Abstract
Background
DNA methylation is a crucial epigenetic alteration involved in diverse biological processes and diseases. Nevertheless, the precise role of DNA methylation in chemotherapeutic drug-induced alopecia remains unclear. This study examined the role and novel processes of DNA methylation in regulating of chemotherapeutic drug-induced alopecia.
Methods
A mouse model of cyclophosphamide (CTX)-induced alopecia was established. Hematoxylin-eosin staining and immunohistochemical staining for the Ki67 proportion and a mitochondrial membrane potential assay (JC-1) were performed to assess the structural integrity and proliferative efficiency of the hair follicle stem cells (HFSCs). Immunofluorescence staining and real-time fluorescence quantitative PCR (RT-qPCR) were performed to determine the expression levels of key HFSC markers, namely Lgr5, CD49f, Sox9, CD200, and FZD10. Differential DNA methylation levels between the normal and CTX-induced model groups were determined through simple methylation sequencing and analyzed using bioinformatics tools. The expression levels of miR-365-1, apoptosis markers, and DAP3 were detected through RT-qPCR and western blotting. In parallel, primary mouse HFSCs were extracted and used as a cell model, which was constructed using 4-hydroperoxycyclophosphamide. The luciferase reporter gene assay was conducted to confirm miR-365-1 binding to DAP3. To measure the expression of relevant indicators, superoxide dismutase (SOD) and malondialdehyde (MDA) kits were used. Methylation-specific PCR (MS-PCR) was performed to determine DNA methylation levels. The regulatory relationship within HFSCs was confirmed through plasmid overexpression of miR-365-1 and DAP3.
Result
In the alopecia areata model, a substantial number of apoptotic cells were observed within the hair follicles on the mouse backs. Immunofluorescence staining revealed that the expression of HFSC markers significantly reduced in the CTX group. Both RT-qPCR and western blotting demonstrated a noteworthy difference in DNA methyltransferase expression. Simple methylation sequencing unveiled that DNA methylation substantially increased within the dorsal skin of the CTX group. Subsequent screening identified miR-365-1 as the most differentially expressed miRNA. miR-365-1 was predicted and confirmed to bind to the target gene DAP3. In the CTX group, SOD and ATP expression markedly reduced, whereas MDA levels were significantly elevated. Cellular investigations revealed 4-HC-induced cell cycle arrest and decreased expression of HFSC markers. MS-PCR indicated hypermethylation modification of miR-365-1 in the 4-HC-induced HFSCs. The luciferase reporter gene experiment confirmed the binding of miR-365-1 to the DAP3 promoter region. miR-365-1 overexpression dramatically reduced apoptotic protein expression in the HFSCs. However, this effect was slightly reversed after DAP3 overexpression in lentivirus.
Conclusion
This study explored the occurrence of miR-365-1 DNA methylation in chemotherapeutic drug-induced alopecia. The results unveiled that miR-365-1 reduces cell apoptosis by targeting DAP3 in HFSCs, thereby revealing the role of DNA methylation of the miR-365-1 promoter in chemotherapeutic drug-induced alopecia.
Keywords: Chemotherapy-induced alopecia, Hair follicle stem cells, RRBS, MiRNA
1. Introduction
Hair loss is a prevalent clinical concern affecting the appearance and psychological well-being of numerous individuals. According to clinical studies, the global prevalence of total baldness, alopecia areata, and common baldness is 0.08%, 0.02%, and 0.03%, respectively [1]. Hair loss is typically multifactorial and is caused by medications and the genetic, environmental, and lifestyle factors [2]. Chemotherapeutic drug-induced hair loss is a common adverse effect during cancer treatment. It affects the physical appearance and potentially leads to decreased self-esteem and quality of life of patients [3,4]. Although research on chemotherapy-induced hair loss has progressed to some extent, the understanding of its underlying mechanisms remains relatively limited [5,6].
DNA methylation is an epigenetic modification in which methyltransferases add methyl groups to the cytosine rings of DNA molecules [7]. This process plays a crucial regulatory role in controlling gene expression, thus enabling gene activity modulation by altering the methylation status of certain genes [8]. DNA methylation is associated with various biological processes and diseases, including cancer, neurological disorders, and cardiovascular conditions and, has been extensively investigated [[9], [10], [11]]. However, the role of DNA methylation in hair loss and its exact role in the mechanism underpinning chemotherapy-induced hair loss remain largely unknown.
This study investigated the potential regulatory roles of DNA methylation in chemotherapy-induced hair loss and the underlying mechanisms. Using both mouse and cell models, we uncovered a hair loss-associated molecular mechanism, specifically focusing on DNA methylation-induced miR-365-1 modification and the interaction of miR-365-1 with DAP3. By comprehensively exploring the role of DNA methylation, we aspire to offer renewed insights into hair loss prevention and treatment, thereby laying the foundation for developing more effective intervention strategies. This research holds significance for improving quality of life of individuals experiencing hair loss, in addition to providing novel insights into the role of DNA methylation in other diseases and physiological processes. Consequently, this study potentially paves the way for extensive research in this field.
2. Method
2.1. Animal experiments
Male C57BL/6 mice aged 6–8 weeks were procured from Shanghai Jihui Laboratory Animal Co. Ltd. (Shanghai, China; license number: SYXK (Hu) 2018-0040). The mice were raised and maintained under standard conditions of room temperature (20°C–26°C) and a relative humidity of 40%–70%. They were exposed to the 12-h light/dark cycle and provided with regular diets. One week after acclimation, 10 mice were randomized into two groups (n = 5 per group): the control group (WT) and the cyclophosphamide (CTX) model group (CTX). The CTX group was intraperitoneally injected with CTX (70 mg/kg) from day 0 to day 15. On day 3, all mice were depilated on the back by using a rosin/wax (1:1) mixture after anesthetizing them with 4% isoflurane inhalation, and photographs were captured. On day 15, the mice were euthanized through cervical dislocation, and skin tissues from the depilated area of each mouse were promptly collected [12]. All animal experiments were conducted strictly according to the Guideline for Ethical Review of Laboratory Animal Welfare of the General Administration of Quality Supervision, Inspection and Quarantine of the People's Republic of China. The animal protocol was approved by Institutional Animal Care and Use Committee (at Yueyang Integrated Chinese and Western Medicine Hospital (Assurance Number: YYLAC-2022-164-4).
2.2. Isolating, sorting, and culturing primary CD49f+/CD200+ mouse hair follicle stem cells (mHFSCs)
Under sterile conditions, the mouse dorsal skin was isolated, minced, and subsequently digested at 37°C for 50 min in a 1.0% trypsin solution with a volume 5 times that of the tissue. The tissue suspension was filtered through a 200-mesh cell strainer, followed by centrifugation at 1500 rpm for 5 min. The cell pellet was resuspended in 200 μL of ice-cold sterile PBS and incubated in the dark at 4°C for 30 min with FITC-labeled anti-mouse CD49f antibody and PE-labeled anti-mouse CD200 antibody.
