Abstract
Attachment of an adenovirus (Ad) to a cell is mediated by the capsid fiber protein. To date, only the cellular fiber receptor for subgroup C serotypes 2 and 5, the so-called coxsackievirus-adenovirus receptor (CAR) protein, has been identified and cloned. Previous data suggested that the fiber of the subgroup D serotype Ad9 also recognizes CAR, since Ad9 and Ad2 fiber knobs cross-blocked each other’s cellular binding. Recombinant fiber knobs and 3H-labeled Ad virions from serotypes representing all six subgroups (A to F) were used to determine whether the knobs cross-blocked the binding of virions from different subgroups. With the exception of subgroup B, all subgroup representatives cross-competed, suggesting that they use CAR as a cellular fiber receptor as well. This result was confirmed by showing that CAR, produced in a soluble recombinant form (sCAR), bound to nitrocellulose-immobilized virions from the different subgroups except subgroup B. Similar results were found for blotted fiber knob proteins. The subgroup F virus Ad41 has both short and long fibers, but only the long fiber bound sCAR. The sCAR protein blocked the attachment of all virus serotypes that bound CAR. Moreover, CHO cells expressing human CAR, in contrast to untransformed CHO cells, all specifically bound the sCAR-binding serotypes. We conclude therefore that Ad serotypes from subgroups A, C, D, E, and F all use CAR as a cellular fiber receptor.
Human adenoviruses (Ads) are associated with a wide range of tropisms (33, 50, 51). They have been classified into six distinct subgroups A to F, with at least 49 serotypes (21, 41), on the basis of their genetic variability, oncogenic potential, and G+C content of their DNA (21, 50–52). A further subclassification of subgroups B (BI and BII) and D (DI, DII, and DIII) has been made on the basis of differential hemagglutination patterns (12, 18, 32, 33, 40, 50). Ads have the ability to infect a wide range of different tissues and have been identified as causative agents of widely different diseases (21, 22, 50, 51). For example, serotypes Ad2 and Ad5 (subgroup C) are associated with upper-airway infections, as is serotype Ad3 (subgroup B), although the latter appears to infect an anatomically distinct region of the airway (22, 50, 51). Serotypes Ad8 and Ad9 (subgroup D) are associated with epidemic keratoconjunctivitis, Ad4 (subgroup E) is associated with pneumonia, Ad12 (subgroup A) is associated with cryptic enteric infection (33), and Ad40 and Ad41 (subgroup F) are associated with gastroenteritis (references 22, 50, and 51 and references therein). Detailed phylogenetic analysis of diverse Ad serotypes has yielded two phenotypic clusters; the gastrointestinal cluster, with subgroups A and F, and the respiratory cluster, with subgroups B, C, and E (3).
It has been suggested that the apparent tropism of different serotypes in different tissues results from virus interactions with distinct cellular receptors (50, 51). Indeed, it has been shown convincingly that the Ad2/5 and Ad3 fiber proteins recognize different cellular receptor proteins (9, 10, 38, 45). It has also been demonstrated that the subgroup B serotype Ad35 recognizes a receptor other than CAR (4). This has led to the construction of Ad5/fiber 3 chimeras (27, 44) and an Ad5/fiber 7 chimera (14) that have an altered fiber receptor tropism and thus can be used to target tissues that display differential fiber 2/fiber 3 receptor levels like the THP1 monocytes (44).
Attachment and uptake into cells of subgroup C Ads occur by separate but cooperative events that result from the interaction of the fiber protein with a receptor for attachment and the penton base protein with a receptor for internalization (55). The 46-kDa coxsackievirus-adenovirus receptor (CAR) protein mediates fiber-dependent attachment of subgroup C Ad2 and Ad5 (4, 49). The C-terminal knob of the fiber protein confers the specificity of the cellular receptor recognition (13, 16, 27, 29, 38, 45). Analysis of the fiber 5 knob at 1.7 Å by X-ray crystallography has yielded a model for the structure of this protein and has identified several exposed amino acid residues and structural loops that have been theorized to be involved in cellular receptor recognition (56). Next, in a process that has been shown to be independent of fiber-cell recognition (11, 27, 55), the viral penton base protein binds to cellular αv-integrins through the RGD loop, a tripeptide motif that protrudes out of the tertiary peptide structure of the penton base, resulting in rapid internalization of the virus particle (30, 45, 46, 52, 54, 55).
Apart from the identification of CAR as the cellular fiber receptor for Ad2 and Ad5 (4, 49), progress has been made in elucidating the molecular mechanisms that underlie cell recognition and attachment. Through cross-competition experiments, it has been shown that the fibers of Ad2, Ad5, and Ad9, despite their distinct tropisms and classification into the different subgroups C and D (21, 33, 50), recognize the same cellular fiber receptor (38).
The observation that the knobs from Ad9 (subgroup D) and Ad2 (subgroup C) cross-compete for binding suggested that Ad serotypes from other subgroups might likewise recognize the same receptor. As a first step, we constructed baculovirus expression clones of eight fiber knob proteins derived from serotypes representing the six subgroups. In competition experiments we found that with the exception of Ad3, all serotypes representing the five subgroups A and C through F, cross-competed with CAR. To allow detailed analysis of this phenomenon, we produced a baculovirus clone that expressed a soluble form of the CAR protein (sCAR) in insect cells. The purified sCAR protein was shown to bind directly to viral capsids and fiber knobs from five of the six subgroups. In addition, preincubation of the CAR binding serotypes with sCAR was shown to block their binding to CAR-expressing cells. Finally, binding experiments with Chinese hamster ovary (CHO) cells that had been transformed with the cDNA of the human CAR gene (4) verified that the CAR protein can be used as a cellular fiber receptor by serotypes from all Ad subgroups except subgroup B.
