Abstract
Schistosomiasis is a neglected tropical disease (NTD) caused by infection with parasitic trematodes of the genus Schistosoma that can lead to debilitating morbidity and mortality. The World Health Organization recommend molecular xenomonitoring of Biomphalaria spp. freshwater snail intermediate hosts of Schistosoma mansoni to identify highly focal intestinal schistosomiasis transmission sites and monitor disease transmission, particularly in low-endemicity areas. A standardised protocol to do this, however, is needed. Here, two previously published primer sets were selected to develop and validate a multiplex molecular xenomonitoring end-point PCR assay capable of detecting S. mansoni infections within individual Biomphalaria spp. missed by cercarial shedding. The assay proved highly sensitive and highly specific in detecting and amplifying S. mansoni DNA and also proved highly sensitive in detecting and amplifying non-S. mansoni trematode DNA. The optimised assay was then used to screen Biomphalaria spp. collected from a S. mansoni-endemic area for infection and successfully detected S. mansoni infections missed by cercarial shedding as well as infections with non-S. mansoni trematodes. The continued development and use of molecular xenomonitoring assays such as this will aid in improving disease control efforts, significantly reducing disease-related morbidities experienced by those in schistosomiasis-endemic areas.
Keywords: Intestinal schistosomiasis, Biomphalaria, Molecular xenomonitoring, Molecular epidemiology, Disease transmission monitoring, Disease control and elimination
Graphical abstract
Highlights
-
•
High throughput, easily interpreted, and reliable multiplex end-point PCR molecular xenomonitoring assay developed.
-
•
This assay enables detection of Schistosoma mansoni infections in Biomphalaria spp. missed by cercarial shedding.
-
•
The assay is also able to detect and identify non-S. mansoni trematode infections in Biomphalaria spp.
-
•
Through the use of this assay, sites of active intestinal schistosomiasis transmission can be identified.
1. Introduction
Schistosomiasis is a neglected tropical disease (NTD) caused by infection with parasitic trematodes of the genus Schistosoma that can lead to debilitating morbidity and mortality (Colley et al., 2014). Whilst it is estimated that over 230 million people are currently infected globally, over 90% of all cases occur within sub-Saharan Africa (McManus et al., 2018). Of these, around one-third are deemed intestinal schistosomiasis, caused predominantly by infection with Schistosoma mansoni but also less commonly by infection with Schistosoma intercalatum and Schistosoma guineensis in some restricted areas of central Africa (Tchuem Tchuenté et al., 2017). Schistosoma mansoni is transmitted via obligate freshwater snail intermediate hosts of the genus Biomphalaria (Gastropoda: Planorbidae) (Morgan et al., 2001). Human infection occurs through contact with contaminated bodies of freshwater, where infectious S. mansoni larval stages (cercariae) shed from infected Biomphalaria spp. hosts contact their definitive human host and penetrate the skin. As such, intestinal schistosomiasis is often highly prevalent in rural areas lacking adequate water, sanitation, and hygiene (WASH) infrastructure, where entire communities rely on bodies of freshwater as a source of drinking water, food (via fishing), a place to bathe, a place to tend livestock, and for recreation (Grimes et al., 2015).
Intestinal schistosomiasis is typically diagnosed using Kato-Katz faecal-egg microscopy (Katz et al., 1972; WHO, 2022). Whilst this method is relatively inexpensive and can be carried out at the point-of-care, it is considered low-throughput and lacks sensitivity, particularly when attempting to diagnose individuals harbouring low-intensity infections (Utzinger et al., 2015). Reliably monitoring disease transmission in areas of low endemicity, such as those that have undergone repeated annual or bi-annual mass drug administration (MDA) with the anthelmintic drug praziquantel (Knopp et al., 2011) or in areas where a recent outbreak of infections has occurred (Kayuni et al., 2020), can therefore be extremely difficult using this method alone. Because of these difficulties, detecting and monitoring the transmission of intestinal schistosomiasis is often also carried out through malacological surveillance of Biomphalaria spp. freshwater snail intermediate hosts followed by cercarial shedding analysis to identify sites of active transmission. This method, however, is also extremely insensitive, primarily because not all freshwater snails harbouring mature, or patent, infections will be actively shedding cercariae and because freshwater snails harbouring prepatent infections will also not shed cercariae (Tavalire et al., 2016; Weerakoon et al., 2018; Kamel et al., 2021). In addition, cercarial shedding can often be misleading owing to morphological similarities between human-infecting and non-human-infecting trematode cercariae. For example, S. mansoni and Schistosoma rodhaini (a morphologically indistinguishable rodent-infecting sister species of S. mansoni) are both transmitted by Biomphalaria spp., and so molecular methods of differentiating these species are needed (Norton et al., 2008; Lu et al., 2016).
An alternative approach used to detect vector-borne pathogens within their host vectors is through molecular xenomonitoring. This method involves the detection and amplification of pathogen DNA from within host vectors typically through some variety of polymerase chain reaction (PCR) (Lu et al., 2016; Schols et al., 2019; Pennance et al., 2020), but also through more novel isothermal nucleic acid amplification technologies such as loop-mediated isothermal amplification (LAMP) (García-Bernalt Diego et al., 2021) or recombinase polymerase/aided amplification (RPA/RAA) (Mesquita et al., 2022). Molecular xenomonitoring has been successfully used to detect and monitor the transmission of several vector-borne parasitic diseases, including malaria (Cameron and Ramesh, 2021), lymphatic filariasis and onchocerciasis (Pilotte et al., 2017), trypanosomiasis (Grébaut et al., 2016), fascioliasis (Rathinasamy et al., 2018), urogenital schistosomiasis (Schols et al., 2019; Pennance et al., 2020) and intestinal schistosomiasis (Lu et al., 2016; Andrus et al., 2023). As such, the World Health Organisation (WHO) has recommended the continued development and implementation of molecular xenomonitoring approaches to identify highly focal sites of ongoing schistosomiasis transmission, particularly in low-transmission areas nearing disease elimination (WHO, 2022). In doing so, more impactful disease control measures can be implemented, such as ceasing costly MDA programmes in areas that would provide no health benefit and concentrating resources in areas where ongoing transmission has been identified.
However, molecular xenomonitoring approaches can be expensive and require sophisticated laboratory infrastructure as well as specialised personnel. In addition, as so few (usually < 5%) of collected freshwater snails are typically found to be harbouring Schistosoma spp. infections when using molecular xenomonitoring approaches, many freshwater snails (ideally > 50 from each malacological surveillance site) is required to thoroughly assess any given location for evidence of schistosomiasis transmission. When developing molecular xenomonitoring methods, the overall assay cost, ability to be implemented in endemic areas, throughput, ease-of-use, speed, reliability, and ease of interpretation should therefore be carefully considered (Kamel et al., 2021). This includes consideration towards the essential preliminary steps needed to isolate DNA from freshwater snail tissues (Adema, 2021).
Here, we aimed to develop a high-throughput, easily interpreted, and reliable molecular xenomonitoring assay for the detection of S. mansoni and other trematode species within Biomphalaria spp. freshwater snail hosts. Whilst previous molecular xenomonitoring assays have been used to detect S. mansoni within Biomphalaria spp. hosts (Lu et al., 2016), these assays do not detect other species of trematode which may influence S. mansoni development within Biomphalaria spp., therefore impacting disease transmission, nor do they use an internal DNA extraction/PCR control, meaning a negative result may be interpreted as non-infection rather than DNA extraction or PCR reaction failure (Pennance et al., 2020). We therefore opted for a multiplex PCR approach capable of also detecting non-S. mansoni trematodes whilst also incorporating an internal reaction control (a Biomphalaria DNA locus). In addition, we used a rapid, automated, and high-throughput DNA extraction methodology together with a high-throughput and easily interpreted end-point PCR approach.