After 1 mL of frozen sterile PBS was added for washing, the cells were centrifuged and the cell mass was collected. Subsequently, the cells were reconstituted to 500 μL of frozen sterile PBS, and CD49f+/CD200+ mouse hair follicle stem cells (mHFSCs) were classified through enzymatic digestion and magnetic bead sorting. These classified cells were cultured in a medium containing DMEM:F12 supplemented with 5 μg/mL insulin, 10 ng/mL EGF, 10 ng/mL bFGF, 10 μg/mL transferrin, 0.36 μg/mL hydrocortisone, 2 mM glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, 15% bovine serum, 50 μg/mL vitamin C, 5 μM Y27632, and 20 ng/mL VEGF-A. The cells were then cultured at 37°C under 5% CO2. The next day, the medium was replaced with fresh medium, and cultivation was continued [13].
2.3. Hematoxylin and eosin staining (H&E staining)
The histopathological morphology of the skin was assessed through hematoxylin and eosin (HE) staining, following a protocol similar to the one previously described. The slides were examined under a BX-51 microscope (Olympus, Japan) and captured using a DP-70 digital camera (Olympus, Japan).
2.4. Immunohistochemical staining
Ki67 immunohistochemical (IHC) expression was determined as follows: first, 4-μm skin sections were baked at 65°C for 120 min. Subsequently, the sections were dewaxed with xylene and subjected to graduated rehydration with alcohols. Following two washes (5 min each) with PBS, the skin sections were immersed in the sodium citrate antigen retrieval solution. Antigen retrieval was achieved by boiling the solution for 20 min. Following three additional washes (5 min each) with PBS, an endogenous peroxidase blocking agent was added to the sections. The sections were then incubated at room temperature for 10 min. After cooling, the sections were washed thrice with PBS (5 min each).
The skin sections were then covered with a 5% bovine serum albumin (BSA) solution and left for 30 min at room temperature. Each section was treated with the primary antibody (approximately 100 μL/section, Abcam, Cambridge, UK), which was dissolved in a 5% BSA solution, and the mixture was incubated overnight at 4°C. After the mixture returned to the room temperature, the sections were washed three times with PBS (5 min each), exposed to the secondary antibody solution for 90 min, and washed three times with PBS (2 min each). The sections were treated with the DAB solution and counterstained with hematoxylin. The sections were thoroughly washed, rehydrated, mounted, and prepared for analysis.
2.5. Immunofluorescence staining
The skin tissues were immersed in 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA) for 30 min, subjected to gradient dehydration with ethanol, and embedded in paraffin. The resulting 6-μm skin sections were dewaxed with xylene, blocked with an IHC blocking buffer (Beyotime Biotechnology, Zhejiang, China) at 37°C for 30 min, and washed (three times, 5 min each, at room temperature) with an IHC washing buffer (Beyotime Biotechnology, China).
Next, the sections were incubated at 37°C for 45 min with primary antibodies against CD200 (ab244560), CD49F (ab181551), SOX-9 (ab185966), CK15 (ab52816), and FZD10 (ab137491) (Abcam, Cambridge, UK). Following another round of washing with an IHC washing solution (three times, 5 min each, at room temperature), the sections were incubated at 37°C for 45 min with the respective secondary antibodies. After the sections were washed thoroughly, they were blocked using an immunofluorescence (IF) blocking buffer (Sigma-Aldrich, USA).
2.6. TUNEL staining
Briefly, the skin sections were stained with the TUNEL reaction solution and incubated in a damp dark room at 37° C for 1 h. Factory plays PBS, equipped with ProLong diamond anti-fading loader and DAPI anti-fading (P36962, Invitrogen) imaging focusing microscope. The SP8 (Leica) apoptosis index was determined by calculating the median values of five TUNEL-positive cells and DAPI-positive cells at the airport.
2.7. Measurement of mitochondrial membrane potential (MMP)
The JC-1 reagent (Beyotime, China) was used to assess the mitochondrial membrane potential (MMP) of epidermal cell. Following the manufacturer's instructions, the skin sections were treated with the JC-1 solution at 37°C for 30 min, and DAPI solution (Sigma-Aldrich, USA) was used to stain the nuclei. Subsequently, changes in JC-1 fluorescence were observed and documented under a fluorescence microscope (Olympus BX53, Japan). The transition of JC-1 aggregates (red fluorescence) to JC-1 monomers (green fluorescence) indicated a decrease in MMP.
2.8. Reduced representation bisulfite sequencing (RRBS)
Genomic DNA extracted from the skin tissues was subjected to digestion with MspI. The ends of the MspI-digested DNA were subsequently repaired, followed by the addition of dA-tails. DNA adapter-linked DNA was treated with disulphite by using the EZ-DNA Methylation Gold Kit (Zymo Research, Orange, CA, USA). Subsequently, disulphite-treated DNA was amplified through PCR. The library DNA concentration was analyzed using Qubit 2.0 (Thermo Fisher Scientific, CA, USA) and readjusted to 1 ng/μL.
Subsequently, the fragment size range and molarity of each library were assessed using Bioanalyzer 2100 (Agilent, CA, USA). The library concentration was recalculated based on the index ratios. After obtaining a sufficient pool of reduced representation bisulfite sequencing (RRBS) libraries for sequencing, comprehensive sequencing was performed on the HiSeq platform (Illumina, USA) [14].
2.8.1. Flow cytometry analysis
mHFSCs from each group were suspended (1 × 106 cells/mL) and stained with primary antibodies on ice in Dulbecco's phosphate-buffered saline containing 10% BSA. The cells were stained with an isotype control antibody (mouse IgG1-FITC, mouse IgG1-PE; Invitrogen, eBioscience™, Shanghai, China) to correct for non-specific binding. Antibody staining was analyzed through fluorescence correlation microscopy by using FACS Aria (Quanta SC, Beckman Coulter INC) [15]. The primary antibodies used were FZD10 (ab137491), CK15 (ab52816), Sox9 (ab185966), and CD200 (ab244560)
2.9. Methylation-specific PCR (MS-PCR)
The Wizard Genomic DNA Purification Kit (Promega) was used to extract genomic DNA from cell chains or clinical samples. Then, DNA was modified with sodium bisulfite by using the EZ-DNA Methylation Gold Kit (Zymo Research, USA) according to the instructions. MS-PCR primers were designed using MethPrimer software, followed by amplification and determination of specific methylation regions on the genomic DNA by using DNA MyTaq TMHS mixed reaction buffer (Bioline, London, UK) [16].
2.10. Transfection of mHFSCs
To generate DAP3- and miR-365-1-5p-overexpressing lentiviral vectors, DNA fragments corresponding to DAP3 and miR-365-1-5p were amplified through PCR by using the genomic DNA of mouse SGC7901 cells. These PCR-amplified segments were integrated into the lentiviral vector pLV-EF1α-IRES-Puro (pLV-ctrl), which resulted in the creation of the viral vectors pLV-DAP3 and pLV-miR-365-1-5p. These vectors were transfected into HEK2937 cells. The culture medium containing lentivirus (pLV-DAP3, pLV-miR-365-1-5p, and pLV-ctrl) was collected at 24-h intervals for three times in total and purified through ultracentrifugation. Full-length cDNA encoding mouse DAP3 and miR-365-1-5p was amplified through PCR. These PCR products were subcloned into the pBOBI and pCMV-HA vectors to obtain overexpression plasmids for DAP3 and miR-365-1-5p, respectively [17].
