Abstract
This article describes the nucleotide sequence of a porcine circovirus (PCV) which possesses a high degree of association with postweaning multisystemic wasting syndrome (PMWS), a newly described disease of young pigs. The DNA sequence of this PMWS-associated PCV (pmws PCV) has 68% homology with that of a previously published nonpathogenic strain of PCV. The strains appear to be closely related yet distinct from one another.
A variety of circoviruses have been identified in a range of animal species (porcine circovirus [PCV] [23], psittacine beak and feather disease virus [21], and chicken anemia virus [25]) and plant species (subterranean clover stunt virus [SCSV] [4], coconut foliar decay virus [CFDV] [22], and banana bunch top virus [BBTV] [12]). Even though all circoviruses have circular single-stranded DNA genomes and small isometric virions, there are very limited similarities among them. The animal circoviruses have insignificant similarity at the nucleotide sequence or protein level with one another and with the plant circoviruses (1, 26, 27). On the other hand, the plant circoviruses have limited similarity with one another at the nucleotide sequence and protein levels (3, 12). Prior to the present study, the only reported nucleotide sequence of porcine circovirus has been for the nonpathogenic (np PCV) strain, which is commonly associated with cultured porcine kidney (PK-15) cells (17). The np PCV was found to have limited protein similarity with only some plant circoviruses (BBTV, CFDV, and SCSV), whereas it has insignificant nucleic acid sequence and protein homology with animal circoviruses (psittacine beak and feather disease virus and chicken anemia virus) (17).
Postweaning multisystemic wasting syndrome (PMWS) is a recently recognized disease of young pigs. Typical clinical signs of PMWS include progressive wasting, dyspnea, tachypnea, occasionally, icterus and, in rare cases, jaundice (5, 11). Postmortem examinations reveal a wide range of lesions; the most common include interstitial pneumonia, lymphadenopathy, and occasionally nephritis and hepatitis (5, 11). Two earlier studies reported that a circovirus appears to be common in swine populations, based upon the prevalence of circovirus antibodies (7, 14). Microscopic examination of hematoxylin-and-eosin-stained tissue sections reveals that PMWS distinctively exhibits intensely basophilic staining inclusion bodies mostly in lymph nodes, tonsils, and Peyer’s patches of the ileum (11). A more recent study on PMWS-affected animals demonstrated the presence of a circovirus by electron microscopy, virus isolation by cell culture, in situ hybridization with a cloned PCV plasmid probe, and immunohistochemical staining with porcine and rabbit immune serum (8). However, in those studies a PCV was used that was derived from persistently infected porcine kidney (PK-15) cell lines (ATCC CCL-33) and was nonpathogenic for experimentally infected pigs (24). In previous work in our laboratory (18), it was reported that PCR was used to detect a characteristic PCV associated with PMWS, pmws PCV. Pigs affected by the disease were always found to contain pmws PCV but not np PCV. The oligonucleotide primers used in that PCR assay were designed from the nucleotide sequence of an np PCV. The pmws PCV and np PCV amplification products were readily distinguishable from one another by restriction endonuclease fragment length polymorphism (RFLP). The amplification products obtained from all PCR-positive clinical tissue specimens exhibited RFLP profiles which were unique for pmws PCV and quite distinct from that of np PCV (18).
The nucleotide sequences of np PCV, derived from persistently infected PK-15 cell lines, were previously reported by two groups of researchers, one based in Ireland (GenBank accession no. U49186 [17]) and the other in Germany (GenBank accession no. Y09921 [16]). These sequences have small (1,759-nucleotide [nt]) circular, single-stranded DNA genomes and over 99% nucleotide sequence homology. We compared the np PCV genome described by the Irish group with pmws PCV.
DNA was extracted from the lungs, lymph nodes, spleens, and tonsils of 100 pigs with PMWS from field cases which were submitted to our facility from several provinces across Canada (most were from Manitoba, but some were from Alberta, Ontario, Prince Edward Island, and Saskatchewan) by methods described elsewhere (10, 10a, 18). We screened DNA samples from these pig tissues by a PCR assay for pmws PCV described elsewhere (10a, 18). Amplification products from all 100 PMWS pigs were analyzed by RFLP. We observed that all PCR positives exhibited RFLP profiles that were unique to pmws PCV yet not identical to one another (10a). We randomly chose to use the tissues from a single PMWS case for PCR and DNA sequencing. Another laboratory (Western College of Veterinary Medicine, Saskatoon, Saskatchewan, Canada) confirmed evidence for PMWS and the presence of PCV in tissues from this random sample by immunohistochemical staining with porcine and rabbit immune serum (see reference 8 for the details about methods).