2. Materials and methods
2.1. Biomphalaria spp. freshwater snails and Schistosoma spp. trematodes used during assay development, validation, and pilot application
Biomphalaria spp. freshwater snails and Schistosoma spp. trematodes (adult worms, miracidia, and ova) used during assay development and validation were provided by the Schistosome and Snail Resource (SSR) (SSR, 2024), the Schistosomiasis Collection at the Natural History Museum (SCAN) archive (Emery et al., 2012) and the Biomedical Research Institute (BRI) (BRI, 2024a, BRI, 2024b). In addition, 92 Biomphalaria spp. snails used during assay pilot application had been previously collected from a single malacological surveillance site situated on the southern shoreline of Lake Malawi, Mangochi District, Malawi (an area endemic for S. mansoni transmission; Kayuni et al., 2020) as part of the Hybridisations in UroGenital Schistosomiasis (HUGS) project (Archer et al., unpublished). Details of all Biomphalaria and Schistosoma material including sample IDs, collection location or laboratory strain, and DNA extraction method are outlined in Table 1.
Table 1.
Biomphalaria and Schistosoma material used during development, validation, and pilot application of the molecular xenomonitoring end-point PCR assay.
| Sample ID | Specimen | Collection location or laboratory strain | S. mansoni infection status | DNA extraction method | Provided by |
|---|---|---|---|---|---|
| Bp | B. pfeifferi (n = 1) | Collected: Lake Victoria, Uganda | Non-infected | DNeasy blood and tissue kit (Qiagen, UK) using standard protocol (Pennance et al., 2018) with minor modificationsa | SSR |
| Bs | B. sudanica (n = 1) | Collected: Lake Victoria, Tanzania | Non-infected | SCAN | |
| Bc | B. choanomphala (n = 1) | Collected: Lake Victoria, Tanzania | Non-infected | SCAN | |
| Sm | S. mansoni adult worms (n = 3, collectively extracted) | Laboratory passaged: Puerto Rico strain | Not applicable | DNeasy blood and tissue kit (Qiagen, UK) using standard protocol (Webster et al., 2012) | SSR |
| Sr | S. rodhaini adult worms (n = 3, collectively extracted) | Laboratory passaged: Uganda strain | Not applicable | SSR | |
| Sh | S. haematobium adult worms (n = 3, collectively extracted) | Laboratory passaged: Senegal strain | Not applicable | SCAN | |
| Sm-mir(1) | S. mansoni miracidia (n = 6, individually extracted) | Laboratory passaged: Puerto Rico strain | Not applicable | DNeasy blood and tissue kit (Qiagen, UK) using standard protocol (Webster et al., 2012) with minor modificationsb | BRI |
| Bg+Sm(1) | B. glabrata exposed to one S. mansoni miracidium (n = 5, individually extracted) | Laboratory-bred: Brazil (BB02) strain, originally collected in 2002 | Infected: Control laboratory infection with one S. mansoni miracidium (Puerto Rico strain) (Section 2.2.4.1) | DNeasy blood and tissue kit (Qiagen, UK), 24 h post miracidia exposure using standard protocol (Pennance et al., 2018) with minor modificationsa | B. glabrata provided by SSR; S. mansoni miracidia provided by BRI |
| Bg+Sm(5) | B. glabrata exposed to five S. mansoni miracidia (n = 5, individually extracted) | Infected: Control laboratory infection with 5 S. mansoni miracidia (Puerto Rico strain) (Section 2.2.4.1) | |||
| Bg | B. glabrata (n = 2 individually extracted) | Non-infected (Section 2.2.4.1) | SSR | ||
| Biomph91(mal) | Biomphalaria spp. (n = 91, individually extracted) | Collected: Lake Malawi, Malawi (Section 2.3.1) | Unknown: Not shedding S. mansoni cercariae | BioSprint 96 workstation and BioSprint 96 DNA Blood Kit (Qiagen, UK) using standard protocol (Pennance et al., 2020) | Not applicable |
| Bp+Sm(mal) | B. pfeifferi infected with S. mansoni (n = 1) | Collected: Lake Malawi, Malawi (Section 2.3.1) | Infected: Natural infection, shedding S. mansoni cercariae | Not applicable |
Abbreviations: SSR, Schistosome and Snail Resource, London, UK (SSR, 2024); SCAN, Schistosome Collections at Natural History Museum, London, UK (Emery et al., 2012); BRI, Biomedical Research Institute, Maryland, USA.
Double volume cell lysis buffers used.
Half volume cell lysis buffers and elution buffer used.
2.2. Assay development and validation
2.2.1. Target DNA loci and primer selection
Two previously published primer sets were selected for development of the multiplex molecular xenomonitoring end-point PCR assay. The commonly used ETTS2 forward and ETTS1 reverse primers were selected to target and amplify the complete nuclear internal transcribed spacer (ITS) region of members of both Gastrapoda (∼1250 bp) and Trematoda (∼1005 bp) (Kane and Rollinson, 1994). This primer set has been used previously within a multiplex molecular xenomonitoring end-point PCR assay to detect trematode infections (including Schistosoma spp.) within Bulinus spp. freshwater snail hosts (Pennance et al., 2020). In addition, S. mansoni NADH dehydrogenase subunit 5 (ND5) forward and reverse primers were selected to target and amplify a 305-bp region of the S. mansoni mitochondrial ND5 gene (Lu et al., 2016). Details of all primer oligonucleotide sequences are provided in Table 2.
Table 2.
Primer sequences used during development, validation, and pilot application of the molecular xenomonitoring end-point PCR assay.
| Name | Target (amplicon length) | Oligonucleotide sequence (5′–3′) | Reference |
|---|---|---|---|
| ETTS2 (F) | Biomphalaria ITS (∼1250 bp) & Schistosoma ITS (∼1005 bp) | TGCTTAAGTTCAGCGGG | Kane and Rollinson (1994); Pennance et al. (2020) |
| ETTS1 (R) | TAACAAGGTTTCCGTAGGTGA | ||
| ND52 (F) | S. mansoni ND5 (305 bp) | ATTAGAGGCAATGCGTGCTC | Lu et al. (2016) |
| ND52 (R) | ATTGAACCAACCCCAAATCA |
Abbreviations: F, forward primer; R, reverse primer.
2.2.2. Specificity testing and multiplex assay optimisation
2.2.2.1. Singleplex Biomphalaria ITS and Schistosoma ITS PCR testing
To confirm amplification of the target Gastrapoda (Biomphalaria) ITS and Trematoda (Schistosoma) ITS loci, singleplex PCR reactions were performed using ETTS2 and ETTS1 primers and B. pfeifferi (Bp), B. sudanica (Bs), B. choanomphala (Bc), S. mansoni (Sm), S. rodhaini (Sr), S. haematobium (Sh) and B. pfeifferi infected with S. mansoni (Bp+Sm(mal)) sample DNA. These were carried out using 25 μl PCR reactions made up of 2 μl template DNA (2 ng/μl) from each DNA sample, 10 pmol of each primer, 21 μl ddH2O and one Illustra PuReTaq ready-to-go PCR bead (Sigma-Aldrich, Dorset, UK). One no-template negative control using 2 μl ddH2O in place of DNA was also used. Singleplex PCR reactions were performed using the following cycling conditions: 5 min at 95 °C; 40 cycles of 30 s at 95 °C, 30 s at 58 °C and 90 s at 72 °C; and 10 min at 72 °C. This was then repeated using an annealing temperature of 60 °C (rather than 58 °C) to check for improved target detection and amplification. Amplicons were visualised by running 7.5 μl PCR product mixed with 2 μl 5× loading buffer blue (Bioline, Essex, UK) stained with GelRed on a 2% agarose gel.
PCR products generated using Bp, Bs, Bc, Sm, Sr and Sh sample DNA were purified using the QIAquick PCR purification kit (Qiagen, Manchester, UK) according to the manufacturer’s instructions and Sanger sequenced in the forward direction using a dilution of the ETTS2 forward primer. PCR products generated using Bp+Sm(mal) sample DNA were visualised under UV light and both ∼1250-bp Biomphalaria and ∼1005-bp Schistosoma bands were individually excised using a fresh scalpel. Excised gel bands were individually purified using the QIAquick Gel purification kit (Qiagen) according to the manufacturer’s instructions and were then purified as described above prior to Sanger sequencing in the forward direction again using a dilution of the ETTS2 forward primer. Sequence data were visualised, trimmed, and edited using Geneious Prime version 2023.01 (Biomatters, LTD) and identified using the Basic Local Alignment Search Tool (BLAST) algorithm within the NCBI database (Geer et al., 2009).