The Inhibitor of DNA methyltransferases 5-Azacytidine(5-AzaC) (19–143, Sigma-Aldrich, USA) was also for HFSC treatment. Subsequent experiments were conducted 48 h after transfection [18].
2.11. RNA isolation and qRT-PCR
Total RNA was extracted from the skin tissues by using TRIzol (Thermo Fisher Scientific, USA). RNA purity was examined using Nanodrop 1000 (Thermo Fisher Scientific, USA). The extracted total RNA was transcribed into cDNA by using M-MLV reverse transcriptase (Promega, Madison, USA) and the ribonuclease inhibitor (Sigma-Aldrich, USA). Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green Real-time PCR Master Mix (Toyobo, Shanghai, China) and the qRT-PCR detection kit (Eppendorf, Hamburg, Germany) for a total of 40 cycles (95°C for 15 s, 58°C for 30 s, and 72°C for 42 s). With 18 rRNA as an internal reference gene, mRNA expression values were calculated using the 2−ΔΔCT method. The specific PCR primers are listed in Table S1.
2.12. Cell cycle analysis
mHFSCs were vaccinated at a density of 1.5 × 106 rooms per table culture and allowed to grow at 6 cm for 24 h. Then, the middle of the serum-free medium, and after 16–18 h of shortage of speech, the rooms enter the G0 phase. Subsequently, several experimental reagents were introduced into serum-free culture medium. The cells (1 × 106) were harvested through pancreatic enzyme-specific digestion, washed two times with cold PBS, and maintained in ice cubes in 70% alcohol for 30 min.
After three rounds of washing with cold PBS, the cells were resuspended in 1 mL Krishan's chromosome solution at 4°C overnight. These cells were filtered through 96-μm-long nylon wells , and 10,000 colored nuclei were analyzed through flow cytometry (BD Biosciences, CA, USA). M-phase DNA histograms of the G0–G1, S and G2 cell cycles were corrected using the Modfit Analytics (BD Biosciences, USA) program. Each condition was repeated three times.
2.13. Western blotting analysis
Total protein was extracted using 1 × RIPA50 lysis buffer (Sigma-Aldrich, USA) for protease inhibitors. Then, the extracted total protein was separated through sodium dodecyl-sulfate polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Bio Rad, CA, USA). The membrane was incubated with primary antibodies and appropriate secondary antibodies overnight at 4°C. The primary antibodies used were anti-caspase-3 rabbit pAb (GB11532-100), anti-cysteine protease-3 rabbit pAb (GB115600-100), anti-Bcl-2 mouse mAb (GB124830-100), anti-Bax mouse mAb, and anti-β-actin mouse mAb (all sourced from Servicebio, Wuhan, China). The DAP3 pAb (Cat. No. 10276-1-AP) was purchased from Proteintech (Wuhan, China).
2.14. Luciferase reporter assay
Luciferase reporter experiments were conducted by referring to a previous study [18]. On the first day, 1.5 × 104 mHFSCs were seeded into a 96-well whiteboard. On the second day, the report constructs were transfected with Lipofectamine 3000. Luciferase activity was evaluated 48 h after transfection. The 3' untranslated region (3' UTR) sequence of the wild-type (WT) or mutant DAP3 was inserted downstream of the firefly luciferase reporter gene. Its expression in the mammalian cells was regulated by adding the SV40 enhancer. No oligonucleotides were inserted into the control vector (GeneCopoeia, Rockville, MD, USA). Renella luciferase is an indicator of successful transfection. Luciferase activity was measured using the LightSwitch Assay Agent (SwitchGear Genomics) according to the manufacturer's instructions.[19].
2.15. Statistical analysis
Data were analyzed using GraphPad Prism 8.0 (GraphPad Software, CA, USA), and the results are expressed as the mean ± standard deviation. The statistical significance between multiple sets of independent data was assessed using one-way analysis of variance, followed by Bonferroni post-hoc tests. A p value of <0.05 was considered statistically significant. To obtain more genes from RRBS for validation, we considered unadjusted p values of <0.05 and fold change of ≥0.4 as qualifying values.
