Abstract
Previous work demonstrated that the adenovirus L1 52/55-kDa protein is required for assembly of viral particles, although its exact role in the assembly process is unclear. The 52/55-kDa protein’s early expression, however, suggests that it might have other roles at earlier times during infection. To uncover any role the 52/55-kDa protein might have at early times and to better characterize its role in assembly, a mutant adenovirus incapable of expressing the 52/55-kDa protein was constructed (H5pm8001). Analysis of the onset and extent of DNA replication and late protein synthesis revealed that H5pm8001-infected 293 cells entered the late stage of infection at the same time as did adenovirus type 5 (Ad5)-infected cells. Interestingly, H5pm8001-infected cells displayed slightly lower levels of replicated viral DNA and late proteins, suggesting that although not required, the 52/55-kDa protein does augment these activities during infection. Analysis of transcripts produced from the major late and IVa2 promoters indicated a slight reduction in H5pm8001-infected compared to Ad5-infected cells at 18 h postinfection that was not apparent at later times. Analysis of particles formed in H5pm8001 cells revealed that empty capsids could form, suggesting that the 52/55-kDa protein does not function as a scaffolding protein. Subsequent characterization of these particles demonstrated that they lacked any associated viral DNA. These findings indicate that the 52/55 kDa-protein is required to mediate stable association between the viral DNA and empty capsid and suggest that it functions in the DNA encapsidation process.
At late times during adenovirus infection, two abundant particles are formed that can be separated by CsCl equilibrium centrifugation (39). The heavier of these particles is the mature virus, while the lighter particles are empty capsids. Analysis of the protein composition of empty capsids shows that although they lack all core components, they contain hexon, penton base, fiber, and the precursor forms of proteins VI and VIII (29, 39, 51, 58). In addition, several other proteins that are not found in the mature virus are found in empty capsids and may function as scaffolding proteins during the assembly process (29, 51, 55, 58). Pulse-chase experiments combined with the analysis of defective particles formed during infection of cells with temperature-sensitive mutants revealed a third, less-abundant class of particles known as assembly intermediates (14, 15). Further characterization of these particles by reversible cross-linking revealed that they could be separated into two components, termed heavy and light intermediates. Light intermediates have the same protein composition as empty capsids but are associated with a small fragment of the viral genome. The heavy intermediates contain the full-length viral genome and lack all scaffolding proteins. A precursor/product relationship between assembly intermediates and mature virions was suggested by kinetic analyses showing that radiolabel incorporated into assembly intermediates could be chased into mature virions (14, 15). A fourth type of particle known as the young virion was identified upon analysis of H2ts1, which contains a temperature-sensitive mutation in the viral protease gene (29, 63, 64). Cells infected with H2ts1 at the nonpermissive temperature accumulate viral particles that contain a full-length viral genome associated with core proteins V and VII. Young virions are identical to mature virions except that several viral proteins are present in a precursor form (IIIa, VI, VII, VIII, and terminal proteins) and proteins X, XI, and XII are absent. Overall, these findings suggest that the first step in viral morphogenesis is association of viral proteins (some in precursor form) with scaffolding proteins to form the empty capsid. The association of viral DNA is the next detectable step and results in the formation of light intermediates. The DNA is then encapsidated, and the scaffolding proteins are degraded or released to produce the heavy intermediate. Young virions are formed by the incorporation of viral core proteins, and the final step is the cleavage of precursor proteins by the viral protease to produce the mature virion.
Characterization of an adenovirus harboring a temperature-sensitive mutation in the L1 52/55-kDa protein (H5ts369) revealed that this protein is required for viral assembly (23). When HeLa cells were infected with H5ts369 at the nonpermissive temperature, light intermediates accumulated. Analysis of these intermediates indicated that they were associated with the left end of the viral genome, suggesting that the 52/55-kDa protein has a role in DNA encapsidation. Later findings indicated that early assembly intermediates have many copies of the 52/55-kDa protein and that these structures gradually lose the 52/55-kDa protein as they mature into virions (22). This led Hasson et al. (22) to suggest that the 52/55-kDa protein may act as a scaffolding protein in a manner similar to that shown for several bacteriophage assembly pathways (reviewed in reference 5).
Despite its clearly demonstrated role in viral assembly, other observations suggested that the 52/55-kDa protein might have additional functions at early times during infection. Unlike other members of the late families of gene products, mRNAs encoding the 52/55-kDa protein are detected very early after infection has commenced (9, 57). Subsequent analysis has revealed the presence of distinct regulatory mechanisms that ensure expression of the 52/55-kDa protein at early times. First, unlike what is seen at late times during infection, when transcription from the major late promoter (MLP) proceeds through to the right end of the genome (1, 17, 65), transcription at early times terminates downstream of the L3 poly(A) site (30, 47). Second, polyadenylation at the L1 poly(A) site was shown to be activated by flanking sequences that facilitate recruitment of processing factors to this site at early times (12, 16, 18). Third, the presence of an intronic splicing repressor immediately upstream of the IIIa splice acceptor results in almost exclusive splicing to the 52/55-kDa protein exon at early times (33). The appearance of the 52/55-kDa protein before viral DNA replication has begun, or before any viral structural proteins have been produced, suggests that it may be responsible for some activity early in the infectious process.
Previously, we reported that the 52/55-kDa protein interacts with the IVa2 protein during infection (21). Since the IVa2 protein is a late-stage-specific transcriptional activator of the MLP (62), this suggested that a possible early role for the 52/55-kDa protein might be to regulate proper temporal activation of late gene expression by interacting with the IVa2 protein. Analysis of assembly intermediates and mature virions, however, revealed that the IVa2 protein was a component of these particles (67), making it possible that interaction with the 52/55-kDa protein was important for viral assembly. Although the phenotype of H5ts369 did not suggest a role for the 52/55-kDa protein at early times, it was possible that the temperature-sensitive mutation did not affect any putative function of the 52/55-kDa protein required prior to assembly of the viral particle. Additionally, the analysis of H5ts369 could not differentiate between a role for the 52/55-kDa protein as a scaffolding protein or in DNA encapsidation (22). To determine if the 52/55-kDa protein has an important role at early times, as well as to further characterize the role of the 52/55-kDa protein in viral assembly, we constructed a mutant adenovirus incapable of expressing the 52/55-kDa protein. The analysis of this mutant virus reveals that the 52/55-kDa protein is not required for entry into the late stage of infection, indicating that it does not provide a necessary function at early times. Characterization of assembly intermediates that form in the absence of the 52/55-kDa protein indicates that it does not function as a scaffolding protein but instead mediates encapsidation of the viral genome.
MATERIALS AND METHODS
Plasmid constructs.
Adenovirus type 5 (Ad5) nucleotide numbering is from the published sequence (10). pTG3602 contains a full-length Ad5 genomic clone and has been described previously (6). The 52/55-kDa open reading frame (ORF) from amino acids 2 to 416 was constructed by cloning the NsiI/HindIII fragment (nucleotides [nt] 11054 to 11565) and the HindIII/SmaI fragment (nt 11565 to 13065) into the PstI and HincII sites of pGEM-3Zf(−) (Promega Corp., Madison, Wis.). The resulting vector was digested with SphI, blunt ended with T4 DNA polymerase, ligated to BamHI linkers (8-mers; Promega Corp.) and digested with BamHI. The resulting 2-kb BamHI fragment containing the 52/55-kDa ORF was then cloned into the BamHI site of pGEM-3Zf(−) to create pGL1.
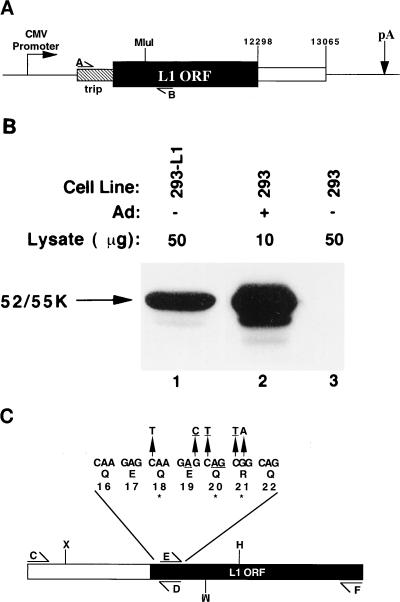
pBK-tripL1 was generated through a series of cloning steps, the details of which we will gladly provide upon request. Briefly, we amplified the 5′ end of a cDNA encoding the 52/55-kDa protein from a late stage adenovirus-infected 293 cell cDNA library (21), using PCR. The primers used corresponded to a 5′ primer (A) complementary to the first exon of the tripartite leader (nt 6051 to 6069) and a 3′ primer (B) complementary to the 52/55-kDa protein ORF (nt 11367 to 11350), and the amplified product therefore contained the tripartite leader and extended to nt 11367 in the 52/55-kDa protein ORF (Fig. 1A). This product was combined with the rest of the 52/55-kDa ORF and cloned into the NheI and XbaI sites of pBK-CMV (Stratagene Cloning Systems, La Jolla, Calif.) to generate pBK-tripL1 (Fig. 1A).