2.2.2.2. Singleplex S. mansoni ND5 PCR testing
To confirm amplification of the target S. mansoni ND5 locus, the above singleplex PCRs were repeated using the S. mansoni ND5 forward and reverse primers in place of ETTS2 and ETTS1 primers. PCRs were again initially performed using an annealing temperature of 58 °C and then repeated using an annealing temperature of 60 °C to check for improved target detection and amplification. Amplicons were visualised as described above. All S. mansoni ND5 PCR products were then purified as described above, and Sanger sequenced in the forward direction using a dilution of the ND52 forward primer. Sequence data were visualised, trimmed, edited, and identified as described above.
2.2.2.3. Multiplex Biomphalaria ITS, Schistosoma ITS and S. mansoni ND5 PCR testing
Multiplex PCR reactions were then performed incorporating both ITS and S. mansoni ND5 primer sets using Bp, Sm, Sr, Sh and Bp+Sm(mal) sample DNA. Bp+Sm(mal) reactions were performed in triplicate. These were carried out using 25 μl PCR reactions made up of 2 μl template DNA (2 ng/μl) from each DNA sample, 10 pmol of each primer, 19 μl ddH2O and one Illustra PuReTaq ready-to-go PCR bead (Sigma-Aldrich). One no-template negative control using 2 μl ddH2O in place of DNA was also used. Multiplex PCR reactions were again initially performed using the following cycling conditions: 5 min at 95 °C; 40 cycles of 30 s at 95 °C, 30 s at 58 °C and 90 s at 72 °C; and 10 min at 72 °C, and then repeated using an annealing temperature of 60 °C (rather than 58 °C) to check for improved target detection and amplification. Amplicons were visualised as described above.
2.2.3. Multiplex assay sensitivity testing
2.2.3.1. Multiplex assay using a single S. mansoni miracidium
The multiplex PCR was performed as described above (annealing temperature 60 °C) using 2 μl S. mansoni miracidia (Sm-mir(1)) template DNA (2 ng/μl) extracted from one single S. mansoni miracidium provided by the BRI. This was carried out using all six individual Sm-mir(1) samples. The PCR assay included one positive control using 2 μl Sm sample DNA (2 ng/μl) and one no-template negative control using 2 μl ddH2O in place of DNA. Amplicons were visualised as described above.
2.2.3.2. Analytical limit-of-detection testing
Sm and Sr sample DNA was normalised to a concentration of 2 ng/μl and then serially diluted to 0.2 ng/μl, 0.02 ng/μl, and 2 pg/μl using ddH2O. Multiplex PCRs were then performed as described above (annealing temperature 60 °C) using 1 μl of each DNA concentration together with 1 μl Bp sample DNA (2 ng/μl). All reactions were performed in triplicate. The PCR assay included four positive controls: two using 2 μl Bp sample DNA (2 ng/μl), one using 2 μl Sm sample DNA (2 ng/μl) and one using 2 μl Sr sample DNA (2 ng/μl). Two no-template negative controls using 2 μl ddH2O in place of DNA were also used. Amplicons were visualised as described above.
2.2.4. Multiplex assay validation
2.2.4.1. Controlled laboratory infection of B. glabrata with S. mansoni miracidia
Twelve laboratory-bred B. glabrata freshwater snails (all ∼3 months old with a discoid shell height of ∼8 mm) were individually placed within plastic pots containing 30 ml ddH2O. A single S. mansoni miracidium was then pipetted into five of the pots. Five S. mansoni miracidia were then pipetted into a further five pots. Miracidia were not added to the two remaining pots. All twelve pots were then covered to ensure snail submersion and stored at 27 °C for 24 h, after which DNA was isolated from all B. glabrata whole tissues. All pots were examined under dissecting microscope to check for the presence or absence of miracidia immediately after B. glabrata were removed.
2.2.4.2. Molecular xenomonitoring PCR
The multiplex molecular xenomonitoring PCR was performed as described above (annealing temperature 60 °C) using all 12 B. glabrata DNA isolates. The PCR assay included two positive controls using 2 μl Bp sample DNA (2 ng/μl) and 2 μl Sm sample DNA (2 ng/μl). One no-template negative control using 2 μl ddH2O in place of DNA was also used. Amplicons were visualised as described above.
2.3. Assay pilot application
2.3.1. Collection and cercarial shedding of Biomphalaria from a S. mansoni-endemic area
Biomphalaria spp. previously collected from a single malacological surveillance site (October 2022) situated on the southern shoreline of Lake Malawi, Mangochi District, Malawi (an area endemic for S. mansoni transmission) were used during assay pilot application. All freshwater snails were collected according to a standard protocol and screened for patent Schistosoma spp. infection by cercarial shedding also according to a standard protocol (Pennance et al., 2022). All necessary personal protective equipment needed to prevent skin contact with contaminated freshwater during malacological collections and cercarial shedding was used at all times.
One of the collected specimens of Biomphalaria (0.7%) was found to be actively shedding Schistosoma spp. cercariae, and so this specimen was individually preserved fully submerged in 100% ethanol within a 2 ml screwcap tube. All remaining specimens of Biomphalaria were collectively preserved fully submerged in 100% ethanol within three 20 ml glass screwcap tubes and transported to the Natural History Museum (NHM), London, UK, under ambient conditions for DNA extraction (June 2023) and molecular analyses. DNA was isolated from each ethanol-preserved specimen of Biomphalaria in batches of 92 samples using the BioSprint 96 workstation and BioSprint 96 DNA Blood Kit (Qiagen) according to a previously outlined protocol (Pennance et al., 2020). This magnetic bead-based DNA extraction protocol was chosen as it allows for rapid and high-throughput multiple-sample processing using a 96-well plate format.
The shedding specimen of Biomphalaria was confirmed as B. pfeifferi through genotyping of the mitochondrial cox1 region according to a standard protocol (Standley et al., 2011; GenBank: OR880348) and the Schistosoma spp. cercariae shed from this specimen were confirmed as S. mansoni through genotyping of the mitochondrial cox1 and nuclear ITS DNA regions also according to a standard protocol (Webster et al., 2013; GenBank: PP390203 and PP388909, respectively).
2.3.2. Molecular xenomonitoring PCR
The multiplex molecular xenomonitoring PCR was used to screen a subset of the collected Biomphalaria DNA isolates for infection with S. mansoni and other trematodes. This subset was comprised of 92 Biomphalaria DNA isolates contained within a single 96-well plate that included the one Biomphalaria (B. pfeifferi) actively shedding S. mansoni cercariae (Bp+Sm(mal) sample DNA; Table 1) as well as 91 non-shedding Biomphalaria (Biomph91(mal) DNA samples; Table 1). The PCR was performed as described above (annealing temperature 60 °C) and included three positive controls using 2 μl Bp sample DNA (2 ng/μl), 2 μl Sm sample DNA (2 ng/μl), and 2 μl Sr sample DNA (2 ng/μl). One no-template negative control using 2 μl ddH2O in place of DNA was also used. Amplicons were visualised as described above.
Samples that amplified only the Biomphalaria ITS locus (considered an internal PCR control) were considered Biomphalaria negative for trematode infection of any sort, including S. mansoni. Samples that successfully amplified all three target loci were considered Biomphalaria positive for S. mansoni infection, as well as potentially other species of the Trematoda. Samples that amplified only the Biomphalaria and Trematoda ITS loci were considered Biomphalaria positive for trematode infection, excluding S. mansoni. Samples that failed to amplify the Biomphalaria ITS locus (regardless of Trematoda ITS and S. mansoni ND5 amplification outcome), were considered to have failed PCR and were repeated. Samples that failed to amplify this locus during the repeat screen were considered to have failed DNA extraction and were omitted from any further analysis.
2.3.3. Confirmatory S. mansoni ND5 PCR and genotyping
To confirm infection with S. mansoni and to validate our protocol, all samples that successfully amplified all three target loci were subjected to an additional singleplex PCR to again amplify the S. mansoni ND5 locus, as described above (Section 2.2.2.2; annealing temperature of 60 °C). Amplicons were visualised, purified, Sanger sequenced and identified as described above (Section 2.2.2.2).