3. Results
3.1. CTX-induced hair loss on the backs of C57 mice
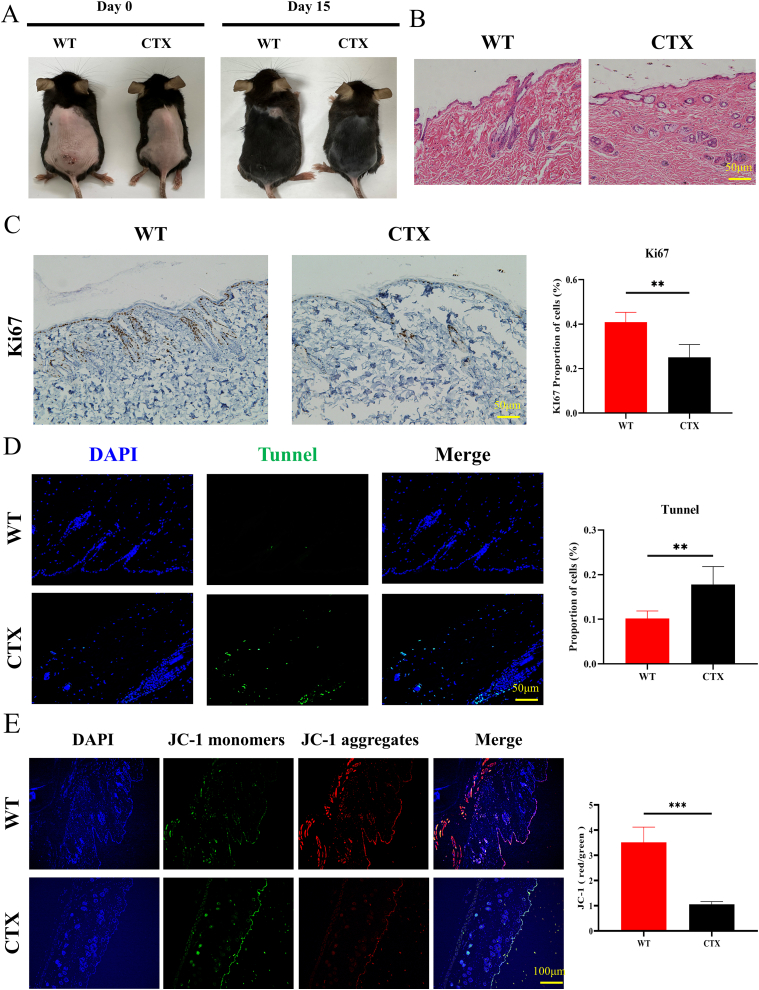
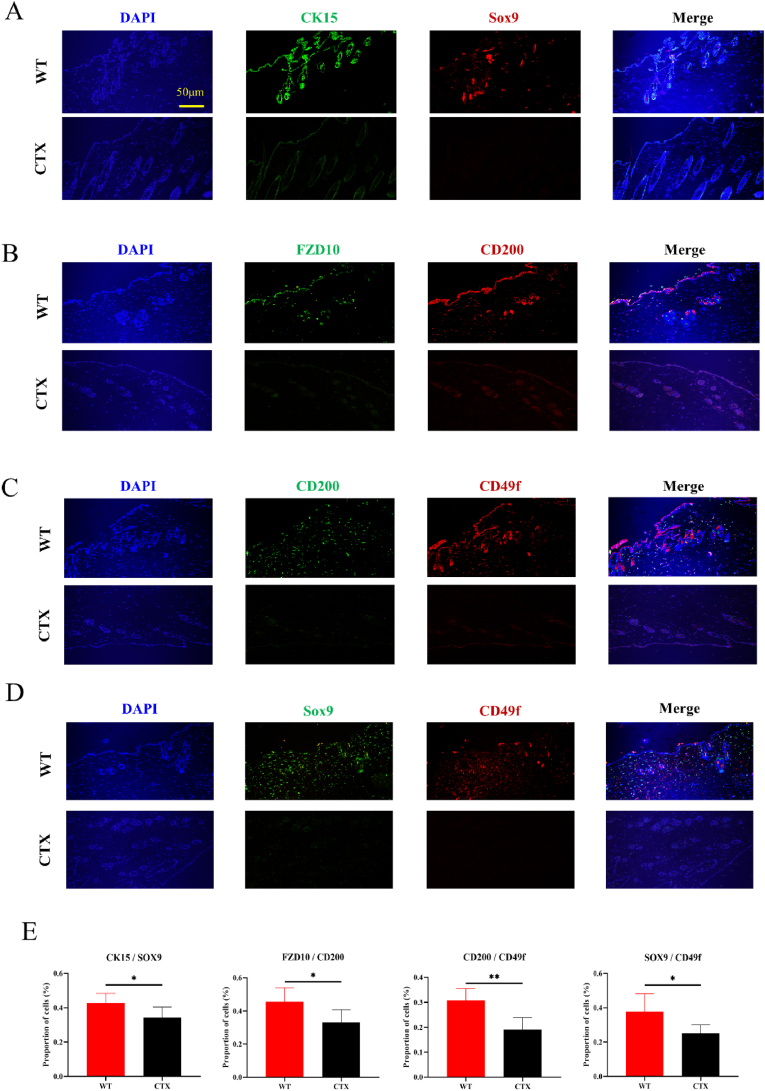
Using CTX, we successfully established a mouse hair loss model. In the CTX group, clear signs of hair loss from the hair follicles were noted on the mouse back. The results were significantly contradictory to those observed in the normal control group. On day 0, we depilated the mice and photographed them for documentation. Fifteen days after CTX was intraperitoneally injected for modeling, the hair on the backs of the mice became sparse and the skin appeared white, which contrasted with the reddish coloration on the backs of the mice in the WT group. This was due to the drug toxicity of CTX (Fig. 1A). HE staining unveiled a significant increase in empty hair follicles in the skin on the mouse backs (Fig. 1B). IHC staining revealed reduced Ki67 expression in the CTX-induced mouse back hair , which indicated suppressed hair proliferation (Fig. 1C). According to TUNEL staining, substantial cell apoptosis occurred in the skin tissue of the hair loss area, which indicated a close association between CTX-induced hair loss and follicular structure damage (Fig. 1D). JC-1 staining demonstrated a significant reduction in mitochondrial membrane potential in CTX-treated mice, suggesting the presence of extensive cell apoptosis in the epidermis (Fig. 1E). Subsequently, IF staining unveiled that the expression of HFSC markers, including CK15, Sox9 (marker for hair growth and development), FZD10, CD200, and CD49f, significantly reduced in the CTX model group (Fig. 2A–D). This result indicated that the chemotherapeutic drug CTX exerted irreversible inhibitory effects on HFSC activity (Fig. 2E).
Fig. 1.
Establishment of the hair loss model induced using the chemotherapeutic drug cyclophosphamide (CTX). (A) Photographs of hairs on the mouse back on day 0 and day 15. (B) HE staining results indicating more empty follicles in the back skin of CTX-treated mice. (C) IHC staining result showing a significant decrease in KI67 proportion after CTX induction. (D) Tunnel staining results demonstrating more cell apoptosis in the CTX group. (E) JC-1 staining results revealing abnormal MMP in the back skin of the CTX group. ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
Fig. 2.
Decreased expression of hair follicle stem cells (HFSCs) after CTX induction. (A) IF images of the HFSC markers CK15, Sox9, FZD10, CD200, and CD49f in the back skin of the WT and CTX groups. (B) Positive area ratio of IF staining. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
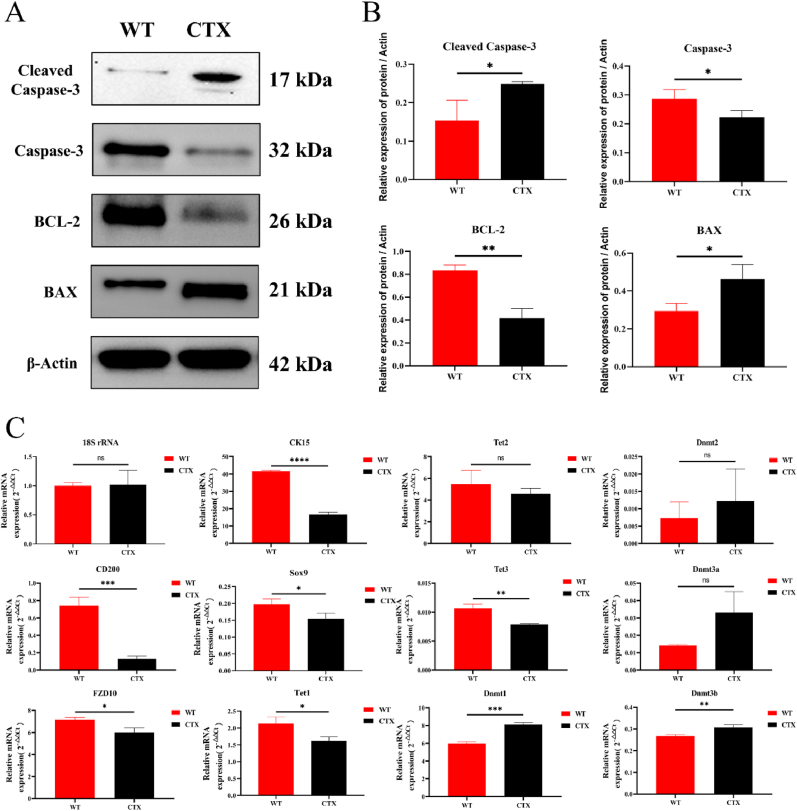
Through western blotting, the expression of mitochondrial apoptosis-related proteins, namely cleaved Caspase-3, Caspase-3, Bcl-2, and BAX, was determined. Mitochondrial damage and cell apoptosis were significant in the CTX-induced back skin (Fig. 3A and B). Subsequently, qRT-PCR was performed to examine mRNA expression of the HFSC markers CD200, CK15, Sox9, and FZD10, all of which showed significant decreased expressions under CTX induction. To investigate whether DNA methylation occurs in the CTX-induced hair loss area in the mouse back skin, qRT-PCR was also performed to determine the expression of DNA methylation- and hydroxymethylation-related genes. The results indicated that the DNA methylation-related genes Dnmt1 and Dnmt3b were significantly upregulated, whereas the DNA hydroxymethylation-related genes Tet1 and Tet3 were significantly downregulated. These findings suggest that CTX-induced mouse back skin undergoes changes in DNA methylation (Fig. 3C).
