Abstract
Human immunodeficiency virus (HIV) envelope binds CD4 and a chemokine receptor in sequence, releasing hydrophobic viral gp41 residues into the target membrane. HIV entry required actin-dependent concentration of coreceptors, which could be disrupted by cytochalasin D (CytoD) without an effect on cell viability or mitosis. Pretreatment of peripheral blood mononuclear cells, but not virus, inhibited entry and infection. Immunofluorescent confocal microscopy of activated cells revealed CD4 and CXCR4 in nonoverlapping patterns. Addition of gp120 caused polarized cocapping of both molecules with subsequent pseudopod formation, while CytoD pretreatment blocked these membrane changes completely.
Elucidation of the mechanism of virus selection of, and entry into, suitable host cells is a key to understanding human immunodeficiency virus (HIV) transmission and pathogenesis. Recent major advances have been the identification of fusin (CXCR4) and various β-chemokine receptors (CKRs [CCR5 and CCR3]) as coreceptors for T-cell line-adapted (TCLA) and macrophage-tropic strains of HIV-1, respectively (2, 4, 5, 7, 10–12, 15, 23). Additional cellular and herpesvirus-encoded CKR-like molecules supporting dual tropism have recently been described (21, 24). Regardless of the coreceptor utilized, sequential binding of gp120 to CD4, followed by interaction of newly exposed and/or stabilized envelope regions with the CKR (28), is postulated to cause conformational changes allowing cryptic regions of transmembrane gp41 to insert into target membranes and mediate fusion with virus or infected cells.
CXC and CC family CKRs are 7-transmembrane domain G protein-coupled molecules capable of transducing activation signals. Recognition of this fact led to investigation of the role of receptor signaling in HIV infection. Results with cytoplasmic tail truncation and cytoplasmic domain mutants (22) suggested that the signaling function of coreceptors is dispensable with respect to viral entry. However, a role for energy-dependent homotypic and/or heterotypic clustering of receptors was not ruled out. Furthermore, those experiments were carried out with tumor cell lines that are constitutively activated and may not fully reflect the entry requirements for normal lymphocytes. We therefore examined the role of energy-dependent, actin-dependent cell surface aggregation of HIV receptors as a determinant of virion entry into CD4+ CKR-expressing peripheral blood mononuclear cells (PBMCs).
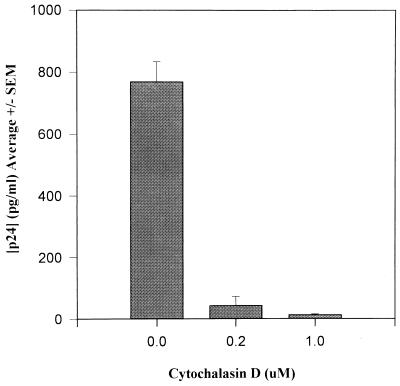
Phytohemagglutinin (PHA)-activated PBMCs were exposed to 0.2 and 1.0 μM cytochalasin D (CytoD; Aldrich, Milwaukee, Wis.) for 1 h and washed free of drug prior to exposure to 104 50% tissue culture infective doses (TCID50) of TCLA HIV-1MN (Advanced Biotechnologies, Inc., Columbia, Md.). CytoD specifically impairs F-actin polymerization in the cytoskeleton (30). While higher concentrations (20 to 40 μM) of CytoD have been used to inhibit antigen induced actin-mediated surface immunoglobulin (Ig) patching and capping on B cells (26), lower concentrations are adequate to inhibit membrane ruffling (8, 31). In our experiments, 0.2 to 1.0 μM CytoD, continuously present, had no effect on PBMC viability or proliferation (not shown). Nevertheless, 1 h of pretreatment with 0.2 and 1.0 μM CytoD resulted in, respectively, 95 and 98% inhibition of viral growth 7 days after infection (Fig. 1). Similar results were obtained with TCLA HIV-1IIIB and macrophage-tropic HIV-1BaL (not shown). HIV-1MN infection is completely blocked in this system by the anti-CD4 monoclonal antibody (MAb) SIM7 developed in our laboratory, as shown previously (18).