2.3.4. Conformation and identification of Biomphalaria infections with other trematode species
To confirm infection with and identify non-S. mansoni trematode species infections, and to validate our protocol, all samples that successfully amplified both Biomphalaria and Trematoda ITS loci only were subjected to an additional singleplex PCR to again amplify the Biomphalaria ITS and Trematoda ITS loci, as described above (Section 2.2.2.1; annealing temperature of 60 °C). Agarose gel bands were then visualised under UV light and the ∼1005-bp Trematoda gel band was excised using a fresh scalpel. Excised gel bands were purified using the QIAquick Gel purification kit (Qiagen) according to the manufacturer’s instructions and were then purified as described above prior to Sanger sequencing in the forward direction (Section 2.2.2.1). Sequence data were visualised, trimmed, edited, and identified as described above (Section 2.2.2.1).
2.4. Ethical considerations
Live B. glabrata (Brazil BB02 strain; routinely maintained according to previously outlined standard operating procedures (BRI, 2024b) were provided by the Schistosome and Snail Resource (SSR, 2024) based across both the Natural History Museum (NHM), London, UK, and the London School of Hygiene and Tropical Medicine (LSHTM), London, UK (Wellcome Trust Biomedical Resource Grant 221368/Z/20/Z (2021–2026)). Controlled laboratory infection of B. glabrata with S. mansoni was performed at the NHM using S. mansoni (Puerto Rico strain) miracidia hatched from ova provided by the Biomedical Research Institute (BRI, 2024a), Rockville, Maryland, USA (Section 2.2.4.1). Ova were isolated and collected from the liver of infected mice following percutaneous tail exposure to S. mansoni cercariae (BRI, 2024a). Animal use was approved the Institutional Animal Care and Use Committee (IACUC) of the Biomedical Research Institute for the Animal Use Protocol, #18-01. Ethical approval and research authorisations for the collection of Biomphalaria spp. from the southern shoreline of Lake Malawi, Malawi, were approved in the UK by the Liverpool School of Tropical Medicine (LSTM) Research Ethics Committee (application 17-018) and in Malawi by the National Health Sciences Research Committee (1805).
3. Results
3.1. Assay development
3.1.1. Specificity testing and multiplex assay optimisation
3.1.1.1. Singleplex Biomphalaria ITS and Schistosoma ITS PCR testing
All Biomphalaria ITS and Schistosoma ITS singleplex reactions performed as anticipated with no non-target amplification. A single ∼1250-bp amplicon was produced when using Bp, Bs and Bc sample DNA, a single ∼1005-bp amplicon was produced when using Sm, Sr and Sh sample DNA, and two amplicons were produced when using Bp+Sm(mal) sample DNA (∼1250 bp and ∼1005 bp in length). Increasing the PCR annealing temperature to 60 °C did not appear to have any observable effect on PCR performance when compared to 58 °C. All Biomphalaria and Schistosoma ITS amplicons (using 60 °C annealing temperature) were successfully confirmed using Sanger sequencing.
3.1.1.2. Singleplex S. mansoni ND5 PCR testing
All S. mansoni ND5 singleplex reactions also performed as anticipated with no non-target amplification. A single 305-bp amplicon was produced when using Sm sample DNA and when using Bp+Sm(mal) sample DNA. No amplicons were produced when using Bp, Bs, Bc, Sr and Sh sample DNA. When using an annealing temperature of 58 °C, the 305-bp S. mansoni ND5 agarose gel bands were present but faint. By comparison, when increasing the annealing temperature to 60 °C, these agarose gel bands were much stronger. Both Sm and Bp+Sm(mal) ND5 amplicons (using an annealing temperature of 60 °C) were successfully confirmed using Sanger sequencing.
3.1.1.3. Multiplex Biomphalaria ITS, Schistosoma ITS and S. mansoni ND5 PCR testing
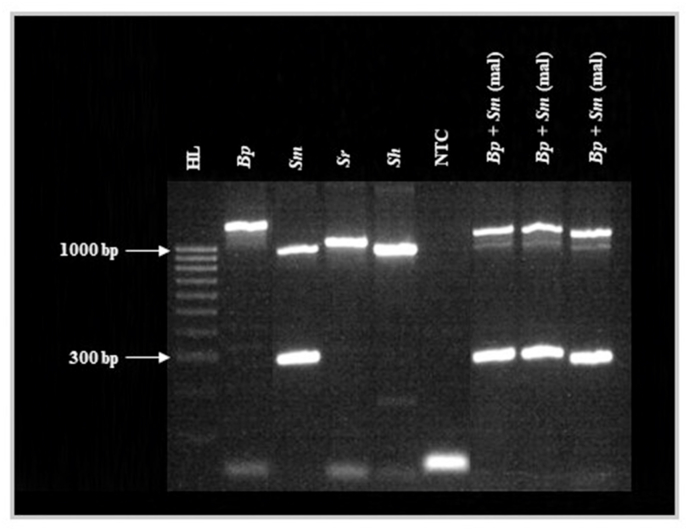
All multiplex reactions performed as anticipated with no non-target amplification. A single ∼1250-bp amplicon was produced when using Bp sample DNA. Two amplicons were produced when using Sm sample DNA (∼1005-bp and 305-bp in length). A single ∼1005-bp amplicon was produced when using Sr and Sh sample DNA. All three target amplicons were produced when using Bp+Sm(mal) sample DNA in all three replicates (∼1250 bp, ∼1005 bp and 305 bp in length; Fig. 1). Increasing the annealing temperature to 60 °C again improved the strength of the 305-bp S. mansoni ND5 agarose gel band in all reactions where S. mansoni DNA was present when compared to an annealing temperature of 58 °C, whilst having no observable effect on Biomphalaria ITS or Schistosoma ITS amplicons. When using both annealing temperatures, the Schistosoma ITS gel band was less strong in multiplex reactions using Bp+Sm(mal) sample DNA when compared to Biomphalaria ITS and Schistosoma ITS singleplex reactions using the same DNA and when using Sm, Sr and Sh sample DNA in multiplex reactions.
Fig. 1.
Agarose gel image of the multiplex molecular xenomonitoring PCR performed using an annealing temperature of 60 °C. Abbreviations: HL, HyperLadder 100 bp (BioLine, UK); Bp, Biomphalaria pfeifferi sample DNA; Sm, Schistosoma mansoni sample DNA; Sr, S. rodhaini sample DNA; Sh, S. haematobium sample DNA; NTC, no template ddH2O negative control; Bp+Sm(mal): B. pfeifferi infected with S. mansoni (collected in Malawi) sample DNA. All Biomphalaria ITS, Schistosoma ITS and S. mansoni ND5 multiplex reactions performed as anticipated with no non-target amplification. GelRed loading buffer stain may affect PCR amplicon migration.
3.1.2. Multiplex assay sensitivity testing
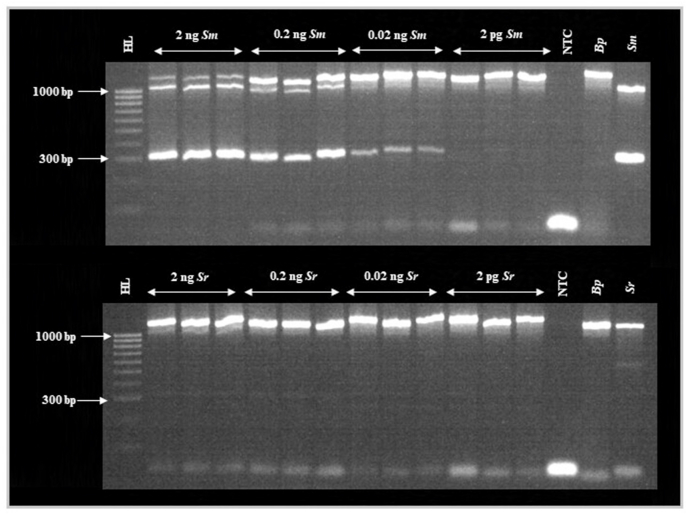
Both ∼1005-bp Schistosoma ITS and 305-bp S. mansoni ND5 amplicons were produced when using a single S. mansoni miracidium in all six Sm-mir(1) DNA samples. Furthermore, the multiplex assay proved capable of detecting S. mansoni DNA with a limit-of-detection of between 0.2 ng and 0.02 ng (Fig. 2). Whilst the strength of the Biomphalaria ITS agarose gel band remained constant in all reactions, the strength of the Schistosoma ITS gel band and the S. mansoni ND5 gel band decreased as S. mansoni DNA concentration decreased. The multiplex assay also proved capable of detecting S. rodhaini DNA with a limit-of-detection of between 2 ng and 0.2 ng (Fig. 2). Again, whilst the strength of the Biomphalaria ITS agarose gel band remained constant in all reactions, the strength of the Schistosoma ITS gel band decreased as S. rodhaini DNA concentration decreased. All control reactions performed as anticipated.