Fig. 3.
Changes in hair follicle stem cell (HFSC) genes, methylation-related genes, and apoptosis proteins in the back skin of the CTX-induced mice. (A) Western blotting detection of the expression of apoptosis-related genes, namely cleaved Caspase-3, Caspase-3, BCL-2, and BAX. (B) Statistical representation of western blotting results. (C) qRT-PCR detection of the expression of HFSC marker-, DNA methylation-, and hydroxymethylation-related genes. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
3.2. 4-HC induces oxidative stress and apoptosis in primary mHFSCs
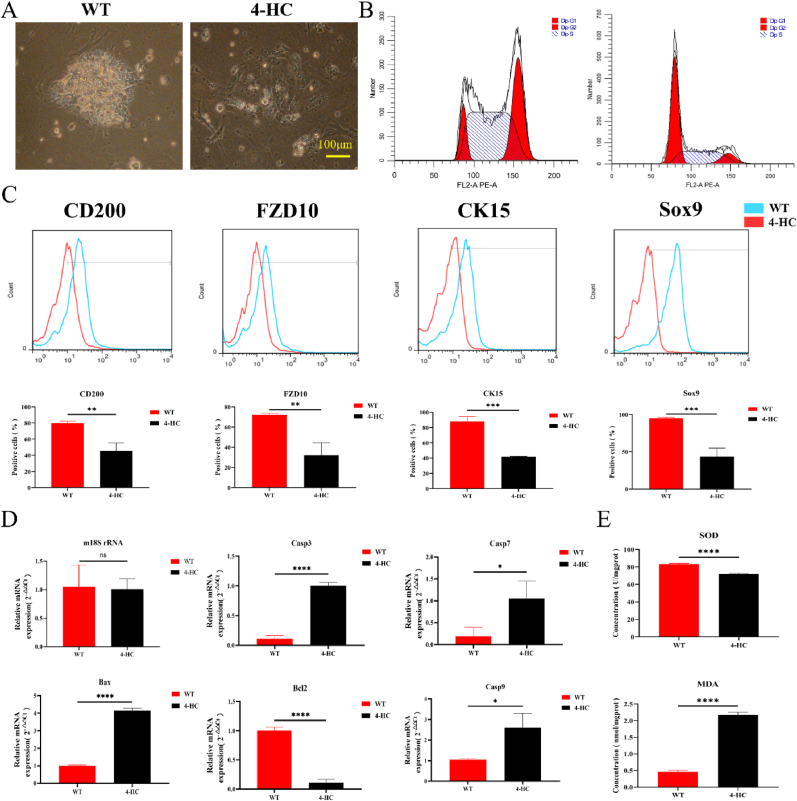
Enzymatic digestion and magnetic bead sorting were used to isolate and enrich CD49f+/CD200+ mHFSCs from the mouse subcutaneous tissue according to the methods used in previous studies [20,21]. The cells were then subjected to primary culture, and mHFSCs from the first generation were used for experimental modeling. In the WT group, mHFSCs had a clonal growth morphology. With passaging, they gradually differentiated and developed a stable cubic pavement stone structure, with a small cell volume, large nuclear/cytoplasmic ratio, smooth surface, minimal wrinkles, and no serrations. After the cell model was induced with 4-HC, cell volume significantly increased, and the losing smooth edges appeared. Cell morphology became disorderly, cell proliferation significantly decreased, and signs of aging were evident (Fig. 4A). Flow cytometry revealed that 4-HC significantly inhibited the cell cycle of HFSCs, displaying a noticeable arrest at the G1 phase (Fig. 4B). The expression of the mHFSC markers, including CD200, FZD10, CK15, and Sox9, decreased significantly (Fig. 4C). According to the qRT-PCR results, the expression of apoptosis-related genes was significantly increased in the 4-HC intervention group (Fig. 4D). SOD and MDA levels suggested that 4-HC increases the oxidative stress levels in the HFSCs (Fig. 4E). Collectively, the results of in vitro cell experiments indicated that 4-HC intervention causes a gradual loss of proliferative capacity and "stemness" characteristics in the HFSCs.
Fig. 4.
Isolation and identification of the primary hair follicle stem cells (HFSCs) in vitro. (A) Photographs of HFSCs from the WT group and the 4-HC group. (B) Cell cycle results for the WT group and the 4-HC group. (C) Expression level changes of mHFSC markers. (D) qRT-PCR detection of the expression of apoptosis-related genes in the WT group and the 4-HC group. (E) SOD and MDA detection reflecting changes in oxidative stress in the WT group and the 4-HC group. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
3.3. Simplified methylation sequencing analysis
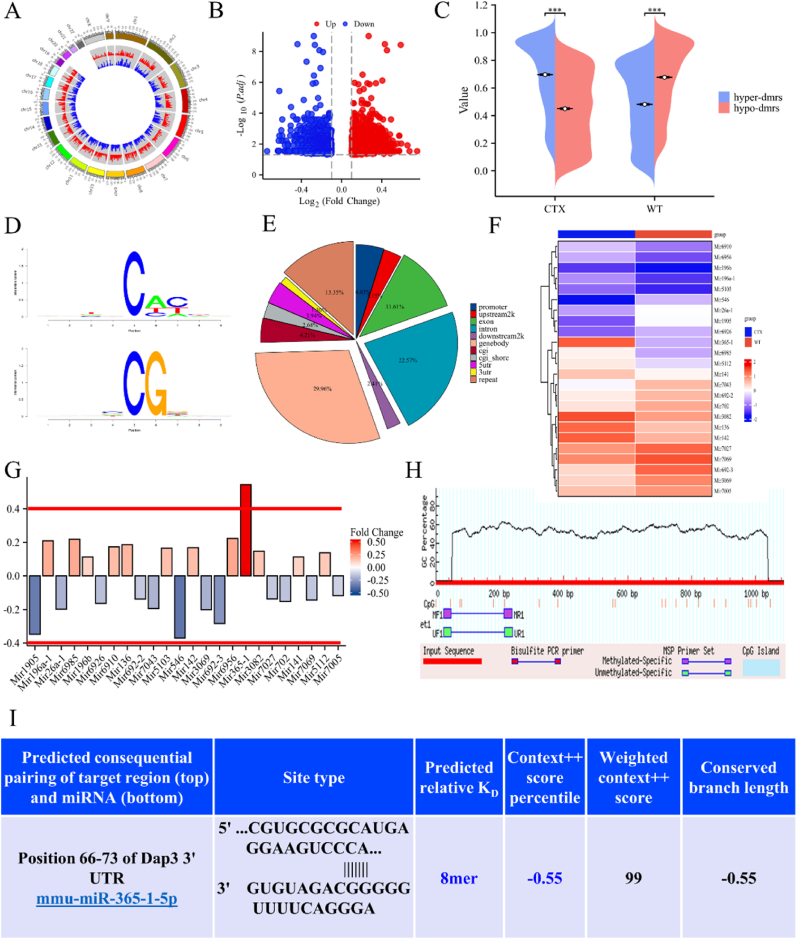
The differences in DNA methylation between the WT and CTX groups were analyzed at the chromosome level through RRBS (Fig. 5A). Fig. 5B presents the different methylated genes in the WT and CTX groups, including 122 hypermethylated genes (logFC ≥ 0.4) and 145 hypomethylated genes (logFC ≤ 0.4). The occurrence of DNA hypermethylation was more in the CTX group, whereas DNA hydroxymethylation was mainly noted in the WT group (Fig. 5C). Methylation site loci were identified by analyzing the characteristics of upstream and downstream sequences of the methylated cytosines in the CG, CHG and CHH backgrounds. Some commonality was noted between the two sample groups, with CG having a CG motif, CHG having a C (C/A/T) GG (A/C) motif, and CHH having a C (A/C/T) (C/T/A) motif (Fig. 