FIG. 1.
Effect of transient treatment of CytoD on infection of PHA-activated PBMCs by HIV-1MN T-cell-tropic virus. Pretreatment was for 1 h at CytoD concentrations of 0.2 and 1 μM and exposed to 104 TCID50 of HIV-1MN for 1 h at 37°C. The cells were washed free of virus and drug and incubated at 37°C. Culture supernatants were assayed for p24 by EIA (Organon Teknika, Durham, N.C.) on day 7. Results represent the mean ± standard error (SEM) of three independent experiments.
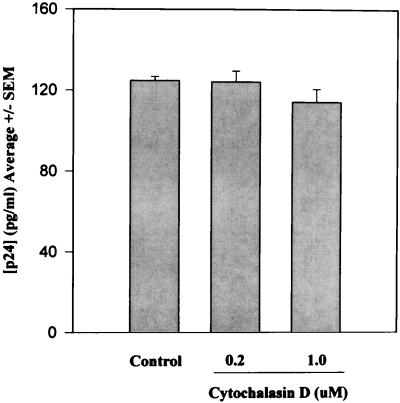
To rule out an additional effect of CytoD on virus budding, activated PBMCs were exposed to 104 TCID50 of HIV-1MN for 24 h and then treated with 10 μM zidovudine (AZT) to prevent subsequent rounds of infection. Cells washed free of virus after an additional 24 h and maintained for 7 days in 10 μM AZT plus CytoD (0.2 or 1.0 μM) showed no significant reduction in virion budding (Fig. 2). One hour of pretreatment of 106 TCID50 HIV-1MN with 5.0 μM CytoD had no antiviral effect when virus and drug were subsequently diluted 100-fold for use in culture (not shown).
FIG. 2.
Effect of continuous treatment of CytoD on virus budding from PHA-activated PBMCs. Cells exposed to 104 TCID50 of HIV-1MN for 24 h at 37°C were then treated with 10 μM AZT for an additional 24 h, washed, and maintained in medium containing AZT and CytoD at 37°C. Supernatants were assayed for p24 on day 7. Results represent the mean ± standard error (SEM) of three independent experiments.
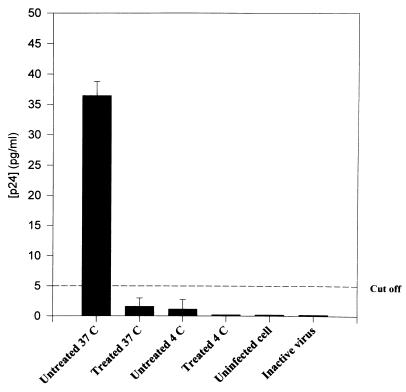
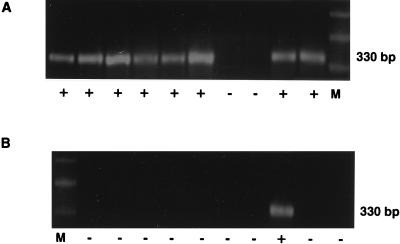
Blockage of HIV infection in CytoD-treated cells appeared to be at the level of virion entry following CD4 binding, as demonstrated by decreased internalized p24 protein within 3 h of exposure to virus (Fig. 3) and decreased frequency of DNA PCR env signal in replicate 12,500-cell aliquots (8 of 10 versus 1 of 10, P < 0.01) at 16 h (Fig. 4). In these experiments, noninternalized virions were removed by trypsinization (19) prior to DNA PCR and the p24 enzyme immunoassay (EIA). To control for incomplete trypsinization of noninternalized virus giving a false-positive signal, PHA-activated cells incubated in medium alone or 1 μM CytoD for 1 h were exposed to virus at 4°C to allow for virus binding without internalization. In some cases, cells were also pretreated with 25 μM AZT overnight to prevent reverse transcription of virus that may have been internalized even at 4°C. To rule out nonspecific sticking of virus to cells, additional controls included cells exposed to heat-inactivated virus (56°C for 3 h). All controls were negative for internalized p24 (Fig. 3) and for env signal in DNA PCR. HLA-DR was successfully amplified in parallel for all aliquots to confirm the presence of template (not shown).