Fig. 2.
Agarose gel image of the multiplex molecular xenomonitoring PCR during sensitivity (analytical limit-of-detection) testing. Abbreviations: HL, HyperLadder 100 bp (BioLine, UK); Sm, Schistosoma mansoni sample DNA; Sr, S. rodhaini sample DNA; NTC, no template ddH2O negative control; Bp, Biomphalaria pfeifferi sample DNA. Whilst the strength of the Biomphalaria ITS agarose gel band remained constant in all reactions, as S. mansoni and S. rodhaini DNA concentration decreased, the strength of the Schistosoma ITS gel bands (both S. mansoni and S. rodhaini) and the S. mansoni ND5 gel band decreased. The multiplex assay proved capable of detecting S. mansoni DNA with a limit-of-detection of between 0.2 ng and 0.02 ng and proved capable of detecting S. rodhaini DNA with a limit-of-detection of between 2 ng and 0.2 ng. GelRed loading buffer stain may affect PCR amplicon migration.
3.1.3. Multiplex assay validation
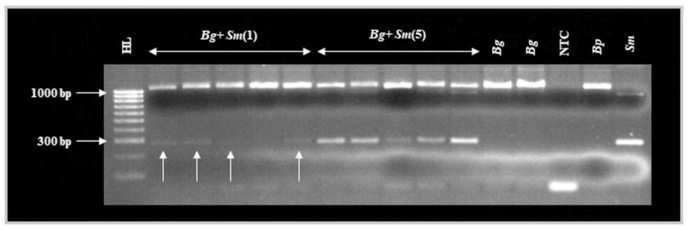
Upon microscopic examination, no miracidia appeared present in any pots immediately after removal of B. glabrata. The multiplex assay proved capable of detecting S. mansoni DNA within Biomphalaria exposed to a single miracidium 24 h post-exposure (Fig. 3). The S. mansoni ND5 locus was detected and visibly amplified (albeit, faintly) in four out of five B. glabrata exposed to a single miracidium. However, the Schistosoma ITS locus was not visibly amplified in any of these samples. The S. mansoni ND5 locus was detected and amplified in all five B. glabrata exposed to five miracidia 24 h post-exposure and the strength of these gel bands appeared stronger than those generated after exposure to a single miracidium 24 h post-exposure (Fig. 3). However, the Schistosoma ITS locus was detected and visibly amplified (albeit, faintly) in only three of these five samples. The Biomphalaria ITS locus was amplified in all 12 B. glabrata samples. All control reactions performed as anticipated.
Fig. 3.
Agarose gel image of the multiplex molecular xenomonitoring PCR during assay validation (controlled laboratory infection of B. glabrata with S. mansoni miracidia). Abbreviations: HL, HyperLadder 100 bp (BioLine, UK); Bg+Sm(1), sample DNA isolated from individual Biomphalaria glabrata exposed to a single miracidium of Schistosoma mansoni; Bg+Sm(5), sample DNA isolated from individual B. glabrata exposed to five miracidia of S. mansoni; Bg, B. glabrata sample DNA isolated from individual B. glabrata not exposed to miracidia of S. mansoni; NTC, no template ddH2O negative control; Bp, B. pfeifferi sample DNA; Sm, S. mansoni sample DNA. Schistosoma mansoni DNA was detected in four of all five Bg+Sm(1) samples (highlighted by upward-facing white arrows). Schistosoma mansoni DNA was also detected in all five Bg+Sm(5) samples. GelRed loading buffer stain may affect PCR amplicon migration.
3.2. Assay pilot application
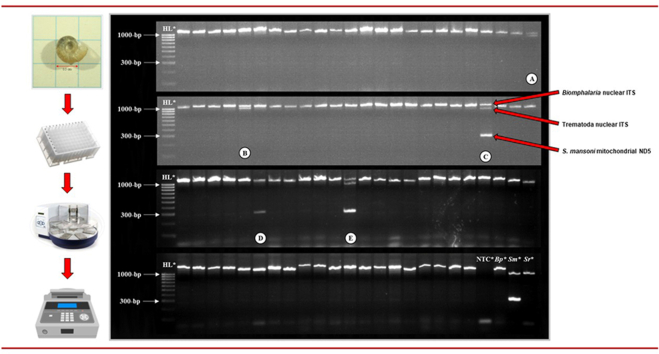
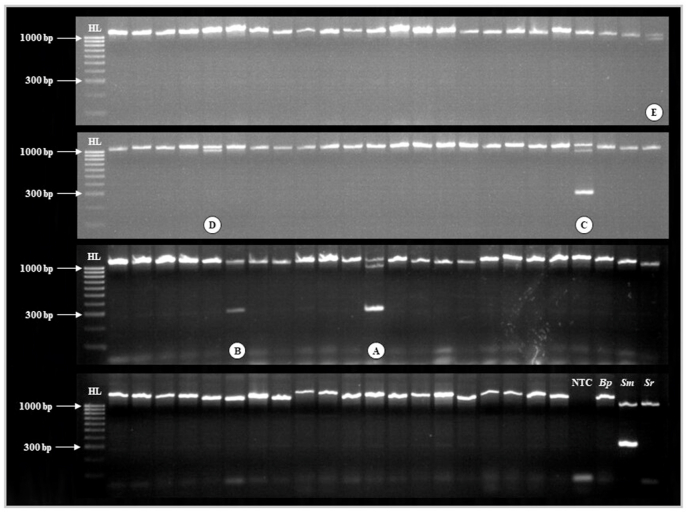
The Biomphalaria ITS locus was amplified in all Biomph91(mal) DNA samples as well as in the Bp+Sm(mal) DNA sample, and so DNA extraction and all PCR reactions were deemed successful (Fig. 4). All three target loci were amplified in the Bg+Sm(mal) sample DNA, as well as two Biomph91(mal) DNA samples, indicating infection with S. mansoni in three of all 92 (3.3%) specimens of Biomphalaria. In one of these Biompha91(mal) samples, the Schistosoma ITS locus was amplified only faintly (Fig. 4, sample B). In addition, both the Biomphalaria ITS and Trematoda ITS loci only were amplified in two Biomph91(mal) DNA samples, indicating infection with non-S. mansoni trematodes in two of all 92 (2.2%) specimens of Biomphalaria (Fig. 4). All control reactions performed as anticipated.
Fig. 4.
Agarose gel image of the high throughput multiplex molecular xenomonitoring PCR assay during pilot application. Abbreviations: HL, HyperLadder 100 bp (BioLine, UK); NTC, no template ddH2O negative control; Bp, Biomphalaria pfeifferi sample DNA; Sm, Schistosoma mansoni sample DNA; Sr, S. rodhaini sample DNA. A: Bp+Sm(mal) sample infected with S. mansoni (detected during cercarial shedding); B and C: Two Biomph91(mal) samples infected with S. mansoni (not detected during cercarial shedding); D and E: Two Biomph91(mal) samples infected with Uvulifer spp. All remaining samples are Biomph91(mal) samples not found to be infected with any trematode species, including S. mansoni. GelRed loading buffer stain may affect PCR amplicon migration.