5D). On investigating the differential methylation regions (DMRs) in genomic regions, we noted that compared with the WT group, differential methylation of genomes accounted for 29.96% of the total methylation in the CTX group, DMR methylation of introns accounted for 22.57%, and the proportion of DMRs in the remaining regions ranged from 2% to 20% (Fig. 5E). An increasing number of studies have reported microRNAs regulate hair follicle cell development either through direct regulation by the transcription factor AR or by blocking cell senescence and death. By contrast, relatively few studies have examined the association between DNA methylation and microRNAs in hair loss [[22], [23], [24]]. As shown in Fig. 5F, all miRNAs that underwent DNA methylation were screened, and mir-365-1 was identified as the most differentially expressed miRNA that underwent the highest degree of DNA methylation (Fig. 5G). We referred to the database MethPrimer to predict the CpG island of mir-365-1 with a 214-bp promoter length (Fig. 5H). The target DAP3 of mir-365-1 was then predicted using the Targetscan website, and its binding sequences were noted (Fig. 5I).
Fig. 5.
Simple methylation sequencing of mouse dorsal skin. (A) The Circos plot illustrating significant differences in DMRs among the groups. (B) The volcano plot of differentially methylated genes between the WT and CTX groups. (C) The Circos plot illustrates significant differences in DMRs between the WT and CTX groups. (D) Identification of motifs at methylation sites. (E) Distribution of differential methylation levels of DMRs in genomic regions for each group. (F) The cluster heatmap displays the miRNA methylation levels in each group. (G) Logarithmic fold change differences in the levels of all miRNAs. (H) The MethPrimer website predicts the methylation sites and promoter sequence of mir-365-1. (I) The Targetscan website predicts the target gene DAP3 of mir-365-1 and its binding sites.
3.4. High methylation of the miR-365-1 promoter region promotes DAP3 expression
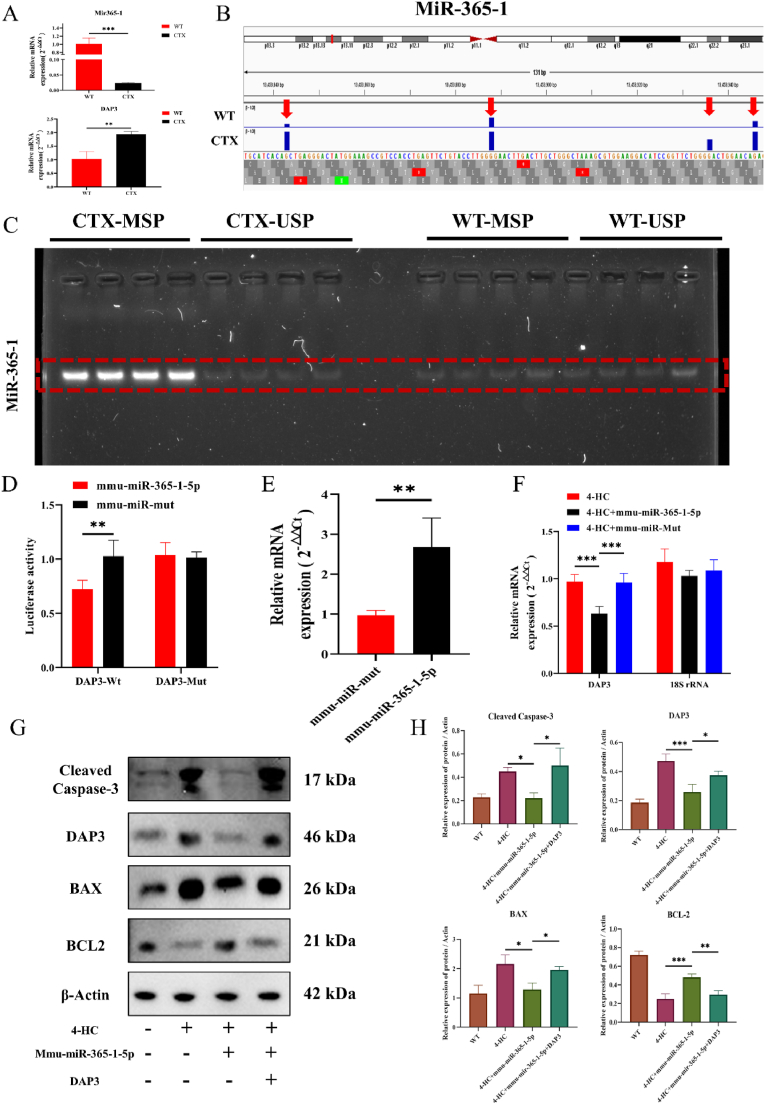
Based on the bioinformatics analysis results, we first confirmed that miR-365-1 expression was downregulated and DAP3 expression was upregulated in the CTX-induced mouse back skin (Fig. 6A). Integrative Genomics Viewer (IGV) analysis revealed multiple highly methylated sites in the miR-365-1 promoter region in the CTX group (Fig. 6B). Subsequently, methylation-specific PCR (MS-PCR) validation confirmed a significant increase in methylation of miR-365-1 in the back skin of CTX-treated mice (Fig. 6C). Moreover, luciferase reporter experiments unveiled a regulatory relationship between miR-365-1 and DAP3 (Fig. 6D). miR-365-1 overexpression was confirmed through qRT-PCR (Fig. 6E). After miR-365-1 was overexpressed, qRT-PCR revealed that DAP3 expression significantly downregulated, suggesting a regulatory relationship between DAP3 and miR-365-1 (Fig. 6F). Subsequently, a DAP3-overexpressing plasmid was constructed to validate the impact of DAP3 and miR-365-1 on HFSC apoptosis (Fig. 6G). According to western blotting results, the expression of apoptosis-related proteins in mHSFCs significantly decreased after miR-365-1 overexpression. However, this effect was reversed with the addition of the DAP3 overexpression vector (Fig. 6H).Therefore, these results indicate a direct regulatory relationship between miR-365-1 and DAP3, suggesting that they have a certain regulatory effect on the phenotype of HSFC apoptosis.