FIG. 3.
Viral entry assay of CytoD-treated (1.0 μM) or untreated PHA-activated PBMCs after exposure to 105 TCID50 of HIV-1MN. After this initial incubation of 1 h at 37°C, cells were washed free of virus and drug, incubated for an additional 2 h, and treated with 0.25% trypsin for 5 min at 37°C. Internalized p24 in cell lysates was measured by EIA. Control cells were mock infected, exposed to virus at 4°C to control for nontrypsinized virus on cell surface, or exposed to heat-inactivated virus.
FIG. 4.
PCR-based limiting dilution analysis of HIV DNA in CytoD-treated or untreated PBMCs. Cells (10 × 106) pretreated with complete medium (A) or 1 μM CytoD (B) were exposed to 105 TCID50 of HIV-1MN for 1 h at 37°C, washed, and incubated at 37°C for 3 h to allow for virus internalization. Noninternalized virus was trypsinized. Cells were treated with 100 U of DNase (Boehringer Mannheim, Indianapolis, Ind.) per ml in the presence of 10 mM MgCl2 for 30 min at 37°C, washed, and maintained in complete medium at 37°C for an additional 8 h. Twofold-serially-diluted aliquots were amplified with nested primer pairs for the HIV-1 env region as described previously (27), and 10 replicates at each dilution were analyzed for signal. The endpoints for untreated and CytoD-treated cells were 3,000 and 12,500 cells, respectively. The presence (+) or absence (−) of amplified signal (HIV-1MN) from 12,500 cells is indicated under each aliquot lane. M, marker DNA.
Confocal immunofluorescence microscopy was used to visualize the effect of 5 μg of HIV-1IIIB gp120 envelope protein (Genentech) per ml on cell surface CXCR4 and CD4 in the presence or absence of CytoD. Two MAbs, FSN-M2 (IgG1 [J. E. K. Hildreth]) and 12G5 (IgG2a [14]), recognizing CXCR4 were used with similar results. SIM7, an anti-CD4 MAb that does not compete with gp120 for binding to CD4, was used. Different isotype specificities and preadsorption of secondary antibodies minimized cross-reactivity. All primary antibodies were used at 2.5 μg/ml, and fluorescein isothiocyanate- or Texas red-conjugated secondary antibodies from Southern Biotechnology Associates (Birmingham, Ala.) were used at 10 μg/ml. Specificity of labeling and absence of signal crossover were established by examination of single-labeled control samples.