FIG. 1.
Helper cell line isolation and construction of the mutant 52/55-kDa ORF. (A) Schematic illustration of the 52/55-kDa protein expression vector (pBK-tripL1) used to generate 293-L1 cells. The location of primers A and B used in cloning are indicated, as well as that of the MluI site at nt 11312. Abbreviations: trip, tripartite leader cDNA; L1 ORF, 52/55-kDa ORF; pA, polyadenylation site; CMV, cytomegalovirus. The open box indicates adenovirus sequences located downstream of the 52/55-kDa ORF. (B) Immunoblot analysis of 293-L1 cells. Whole-cell lysates prepared from 293-L1 or 293 cells were examined by using a polyclonal rabbit antibody to the 52/55-kDa protein. Ad: +, lysate was prepared from adenovirus-infected 293 cells at 20 h postinfection; −, uninfected cell lysate. Lysate (μg), amount of lysate loaded. (C) Schematic representation of the mutant 52/55-kDa ORF. The locations of primer binding sites (C, D, E, and F) and restriction endonuclease cleavage sites (M, H, and X) used in the construction of the mutant are indicated at the bottom. M, MluI; H, HindIII; X, XbaI. The upper portion of the figure displays the nucleotide sequence of the N terminus of the 52/55-kDa protein ORF (nt 11096 to 11116). The encoded amino acid is indicated below each triplet, and the codon position within the 52/55-kDa ORF is indicated below that. Arrows indicate point mutations that were introduced into this region. Underlined nucleotides correspond to the SpeI site that is created by these mutations. ∗, position of stop codons created by these manipulations.
A PCR strategy was employed to mutate the 52/55-kDa protein ORF (Fig. 1C). A 5′ fragment (CD) extending from nt 10321 to 11115 was synthesized by using primers C (5′-CAGGTACTGGTATCCCACC-3′) and D (5′-CTACTAGTCTTACTCTTGCCGCTGCTG-3′). A 3′ fragment (EF) extending from nt 11100 to 12298 was synthesized by using primers E (5′-GTAAGACTAGTAGCAGACATGCAGGGC-3′) and F (5′-CTTAGTACTCGCCGTCCTC-3′). Primers D and E are complementary at their 5′ ends and alter the sequence between nt 11102 and 11113 from CAA GAG CAG CGC to TAA GAC TAG TAG. Bold nucleotides represent mutations introduced by these primers, and underlined nucleotides indicate a SpeI site that was created in the mutant. Fragments CD and EF were digested with SpeI, ligated, and PCR amplified, using primers C and F to generate the ΔL1 fragment (Fig. 1). The XbaI/HindIII fragment (nt 10589 to 11565) was isolated from the ΔL1 fragment and used to replace the corresponding region of the wild-type gene in a clone of the viral KpnB fragment (nt 8537 to 14290) to generate pKpnBΔL1.
For the synthesis of antisense riboprobes, different regions of the adenovirus genome were cloned into pGEM-3Zf(−). pGL3pA contains Ad2 sequences that span the L3 poly(A) site (nt 22237 to 22667) from pGEML1 (12). pGL3pArev contains the same sequences in the reverse orientation. pGHwt contains Ad2 sequences spanning the L1 poly(A) site (nt 13976 to 14146). pGE53/Pst contains Ad5 sequences spanning the IVa2 coding region (nt 4060 to 4245) isolated from clone E53 (21). Transcription of pGL3pA, pGL3pArev, pGHwt, and pGE53/Pst by T7 RNA polymerase produces transcripts complementary to the L3, E2A, L1, and IVa2/E2B mRNAs, respectively.
Cell lines.
Low-passage 293 cells (20) were maintained as described previously (21). A cell line that stably expresses the 52/55-kDa protein was generated by transfecting pBK-tripL1 into 293 cells. Approximately 5 × 105 293 cells were plated in a 6-cm-diameter dish 48 h prior to transfection. Four to 6 h prior to transfection, cells were fed with 4.5 ml of complete medium. Calcium phosphate-DNA precipitates were prepared as described previously, using 10 μg of pBK-tripL1 and 10 μg of pGEM DNA per ml of precipitate (34). Precipitate (0.5 ml) was added to the cells, and 16 h later the cells were washed twice with phosphate-buffered saline (PBS) and fed with fresh medium. Twenty-four hours later, the cells were trypsinized and plated at increasing dilutions to allow for the outgrowth of individual colonies in the presence of G418, which was added the following day at 0.5 mg/ml. Cells were fed every 3 to 4 days with fresh medium containing G418 until colonies could be detected. Colonies were picked, expanded, and analyzed by immunoblotting for expression of the 52/55-kDa protein. Following isolation, one positive cell line that expressed the highest level of the 52/55-kDa protein was subcloned by limiting dilution and designated 293-L1.
Viruses.
Ad5 stocks were prepared on 293 cells as described previously (19). H5ts369 contains a temperature-sensitive mutation in the 52/55-kDa protein and was grown on 293 monolayers at 32°C (23). Plaque assays were performed at 37°C, except for H5ts369 assays which were incubated at either 32 or 39.5°C.
To construct H5pm8001, we employed the bacterial recombination protocol described by Chartier et al. (6). pKpnBΔL1 was digested with KpnI, and the KpnBΔL1 fragment (nt 8537 to 14290) was isolated. Ten nanograms of the purified KpnBΔL1 fragment and 10 ng of pTG3602 that had been linearized at the PmeI site (nt 13258) were cotransformed into Escherichia coli BJ5183 and plated on medium containing ampicillin. Since the yield of plasmid DNA from BJ5183 was too low to analyze directly, it was used to transform DH5α cells. Restriction analysis of a plasmid isolated from this transformation (pTG3602ΔL1) indicated that a SpeI site had been introduced into the 52/55-kDa protein ORF and that no rearrangements had occurred. 293-L1 cells were transfected with 1 μg of PacI-digested pTG3602ΔL1 and monitored for the development of cytopathic effect (CPE). CPE became evident 11 days later, at which time a viral lysate was prepared and plaqued on 293-L1 and 293 cells. No plaques were detected on 293 cells. Two isolated plaques were picked from 293-L1 cells and replaqued on both cell lines. Again, no plaques were detected on 293 cells, and several isolated plaques picked from 293-L1 cells were used as master stocks for the growth of H5pm8001. DNA was prepared from plaque lysates by using a modification of the procedure described by Hirt (26). Plaque lysates were adjusted to 0.6% sodium dodecyl sulfate (SDS)–10 mM EDTA and incubated at room temperature for 15 min, followed by the addition of NaCl to 1.5 M and incubation overnight at 4°C. Samples were spun at top speed in a microcentrifuge for 10 min. Ten microliters containing 50 mg of predigested pronase/ml and 5 μl containing 10 mg of RNase A/ml were added to the supernatants, and the samples were incubated at 37°C for 1 h, extracted twice with phenol and once with chloroform-isoamyl alcohol (24:1), precipitated, washed, and resuspended in 100 μl TE (10 mM Tris, 1 mM EDTA, pH 8). Protein samples were prepared in E1A lysis buffer and analyzed by immunoblot as described previously (21).
Viral growth assays.
One million cells were plated on a 35-mm-diameter plate and allowed to attach overnight. Cells were infected the next day by aspirating the medium and adding virus at a multiplicity of infection (MOI) of 0.1 in 0.5 ml of Dulbecco modified Eagle medium (DMEM)–2% fetal bovine serum (FBS). Following adsorption of virus for 90 min, 2 ml of complete medium was added. Viral lysates were prepared by washing cells twice with PBS and resuspending in 0.25 ml of DMEM–2% FBS, followed by three cycles of freeze-thaw. Lysates were plaqued on 293-L1 or 293 cells as described above.
Infection time course analysis.
Infections for the analysis of RNA used 106 cells, whereas infections for the isolation of DNA and protein used 5 × 105 cells. Cells were trypsinized, spun at 250 × g for 5 min, resuspended in complete medium, and counted in a hemocytometer. Cells were again pelleted at 250 × g for 5 min and resuspended at 107 cells/ml in DMEM–2% FBS containing virus at an MOI of 10. Virus was allowed to adsorb for 20 min at 37°C with occasional mixing, followed by the addition of 1 ml of complete medium and plating. At the indicated times postinfection, cells were washed twice with PBS and harvested. For immunoblot analysis, whole-cell lysates were prepared by lysing the cells in 25 μl of E1A lysis buffer as described previously (21). The preparation of rabbit polyclonal antiserum that recognizes the 52/55-kDa protein, as well as the mouse monoclonal antibodies to the IVa2 and 72-kDa DNA binding protein, have been described previously (21, 37, 54). Polyclonal rabbit antibodies to the fiber protein were the kind gifts of P. Hearing and C. Anderson.