Fig. 6.
Methylation identification of miR-365-1 and its direct regulatory relationship with DAP3. (A) qRT-PCR detection of differences in the expression of mir-365-1 and DAP3 in the dorsal skin of mice from the WT and CTX groups. (B) IGV software identification of the methylation sites of mir-365-1. (C) MS-PCR identification of the highly methylated state in the dorsal skin of mice from the CTX group. (D) The luciferase reporter gene experiment identifying the binding relationship between mir-365-1 and DAP3. (E) qRT-PCR reveals that mmu-miR-365-1 can successfully overexpress mir-365-1. (F) qRT-PCR unveils that mir-365-1 overexpression significantly reduces DAP3 expression. (G) Western blot detection of changes in the expression of apoptosis-related genes under the influence of 4-HC, mmu-miR-365-1, and DAP3-overexpressing plasmids. (H) Statistical chart of protein expression determined through western blotting. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001.
4. Discussion
Hair loss is a common clinical problem, mostly among cancer patients receiving chemotherapy [25,26]. Although some studies have investigated about hair loss, the specific molecular mechanisms underlying this problem remain unclear. Therefore, this study was conducted to gain a deeper understanding of the role of DNA methylation in hair loss and the underlying potential mechanisms. By establishing the mouse and cell models, we achieved a series of crucial experimental results.
Studies on chemotherapy-induced hair loss have predominantly focused on the mechanisms such as cell apoptosis and stem cell damage. For instance, Purba TS et al. found that CDK4/6 inhibitors could reduce stem cell damage in a paclitaxel-induced hair loss model [27]. Piccini et al. reported the protective effects of peroxisome proliferation-activated receptor γ signal transduction on HFSCs against chemotherapy, thereby preventing cell apoptosis and epithelial–mesenchymal transition [28]. Various targeted compounds have been designed against different targets. For example, Yang et al. designed a TLR7 agonist for the treatment of chemotherapy-induced hair loss [29]. However, no current research specifically addresses the relationship between chemotherapy-induced hair loss and DNA methylation.
According to our results, DNA methylation may be among the crucial regulatory factors for hair loss. It is an epigenetic modification process widely involved in regulating gene expression, and its role in hair loss may be associated with the regulation of multiple key genes. We also identified the regulatory relationship between miR-365-1 and DAP3. RRBS results revealed miR-365-1 as the most significantly differentially expressed miRNAs and DAP3 as its target gene. This association suggests that the miR-365-1/DAP3 pathway plays a critical role in hair loss. DAP3 plays a major role in mitochondrial function and is also associated with the functional changes observed in the mitochondria. Our results indicate signs of oxidative stress in the CTX-induced hair loss model. SOD and MDA are vital biomolecules involved in cellular oxidative stress and free radical reactions. SOD targets superoxide radicals, catalyzes the reduction reaction of these radicals, and converts them to oxygen molecules and hydrogen peroxide, thereby protecting cells from superoxide radical damage and oxidative stress [30]. MDA, on the other hand, is a lipid peroxide and is often considered a hallmark of free radical-induced lipid oxidation [31]. The decreased SOD levels and the increased MDA levels suggest that oxidative stress is involved in hair loss. Oxidative stress-induced changes in the mitochondrial function is also a key finding of our study. The decreased MMP and the increased mitochondrial ROS levels may induce oxidative damage and apoptosis, which possibly contribute eventually to hair loss.
This study enhances our understanding of the hair loss mechanism and provides crucial clues for future hair loss treatment strategies. First, DNA methylation may be a promising target for hair loss intervention. By regulating the DNA methylation levels, the expression of affected genes may be restored. Second, the miR-365-1/DAP3 pathway and mitochondrial function may serve as key therapeutic targets. By modulating the activity of these pathways, hair loss severity may be alleviated. However, our study also has certain flaws; for example, we only conducted luciferase experiments to validate the relationship between miR-365-1-5p and DAP3 and did not validate the knockdown for their binding sites. For DNA methyltransferases specifically regulating DNA methylation of miR-365-1-5p, only 5-AzaC, a universal inhibitor of DNA methyltransferases, was used for blocking validation. We failed to target specific proteins. Our subsequent studies will remedy these deficiencies.
In summary, our study reveals the roles of multiple factors, including DNA methylation, the miR-365-1/DAP3 pathway, and oxidative stress, in hair loss, thereby providing strong scientific evidence for further exploring the mechanisms underlying hair loss and developing treatment strategies. These findings are expected to offer more effective treatment options for hair loss patients and open up new research avenues on DNA methylation- and mitochondrial function-related diseases. Future studies must explore the detailed regulatory networks of these mechanisms and the development of potential therapeutic drugs and interventions.
Funding
Innovative Team Projects of Shanghai Municipal Commission of Health (2022CX011), Young Qi-Huang Scholar of National Administration of TCM, Shanghai Municipal Health Commission (20214Y0175), Evidence-Based Capacity Building for TCM Specialty Therapies for Skin Diseases of National Administration of TCM, High-level Chinese Medicine Key Discipline Construction Project (Integrative Chinese and Western Medicine Clinic) of National Administration of TCM (zyyzdxk-2023065), 2023 Shanghai University of Traditional Chinese Medicine "Cultivation Program for Outstanding Doctoral Students in Key Areas" (GJ2023028), Sailing Program of Shanghai Rising-Star Program (22YF1449700), and National Natural Science Foundation of China (82074428).
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability
The full complement of data accumulated for these studies is available upon request.
CRediT authorship contribution statement
Xin Liu: Writing – review & editing, Writing – original draft, Resources, Data curation, Conceptualization. Ruofan Xi: Data curation. Xinran Du: Conceptualization. Yi Wang: Data curation. Linyan Cheng: Resources. Ge Yan: Writing – original draft. Jianyong Zhu: Writing – review & editing. Te Liu: Writing – review & editing. Fulun Li: Writing – review & editing, Funding acquisition.
Acknowledgments
We acknowledge the support of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine and Shanghai Geriatric Institute of Chinese Medicine.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2024.03.001.
Contributor Information
Te Liu, Email: liute1979@shutcm.edu.cn.