Sixteen primers suitable for PCR were selected from a published np PCV sequence (GenBank accession no. U49186 [17]) with the Primer computer program (15). All of the appropriate primers were selected for use in several separate PCRs which would yield several fragments overlapping, overall covering the entire pmws PCV genome (based upon the assumption that the pmws PCV genome should at least be similar to that of the np PCV genome). The sequences of these fragments (5′ to 3′) were as follows: 1F (nt 24 to 43), GCACCTCGGCAGCGTCAGTG; 2F (nt 378 to 399), GGAAGCGCAGCGACCTGTCTAC; 3F (nt 426 to 451), GGTCTTTGGTGACTGTAGCCGAGCAG; 4F (nt 888 to 914), GGAAGACTGCTGGAGAACAATCCACGG; 5F (nt 904 to 927), ACAATCCACGGAGGTACCCGAAGG; 6F (nt 947 to 972), CCACCCTGTGCCCTTTTCCCATATAA; 7F (nt 1231 to 1253), TGGGGGTGAAGTACCTGGAGTGG; 8F (nt 1682 to 1704), GCGGGTCCTTCTTCTGCGGTAAC; 9R (nt 43 to 24), CACTGACGCTGCCGAGGTGC; 10R (nt 399 to 378), GTAGACAGGTCGCTGCGCTTCC; 11R (nt 451 to 426), CTGCTCGGCTACAGTCACCAAAGACC; 12R (nt 914 to 888), CCGTGGATTGTTCTCCAGCAGTCTTCC; 13R (nt 927 to 904), CCTTCGGGTACCTCCGTGGATTGT; 14R (nt 972 to 947), TTATATGGGAAAAGGGCACAGGGTGG; 15R (nt 1253 to 1231), CCACTCCAGGTACTTCACCCCCA; and 16R (nt 1704 to 1682), GTTACCGCAGAAGAAGGACCCGC.
The PCR was performed as described elsewhere (10) except that Taq DNA polymerase was the only enzyme used in the reactions. After thermocycling was complete, PCR products were analyzed by gel electrophoresis as previously described (10). A PCR-positive sample was randomly chosen for DNA sequencing.
Before sequencing, 10 μg of each PCR product for pmws PCV was purified and concentrated with Microcon-100 (Fisher Scientific) 100,000-molecular-weight-cutoff microcentrifuge filter units according to the manufacturer’s recommendations. Both strands of the purified PCR products were sequenced with their corresponding PCR primers at a commercial facility (SeqWright, Houston, Tex.) by the Applied Biosystems Prism dye-terminator dideoxy system.
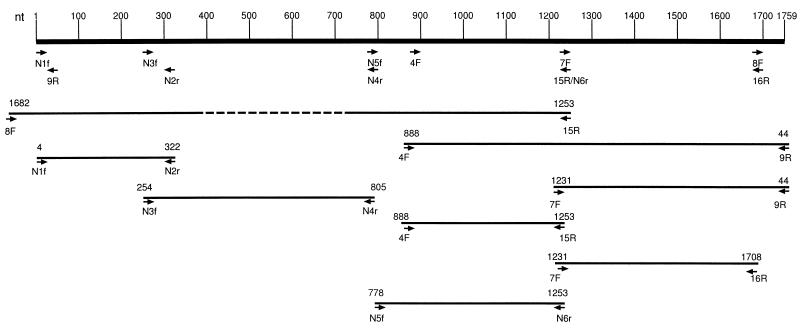
All 16 primers generated PCR amplification products of the expected sizes from DNA of the PK-15 cell line infected with np PCV; however, only six of these primers (4F, 7F, 8F, 9R, 15R, and 16R) were also able to produce PCR amplification products from DNA of PMWS-affected pigs (results not shown). These six primers were used in five different combinations of pairs to produce the following PCRs: 4F/15R (366 nt), 7F/16R (474 nt), 7F/9R (572 nt), 4F/9R (915 nt), and 8F/15R (1,331 nt). The DNA from a single randomly chosen PMWS-affected pig was subjected to these five different PCRs, and both strands of each amplification product were sequenced (Fig. 1). The raw sequencing data obtained from the three smaller PCRs (4F/15R, 7F/16R, and 7F/9R) were clearly readable in their entirety for both strands. About 850 nt in each of the two larger PCRs (8F/15R and 4F/9R) could be read with certainty (a maximum of about 425 nt from each strand). Raw sequence data for a total of 4,120 nt of pmws PCV were obtained from these five pairs of PCRs.