Viral DNA was prepared by using the modified Hirt procedure described above and analyzed by Southern blotting. Samples were digested with KpnI and SpeI, electrophoresed on a 0.8% agarose gel, transferred to Zetaprobe membrane (Bio-Rad Laboratories, Hercules, Calif.), and prehybridized in 0.5 M NaHPO4–1 mM EDTA–7% SDS (pH 7.2) at 65°C for 1 h. A radiolabeled, Ad5-specific probe was synthesized using pTG3602 and the Random Primer Labeling kit (Life Technologies Inc., Gaithersburg, Md.) and boiled immediately before use. The prehybridization solution was removed, and fresh solution containing probe (106 cpm/ml) was added to the membrane and incubated overnight at 65°C. The membrane was rinsed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature, twice with 40 mM NaHPO4 (pH 7.2)–1 mM EDTA–5% SDS at 65°C for 30 min, twice with 40 mM NaHPO4 (pH 7.2)–1 mM EDTA–1% SDS at 65°C for 30 min, blotted dry, and exposed to film. The amount of bound probe was quantitated in a PhosphorImager (Molecular Dynamics Inc., Sunnyvale, Calif.).
RNA was isolated by the method of Chomczynski and Sacchi (8), except that all volumes were reduced by 75%. The RNA was precipitated, resuspended in 0.1 ml of TE, and quantitated by measuring the optical density at 260 nm. Poly(A)+ RNA was purified from 16 μg of total RNA as described previously (13). One-fourth of the poly(A)+ RNA was electrophoresed on a formaldehyde-denaturing gel and transferred to a Zetaprobe nylon membrane. Prehybridization, hybridization, and washing were the same as described above for Southern analysis. Plasmids used for the synthesis of riboprobes were linearized with HindIII, extracted once with phenol, once with chloroform-isoamyl alcohol (24:1), precipitated, and resuspended in TE at 1 mg/ml. One microliter of linearized plasmid was incubated at 37°C for 1 h with 1 U of T7 RNA polymerase (Life Technologies Inc.) in the manufacturer’s recommended buffer supplemented with 50 μCi of [32P]UTP, 10 μM UTP, and 0.5 mM (each) ATP, CTP, and GTP, followed by the addition of 1 U of RNase-free DNase I and incubation at 37°C for another 15 min. Riboprobes were extracted once with phenol and once with chloroform-isoamyl alcohol (24:1) and were purified in a G50 quickspin column (Boehringer Mannheim Corp., Indianapolis, Ind.) before being added to membranes.
Transmission electron microscopy.
Infections for analysis by electron microscopy were carried out as described above for the time course analysis. At 40 h postinfection, cells were washed twice with PBS, fixed for 1 h in 2% buffered glutaraldehyde, postfixed for 1 h with buffered 1% osmium tetroxide, dehydrated, and infiltrated with Epon epoxy resin. The resulting block was then sectioned, and grids containing the ultrathin sections were double stained with lead citrate and uranyl acetate and examined by using a Philips CM-100 transmission electron microscope operated at 60 kV.
Purification of assembly intermediates.
H5pm8001 infections were carried out at an MOI of 5; all other infections were at an MOI of 10. H5ts369 infections were carried out at 39.5°C; all others were at 37°C. Forty-eight hours postinfection, 2 × 108 cells were centrifuged at 250 × g, washed with PBS, and resuspended in 5 ml of 10 mM Tris, pH 8. The cells were lysed by three cycles of freezing and thawing and centrifuged at 1,500 × g for 15 min, and the supernatant was layered onto a 28-ml step gradient prepared by underlaying 14 ml of a 1.45-g/cm3 CsCl solution beneath 14 ml of a 1.2-g/cm3 CsCl solution. The virus was centrifuged in an SW28 rotor at 72,000 × g for 2 h, harvested, and layered onto a preformed (1.2 to 1.45 g/cm3) CsCl gradient and centrifuged in an SW41 rotor at 68,500 × g for 16 h. After centrifugation, fractions were collected from the gradient and analyzed for protein content by using the Bio-Rad protein assay kit to determine the location of viral particles. The density of individual fractions was determined with a refractometer.
For analysis of protein composition, particles were added directly to sample release buffer (21), electrophoresed on a 10% SDS-polyacrylamide gel, and visualized directly by using the Gel Code silver stain kit (Pierce Chemical Company, Rockford, Ill.) or by immunoblotting. DNA was isolated from viral particles by diluting into 0.3 ml of pronase digestion buffer (2 mg of predigested pronase/ml, 50 mM Tris, 1 mM EDTA, 0.5% SDS, pH 7.5) and incubating at 37°C for 1 h with occasional mixing. Samples were extracted twice with phenol, once with chloroform-isoamyl alcohol (24:1), precipitated, washed with 70% ethanol, dried, and resuspended in water. Southern analysis was as described above.
RESULTS
Construction of 293-L1 helper cell line.
Previous work had demonstrated that the adenovirus 52/55-kDa protein is required for the assembly of viral particles (23), indicating that propagation of a 52/55-kDa protein null mutant virus would require the isolation of a complementing cell line. To generate a cell line that expressed the 52/55-kDa protein, the construct shown in Fig. 1A was transfected into 293 cells. We included a cDNA for the adenovirus tripartite leader at the 5′ end of the ORF in an effort to increase expression of the 52/55-kDa protein during adenovirus infection. The tripartite leader is a 210-bp sequence spliced to the 5′ end of all mRNAs produced from the MLP at late times during infection and has been shown to enhance transport of these mRNAs out of the nucleus and their subsequent translation (2, 28, 36). Figure 1B shows that the 52/55-kDa protein was detectable in lysates prepared from 293-L1 cells, a clonal cell line isolated after transfection of 293 cells with pBK-tripL1, while it was not detected in untransfected cells. Comparison with late-stage Ad5-infected 293 cell lysates indicated that the level of 52/55-kDa protein expressed in this cell line was at least 10% of that seen during a normal adenovirus infection. To confirm that the level of 52/55-kDa protein expression in 293-L1 cells was sufficient to complement a virus with a defective 52/55-kDa protein, we examined the ability of H5ts369 to grow on 293-L1 cells at the nonpermissive temperature. While replication of H5ts369 in 293 cells was strictly temperature dependent, H5ts369 grew nearly as well in 293-L1 cells at the nonpermissive temperature as at the permissive temperature (Table 1). These results demonstrate that 293-L1 cells express the 52/55-kDa protein at levels approaching that seen during adenovirus infection and this is sufficient for complementation of a virus expressing a defective 52/55-kDa protein.
TABLE 1.
Plaquing efficiency of H5ts369 on 293-L1 cells
| Cell line | Titer (PFU/ml)a
|
|
|---|---|---|
| 32°C | 39.5°C | |
| 293-L1 | 1.3 × 1010 | 1.1 × 1010 |
| 293 | 1.5 × 1010 | 6.3 × 105 |
H5ts369 was plaqued on 293 or 293-L1 cells at the permissive (32°C) or nonpermissive (39.5°C) temperature.
Isolation of pm8001.
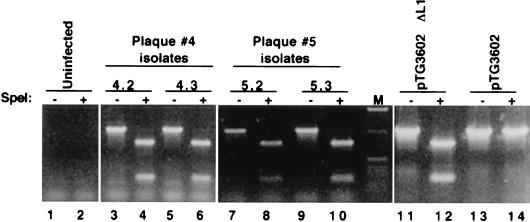
We employed the bacterial recombination system described by Chartier et al. (6) to generate a recombinant Ad5 genome containing several point mutations at the 5′ end of the 52/55-kDa protein ORF (pTG3602ΔL1, Fig. 1C). In addition to introducing stop codons at positions 18, 20, and 21, these mutations create a SpeI site. We confirmed that these mutations block expression of the 52/55-kDa protein by cloning the mutant ORF into the pBK-CMV expression vector and transfecting it into 293 cells. Analysis of transfected cell lysates did not reveal any detectable 52/55-kDa protein, while cells transfected with the wild-type ORF produced readily detectable levels (data not shown). The mutant adenovirus was generated by digesting pTG3602ΔL1 with PacI and transfecting it into 293-L1 cells. Following two rounds of plaque purification, viral DNA was prepared from several isolated plaques and analyzed by PCR for the presence of a SpeI site in the 52/55-kDa protein ORF (Fig. 2). Amplification of pTG3602 or pTG3602ΔL1 gives rise to the expected 1-kb fragment spanning the 5′ end of the 52/55-kDa protein ORF, but only the product amplified from pTG3602ΔL1 is digestible with SpeI (compare lanes 12 and 14). Analysis of a number of different plaques from two independent isolates indicated that all had the expected SpeI site in the 52/55-kDa protein ORF, confirming that a clonal population of recombinant adenovirus had been isolated. These results demonstrated that a mutant adenovirus containing the expected mutations in the 52/55-kDa protein ORF, designated H5pm8001, had been isolated.
FIG. 2.
PCR analysis of plaque isolates. Viral DNA was prepared from plaque isolates and analyzed by PCR using primers B and C (Fig. 1). Samples were either analyzed directly (lanes −) or were adjusted to 5 mM MgCl2 and digested with 0.5 U of SpeI (lanes +) prior to electrophoresis on a 1.2% agarose gel. pTG3602ΔL1 and pTG3602 indicate amplification from the mutant or wild-type plasmid, respectively. Uninfected, amplification of DNA prepared from uninfected 293-L1 cells. Lane M, 1-kb molecular size marker.