Fulun Li, Email: drlifulun@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Lee H.H., et al. Epidemiology of alopecia areata, ophiasis, totalis, and universalis: a systematic review and meta-analysis. J. Am. Acad. Dermatol. 2020;82(3):675–682. doi: 10.1016/j.jaad.2019.08.032. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg L.J. Alopecia - new building blocks. J. Am. Acad. Dermatol. 2023;89(2S):S1–S2. doi: 10.1016/j.jaad.2023.05.048. [DOI] [PubMed] [Google Scholar]
- 3.Alhanshali L., et al. Medication-induced hair loss: an update. J. Am. Acad. Dermatol. 2023;89(2S):S20–S28. doi: 10.1016/j.jaad.2023.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Sikora M., Rudnicka L. Chemotherapy-induced alopecia - the urgent need for treatment options. J. Eur. Acad. Dermatol. Venereol. 2019;33(2):e69–e70. doi: 10.1111/jdv.15207. [DOI] [PubMed] [Google Scholar]
- 5.Yin L., et al. Dermatologist awareness of scalp cooling for chemotherapy-induced alopecia. J. Am. Acad. Dermatol. 2023;88(1):176–179. doi: 10.1016/j.jaad.2022.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Basmanav F.B., Nothen M.M., Betz R.C. Insights into the biology of persistent chemotherapy-induced alopecia via genomic approaches-an avenue to clinical translation? JAMA Dermatol. 2020;156(9):947–948. doi: 10.1001/jamadermatol.2020.1866. [DOI] [PubMed] [Google Scholar]
- 7.Marx V. Genetics: profiling DNA methylation and beyond. Nat. Methods. 2016;13(2):119–122. doi: 10.1038/nmeth.3736. [DOI] [PubMed] [Google Scholar]
- 8.DNA methylation provides molecular links underlying complex traits. Nat. Genet. 2023;55(1):12–13. doi: 10.1038/s41588-022-01249-y. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Relevance of DNA methylation in the management of cancer. Lancet Oncol. 2003;4(6):351–358. doi: 10.1016/s1470-2045(03)01115-x. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y., et al. Epigenetic regulation in cardiovascular disease: mechanisms and advances in clinical trials. Signal Transduct. Targeted Ther. 2022;7(1):200. doi: 10.1038/s41392-022-01055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood H. Neurodegenerative disease: altered DNA methylation and RNA splicing could be key mechanisms in Huntington disease. Nat. Rev. Neurol. 2013;9(3):119. doi: 10.1038/nrneurol.2013.23. [DOI] [PubMed] [Google Scholar]
- 12.Chen G., et al. Protection against cyclophosphamide-induced alopecia and inhibition of mammary tumor growth by topical 1,25-dihydroxyvitamin D3 in mice. Int. J. Cancer. 1998;75(2):303–309. doi: 10.1002/(sici)1097-0215(19980119)75:2<303::aid-ijc21>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 13.Kim C.S., et al. Glutamine metabolism controls stem cell fate reversibility and long-term maintenance in the hair follicle. Cell Metabol. 2020;32(4):629–642 e8. doi: 10.1016/j.cmet.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Steele C.D., et al. Signatures of copy number alterations in human cancer. Nature. 2022;606(7916):984–991. doi: 10.1038/s41586-022-04738-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai L., et al. Bovine serum albumin aggravates macrophage M1 activation and kidney injury in heterozygous Klotho-deficient mice via the gut microbiota-immune axis. Int. J. Biol. Sci. 2021;17(3):742–755. doi: 10.7150/ijbs.56424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L., et al. ACSS3 represses prostate cancer progression through downregulating lipid droplet-associated protein PLIN3. Theranostics. 2021;11(2):841–860. doi: 10.7150/thno.49384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mokabber H., Najafzadeh N., Mohammadzadeh Vardin M. miR-124 promotes neural differentiation in mouse bulge stem cells by repressing Ptbp1 and Sox9. J. Cell. Physiol. 2019;234(6):8941–8950. doi: 10.1002/jcp.27563. [DOI] [PubMed] [Google Scholar]
- 18.Du K.T., et al. MiR-214 regulates the human hair follicle stem cell proliferation and differentiation by targeting EZH2 and wnt/beta-catenin signaling way in vitro. Tissue Eng Regen Med. 2018;15(3):341–350. doi: 10.1007/s13770-018-0118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye M., et al. LncRNA NALT1 promotes colorectal cancer progression via targeting PEG10 by sponging microRNA-574-5p. Cell Death Dis. 2022;13(11):960. doi: 10.1038/s41419-022-05404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue K., et al. Differential expression of stem-cell-associated markers in human hair follicle epithelial cells. Lab. Invest. 2009;89(8):844–856. doi: 10.1038/labinvest.2009.48. [DOI] [PubMed] [Google Scholar]
- 21.Kloepper J.E., et al. Immunophenotyping of the human bulge region: the quest to define useful in situ markers for human epithelial hair follicle stem cells and their niche. Exp. Dermatol. 2008;17(7):592–609. doi: 10.1111/j.1600-0625.2008.00720.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., et al. MiR-199a-3p regulates the PTPRF/beta-Catenin Axis in hair follicle development: insights into the pathogenic mechanism of alopecia areata. Int. J. Mol. Sci. 2023;24(24) doi: 10.3390/ijms242417632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li K., et al. The AR/miR-221/IGF-1 pathway mediates the pathogenesis of androgenetic alopecia. Int. J. Biol. Sci. 2023;19(11):3307–3323. doi: 10.7150/ijbs.80481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae S., et al. Arctiin blocks hydrogen peroxide-induced senescence and cell death though microRNA expression changes in human dermal papilla cells. Biol. Res. 2014;47(1):50. doi: 10.1186/0717-6287-47-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haque E., et al. Management of chemotherapy-induced alopecia (CIA): a comprehensive review and future directions. Crit. Rev. Oncol. Hematol. 2020;156 doi: 10.1016/j.critrevonc.2020.103093. [DOI] [PubMed] [Google Scholar]
- 26.Dunnill C.J., et al. A clinical and biological guide for understanding chemotherapy-induced alopecia and its prevention. Oncol. 2018;23(1):84–96. doi: 10.1634/theoncologist.2017-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purba T.S., et al. CDK4/6 inhibition mitigates stem cell damage in a novel model for taxane-induced alopecia. EMBO Mol. Med. 2019;11(10) doi: 10.15252/emmm.201911031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piccini I., et al. Peroxisome proliferator-activated receptor-gamma signalling protects hair follicle stem cells from chemotherapy-induced apoptosis and epithelial-mesenchymal transition. Br. J. Dermatol. 2022;186(1):129–141. doi: 10.1111/bjd.20745. [DOI] [PubMed] [Google Scholar]
- 29.Yang J., et al. Design, synthesis, and biological activity of TLR7-based compounds for chemotherapy-induced alopecia. Invest. N. Drugs. 2020;38(1):79–91. doi: 10.1007/s10637-019-00793-5. [DOI] [PubMed] [Google Scholar]
- 30.Sheng Y., et al. Superoxide dismutases and superoxide reductases. Chem Rev. 2014;114(7):3854–3918. doi: 10.1021/cr4005296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams A., De Kimpe N., van Boekel M.A. Modification of casein by the lipid oxidation product malondialdehyde. J. Agric. Food Chem. 2008;56(5):1713–1719. doi: 10.1021/jf072385b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full complement of data accumulated for these studies is available upon request.