FIG. 1.
Map depicting the eight overlapping fragments of pmws PCV genomic DNA that were PCR amplified and sequenced in this study. The scale at the top represents nucleotide position numbers derived from a published np PCV sequence (17). This is a circular genome; therefore, nucleotide position 1759 abuts nucleotide position 1. Arrows denote location and orientation of primers that were used for PCR and sequencing. The eight overlapping horizontal lines below the graduated scale represent the eight portions of pmws PCV genomic DNA that were PCR amplified and sequenced. The dashed line in the 8F/15R PCR product indicates where legible sequence ended because it was too far from either primer. The nucleotide sequences from all eight PCR products result in an accumulated total of 6,480 nt. After all of the sequences were aligned, we observed that approximately 1,450 nt were sequenced at least three times and that 320 nt were sequenced twice.
The raw sequence data were aligned, creating a preliminary 1,360-nt contiguous sequence of pmws PCV which consisted of a region of approximately 930 nt that was sequenced four times and separate 360- and 70-nt regions that were sequenced once.
Six new primers were designed based upon this preliminary pmws PCV sequence. Their sequences (5′ to 3′) were as follows (nucleotide position numbers are based upon the completed pmws PCV sequence given in Fig. 2): N1f (nt 4 to 22), AGCGCACTTCGGCAGCGGC; N2r (nt 337 to 306), TATTCTTTATTCTGCTGATCAGTTCCTTTGGC; N3f (nt 267 to 292), GTGAAGTGGTATTTGGGTGCCCGCTG; N4r (nt 817 to 791), ATTGCTGGTAATCAAAATACTGCGGGCC; N5f (nt 790 to 819), TGGCCCGCAGTATTCTGATTACCAGCAATC; and N6r (nt 1242 to 1268), CCACTCCCGTTAATTCACACCCAAACC. These six new primers derived from the preliminary pmws PCV sequence were used to produce the following three different PCRs: N1f/N2r (334 nt), N3f/N4r (551 nt), and N5f/N6r (469 nt). The three new PCRs were performed on DNA from the same PMWS-affected pig, and both strands of each amplification product were sequenced (Fig. 1). A total of about 2,360 nt of raw sequencing data was obtained from these three new PCR products.
FIG. 2.
Nucleotide sequence of pmws PCV (GenBank accession no. AF027217 [this study]) aligned with np PCV (GenBank accession no. U49186 [17]). Numbering used here is based upon numbering that was used for np PCV, where nt position 1 is the first A residue immediately downstream of the putative nick site in the nonanucleotide motif. Total genome sizes are 1,768 nt for pmws PCV and 1,759 nt for np PCV. Homologous nucleotides are indicated by asterisks. Potential polyadenylation sites are overlined in the putative viral strand and underlined in the complementary strand.
The nucleotide sequences from all eight overlapping PCR products for pmws PCV were aligned (an accumulated total of 6,480 nt), generating a final 1,768-nt contiguous consensus sequence for pmws PCV (Fig. 2). Overall, approximately 1,450 nt were sequenced at least three times, and approximately 320 nt were sequenced twice.
The nucleotide sequences were analyzed with the computer programs Align (20), Basic Local Alignment Search Tool (BLAST, available on the Internet from the National Center for Biotechnology Information at http://www.ncbi.nlm.nih.gov [2]), ClustalV (13), and Numseq (9). Analysis of the DNA and predicted protein sequences of pmws PCV with BLASTn and BLASTx, respectively, detected any considerable homology only for np PCV. BLASTx detected scant homology between the Rep protein of pmws PCV and BBTV, CFDV, and SCSV, similarly to what was previously reported for np PCV (17).
The DNA genome of pmws PCV is 9 nt larger than that of np PCV (Fig. 2). Overall, these two genomes have 69% sequence homology, with their first halves (nt position 1 to 900) having over 82% sequence homology and their second halves (nt 901 to 1768/1759) having 62% homology. The genome of pmws PCV was determined to be circular, based upon reiteration of sequences flanking the end nucleotide positions (region between nt 1730 to 1768 and 1 to 30), from several PCR product sequences (4F/9R, 7F/9R, 8F/15R, and N1f/N2r).