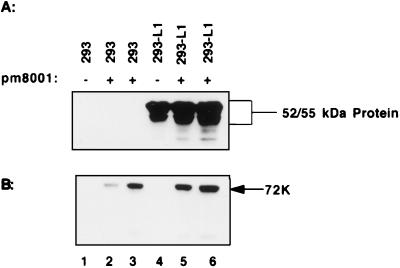
To confirm that the mutations on the viral genome blocked expression of the 52/55-kDa protein, we infected two H5pm8001 isolates into 293 or 293-L1 cells and examined whole-cell lysates for the presence of the 52/55-kDa protein. Figure 3A shows that, as expected, the 52/55-kDa protein was detected in all 293-L1 cell lysates tested. When 293 cells were infected with either isolate of H5pm8001, however, no 52/55-kDa protein was detected (lanes 2 and 3), confirming that the point mutations in pm8001 did block expression of the 52/55-kDa protein. To prove that H5pm8001 was indeed infecting the cells, the lysates were probed for the viral E2A 72-kDa DNA binding protein (72K protein). Figure 3B shows that all infected cell lysates were positive for the 72K protein, confirming a specific block in expression of the 52/55-kDa protein.
FIG. 3.
Immunoblot analysis of 293 and 293-L1 cells infected with H5pm8001. Whole-cell lysates were prepared from uninfected (−) or H5pm8001-infected (+) 293 and 293-L1 cells. Fifty micrograms of each lysate was analyzed by immunoblot with the indicated antibody. (A) Analysis with antibodies to the 52/55-kDa protein. (B) The blot in panel A was stripped and reprobed with antibodies to the E2A 72-kDa DNA binding protein (72K).
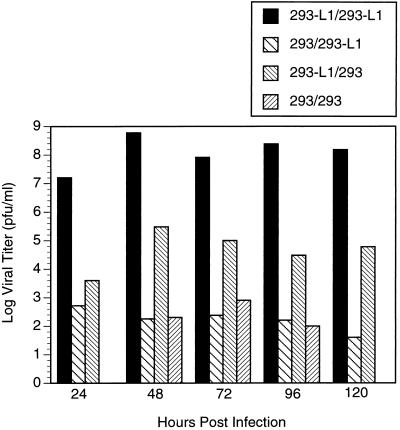
Analysis of the growth characteristics of H5pm8001 revealed a clear dependence on coexpression of the 52/55-kDa protein for viral growth. Figure 4 shows that when 293 cells were infected with H5pm8001 the viral yield did not exceed 103 PFU/ml, even when infections were allowed to proceed for 5 days. In contrast, when 293-L1 cells were infected with H5pm8001, yields of 108 PFU/ml were obtained after only 48 h. When the plaquing efficiency of H5pm8001 grown in 293-L1 cells was compared on 293 and 293-L1 cells, a 3- to 4-log difference was observed. The smaller difference observed when comparing plaquing efficiency was most likely due to the appearance of wild-type revertants that arose due to recombination with the 52/55-kDa protein coding sequences present in 293-L1 cells. In support of this, PCR analysis of H5pm8001 lysates prepared after extended passage in 293-L1 cells indicated that a fraction of the virus in these lysates had lost the SpeI site located in the 52/55-kDa ORF and the 52/55-kDa protein could be detected by immunoblot when these lysates were used to infect 293 cells (data not shown). All H5pm8001 stocks used for the experiments described below tested negative for wild-type adenovirus by PCR, exhibited at least a 4-log-lower titer on 293 cells than on 293-L1 cells, and did not produce detectable 52/55-kDa protein in 293 cells.
FIG. 4.
Growth characteristics of H5pm8001. 293 or 293-L1 cells were infected with H5pm8001, and viral lysates were prepared at 24-h intervals for 5 days. The titers of these lysates were then determined on both 293 and 293-L1 cells. Labeling of the bars corresponds to the cell line from which the lysate was prepared, followed by the cell line in which it was titered. The 293/293 sample was not assayed at 24 or 120 h.
Viral gene expression and DNA replication in pm8001-infected cells.
As mentioned above, the 52/55-kDa protein’s early expression during infection suggests that it might have other functions in addition to its demonstrated role in assembly. In particular, because the IVa2 protein is a transcriptional activator of the MLP (62), interaction between the 52/55-kDa protein and the IVa2 protein might be important for proper regulation of gene expression during infection. Since DNA replication is a prerequisite for entry into the late stage of infection and activation of the MLP (60), we compared the onset and extent of viral DNA replication in 293 cells infected with H5pm8001 or Ad5. Figure 5 reveals that replicated viral DNA was detected by 12 h postinfection in both Ad5 and H5pm8001 infections. Although in this experiment H5pm8001 produced more DNA at 12 h postinfection than did Ad5, this difference was not apparent in other experiments. Despite the relative abundance of H5pm8001 DNA at 12 h postinfection, by 18 h postinfection the Ad5 infections consistently contained more viral DNA (compare lanes 9 and 12). Quantitation of the amount of viral DNA at 18 h postinfection in two independent experiments indicated that an average of four times more DNA accumulated in Ad5- versus H5pm8001-infected 293 cells. This difference was not apparent when 293-L1 cells were infected with H5pm8001 (compare lanes 1 to 3 with lanes 7 to 9). These results show that although the onset of viral DNA replication is not affected in H5pm8001-infected 293 cells, viral DNA does not accumulate to the same levels as those seen in wild-type Ad5 infections.
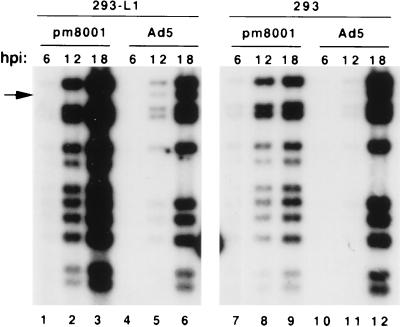
FIG. 5.
Analysis of viral DNA replication. 293 or 293-L1 cells were infected with H5pm8001 or Ad5, and viral DNA was prepared at the indicated hours postinfection (hpi). The DNA was digested with KpnI and SpeI and analyzed by Southern using a radiolabeled Ad5-specific probe. The arrow indicates the location of the KpnB fragment (nt 8527 to 11311) that is cleaved by SpeI in DNA isolated from H5pm8001 infections.
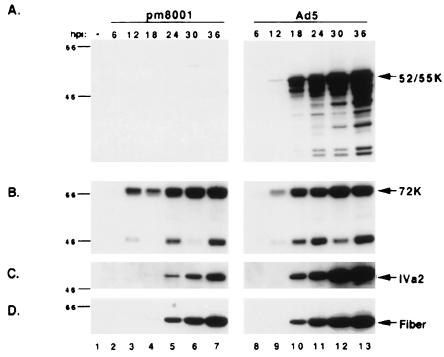
Next, we wanted to determine if the loss of the 52/55-kDa protein resulted in any major defects in viral gene expression. For this experiment, protein lysates prepared from H5pm8001-infected or Ad5-infected 293 cells at various times postinfection were examined by immunoblot for the presence of early and late viral proteins. Figure 6A shows that, as expected, no 52/55-kDa protein was detected in H5pm8001 cell lysates, while it could be detected by 12 h postinfection in Ad5 infections. Analysis of early gene expression using an antibody to the E2A 72K protein revealed no major differences between H5pm8001-infected and Ad5-infected 293 cells (Fig. 6B). Analysis of IVa2 expression revealed that although it was detected at the same time in infections with either virus, significantly less accumulated in H5pm8001-infected cells (Fig. 6C, compare lanes 4 to 7 with lanes 10 to 13). Similarly, the fiber protein could be detected by 18 h postinfection with either virus, but there was consistently less produced in H5pm8001 infections than in Ad5 infections (Fig. 6D, compare lanes 4 to 7 with lanes 10 to 13). These results suggest that the 52/55-kDa protein is not required for expression of late proteins but is important for achieving the level of late protein synthesis seen during a wild-type infection.
FIG. 6.
Expression of viral proteins in H5pm8001-infected 293 cells. Twenty-five micrograms of whole-cell lysates prepared from 293 cells infected with H5pm8001 or Ad5 at the indicated times postinfection were analyzed by immunoblot using antibodies to the 52/55-kDa protein (52/55K) (A), to the E2A 72-kDa DNA binding protein (72K) (B), to the IVa2 protein (C), and to the fiber protein (D). hpi, hours postinfection. The faster-migrating band in panel B is a degradation product of the 72K protein (54). The locations of molecular weight markers are indicated on the left.