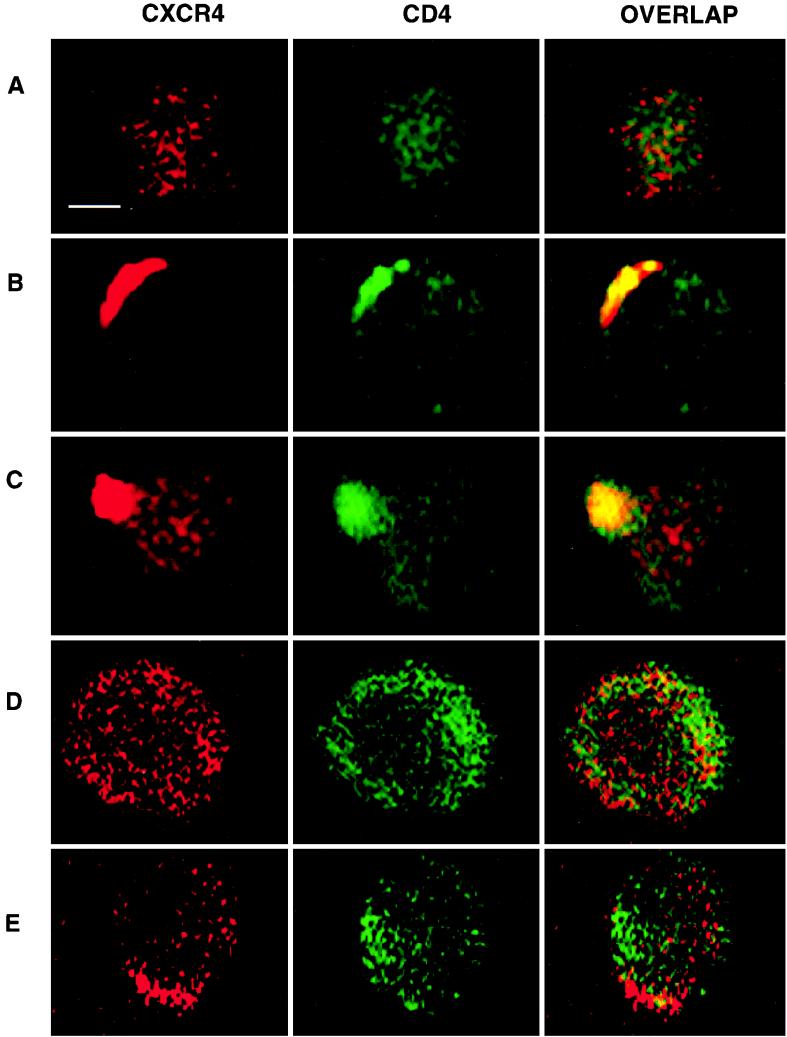
In the absence of gp120, CD4 and CXCR4 maintained a diffusely punctate surface staining pattern with minimal overlap on PHA-activated cells. Addition of gp120 at 37°C resulted in aggregation of both receptors in overlapping patches by 45 min and complete coreceptor colocalization in a polarized pattern at 60 min. Interestingly, with an increased incubation time of 90 min, a pseudopod was seen to form from the region of cells where receptors had polarized. Pretreatment of cells with 1 μM CytoD for 1 h abrogated these gp120-induced surface events (Fig. 5).
FIG. 5.
Actin-dependent colocalization of surface CD4 and CXCR4, visualized by confocal immunofluorescence microscopy. A diffusely punctate distribution of CD4 (green) and CXCR4 (red) is seen 30 min after addition of HIV-1IIIB rgp120 (A). Colocalized coreceptors appear as a false yellow polarized cap by 45 min (B), and by 90 min, a pseudopod has started to form from the area of receptor colocalization (C). Following 1 h of transient pretreatment with CytoD (1 μM), cells exhibit no changes from the baseline distribution of CD4 and CXCR4 in the presence of gp120 over 90 min (D). In the absence of gp120 or CytoD pretreatment, occasional cells may start to form homotypic polarized aggregates of CXCR4 which do not involve CD4 colocalization and produce no yellow overlap (E). Essentially identical patterns were seen with MAb 12G5 (not shown). Scale bar, 2.5 μm.
Cellular adhesion molecules are known to cluster at the surface of pseudopods and play a role in cell migration (9). The emergence of pseudopods from the area in which CD4 and chemokine receptors polarize is suggestive of a chemotactic response to soluble envelope (29) and may reflect additional interactions with cell surface adhesion molecules (19a).