The putative viral single-stranded DNA forms of pmws PCV and np PCV both contain potential polyadenylation addition [poly(A)] signal sequences (AATAA [19]) at conserved positions. Poly(A) sites are found at two places in pmws PCV (nt positions 327 to 332 and 983 to 988) which align with poly(A) sites in np PCV (nt positions 314 to 319 and 973 to 978). The complementary (minus) strand of pmws PCV has only one poly(A) site (nt positions 1022 to 1027), which aligns with one of the minus-strand poly(A) sites of np PCV (nt positions 1015 to 1030). The minus strand of np PCV contains an extra possible poly(A) site (nt positions 1184 to 1189).
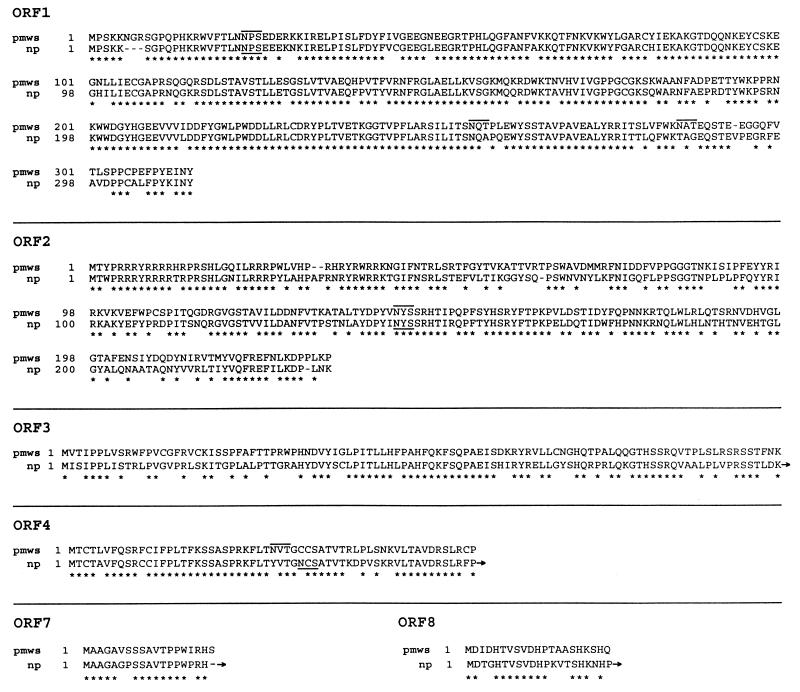
The two types of PCVs, np PCV and pmws PCV, both contain 11 potential open reading frames (ORFs). The locations and orientations of these ORFs are compared (Table 1), as are the shared homologies, sizes, and glycosylation sites for their predicted proteins. The six homologous predicted proteins encoded by ORFs 1, 2, 3, 4, 7, and 8 in pmws PCV and np PCV are aligned for comparison (Fig. 3).
TABLE 1.
Comparison of ORFs in pmws PCV and np PCVa
| ORF | pmws PCV
|
np PCV
|
Homology (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomic DNA
|
Protein
|
Genomic DNA
|
Protein
|
||||||||
| Position (nt) | Strandb | No. of aa | kDa | Glycc | Position (nt) | Strand | No. of aa | kDa | Glyc | ||
| 1 | 51–995 | V | 314 | 35.8 | + | 47–985 | V | 312 | 35.7 | + | 85 |
| 2 | 1735–1034 | C | 233 | 27.8 | + | 1723–1022 | C | 233 | 27.8 | + | 66 |
| 3 | 671–357 | C | 104 | 11.9 | − | 658–38 | C | 206 | 23.2 | − | 62 |
| 4 | 565–386 | C | 59 | 6.5 | + | 552–205 | C | 115 | 13.3 | + | 83 |
| 5 | 1016–1177 | V | 53 | 6.2 | − | 1163–1450 | V | 95 | 9.8 | + | None |
| 6 | 1611–1530 | C | 27 | 2.8 | − | 1518–1330 | C | 62 | 6.7 | + | None |
| 7 | 1682–1741 | V | 19 | 1.9 | − | 1670–81 | V | 56 | 6.0 | − | 79 |
| 8 | 753–688 | C | 21 | 2.3 | − | 740–627 | C | 37 | 4.3 | − | 67 |
| 9 | 92–1732 | C | 42 | 4.6 | − | 968–873 | C | 31 | 3.4 | − | None |
| 10 | 1524–1631 | V | 35 | 4.1 | − | 1642–1755 | V | 3.7 | 3.7 | + | None |
| 11 | 1033–989 | C | 14 | 1.8 | − | 648–719 | V | 23 | 2.8 | − | None |
Traits compared were genomic position and orientation of ORF (which viral nucleic acid strand the ORF encoded), predicted size of ORF-encoded protein (number of amino acids and molecular mass), presence of potential glycosylation sites, and homology between ORFs in pmws PCV and np PCV.