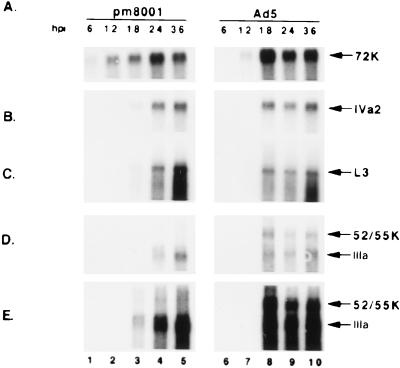
To determine if the decrease in late protein accumulation seen with H5pm8001 correlated with reduced transcription, we examined the steady-state levels of viral RNAs corresponding to the E2A, IVa2, L3, and L1 families of gene products. Analysis of the E2A transcripts in H5pm8001- and Ad5-infected 293 cells revealed no major difference in the time of appearance or accumulation of these mRNAs (Fig. 7A). IVa2, L3, and IIIa mRNAs were detected at 18 h postinfection in H5pm8001- or Ad5-infected cells and accumulated to roughly equivalent levels in both infections (Fig. 7B, C, and D, respectively). Although not dramatic, one difference that was apparent from this analysis was that the late mRNAs were less abundant at the beginning of the late phase in H5pm8001 infections than in Ad5 infections (compare lanes 3 and 8 in Fig. 7B, C, and D). After normalizing to the level of E2A transcripts there was 30 to 40% less late mRNA in H5pm8001- than in Ad5-infected cells at 18 h postinfection, while they accumulated to equal or greater levels at later times. This difference was apparent in multiple independent experiments and may explain the lower level of late proteins observed in H5pm8001-infected 293 cell lysates (Fig. 6). Another difference that was apparent from this analysis was that no mRNA corresponding to the 52/55-kDa protein was detected in H5pm8001-infected 293 cells (Fig. 7D and E). Despite the lack of detectable 52/55-kDa protein mRNA, IIIa transcripts did not appear to be expressed earlier or at higher levels in H5pm8001 infections. These results demonstrate that the 52/55-kDa protein is not essential for correct temporal regulation of viral gene expression but indicate that it does contribute to full activation of late gene expression at the transition from the early to late stage of infection.
FIG. 7.
Analysis of RNA produced in H5pm8001-infected 293 cells. Poly(A)+ RNA was prepared from 293 cells infected with H5pm8001 or Ad5 at the indicated times and analyzed by Northern blotting, using probes specific for E2A mRNA (A), IVa2 mRNA (B), L3 mRNA (C), and L1 mRNA (D). (E) A longer exposure of the blot in panel D to demonstrate the lack of 52/55-kDa protein mRNA in H5pm8001 infections. hpi, hours postinfection.
Analysis of pm8001 assembly intermediates.
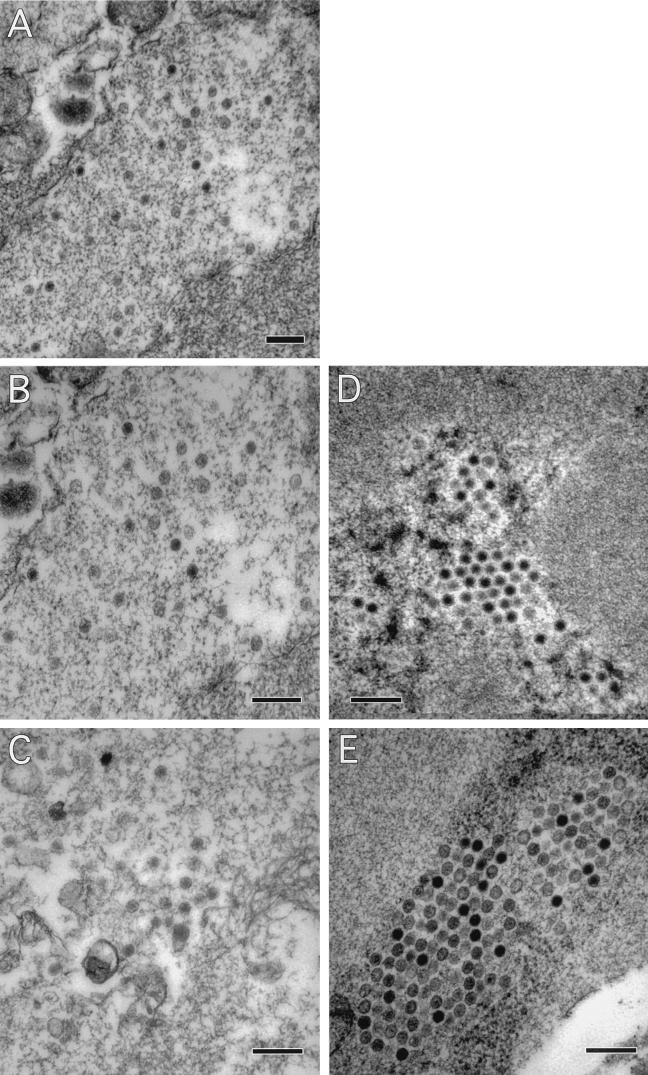
Since H5pm8001 was able to make the transition from the early to late stage of infection and express late proteins, we next used transmission electron microscopy to examine whether any assembly intermediates were formed in H5pm8001-infected 293 cells. Figure 8A shows that capsid-like structures were visible in H5pm8001-infected 293 cells. Unlike the capsids seen in Ad5-infected cells, which contained a darkly staining core, capsids in H5pm8001-infected 293 cells were lightly staining, suggesting that they lacked core components (compare Fig. 8E with B and C). Analysis of H5pm8001-infected 293-L1 cells revealed a larger proportion of darkly staining capsid structures that more closely resembled those seen in Ad5-infected cells (Fig. 8D). The appearance of lightly staining capsid structures in H5pm8001-infected 293 cells was similar to what had been reported previously for H5ts369 (23) and suggested that, like those of the temperature-sensitive mutant, H5pm8001 capsids did not contain a full complement of viral DNA.
FIG. 8.
Transmission electron microscopy of H5pm8001-infected cells. (A, B, and C) 293 cells infected with H5pm8001. (D) 293-L1 cells infected with H5pm8001. (E) 293 cells infected with Ad5. Magnification in panel A, ×25,000. Magnification panels D to E, ×46,000. Bars, 300 nm.
The intermediates were further characterized by using CsCl gradients for purification and density determination. As expected, two distinct populations of viral particles were purified from Ad5-infected 293 cells: the heavier particle (1.34 g/cm3) corresponded to mature virions, while the lighter particle (1.29 g/cm3) represented empty capsids (39, 51, 55). Analysis of H5pm8001-infected 293-L1 cells also revealed particles with densities of 1.34 g/cm3 and 1.29 g/cm3. Characterization of particles from H5pm8001-infected 293 cells, however, revealed only a single population of particles, with a density of 1.29 g/cm3. Examination of H5ts369 intermediates from 293 cells infected at the nonpermissive temperature indicated that, as previously published, these cells accumulated particles that were slightly denser than empty capsids, with a median density of 1.31 g/cm3 (23). No particles with densities greater than 1.29 g/cm3 were detected in H5pm8001-infected 293 cells, indicating a slightly different phenotype from that seen with H5ts369.
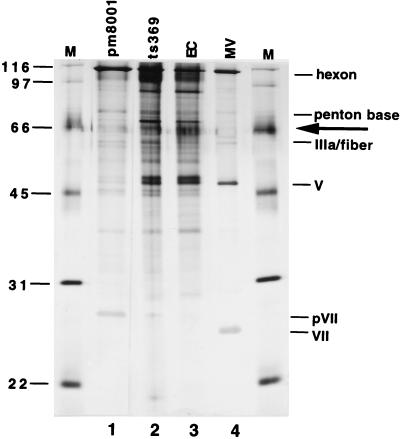
Previous work had demonstrated no appreciable differences in the protein composition of Ad5 empty capsids and H5ts369 intermediates formed at the nonpermissive temperature (23). To determine if intermediates formed in the absence of the 52/55-kDa protein had any major differences with Ad5 empty capsids or H5ts369 intermediates, we analyzed the protein composition of purified particles by silver staining. Analysis of intermediates purified from Ad5-, H5pm8001-, and H5ts369-infected 293 cells revealed that all appeared to contain hexon, penton base, and fiber (Fig. 9). As reported previously, the proteins found in empty capsids and H5ts369 intermediates were very similar. Comparison with H5pm8001 intermediates revealed no major differences in the protein composition of these particles; however, the relative abundance of some of the proteins seemed to be altered. Noticeably, H5pm8001 intermediates consistently contained more pVII than did H5ts369 intermediates. Additionally, a protein with an apparent molecular size of 70 kDa was readily detectable in both H5ts369 and Ad5 intermediates but was much less abundant in H5pm8001 particles. The 90-kDa protein visible in lanes 2 and 3 was not consistently detected in all experiments. These results demonstrate that H5pm8001 intermediates and those formed during infection with either Ad5 or H5ts369 have very similar protein compositions.
FIG. 9.
Analysis of the protein composition of H5pm8001 intermediates. Particles were purified by two rounds of CsCl gradient centrifugation and analyzed by silver staining. Lanes: pm8001, intermediates from H5pm8001-infected 293 cells; MV and EC, mature virions and empty capsids, respectively, prepared from Ad5-infected 293 cells; ts369, particles prepared from H5ts369-infected 293 cells at 39.5°C. The identities of several components of the mature virion are indicated based on their mobility in the gel. The arrow denotes the 70-kDa protein described in the text and molecular weight markers are indicated on the left.