Infection with S. mansoni was confirmed in all three S. mansoni-infected samples through singleplex PCR, Sanger sequencing and BLAST analysis of the S. mansoni ND5 locus, validating our molecular xenomonitoring protocol. The prevalence of S. mansoni infection in these 92 Biomphalaria freshwater snails was therefore increased from 1.1% to 3.3% through use of the multiplex molecular xenomonitoring PCR. Both trematode infections were confirmed as Uvulifer spp. (closest match GenBank: MK604882; mean percent identity score: 96.6; query cover 86.3%) through agarose gel excision, purification, Sanger sequencing and BLAST analysis of the Trematoda ITS locus. This diplostomid trematode has a three-host life-cycle: specific genera of freshwater snails including Biomphalaria (López-Hernández et al., 2019), freshwater sunfish, and piscivorous birds such as kingfishers (highly prevalent along this shoreline of Lake Malawi; Johnston, 1989).
4. Discussion
In areas of low disease endemicity, highly sensitive and reliable methods of detecting S. mansoni transmission are crucial for successful disease monitoring and control. Molecular xenomonitoring is a valuable and extremely sensitive approach that can be used to detect and monitor the transmission of vector-borne pathogens within their host vectors. Here, we aimed to develop a high throughput, easily interpreted, and reliable molecular xenomonitoring assay for the detection of S. mansoni and other trematode species within Biomphalaria spp. freshwater snail hosts.
The molecular xenomonitoring assay was successful in detecting and amplifying the ITS loci of a range of Biomphalaria and Schistosoma species. The Biomphalaria ITS locus was therefore deemed suitable as an internal DNA extraction/PCR reaction control and the ETTS2 forward and ETTS1 reverse primers were deemed capable of detecting trematode species other than S. mansoni, as also shown previously using non-Schistosoma trematode infections in Bulinus spp. freshwater snails (Pennance et al., 2020). In addition, the assay proved highly specific in detecting and amplifying the S. mansoni ND5 locus in both singleplex and multiplex reactions. Of note, the S. mansoni-specific ND5 primers did not cross-react with sister species S. rodhaini adult worm DNA at any point during these analyses.
The molecular xenomonitoring assay also proved highly sensitive during sensitivity testing and during controlled laboratory infection of B. glabrata with S. mansoni miracidia testing. Both ∼1005-bp Schistosoma ITS and 305-bp S. mansoni ND5 amplicons were successfully produced when using a single S. mansoni miracidium, and during analytical limit-of-detection testing, the assay was able to detect extremely low concentrations of S. mansoni DNA (the Schistosoma ITS and S. mansoni ND5 loci were detected and amplified using 0.02 ng template DNA) and S. rodhaini DNA (the Schistosoma ITS locus was detected and amplified using 0.2 ng template DNA). These were both deemed above that necessary to detect DNA from a single miracidium of both S. mansoni and S. rodhaini (∼2.5 ng/μl and ∼2.1 ng/μl, respectively, as measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Massacuhsetts, USA) (Archer et al., unpublished data). Furthermore, during controlled laboratory infection of B. glabrata with S. mansoni miracidia testing, the S. mansoni ND5 locus was detected and visibly amplified (albeit, faintly) in four of all five B. glabrata exposed to just one S. mansoni miracidium 24 h post miracidia exposure. The reason the S. mansoni ND5 locus failed to amplify in one of these B. glabrata samples exposed to one single miracidium is likely because the miracidium failed to penetrate snail soft tissues but was then missed by microscopy following exposure; however, it is not possible to confirm this. Additionally, it should also be noted that in all five of these samples, the Schistosoma ITS locus failed to visibly amplify. The S. mansoni ND5 locus was also detected and visibly amplified in all five B. glabrata exposed to five S. mansoni miracidium 24 h post miracidia exposure. All five of these amplicons were more visible than those produced by B. glabrata infection with just one S. mansoni miracidium. Similarly, however, the Schistosoma ITS locus failed to visibly amplify in two of these five samples.
This is likely caused by two primary factors. First, the ETTS2 forward and ETTS1 reverse primers appear to have lower sensitivity in detecting and amplifying the Schistosoma ITS locus when compared to the sensitivity of the S. mansoni ND5 primer set in detecting and amplifying the S. mansoni ND5 locus. This was demonstrated during analytical sensitivity testing using serially diluted S. mansoni and S. rodhaini adult worm DNA and is, itself, likely caused by the significantly lower copy-number per cell of nuclear DNA when compared to mitochondrial DNA (whilst the Schistosoma nuclear ITS region is tandemly repeated many times, multiple thousand copies of mitochondrial DNA will be present within a cell), as well as by PCR biases for smaller amplicons at reduced DNA concentrations. Secondly, this is likely also caused by ETTS2 forward and ETTS1 reverse primers being depleted during amplification of the Biomphalaria ITS region as a much greater concentration of Biomphalaria DNA will be present within PCR reactions. A similar outcome was found previously when using the same ETTS2 forward and ETTS1 reverse primers to detect trematode infections within Bulinus spp. freshwater snail hosts (Pennance et al., 2020). It should therefore be noted that very early pre-patent (< 24 h post-exposure) Biomphalaria spp. infections with only few non-S. mansoni trematode miracidia may not be detected using this assay.
During assay pilot application, all 92 Biomphalaria DNA isolates collected from a S. mansoni-endemic area successfully detected and amplified the Biomphalaria ITS locus. As such, no reactions were considered to have failed either DNA extraction or PCR and the high-throughput BioSprint 96 DNA extraction protocol was deemed reliable as a means of isolating DNA from freshwater snail tissues, as demonstrated previously (Pennance et al., 2020). The assay also successfully detected the S. mansoni infection in Bg+Sm(mal) sample DNA as well as, importantly, two Biomph91(mal) DNA samples not found to be infected during cercarial shedding. Interestingly, one of these S. mansoni-infected Biomph91(mal) DNA samples (Fig. 4, sample B), only faintly amplified the Schistosoma ITS locus, suggesting that this may be a recently established infection with very few, or just one, S. mansoni miracidia. In addition, the assay successfully detected non-S. mansoni trematode infections in two Biomph91(mal) DNA samples, later confirmed as Uvulifer spp. Whilst there appears to be no available studies investigating whether established Uvulifer spp. infections within Biomphalaria spp. freshwater snails impact S. mansoni development and transmission, other trematode species have been found to influence S. mansoni development within Biomphalaria spp. (Laidemitt et al., 2019), and so this may also be the case for Uvulifer spp. infections. Nevertheless, this finding is of both ecological and zoological interest, and, to our understanding, is the first report of Uvulifer spp. infecting Biomphalaria in this area.
The current study, and molecular xenomonitoring more broadly, does have some limitations. Whilst end-point PCR is relatively inexpensive compared to more sophisticated PCR approaches such as real-time or quantitative PCR (qPCR), this molecular xenomonitoring assay does still require costly reagents and laboratory equipment, as well as specialised personnel. In addition, the assay requires sophisticated laboratory infrastructure seldom available in schistosomiasis-endemic areas. As such, the continued development of more portable, rapid, and easy-to-use nucleic acid amplification technologies that can be carried out at the point-of-need, such as LAMP and RPA/RAA, for molecular xenomonitoring purposes is encouraged here (Mesquita et al., 2022). For example, the development and validation of a duplex RPA/RAA assay capable of detecting both extremely low levels of S. mansoni DNA as well as an internal Biomphalaria control locus. Furthermore, the continued development of DNA extraction technologies capable of isolating DNA from freshwater snail tissues in resource-poor settings is also encouraged (Mesquita et al., 2022).
Whilst the assay proved to be high throughput, pooling multiple collected Biomphalaria specimens (for example, those collected from one individual malacological collection site) to be screened within one PCR reaction may further dramatically increase assay throughput whilst still allowing for the identification of transmission sites, as suggested previously (Pennance et al., 2020). This would, however, require an extremely sensitive molecular xenomonitoring protocol capable of detecting S. mansoni DNA within a much greater concentration of Biomphalaria DNA, for example, potentially real-time PCR. Nevertheless, pooling strategies such as this should be explored.