There are two broad interpretations of these data which are not mutually exclusive. Coclustering of CD4-CKR heterodimers may provide a critical density necessary for each gp120 monomer of the trimeric envelope spike to bind a CKR at the same time. This would be important if conformational changes releasing all three gp41 domains simultaneously were required for high-efficiency entry. This would be analogous to density thresholds demonstrated for adhesion molecule interactions and T-cell receptor triggering (13). Alternatively or additionally, cocapping of receptors may transduce activation signals important for some as yet undefined actin-dependent steps. Cocapping-induced signals may be less crucial for infection of constitutively activated transformed human cells. This would explain the absence of coreceptor polarization in recent studies of CD4+ mink lung cells, although localized copatching was observed (28). Neither of these interpretations requires the assumption that individual CD4-coreceptor heterodimers are completely absent from the cell surface prior to contact with HIV envelope. Previous findings of the temperature dependence of virion entry have generally been interpreted as a reflection of lipid bilayer mobility thermodynamics; however, energy-dependent cytoskeletal movement is also temperature sensitive and could explain decreased viral entry at 4°C (16, 17).
Regardless of the exact mechanisms by which receptor clustering enables viral entry, the dependence on intact cytoskeleton contraction has implications for host cell selection beyond selective coreceptor usage. In order to replicate efficiently, HIV virions must enter mononuclear cells that are sufficiently activated to provide phosphorylated nucleotides for reverse transcription and energy-dependent transport of the preintegration complex into the nucleus. Once integrated, the provirus depends on host cells to produce progeny before their demise or elimination. Greater than 90% of CD4+ cells are quiescent in a healthy individual and therefore are less than ideal hosts for HIV. Moreover, >90% of activated lymphocytes are destined for programmed cell death, with a life span roughly equal to the HIV minimum generation time (1, 32). Also, HIV may replicate in the setting of cell lysis and fragmentation, which releases membrane vesicles which may bear CD4 and CKRs (3).
Faced with a multitude of inappropriate potential host cells and cell fragments, the ability to enter only cells which are viable and activated would be a major selective advantage. Caspase-mediated activation of a p21-activated kinase, PAK2, is implicated in the dysregulation of cytoskeletal actin very early in the course of apoptosis (25). If coreceptor clustering requires both activated cytoskeletal contraction and an intact cell membrane free of early apoptotic changes, it could serve as an indicator of host fitness, which the virus probes interactively prior to entry.
This model makes several experimentally verifiable predictions. (i) Rigorously purified quiescent cells expressing CD4 and CKRs and cells in the early stages of apoptosis will fail to exhibit receptor clustering upon interaction with HIV envelope and will be only minimally permissive for entry of CD4-bound virions. Recently reported studies support the resistance to infection of highly purified resting cells (6). (ii) HIV which does not enter such cells will remain infectious for suitable hosts, including formerly quiescent cells which become activated within a short period of binding virus. (iii) HIV mutants which make use of a single receptor or are otherwise independent of cytoskeletal activation will not be pathogenic or replication competent in vivo unless they are also capable of activating quiescent cells upon binding. Experiments are under way to test these hypotheses. Our results may be generalizable to other viruses. Murine ecotropic retrovirus receptors actively cluster, and Kizhatil and Albritton have recently shown that entry can be blocked by CytoD (20).
Acknowledgments
We thank J. A. Hoxie for 12G5 antibody, J. Mitchell and R. Hampton for technical help, and M. Delannoy for assistance with confocal microscopy.
This work was supported in part by NIH grant 2RO1 AI31806-05 and by American Cancer Society grant IRG 11-36.