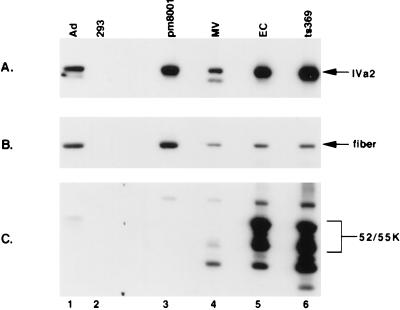
Since we had previously demonstrated that the IVa2 and 52/55-kDa proteins interact during infection (21), we examined whether the IVa2 protein was incorporated into assembly intermediates in H5pm8001-infected 293 cells. Purified particles prepared from H5pm8001-, Ad5-, and H5ts369-infected 293 cells were examined by immunoblot for the presence of the IVa2 protein. Figure 10A shows that the IVa2 protein was detectable in mature virions and Ad5 empty capsids. Analysis of H5ts369 intermediates revealed that the IVa2 protein was incorporated into these particles, indicating that the mutation in H5ts369 did not affect IVa2’s incorporation into virions. Finally, analysis of H5pm8001 intermediates demonstrated that the IVa2 protein was incorporated into capsids in the absence of the 52/55-kDa protein. Interestingly, mature virions displayed a second immunoreactive band that migrated slightly faster than that detected in empty capsids, intermediates, or whole-cell lysates. This suggests that the IVa2 protein is processed during virus assembly and may explain the discrepancy between the previously reported size of the IVa2 protein (56 kDa) and the isolation of a 50-kDa protein from virions that was identified as IVa2 by tryptic peptide mapping (50, 67). To confirm that the processing of IVa2 seen in Ad5 mature virions was not due to general degradation of the purified particles, we analyzed the immunoblot shown in Fig. 10A with antibodies to the fiber protein. Figure 10B shows that no degradation of fiber is seen in any of the particles, confirming that the results shown in Fig. 10A were specific to the IVa2 protein. As expected, analysis of these particles with antibodies to the 52/55-kDa protein confirmed that it was present in Ad5 and H5ts369 intermediates but absent from mature virions and H5pm8001 intermediates (Fig. 10C).
FIG. 10.
Immunoblot analysis of H5pm8001 intermediates. Viral particles were purified and analyzed with the indicated antibodies. Lanes: pm8001, intermediates from H5pm8001-infected 293 cells; MV and EC, mature virions and empty capsids, respectively, prepared from Ad5-infected 293 cells; ts369, particles prepared from 293 cells infected with H5ts369 at 39.5°C. (A) Anti-IVa2 immunoblot. (B) Anti-fiber immunoblot. (C) Anti-52/55-kDa protein immunoblot. Ad, Whole-cell lysate prepared from Ad5-infected 293 cells at 20 h postinfection; 293, uninfected whole-cell lysate.
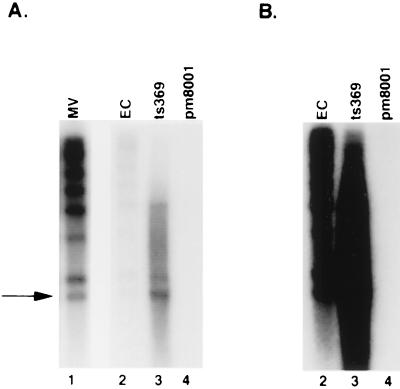
Characterization of H5ts369 intermediates formed at the nonpermissive temperature revealed that these particles differed from empty capsids due to their association with the left end of the viral genome (23). Since the intermediates found in H5pm8001-infected 293 cells appeared to have the same density as empty capsids, we examined whether they were associated with any viral DNA. Viral DNA was prepared from Ad5, H5pm8001, and H5ts369 intermediates, digested with KpnI and ClaI, and analyzed by Southern blotting with an adenovirus-specific probe. ClaI cleaves Ad5 at nt 982, close to the left end of the viral genome. Cleavage of DNA isolated from H5ts369 intermediates revealed a predominant band corresponding to the left end-derived ClaI fragment, confirming association of these particles with the left end of the viral genome (Fig. 11, lane 3). Although empty capsids displayed a small amount of associated viral DNA, the entire genome was represented (lane 2). The detection of DNA in empty capsids is most likely due to contamination of these preparations with a small amount of mature virions. This was confirmed by our ability to detect a small amount of the core proteins pVII and VII in these preparations (data not shown). Analysis of H5pm8001 intermediates failed to reveal any associated viral DNA even after extended exposure of the blot (Fig. 11B), suggesting that the 52/55-kDa protein is required for stable association of the viral genome with the empty capsid.
FIG. 11.
Analysis of viral DNA associated with H5pm8001 intermediates. Viral DNA was prepared from purified particles, digested with KpnI and ClaI, and analyzed by Southern with an Ad5-specific probe. Lanes: pm8001, intermediates from H5pm8001-infected 293 cells; MV and EC, mature virions and empty capsids, respectively, prepared from Ad5-infected 293 cells; ts369, particles prepared from 293 cells infected with H5ts369 at 39.5°C. (B) Longer exposure of the blot shown in panel A, with lane 1 removed. The arrow indicates the left end-specific ClaI fragment.
DISCUSSION
In this report we have described the isolation and characterization of an adenovirus harboring a mutation that blocks expression of the 52/55 kDa-protein, H5pm8001. As expected, successful propagation of H5pm8001 required the generation of a helper cell line capable of providing the 52/55-kDa protein in trans. Our analysis of the ability of H5pm8001 to progress through the viral life cycle in 293 cells has provided several insights into the role of the 52/55-kDa protein during infection. First, the 52/55-kDa protein was not required for entry into the late stage of infection. Second, the 52/55-kDa protein was not required for full activation of the MLP at late times during infection. Third, the 52/55-kDa protein was not required for the formation of empty capsid structures, suggesting that it does not function as a scaffolding protein during assembly. Finally, the inability to detect DNA associated with capsids that form in H5pm8001-infected 293 cells strongly argues that the 52/55-kDa protein is essential for association between the viral genome and the empty capsid or in the encapsidation process itself.
The early appearance of the 52/55-kDa protein has been taken as an indication that it might have additional functions at early times during infection (23). In addition, the identification of a variety of regulatory mechanisms apparently designed to ensure the early expression of the 52/55-kDa protein strengthens this argument (12, 16, 18, 30, 57, 66). Our analysis of H5pm8001-infected 293 cells, however, did not reveal a requirement for the 52/55-kDa protein at early times: there was no effect on the expression of the E2A 72K protein or RNA. Furthermore, as determined by examining the onset of DNA replication and gene expression, H5pm8001-infected 293 cells made the transition from the early to late stage of infection at the same time as did Ad5-infected cells. Analysis of DNA replication and late gene expression, however, did reveal some differences between H5pm8001-infected and Ad5-infected 293 cells. First, H5pm8001-infected 293 cells accumulated less viral DNA as the infection progressed (25% of that of Ad5-infected cells at 18 h postinfection). This difference was either less dramatic or not apparent at earlier times or when H5pm8001 was infected into 293-L1 cells. Second, lower levels of late proteins were synthesized in H5pm8001 infections as determined by immunoblotting with antibodies to the fiber and IVa2 proteins. One possible explanation for this finding would be that the 52/55-kDa protein stabilized proteins produced at late times during infection, thereby allowing them to accumulate to higher levels. Pulse-chase experiments designed to examine this possibility, however, revealed no difference in the half life of the IVa2 protein in H5pm8001-infected or Ad5-infected 293 cells. Third, Northern analysis revealed that although L3, L1, and IVa2 mRNAs could be detected by 18 h postinfection in H5pm8001 and Ad5 infections, they were consistently less abundant in H5pm8001 infections at this time (60 to 70% of wild type levels). Despite the differences apparent at 18 h postinfection, these mRNAs accumulated to similar levels at later times in infections with either virus.
Interestingly, a similar phenotype has been reported for adenoviruses containing mutations in MLP elements. Initial characterization of the MLP had suggested that it was a relatively simple promoter composed of a TATA box and an upstream promoter element (UPE) (3). The UPE is a cis-acting sequence element that is specifically bound by a factor termed USF/MLTF at late times during infection, and is required for full transcriptional activity in constructs that remove the MLP from its natural context in the viral genome (25, 27, 41, 56). Reach et al. described the construction of an adenovirus (H5USF0) containing point mutations in the UPE that abolish binding of USF, and impair transcription from the MLP in vitro (52). Analysis of transcription from the MLP in H5USF0-infected HeLa cells, however, revealed only a twofold decrease at 12 h postinfection, that was no longer apparent by 24 h postinfection (52). Their findings suggested that activation of the MLP involves multiple, partially redundant, cis-acting DNA sequence elements, and subsequent analysis of the MLP has identified a CAAT box, along with MAZ and Sp1 binding sites, and a downstream element (DE) that contribute to activation of the MLP (35, 40, 49, 52, 53).