Additionally, the development and application of species-specific Biomphalaria internal reaction control loci would be of great malacological interest, allowing for the rapid identification of morphologically similar Biomphalaria species that may differ in their ability to transmit S. mansoni (e.g. B. pfeifferi and B. sudanica; Lu et al., 2016; Mutuku et al., 2017), without the need to generate sequence data (Pennance et al., unpublished data). Using species-specific Biomphalaria loci would also mean that any morphologically similar non-Biomphalaria genus of freshwater snails that do not transmit S. mansoni, such as Gyraulus spp., could also be identified and omitted from any analysis. Similarly, whilst S. mansoni sister-species S. rodhaini is not known to infect humans, rare occurrences of natural hybridisations between S. mansoni and S. rodhaini have been identified in Kenya and Tanzania (Leger and Webster, 2017) and so the detection and monitoring of S. rodhaini transmission within Biomphalaria is not only of zoological interest, but also of potential medical importance. As such, the implementation of an additional assay locus, capable of detecting S. rodhaini whilst also differentiating S. rodhaini from S. mansoni within Biomphalaria hosts, is encouraged. Interestingly, the S. mansoni ND5 primer set used here has previously been shown to detect and amplify an ∼800-bp region of the S. rodhaini ND5 gene (Rwanda strain), and so clarification on the specificity of this DNA target is needed (Lu et al., 2016).
5. Conclusions
Here, we developed and validated a high throughput, easily interpreted, and reliable molecular xenomonitoring assay for the detection of S. mansoni and other trematode species within Biomphalaria spp. freshwater snail hosts. Furthermore, we applied this assay to successfully detect S. mansoni and non-S. mansoni trematode infections within Biomphalaria freshwater snails collected from a S. mansoni-endemic area. Molecular xenomonitoring assays such as this can be used to identify highly focal S. mansoni transmission sites, even in areas of low endemicity. Their continued development and use, particularly in areas nearing disease elimination, has been recommended by the WHO and will aid in improving disease control efforts, significantly reducing disease-related morbidities experienced by those in schistosomiasis-endemic areas.
Funding
JA is supported by a Medical Research Council Doctoral Training Programme (MRC-DTP) studentship. JA was also awarded a London Centre for Neglected Tropical Disease Research (LCNTDR) student travel grant. SMY and GG were funded through the London School of Hygiene and Tropical Medicine (LSHTM) MSc Research Project fund (2023 and 2022, respectively). The Hybridisations in UroGenital Schistosomiasis (HUGS) study is funded by a Wellcome Trust Joint Investigator Award 220818/Z/20/Z.
Ethical approval
Animal use was approved by the Institutional Animal Care and Use Committee (IACUC) of the Biomedical Research Institute for the Animal Use Protocol, #18-01. Ethical approval and research authorisations for the collection of Biomphalaria spp. from the southern shoreline of Lake Malawi, Malawi, were approved in the UK by the Liverpool School of Tropical Medicine (LSTM) Research Ethics Committee (application 17-018) and in Malawi by the National Health Sciences Research Committee (1805).
CRediT authorship contribution statement
John Archer: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization, Project administration, Funding acquisition. Shi Min Yeo: Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – review & editing, Visualization, Project administration. Grace Gadd: Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – review & editing, Visualization, Project administration. Tom Pennance: Methodology, Writing – review & editing. Lucas J. Cunningham: Resources, Writing – review & editing. Alexandra Juhàsz: Resources, Writing – review & editing. Sam Jones: Resources, Writing – review & editing. Priscilla Chammudzi: Resources, Writing – review & editing. Donales R. Kapira: Resources, Writing – review & editing. David Lally: Resources, Writing – review & editing. Gladys Namacha: Resources, Writing – review & editing. Bright Mainga: Resources, Writing – review & editing. Peter Makaula: Resources, Writing – review & editing, Project administration. James E. LaCourse: Resources, Writing – review & editing, Project administration. Sekeleghe A. Kayuni: Resources, Writing – review & editing, Project administration. Janelisa Musaya: Resources, Writing – review & editing, Project administration, Funding acquisition. J. Russell Stothard: Resources, Writing – review & editing, Project administration, Funding acquisition. Bonnie L. Webster: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Writing – review & editing, Funding acquisition. All authors read and approved the final manuscript.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the Schistosome and Snail Resource (SSR) (Wellcome Trust Biomedical Resource Grant 221368/Z/20/Z (2021-2026)) for the provision of Bi. glabrata and Schistosoma life-cycle stages, and SSR team members Dr Adam Cieplinski, Dr Fernanda Sales Coelho, Dr Aidan Emery, Dr Vanessa Yardley and Dr Amaya Bustinduy. In addition, we are grateful to the Biomedical Research Institute (BRI), Maryland, USA (https://www.afbr-bri.org), for provision of live S. mansoni ova. The authors also thank the Natural History Museum, London for the material provided via the Schistosomiasis Collection at the Natural History Museum (SCAN) archive (Wellcome Trust Biomedical Resource Grant 104958/Z/14/Z, 2010-2020). We are also grateful to David Oguttu and Fred Besigye for initial Schistosoma rodhaini collection and provision. All samples used here were collected and curated as part of a collaborative effort and we acknowledge the support and generosity of the partnerships involved in the collections, particularly in relation to all endemic countries involved, and past colleagues that maintained the live material. Thanks also to Claire Griffin of the NHM’s Molecular Biology Laboratories for assistance in Sanger sequencing.
Data availability
The data supporting the conclusions of this article are included within the article.
References
- Adema C.M. Sticky problems: Extraction of nucleic acids from molluscs. Philos. Trans. R. Soc. B Biol. Sci. 2021;376 doi: 10.1098/rstb.2020.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrus P.S., Stothard J.R., Wade C.M. Seasonal patterns of Schistosoma mansoni infection within Biomphalaria snails at the Ugandan shorelines of Lake Albert and Lake Victoria. PLoS Negl. Trop. Dis. 2023;17 doi: 10.1371/journal.pntd.0011506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRI . Biomedical Research Institute; Rockville, Maryland, USA: 2024. Isolating Schistosoma spp. eggs from murine liver and gut.https://www.afbr-bri.org/schistosomiasis/standard-operating-procedures/isolating-schistosoma-spp-eggs-from-murine-liver-and-gut/ [Google Scholar]
- BRI . Biomedical Research Institute; Rockville, Maryland, USA: 2024. Procedures common to maintaining intermediate snail hosts of Schistosoma spp.https://www.afbr-bri.org/schistosomiasis/standard-operating-procedures/ [Google Scholar]
- Cameron M.M., Ramesh A. The use of molecular xenomonitoring for surveillance of mosquito-borne diseases. Philos. Trans. R. Soc. B Biol. Sci. 2021;376 doi: 10.1098/rstb.2019.0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley D.G., Bustinduy A.L., Secor W.E., King C.H. Human schistosomiasis. Lancet. 2014;383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery A.M., Allan F.E., Rabone M.E., Rollinson D. Schistosomiasis collection at NHM (SCAN) Parasites Vectors. 2012;5:185. doi: 10.1186/1756-3305-5-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bernalt Diego J., Fernández-Soto P., Febrer-Sendra B., Crego-Vicente B., Muro A. Loop-mediated isothermal amplification in schistosomiasis. J. Clin. Med. 2021;10:511. doi: 10.3390/jcm10030511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer L.Y., Marchler-Bauer A., Geer R.C., Han L., He J., He S., et al. The NCBI BioSystems database. Nucleic Acids Res. 2009;38:492–496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grébaut P., Melachio T., Nyangmang S., Eyenga V.E., Njitchouang G.-R., Ofon E., et al. Xenomonitoring of sleeping sickness transmission in Campo (Cameroon) Parasites Vectors. 2016;9:201. doi: 10.1186/s13071-016-1479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes J.E., Croll D., Harrison W.E., Utzinger J., Freeman M.C., Templeton M.R. The roles of water, sanitation, and hygiene in reducing schistosomiasis: A review. Parasites Vectors. 2015;8:156. doi: 10.1186/s13071-015-0766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston D.W. Feeding ecology of pied kingfishers on Lake Malawi, Africa. Biotropica. 1989;21:275. doi: 10.2307/2388655. [DOI] [Google Scholar]