V, virus strand; C, complementary strand.
Glyc, glycosylation site; +, contains glycosylation signal (asparagine sequon N-X-S or N-X-T); −, does not contain asparagine sequon.
FIG. 3.
Alignments of predicted amino acid sequences of several ORFs in pmws PCV and np PCV. Potential glycosylation signal sequences (asparagine sequons NXS or NXT, where X = any amino acid [6]) are overlined in pmws PCV and underlined in np PCV. The proteins encoded by ORF1 would be 314 aa long and have a molecular mass of 35.8 kDa in pmws PCV, be 312 aa long and have a molecular mass of 35.7 kDa in np PCV, and have 86% homology. The proteins encoded by ORF2 would be 233 aa long and 27.8 kDa in pmws PCV, 233 aa long and 27.8 kDa in np PCV, and have 66% homology. The proteins encoded by ORF3 would be 104 aa and 11.9 kDa in pmws PCV and 206 aa and 23.2 kDa in np PCV and have 62% homology. The proteins encoded by ORF4 would be 59 aa and 6.5 kDa in pmws PCV and 115 aa and 13.3 kDa in np PCV and have 83% homology. The proteins encoded by ORF7 would be 19 aa and 1.9 kDa in pmws PCV and 56 aa and 6.0 kDa in np PCV and have 79% homology. The proteins encoded by ORF8 would be 21 aa and 2.3 kDa in pmws PCV and 37 aa and 4.3 kDa in np PCV and have 67% homology. All amino acid sequence alignments shown involve the full-length proteins in pmws PCV and the homologous portions of appropriate lengths for their counterparts in np PCV. Thus, the arrows shown at the ends of protein sequences encoded by ORFs 3, 4, 7, and 8 in np PCV indicate that these proteins continue further, and their remaining sequences are not shown.
The proteins encoded by ORF1 are of similar sizes in pmws PCV and np PCV. Likewise, the proteins encoded by ORF2 are of similar sizes in both PCVs. Most of the remaining nine ORFs (3 to 11) in pmws PCV are smaller than their counterparts in np PCV, except for ORFs 9 and 10, which are larger in pmws PCV than in np PCV.
Potential glycosylation sequences (also called asparagine sequons NXS or NXT, where X represents any amino acid [6]) are indicated in Figure 3. The proteins encoded by ORFs 1, 2, and 4 in pmws PCV contain sequons. The proteins encoded by ORFs 1, 2, 4, 5, 6, and 10 in np PCV contain sequons. The circoviruses have similar first sequons in their ORF1-encoded proteins, at amino acids (aa) 23 to 25 (NPS) in pmws PCV, and at aa 20 to 22 (NPS) in np PCV. However, the ORF1-encoded protein in pmws PCV has two extra sequons, at aa 256 to 258 (NQT) and aa 286 to 288 (NAT). Both viruses have single sequons in their ORF2-encoded proteins, at aa 143 to 145 (NYS) in pmws PCV and aa 102 to 104 (also NYS) in np PCV. Both viruses lack sequons in their proteins encoded by ORF3, ORF7, ORF8, ORF9, and ORF11. Both have similarly placed sequons in their ORF4-encoded proteins, at aa 30 to 32 (NVT) in pmws PCV and aa 34 to 36 (NCS) in np PCV. There are sequons in the proteins encoded by ORF5 (aa 64 to 66 and 69 to 71), ORF6 (aa 6 to 8, 27 to 29, and 37 to 39) and ORF10 (aa 21 to 23) in np PCV but not in pmws PCV.
Both PCV genomes have nearly identical predicted stem-loop structures and nonanucleotide motif sequences (Fig. 4). This region is known to be required for np PCV genome replication (16). The starting point used for numbering the nucleotide sequence positions in both PCV genomes is located at the right-most A residue within the nonanucleotide sequence motif, 5′-AAGTATTAC-3′ (nt positions 1762 to 2 in pmws PCV and 1753 to 2 in np PCV), which is immediately downstream of the putative nick site in between the rightmost T and A residues (Fig. 3).