REFERENCES
- 1.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC-CKR5: a RANTES, MIP-1 alpha, MIP-1 beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Aupeix K, Hugel B, Martin T, Bischoff P, Lill H, Pasqualli J-L, Freyssinet J-M. The significance of shed membrane particles during programmed cell death in vitro, and in vivo, in HIV-1 infection. J Clin Invest. 1997;99:1546–1554. doi: 10.1172/JCI119317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for lestr/fusin and blocks HIV-1 entry. Nature (London) 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 5.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.Chou C-S, Ramilo O, Vitetta E S. Highly purified CD25(−) resting T cells cannot be infected de novo with HIV-1. Proc Natl Acad Sci USA. 1997;94:1361–1365. doi: 10.1073/pnas.94.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 8.Cooper J A. Effect of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Pozo M A, Sanchez-Mateos P, Nieto M, Sanchez-Madrid F. Chemokines regulate cellular polarization and adhesion receptor redistribution during lymphocyte interaction with endothelium and extracellular matrix. Involvement of cAMP signaling pathway. J Cell Biol. 1995;131:495–508. doi: 10.1083/jcb.131.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 11.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 12.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 13.Dustin M L, Ferguson L M, Chan P-Y, Springer T A, Golan D E. Visualization of CD2 interaction with LFA3 and determination of two-dimensional dissociation constant for adhesion receptors in a contact area. J Cell Biol. 1996;132:465–474. doi: 10.1083/jcb.132.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enders M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenauhaggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Frey S, Marsh M, Günther S, Pelchen-Matthews A, Stephens P, Ortlepp S, Stegmann T. Temperature dependence of cell-cell fusion induced by the envelope glycoprotein of human immunodeficiency virus type 1. J Virol. 1995;69:1462–1472. doi: 10.1128/jvi.69.3.1462-1472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y-K, Hart T K, Jonak Z L, Bugelski P J. Physicochemical dissociation of CD4-mediated syncytium formation and shedding of human immunodeficiency virus type 1 gp120. J Virol. 1993;67:3818–3825. doi: 10.1128/jvi.67.7.3818-3825.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez M B, Hildreth J E K. Antibody to adhesion molecule LFA-1 enhances plasma neutralization of human immunodeficiency virus type 1. J Virol. 1995;69:4628–4632. doi: 10.1128/jvi.69.8.4628-4632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himathongkham S, Luciw P A. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 19a.Iyengar, S., D. H. Schwartz, and J. E. K. Hildreth. Submitted for publication.
- 20.Kizhatil K, Albritton L M. Requirements of different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J Virol. 1997;71:7145–7156. doi: 10.1128/jvi.71.10.7145-7156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Z-H, Berson J F, Chen Y-H, Turner J D, Zhang T-Y, Sharron M, Jenks M H, Wang Z-X, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 24.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 25.Rudel T, Bokoch G M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 26.Schliwa M. Action of cytochalasin D on cytoskeletal networks. J Cell Biol. 1982;92:79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz D H, Sharma U, Busch M, Weinhold K, Matthews T, Lieberman J, Brix D, Farzadegan H, Margolick J, Quinn T, Davis B, Bagasra O, Pomerantz R, Viscidi R. Absence of replicating infectious virus and unique immune response in an asymptomatic HIV+ long-term survivor. AIDS Res Hum Retroviruses. 1994;12:1703–1711. doi: 10.1089/aid.1994.10.1703. [DOI] [PubMed] [Google Scholar]
- 28.Ugolini S, Moulard M, Mondor I, Barois N, Demandolx D, Hoxie J, Brelot A, Alizon M, Davoust J, Sattentau Q J. HIV-1 gp120 induces an association between CD4 and the chemokine receptor CXCR4. J Immunol. 1997;159:3000–3008. [PubMed] [Google Scholar]
- 29.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 30.Wessels N, Spooner B, Ash J, Bradley H, Iuduena M, Taylor E, Wrenn J, Yamada K. Microfilaments in cellular and developmental processes. Science. 1971;171:135–143. doi: 10.1126/science.171.3967.135. [DOI] [PubMed] [Google Scholar]
- 31.Yahara I, Harada F, Sekita S, Yoshihira K, Natori S. Correlation between effects of 24 different cytochalasins on cellular structures and cellular events and those on actin in vivo. J Cell Biol. 1982;92:69–78. doi: 10.1083/jcb.92.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zinkernagel R M. Immunology taught by viruses. Science. 1996;271:173–178. doi: 10.1126/science.271.5246.173. [DOI] [PubMed] [Google Scholar]