- Kamel B., Laidemitt M.R., Lu L., Babbitt C., Weinbaum O.L., Mkoji G.M., Loker E.S. Detecting and identifying Schistosoma infections in snails and aquatic habitats: A systematic review. PLoS Negl. Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane R.A., Rollinson D. Repetitive sequences in the ribosomal DNA internal transcribed spacer of Schistosoma haematobium, Schistosoma intercalatum and Schistosoma mattheei. Mol. Biochem. Parasitol. 1994;63:153–156. doi: 10.1016/0166-6851(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Katz N., Chaves A., Pellegrino J. A simple device for quantitative stool thick-smear technique in schistosomiasis mansoni. Rev. Inst. Med. Trop. Sao Paulo. 1972;14:397–400. [PubMed] [Google Scholar]
- Kayuni S.A., O'Ferrall A.M., Baxter H., Hesketh J., Mainga B., Lally D., et al. An outbreak of intestinal schistosomiasis, alongside increasing urogenital schistosomiasis prevalence, in primary school children on the shoreline of Lake Malawi, Mangochi District, Malawi. Inf. Dis. Poverty. 2020;9:121. doi: 10.1186/s40249-020-00736-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopp S., Speich B., Hattendorf J., Rinaldi L., Mohammed K.A., Khamis I.S., et al. Diagnostic accuracy of Kato-Katz and FLOTAC for assessing anthelmintic drug efficacy. PLoS Negl. Trop. Dis. 2011;5 doi: 10.1371/journal.pntd.0001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidemitt M.R., Anderson L.C., Wearing H.J., Mutuku M.W., Mkoji G.M., Loker E.S. Antagonism between parasites within snail hosts impacts the transmission of human schistosomiasis. eLife. 2019;8 doi: 10.7554/eLife.50095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger E., Webster J.P. Hybridizations within the genus Schistosoma: Implications for evolution, epidemiology and control. Parasitology. 2017;144:65–80. doi: 10.1017/S0031182016001190. [DOI] [PubMed] [Google Scholar]
- López-Hernández D., Locke S.A., De Assis J.C.A., Drago F.B., De Melo A.L., Rabelo É.M.L., Pinto H.A. Molecular, morphological, and experimental-infection studies of cercariae of five species in the superfamily Diplostomoidea (Trematoda: Digenea) infecting Biomphalaria straminea (Mollusca: Planorbidae) in Brazil. Acta Trop. 2019;199 doi: 10.1016/j.actatropica.2019.105082. [DOI] [PubMed] [Google Scholar]
- Lu L., Zhang S.M., Mutuku M.W., Mkoji G.M., Loker E.S. Relative compatibility of Schistosoma mansoni with Biomphalaria sudanica and B. pfeifferi from Kenya as assessed by PCR amplification of the S. mansoni ND5 gene in conjunction with traditional methods. Parasites Vectors. 2016;9:166. doi: 10.1186/s13071-016-1457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus D.P., Dunne D.W., Sacko M., Utzinger J., Vennervald B.J., Zhou X.-N. Schistosomiasis. Nat. Rev. Dis. Prim. 2018;4:13. doi: 10.1038/s41572-018-0013-8. [DOI] [PubMed] [Google Scholar]
- Mesquita S.G., Lugli E.B., Matera G., Fonseca C.T., Caldeira R.L., Webster B. Development of real-time and lateral flow recombinase polymerase amplification assays for rapid detection of Schistosoma mansoni. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1043596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J.A.T., Dejong R.J., Snyder S.D., Mkoji G.M., Loker E.S. Schistosoma mansoni and Biomphalaria: Past history and future trends. Parasitology. 2001;123:211–228. doi: 10.1017/S0031182001007703. [DOI] [PubMed] [Google Scholar]
- Mutuku M.W., Lu L., Otiato F.O., Mwangi I.N., Kinuthia J.M., Maina G.M., et al. A Comparison of Kenyan Biomphalaria pfeifferi and B. sudanica as vectors for Schistosoma mansoni, including a discussion of the need to better understand the effects of snail breeding systems on transmission. J. Parasitol. 2017;103:669–676. doi: 10.1645/17-72. [DOI] [PubMed] [Google Scholar]
- Norton A., Rollinson D., Richards L., Webster J. Simultaneous infection of Schistosoma mansoni and S. rodhaini in Biomphalaria glabrata: Impact on chronobiology and cercarial behaviour. Parasites Vectors. 2008;1:43. doi: 10.1186/1756-3305-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennance T., Ame S.M., Amour A.K., Suleiman K.R., Allan F., Rollinson D., Webster B.L. Occurrence of Schistosoma bovis on Pemba Island, Zanzibar: Implications for urogenital schistosomiasis transmission monitoring. Parasitology. 2018;145:1727–1731. doi: 10.1017/S0031182018001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennance T., Ame S.M., Amour A.K., Suleiman R., Mushin M.T., Kabole F., et al. Transmission and diversity of Schistosoma haematobium and S. bovis and their freshwater intermediate hosts Bulinus globosus and B. nasutus in the Zanzibar Archpelago, United Republic of Tanzania. PLoS Negl. Trop. Dis. 2022;16 doi: 10.1371/journal/pntd.0010585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennance T., Archer J., Lugli E.B., Rostron P., Llanwarne F., Ali S.M., et al. Development of a molecular snail xenomonitoring assay to detect Schistosoma haematobium and Schistosoma bovis infections in their Bulinus snail hosts. Molecules. 2020;25:4011. doi: 10.3390/molecules25174011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotte N., Unnasch T.R., Williams S.A. The current status of molecular xenomonitoring for lymphatic filariasis and onchocerciasis. Trends Parasitol. 2017;33:788–798. doi: 10.1016/j.pt.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Rathinasamy V., Hosking C., Tran L., Kelley J., Williamson G., Swan J., et al. Development of a multiplex quantitative PCR assay for detection and quantification of DNA from Fasciola hepatica and the intermediate snail host, Austropeplea tomentosa, in water samples. Vet. Parasitol. 2018;259:17–24. doi: 10.1016/j.vetpar.2018.06.018. [DOI] [PubMed] [Google Scholar]
- Schols R., Carolus H., Hammoud C., Mulero S., Mudavanhu A., Huyse T. A rapid diagnostic multiplex PCR approach for xenomonitoring of human and animal schistosomiasis in a ‘One Health’ context. Trans. R. Soc. Trop. Med. Hyg. 2019;113:722–729. doi: 10.1093/trstmh/trz067. [DOI] [PubMed] [Google Scholar]
- SSR . 2024. Schistosome and Snail Resource.https://www.nhm.ac.uk/our-science/research/projects/schistosome-snail-resource.htmlhttps://www.lshtm.ac.uk/research/centres-projects-groups/schistosome-and-snail-resource [Google Scholar]
- Standley C.J., Wade C., Stothard J.R. A fresh insight into transmission of schistosomiasis: A misleading tale of Biomphalaria in Lake Victoria. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalire H.F., Blouin M.S., Steinauer M.L. Genotypic variation in host response to infection affects parasite reproductive rate. Int. J. Parasitol. 2016;46:123–131. doi: 10.1016/j.ijpara.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Tchuem Tchuenté L.A., Rollinson D., Stothard J.R., Molyneux D. Moving from control to elimination of schistosomiasis in sub-Saharan Africa: Time to change and adapt strategies. Inf. Dis. Poverty. 2017;6:42. doi: 10.1186/s40249-017-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzinger J., Becker S.L., van Lieshout L., van Dam G.J., Knopp S. New diagnostic tools in schistosomiasis. Clin. Microbiol. Infect. 2015;21:529–542. doi: 10.1016/j.cmi.2015.03.014. [DOI] [PubMed] [Google Scholar]
- Webster B.L., Emery A.M., Webster J.P., Gouvras A., Garba A., Diaw O., et al. Genetic diversity within Schistosoma haematobium: DNA barcoding reveals two distinct groups. PLoS Negl. Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B.L., Webster J.P., Gouvras A.N., Garba A., Lamine M.S., Diaw O.T., et al. DNA ‘barcoding’ of Schistosoma mansoni across sub-Saharan Africa supports substantial within locality diversity and geographical separation of genotypes. Acta Trop. 2013;128:250–260. doi: 10.1016/j.actatropica.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Weerakoon K.G., Gordon C.A., McManus D.P. DNA diagnostics for schistosomiasis control. Trav. Med. Infect. Dis. 2018;3:81. doi: 10.3390/tropicalmed3030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; Geneva, Switzerland: 2022. Guideline on control and elimination of human schistosomiasis.https://www.who.int/publications/i/item/9789240041608 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the conclusions of this article are included within the article.