FIG. 4.
(a) Alignment of the putative DNA replication sequences in pmws PCV and np PCV. Homology is indicated by an asterisk. (b) Comparison of the stem-loop structures predicted from the sequences in panel a. In both panels the nonanucleotide sequence motifs are in bold-faced type, and arrows mark the putative nick sites. The A residues immediately downstream of these nick sites are used as the starting point for numbering these genomes. All sequences are given in the 5′-to-3′ direction.
It is perhaps not surprising that the proteins encoded by ORF1 have the most highly conserved sequence (they have 85% homology) considering that they code for the putative Rep protein, required for genome replication. The proteins encoded by ORFs 2, 3, 4, 7, and 8 in pmws PCV are obviously closely related to their respective counterparts in np PCV. However, their differences are striking, especially for those proteins encoded by ORFs 3, 4, 7, and 8, which in pmws PCV are about half the size of their counterparts in np PCV. Furthermore, ORFs 5, 6, 9, 10, and 11 in these two circoviruses lack any homology with any other ORF. These predicted differences in proteins, encoded in pmws PCV, are possibly the contributing factors for the pathogenesis, clinical signs, and lesions associated with PMWS.
Work needs to be done in identifying and characterizing the proteins that are actually produced by pmws PCV before their functions can be known (by gel electrophoresis and by probing with antibodies raised against purified pmws PCV). It is hoped that such research will extend our understanding of the roles played by these proteins and of their different traits (overall amino acid sequence, hydrophobic, and hydrophilic domains, and glycosylation sites) in relation to the differences in virulence between pmws PCV and np PCV.
The recent epidemic of PMWS in North American pigs has a characteristic PCV, pmws PCV, associated with the disease. The similarity of pmws PCV to np PCV in nucleotide sequence, 6 of 11 protein sequences, genome structure and organization, and host cell preferences demonstrates that they are closely related and may have a common ancestor.
The PCR assay is a useful tool for studying diseases such as PMWS. We had previously reported using PCR and detecting only pmws PCV in pigs affected by PMWS (18). In an upcoming study, we will fully describe a PCR assay with primers based upon the pmws PCV sequence given here and its efficacy for detecting pmws PCV in pigs (10a). Studies involving purified pmws PCV used to experimentally infect pigs can extend our understanding of the development and pathology of PMWS. Such studies can make use of probes and PCR assays that are based upon the pmws PCV nucleotide sequence presented here.
PMWS is an important new disease in pigs. We hope the nucleotide sequence of pmws PCV described here will be useful for improving our understanding of the disease PMWS and developing effective vaccines against the disease and diagnostic procedures such as the PCR for detecting the etiological agent.
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of pmws PCV described in the present study is AF027217.
Acknowledgments
We are most grateful to Cheryl Sachvie for valuable technical assistance.
REFERENCES
- 1.Allan G M, Phenix K V, Todd D, McNulty M S. Some biological and physico-chemical properties of porcine circovirus. J Vet Med Ser B. 1994;41:17–26. doi: 10.1111/j.1439-0450.1994.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 2.Altshul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Burns T M, Harding R M, Dale J L. The genome organization of banana bunchy top virus: analysis of six ssDNA components. J Gen Virol. 1995;76:1471–1482. doi: 10.1099/0022-1317-76-6-1471. [DOI] [PubMed] [Google Scholar]
- 4.Chu P G W, Keese P, Qiu B S, Waterhouse P M, Gerlach W L. VIIIth International Congress of Virology Abstracts. 1990. Novel ssDNA genome organization of a new plant virus, abstr. W82-001; p. 125. [Google Scholar]
- 5.Clark E G. Post-weaning multisystemic wasting syndrome. Proc Am Assoc Swine Pract. 1997;28:499–501. [Google Scholar]
- 6.Darnell J E, Lodish H F, Baltimore D. Molecular cell biology. New York, N.Y: Scientific American Books; 1986. Assembly of organelles: protein glycosylation; p. 961. [Google Scholar]