The DE consists of two DNA sequence elements located downstream of the MLP called DE1 and DE2 that are bound by protein complexes termed DEF-A and DEF-B, respectively (31, 32, 42, 43, 62). Purification of these complexes has indicated that DEF-B consists of IVa2 homodimers, while DEF-A consists of IVa2 and an unknown 40-kDa protein (32, 62). Our demonstration that the IVa2 and 52/55-kDa proteins interact (21), combined with the observation that one of the major degradation products of the 52/55-kDa protein is 40 kDa (22), suggests that this may be the functional partner of IVa2 in the DEF-A complex. Although the downstream elements are required for full activation of the MLP in vitro, analysis of an adenovirus containing a mutated DE1 site did not display any defect in transcription from the MLP, suggesting that the downstream element may be functionally redundant as well (53). It should be noted, however, that this mutant did not disrupt the DE2 site, which also contains a DEF-A binding site, leaving open the possibility that in this mutant DEF-A function is provided through an interaction with the DE2 site (43, 53).
Despite the similarity between H5pm8001 and adenoviruses containing mutations in single MLP elements, some differences exist as well. Although H5USF0 displayed a twofold reduction in DNA replication at 12 h postinfection, at later times, DNA accumulated to wild-type levels (52). H5pm8001 displayed a fourfold drop in DNA replication at 18 h postinfection that was not apparent at earlier times. One possible explanation for the lower level of late proteins seen in H5pm8001-infected cells could be that there are fewer viral templates available for the transcriptional machinery. This explanation could account for the lower level of L3, IVa2, and IIIa transcripts observed at 18 h postinfection but is not consistent with the detection of equivalent amounts of these mRNAs at later times. Also, the levels of E2A 72K protein and mRNA were unaffected at late times in H5pm8001 infections, suggesting that, at least in this case, the accumulation of viral mRNA and protein was independent of template concentration. The most dramatic difference between H5pm8001 and H5USF0 was that the H5USF0 mutation only affected transcription from the MLP (52). Our analysis indicated that in H5pm8001-infected 293 cells, the reduced level of transcription was not restricted to the MLP but extended to the IVa2 promoter as well. The IVa2 transcription start site is separated from the MLP initiation site by 210 bp, and the two are divergently transcribed. In vitro analysis of the IVa2 promoter has indicated a certain degree of overlap with MLP regulatory elements, and mutations that affect MLP activity have been shown to affect IVa2 promoter activity as well (7, 44–46). Interestingly, characterization of the IVa2 promoter has not included sequences that extend to the DE, so a role for these sequences, and hence the 52/55-kDa protein in activation of the IVa2 promoter, cannot be ruled out.
Characterization of intermediates formed in H5pm8001-infected 293 cells revealed that particles formed with a density identical to that of empty capsids (1.29 g/cm3). This is in contrast to what was seen for H5ts369, which accumulated particles with a density of 1.31 g/cm3 (23). Additionally, comparison of the polypeptide composition of H5pm8001 intermediates with those of Ad5 empty capsids and H5ts369 intermediates revealed some subtle differences. Ad5 empty capsids and H5ts369 intermediates contained a polypeptide with an apparent molecular size of 70 kDa that was present at much lower levels in H5pm8001 intermediates. Previous reports demonstrating an interaction of adenovirus capsid components with hsp70 suggest a possible identity for this protein (38, 48). Another difference that was apparent from this analysis was that H5pm8001 intermediates consistently displayed more pVII than did H5ts369 intermediates or empty capsids. D’Halluin et al. reported that although pVII was present in all gradient fractions it could only be cross-linked to young virions, indicating that it was specifically associated with these particles (14). As the density and protein composition of H5pm8001 intermediates were very similar to what has been reported previously for empty capsids (39), this would suggest that the abundance of pVII was due to nonspecific association with the pm8001 intermediates.
Characterization of H5ts369 suggested several possible roles for the 52/55-kDa protein during assembly. The observation that the 52/55-kDa protein was present in assembly intermediates but absent from mature virions suggested that it might function as a scaffolding protein (22). In other viral systems, mutation or elimination of scaffolding proteins results in either the complete lack of capsid formation or the appearance of aberrant capsid structures (5, 11, 59, 61). The finding of normal-appearing capsid structures in H5pm8001-infected 293 cells indicates that the 52/55-kDa protein most likely does not function as a scaffolding protein during capsid assembly. Characterization of the viral DNA associated with intermediates formed in H5ts369-infected HeLa cells at the nonpermissive temperature demonstrated an association with the left end of the viral genome (23). This suggested that the 52/55-kDa protein was involved in either association between the DNA and capsid or in DNA packaging. Previous work analyzing an adenovirus that has a deleted packaging signal (H5dl309-A5) indicated that the viral packaging signal is required for capsid assembly to take place. Cells infected with H5dl309-A5 accumulate normal levels of viral DNA and late proteins, but no empty capsids are produced (22, 24). Presumably, this reflects a requirement for interaction between capsid components and the packaging signal that initiates the assembly process and the formation of empty capsids. The inability to detect DNA associated with empty capsids would indicate that this initial interaction between capsid components and the packaging signal is relatively unstable or short-lived. If this model is correct, then the appearance of empty capsids in H5pm8001-infected 293 cells suggests that the 52/55-kDa protein functions subsequent to the initial interaction between the packaging signal and capsid components.
The results presented above indicate that the 52/55-kDa protein is required for stable association between the viral DNA and capsid. This is consistent with a role for the 52/55-kDa protein in encapsidation of the viral genome. Although very little is known regarding the mechanism by which the adenovirus chromosome is inserted into the empty capsid, this event most likely requires interaction between factors bound at the packaging signal and capsid components (4). We would speculate that the interaction between the IVa2 and 52/55-kDa proteins is involved in packaging the viral genome. It will be interesting to examine if these proteins, alone or in combination, have any affinity for the adenovirus packaging signal.
ACKNOWLEDGMENTS
We thank the members of the Imperiale laboratory for helpful comments and suggestions throughout the course of this work; Erle Robertson for critically reading the manuscript; Tom Shenk for H5ts369; Transgene S.A. for pTG3602; and Arnie Levine, Claude Kedinger, Carl Anderson, and Patrick Hearing for various antibodies.
This work was supported by PHS grant GM34902 to M.J.I. and core grant P30 CA46592 to the University of Michigan Comprehensive Cancer Center. K.E.G. was supported in part by a Horace Rackham Predoctoral Fellowship from the University of Michigan.
