. Lignanamides: Sources, biosynthesis and potential health benefits - a minireview. Crit Rev Food Sci. Nutr. 2021;61:1404–1414. doi: 10.1080/10408398.2020.1759025. [DOI] [PubMed] [Google Scholar]
- 23.Liu S., Jiang J., Ma Z., et al. The role of hydroxycinnamic acid amide pathway in plant immunity. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.922119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeiss D.R., Piater L.A., Dubery I.A. Hydroxycinnamate amides: Intriguing conjugates of plant protective metabolites. Trends Plant Sci. 2021;26:184–195. doi: 10.1016/j.tplants.2020.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Fontanals N., Marcé R.M., Borrull F., et al. Mixed-mode ion-exchange polymeric sorbents: Dual-phase materials that improve selectivity and capacity. Trends Analyt. Chem. 2010;29:765–779. [Google Scholar]
- 26.Gilart N., Cormack P.A.G., Marcé R.M., et al. Selective determination of pharmaceuticals and illicit drugs in wastewaters using a novel strong cation-exchange solid-phase extraction combined with liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 2014;1325:137–146. doi: 10.1016/j.chroma.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Salas D., Borrull F., Marcé R.M., et al. Study of the retention of benzotriazoles, benzothiazoles and benzenesulfonamides in mixed-mode solid-phase extraction in environmental samples. J. Chromatogr. A. 2016;1444:21–31. doi: 10.1016/j.chroma.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 28.Shah P.A., Shrivastav P.S., George A. Mixed-mode solid phase extraction combined with LC-MS/MS for determination of empagliflozin and linagliptin in human plasma. Microchem. J. 2019;145:523–531. [Google Scholar]
- 29.Wu T., Lv H., Wang F., et al. Characterization of polyphenols from Lycium ruthenicum fruit by UPLC-Q-TOF/MSE and their antioxidant activity in Caco-2 cells. J. Agric. Food Chem. 2016;64:2280–2288. doi: 10.1021/acs.jafc.6b00035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.