- 7.Dulac G C, Afshar A. Porcine circovirus antigens in PK-15 cell line (ATCC CCL-33) and evidence of antibodies to circovirus in Canadian pigs. Can J Vet Res. 1989;53:431–433. [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis J, Hassard L, Clark E, Harding J, Allan G, Willson P, Strakappe J, Martin K, McNeilly F, Meehan B, Todd D, Haines D. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J. 1998;39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 9.Fristensky B, Li J T, Wu R. Portable microcomputer software for nucleotide analysis. Nucleic Acids Res. 1982;10:6451–6463. doi: 10.1093/nar/10.20.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamel A L, Wasylyshen M D, Nayar G P S. Rapid detection of bovine viral diarrhea virus by using RNA extracted directly from assorted specimens and a one-tube reverse transcription PCR assay. J Clin Microbiol. 1995;33:287–291. doi: 10.1128/jcm.33.2.287-291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harding J C. Post-weaning multisystemic wasting syndrome (PMWS): preliminary epidemiology and clinical presentation. Proc Am Assoc Swine Pract. 1997;28:503. [Google Scholar]
- 12.Harding R M, Burns T M, Hafner G, Dietzgen R G, Dale J L. Nucleotide sequence of one component of the banana bunchy top virus genome contains a putative replicase gene. J Gen Virol. 1993;74:323–328. doi: 10.1099/0022-1317-74-3-323. [DOI] [PubMed] [Google Scholar]
- 13.Higgin D G, Sharp P. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;45:333–338. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 14.Hines R K, Lukert D. Porcine circovirus: a serological survey of swine in the United States. Swine Health and Production. 1995;3:71–73. [Google Scholar]
- 15.Lincoln S E, Daly M E, Lander E S. Primer: a computer program for automatically selecting PCR primers. Cambridge, Mass: MIT Center for Genome Research and Whitehead Institute for Biomedical Research; 1990. [Google Scholar]
- 16.Mankertz A, Persson F, Mankertz J, Blaess G, Buhk H J. Mapping and characterization of the origin of DNA replication of porcine circovirus. J Gen Virol. 1997;71:2562–2566. doi: 10.1128/jvi.71.3.2562-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meehan B M, Creelan J L, McNulty M S, Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol. 1997;78:221–227. doi: 10.1099/0022-1317-78-1-221. [DOI] [PubMed] [Google Scholar]
- 18.Nayar G P S, Hamel A, Lin L. Detection and characterization of porcine circovirus associated with post-weaning multisystemic wasting syndrome in pigs. Can Vet J. 1997;38:385–386. [PMC free article] [PubMed] [Google Scholar]
- 19.Nevins J R, Darnell J E., Jr Steps in the processing of Ad2 mRNA: poly (A) + nuclear sequences are conserved and poly (A) addition precedes splicing. Cell. 1978;15:1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- 20.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie B W, Niagro F D, Lukert P D, Steffens W L, Latimer K S. Characterization of a new virus from cockatoos with psittacine beak and feather disease virus. Virology. 1989;171:83–88. doi: 10.1016/0042-6822(89)90513-8. [DOI] [PubMed] [Google Scholar]
- 22.Rohde W, Randles J W, Langridge P, Hanold D. Nucleotide sequence of a circular single-stranded DNA associated with coconut foliar decay virus. Virology. 1990;176:643–651. doi: 10.1016/0042-6822(90)90038-s. [DOI] [PubMed] [Google Scholar]
- 23.Tischer I, Gelderblom H, Vettermann W, Koch M A. A very small porcine virus with a circular single-stranded DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 24.Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on the pathogenicity of porcine circovirus. Arch Virol. 1986;91:271–276. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- 25.Todd D, Creelan J E, Mackie D P, Rixon F, McNulty M S. Purification and biochemical characterization of chicken anaemia agent. J Gen Virol. 1990;71:819–823. doi: 10.1099/0022-1317-71-4-819. [DOI] [PubMed] [Google Scholar]
- 26.Todd D, Niagro F D, Ritchie B W, Curran W, Allan G M, Lukert P D, Latimer K S, Steffens W L, McNulty M S. Comparison of three animal viruses with circular single-stranded DNA genomes. Arch Virol. 1991;117:129–135. doi: 10.1007/BF01310498. [DOI] [PubMed] [Google Scholar]
- 27.Todd D, Phenix K V, Allan G M, McNulty M S, Buhk H J, Tischer I. IXth International Congress of Virology Abstracts. 1993. Comparison of chicken anaemia virus and porcine circovirus, members of the vertebrate virus family Circoviridae, abstr. P76-2; p. 367. [Google Scholar]