REFERENCES
- 1.Bachenheimer S, Darnell J E. Adenovirus-2 mRNA is transcribed as part of a high-molecular-weight precursor RNA. Proc Natl Acad Sci USA. 1975;72:4445–4449. doi: 10.1073/pnas.72.11.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berget S M, Moore C, Sharp P A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berk A J. Adenovirus promoters and E1A transactivation. Annu Rev Genet. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- 4.Black L W. DNA packaging in dsDNA bacteriophages. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 321–374. [Google Scholar]
- 5.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 15–92. [Google Scholar]
- 6.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Vinnakota R, Flint S J. Intragenic activating and repressing elements control transcription from the adenovirus IVa2 initiator. Mol Cell Biol. 1994;14:676–685. doi: 10.1128/mcb.14.1.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Chow L T, Broker T R, Lewis J B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979;134:265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- 10.Chroboczek J, Bieber F, Jacrot B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology. 1992;186:280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- 11.Desai P, Watkins S C, Person S. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J Virol. 1994;68:5365–5374. doi: 10.1128/jvi.68.9.5365-5374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeZazzo J D, Falck-Pedersen E, Imperiale M J. Sequences regulating temporal poly(A) site switching in the adenovirus major late transcription unit. Mol Cell Biol. 1991;11:5977–5984. doi: 10.1128/mcb.11.12.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeZazzo J D, Imperiale M J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989;9:4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Halluin J C, Martin G R, Torpier G, Boulanger P A. Adenovirus type 2 assembly analyzed by reversible cross-linking of labile intermediates. J Virol. 1978;26:357–363. doi: 10.1128/jvi.26.2.357-363.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edvardsson B, Everitt E, Jornvall H, Prage L, Philipson L. Intermediates in adenovirus assembly. J Virol. 1976;19:533–547. doi: 10.1128/jvi.19.2.533-547.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falck-Pedersen E, Logan J. Regulation of poly(A) site selection in adenovirus. J Virol. 1989;63:532–541. doi: 10.1128/jvi.63.2.532-541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser N W, Nevins J R, Ziff E, Darnell J E., Jr The major late adenovirus type-2 transcription unit: termination is downstream from the last poly(A) site. J Mol Biol. 1979;129:643–656. doi: 10.1016/0022-2836(79)90474-1. [DOI] [PubMed] [Google Scholar]
- 18.Gilmartin G M, Hung S L, DeZazzo J D, Fleming E S, Imperiale M J. Sequences regulating poly(A) site selection within the adenovirus major late transcription unit influence the interaction of constitutive processing factors with the pre-mRNA. J Virol. 1996;70:1775–1783. doi: 10.1128/jvi.70.3.1775-1783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham F L, Prevec L. Manipulation of adenovirus vectors. Methods Mol Biol. 1991;7:109–128. doi: 10.1385/0-89603-178-0:109. [DOI] [PubMed] [Google Scholar]
- 20.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 21.Gustin K E, Lutz P, Imperiale M J. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J Virol. 1996;70:6463–6467. doi: 10.1128/jvi.70.9.6463-6467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasson T B, Ornelles D A, Shenk T. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J Virol. 1992;66:6133–6142. doi: 10.1128/jvi.66.10.6133-6142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasson T B, Soloway P D, Ornelles D A, Doerfler W, Shenk T. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J Virol. 1989;63:3612–3621. doi: 10.1128/jvi.63.9.3612-3621.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hearing P, Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983;33:695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- 25.Hen R, Sassone-Corsi P, Corden J, Gaub M P, Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci USA. 1982;79:7132–7136. doi: 10.1073/pnas.79.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 27.Hu S L, Manley J L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci USA. 1981;78:820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Flint S J. The tripartite leader sequence of subgroup C adenovirus major late mRNAs can increase the efficiency of mRNA export. J Virol. 1998;72:225–235. doi: 10.1128/jvi.72.1.225-235.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishibashi M, Maizel J V., Jr The polypeptides of adenovirus. V. Young virions, structural intermediate between top components and aged virions. Virology. 1974;57:409–424. doi: 10.1016/0042-6822(74)90181-0. [DOI] [PubMed] [Google Scholar]
- 30.Iwamoto S, Eggerding F, Falck-Pedersen E, Darnell J E., Jr Transcription unit mapping in adenovirus: regions of termination. J Virol. 1986;59:112–119. doi: 10.1128/jvi.59.1.112-119.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen-Durr P, Boeuf H, Kedinger C. Replication-induced stimulation of the major late promoter of adenovirus is correlated to the binding of a factor to sequences in the first intron. Nucleic Acids Res. 1988;16:3771–3786. doi: 10.1093/nar/16.9.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen-Durr P, Mondesert G, Kedinger C. Replication-dependent activation of the adenovirus major late promoter is mediated by the increased binding of a transcription factor to sequences in the first intron. J Virol. 1989;63:5124–5132. doi: 10.1128/jvi.63.12.5124-5132.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 34.Kingston R E, Chen C A, Okayama H, Rose J K. Calcium phosphate transfection. In: Ausubel M F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1995. pp. 9.1.4–9.1.11. [Google Scholar]
- 35.Leong K, Lee W, Berk A J. High-level transcription from the adenovirus major late promoter requires downstream binding sites for late-phase-specific factors. J Virol. 1990;64:51–60. doi: 10.1128/jvi.64.1.51-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logan J, Shenk T. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc Natl Acad Sci USA. 1984;81:3655–3659. doi: 10.1073/pnas.81.12.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutz P, Kedinger C. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J Virol. 1996;70:1396–1405. doi: 10.1128/jvi.70.3.1396-1405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macejak D G, Luftig R B. Association of HSP70 with the adenovirus type 5 fiber protein in infected HEp-2 cells. Virology. 1991;180:120–125. doi: 10.1016/0042-6822(91)90015-4. [DOI] [PubMed] [Google Scholar]
- 39.Maizel J V, Jr, White D O, Scharff M D. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology. 1968;36:126–136. doi: 10.1016/0042-6822(68)90122-0. [DOI] [PubMed] [Google Scholar]
- 40.Mansour S L, Grodzicker T, Tjian R. Downstream sequences affect transcription initiation from the adenovirus major late promoter. Mol Cell Biol. 1986;6:2684–2694. doi: 10.1128/mcb.6.7.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto N G, Moncollin V, Egly J M, Chambon P. Specific interaction between a transcription factor and the upstream element of the adenovirus-2 major late promoter. EMBO J. 1985;4:3563–3570. doi: 10.1002/j.1460-2075.1985.tb04118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mondesert G, Kedinger C. Cooperation between upstream and downstream elements of the adenovirus major late promoter for maximal late phase-specific transcription. Nucleic Acids Res. 1991;19:3221–3228. doi: 10.1093/nar/19.12.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mondesert G, Tribouley C, Kedinger C. Identification of a novel downstream binding protein implicated in late-phase-specific activation of the adenovirus major late promoter. Nucleic Acids Res. 1992;20:3881–3889. doi: 10.1093/nar/20.15.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natarajan V, Madden M J, Salzman N P. Proximal and distal domains that control in vitro transcription of the adenovirus IVa2 gene. Proc Natl Acad Sci USA. 1984;81:6290–6294. doi: 10.1073/pnas.81.20.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Natarajan V, Madden M J, Salzman N P. Positive and negative control sequences within the distal domain of the adenovirus IVa2 promoter overlap with the major late promoter. J Virol. 1985;55:10–15. doi: 10.1128/jvi.55.1.10-15.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natarajan V, Madden M J, Salzman N P. Identification of a transcription factor which interacts with the distal domain of the adenovirus IVa2 promoter. J Virol. 1987;61:646–652. doi: 10.1128/jvi.61.3.646-652.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nevins J R, Wilson M C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981;290:113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- 48.Niewiarowska J, D’Halluin J C, Belin M T. Adenovirus capsid proteins interact with HSP70 proteins after penetration in human or rodent cells. Exp Cell Res. 1992;201:408–416. doi: 10.1016/0014-4827(92)90290-o. [DOI] [PubMed] [Google Scholar]
- 49.Parks C L, Shenk T. Activation of the adenovirus major late promoter by transcription factors MAZ and Sp1. J Virol. 1997;71:9600–9607. doi: 10.1128/jvi.71.12.9600-9607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Persson H, Mathisen B, Philipson L, Pettersson U. A maturation protein in adenovirus morphogenesis. Virology. 1979;93:198–208. doi: 10.1016/0042-6822(79)90287-3. [DOI] [PubMed] [Google Scholar]
- 51.Prage L, Hoglund S, Philipson L. Structural proteins of adenoviruses. 8. Characterization of incomplete particles of adenovirus type 3. Virology. 1972;49:745–757. doi: 10.1016/0042-6822(72)90531-4. [DOI] [PubMed] [Google Scholar]
- 52.Reach M, Babiss L E, Young C S. The upstream factor-binding site is not essential for activation of transcription from the adenovirus major late promoter. J Virol. 1990;64:5851–5860. doi: 10.1128/jvi.64.12.5851-5860.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reach M, Xu L X, Young C S. Transcription from the adenovirus major late promoter uses redundant activating elements. EMBO J. 1991;10:3439–3446. doi: 10.1002/j.1460-2075.1991.tb04908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reich N C, Sarnow P, Duprey E, Levine A J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 55.Rosenwirth B, Tjia S, Westphal M, Doerfler W. Incomplete particles of adenovirus. II. Kinetics of formation and polypeptide composition of adenovirus type 2. Virology. 1974;60:431–437. doi: 10.1016/0042-6822(74)90337-7. [DOI] [PubMed] [Google Scholar]
- 56.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw A R, Ziff E B. Transcripts from the adenovirus-2 major late promoter yield a single early family of 3′ coterminal mRNAs and five late families. Cell. 1980;22:905–916. doi: 10.1016/0092-8674(80)90568-1. [DOI] [PubMed] [Google Scholar]
- 58.Sundquist B, Everitt E, Philipson L, Hoglund S. Assembly of adenoviruses. J Virol. 1973;11:449–459. doi: 10.1128/jvi.11.3.449-459.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 60.Thomas G P, Mathews M B. DNA replication and the early to late transition in adenovirus infection. Cell. 1980;22:523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- 61.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tribouley C, Lutz P, Staub A, Kedinger C. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J Virol. 1994;68:4450–4457. doi: 10.1128/jvi.68.7.4450-4457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber J. Genetic analysis of adenovirus type 2. III. Temperature sensitivity of processing viral proteins. J Virol. 1976;17:462–471. doi: 10.1128/jvi.17.2.462-471.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber J, Begin M, Khittoo G. Genetic analysis of adenovirus type 2. II. Preliminary phenotypic characterization of temperature-sensitive mutants. J Virol. 1975;15:1049–1056. doi: 10.1128/jvi.15.5.1049-1056.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weber J, Jelinek W, Darnell J E., Jr The definition of a large viral transcription unit late in Ad2 infection of HeLa cells: mapping of nascent RNA molecules labeled in isolated nuclei. Cell. 1977;10:611–616. doi: 10.1016/0092-8674(77)90093-9. [DOI] [PubMed] [Google Scholar]
- 66.Wilson M C, Darnell J E., Jr Control of messenger RNA concentration by differential cytoplasmic half-life: adenovirus messenger RNAs from transcription units 1A and 1B. J Mol Biol. 1981;148:231–251. doi: 10.1016/0022-2836(81)90537-4. [DOI] [PubMed] [Google Scholar]
- 67.Winter N, D’Halluin J C. Regulation of the biosynthesis of subgroup C adenovirus protein IVa2. J Virol. 1991;65:5250–5259. doi: 10.1128/jvi.65.10.5250-5259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]