MATERIALS AND METHODS
General methods.
DNA manipulations were performed by standard methods (1). All DNA digests were done in an universal restriction enzyme buffer, which has been described previously (39). Protein analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting was performed as described previously (1, 28).
Viruses and cell lines.
Ad2, Ad3, Ad4, Ad5, Ad7, Ad9, Ad12, Ad15, Ad19, Ad31, and Ad41 were obtained as stocks from the American Type Culture Collection (Rockville, Md.) and were passaged on 293/ORF6 cells (5) in Dulbecco’s modified Eagle’s medium (Gibco BRL, Gaithersburg, Md.) supplemented with 5% fetal calf serum. The cell lines used and their maintenance have been described elsewhere (38). CHO-CAR cells, which express the full-length human CAR gene (4), were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. [methyl-3H]thymidine-labeled Ads were produced as described previously (38).
All virus stocks were subjected to quality control by PCR with sets of primers specifically designed for each serotype. The sense primers were designed to hybridize to the junction of the shaft and the knob region of the fiber gene, the so-called TLWT hinge (47), which is conserved among most serotypes. Specificity for individual serotypes was provided by the antisense primers that were designed against the highly variable HI loop of the fiber knob (56). All virus stocks were evaluated with all primer pairs. No cross-contamination for any of the viruses was observed. The PCR products obtained were digested with enzymes specific for each knob DNA sequence and analyzed on agarose gels.
Fiber gene sequences.
All Ad fiber gene sequences used for similarity studies and expression construct cloning were obtained from the GenBank/EMBL databases. The accession numbers and references are as follows: F2, P03275 (17); F3, M12411 (42); F4, L19194 (15); F5, M18369 (6, 7); F7, P15141 (19); F8, X74660 (36); F9, X74659 (36); F12, X73487 (43); F15, X74658 and X74659 (35a); F19, X94485 (35a); F31, X76548 (37); F40 Long, P18047 (25); F40 Short, M28822 (24); F41 Long, P14267 (35); F41 Short, X17016 (34).
Expression constructs.
The baculovirus expression vector pAcSG2 was used for expression (Pharmingen, San Diego, Calif.) of the sCAR and fiber knob proteins. The sCAR clone was designed to contain amino acids 1 to 236 of the human CAR extracellular domain (4), upstream of a linker sequence and a FLAG tag consisting of the amino acids DYKDDDDK for detection and purification purposes (Eastman Kodak Co., Rochester, N.Y.). Construction of His-tagged versions of the fiber 3 knob (F3K), F5K, and F9K has been described previously (38). All other His-tagged fiber constructs were generated by designing primer pairs for PCR amplification between the hinge sequence TLWT, or similar sequence (8, 47), and the native C terminus. All PCRs were performed with Ultma DNA polymerase (Perkin-Elmer, Foster City, Calif.) by a standard PCR protocol. The DNA sequences of all expression clones were determined with an automated ABI 373 DNA sequencer (Applied Biosystems, Foster City, Calif.). Except for the designed DNA sequence changes, no nucleotide differences were found in comparison with the original sequences reported in the GenBank/EMBL databases. Insect virus expression clones were generated by using the Bsu36 I-digested BacPak DNA and transfection kit (Invitrogen, San Diego, Calif.). Recombinant His-tagged fiber knob proteins were produced in Tn5 B1-4 insect cells and purified as described previously (38, 55). FLAG-tagged sCAR protein was purified on an affinity column with a coupled anti-FLAG M2 antibody as specified by the manufacturer (Eastman Kodak Co.) with one modification: the 500-μl elution fractions, which are at pH 3.5, were neutralized by collection in tubes that contained 30 μl of 1 M Tris (pH 8.0).
Slot-blot experiments.
For the sCAR binding experiments, either virions (2 × 1010 particles) or purified fiber protein (2 μg) was blotted onto a nitrocellulose membrane. After blocking for 1 h in phosphate-buffered saline (PBS) with 5% milk, the blot was incubated overnight in PBS with 0.5% milk and sCAR protein (100 ng/ml) at 4°C. After two washes, the blot was incubated with a 1:10,000 dilution of the anti-FLAG mouse monoclonal antibody M2 (Eastman Kodak Co.) and incubated for 2 h at room temperature (RT). After two washes with PBS–0.5% milk–0.05% Igepal CA 630 (Sigma, St. Louis, Mo.), the blot was incubated for 2 h in the same buffer with a 1:10,000 dilution of a goat-anti mouse secondary antibody conjugated to horseradish peroxidase (Boehringer Mannheim, Indianapolis, Ind.). Detection was carried out with the ECL Western blot detection kit (Amersham Life Sciences, Arlington Heights, Ill.).
Competition experiments.
Assays to evaluate the binding of Ad serotypes were done essentially as described previously (38). Briefly, 106 A549, CHO, or CHO-CAR cells were preincubated at 37°C for 1 h and then chilled at 4°C for 10 min. 3H-labeled virions were added, and the mixture was incubated for 1 h. The cells were washed twice with cold PBS, pelleted, resuspended in 100 μl of PBS, and counted directly in a scintillation counter. Competition experiments with sCAR protein were done at RT with 2 × 106 Ramos cells.
RESULTS
Fiber knob cross-competition.
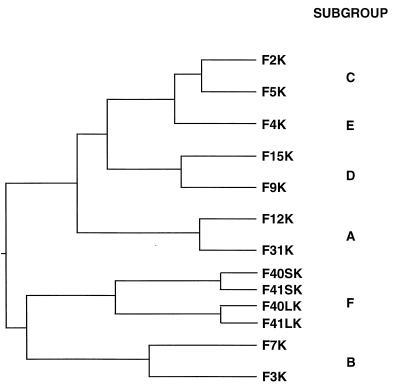
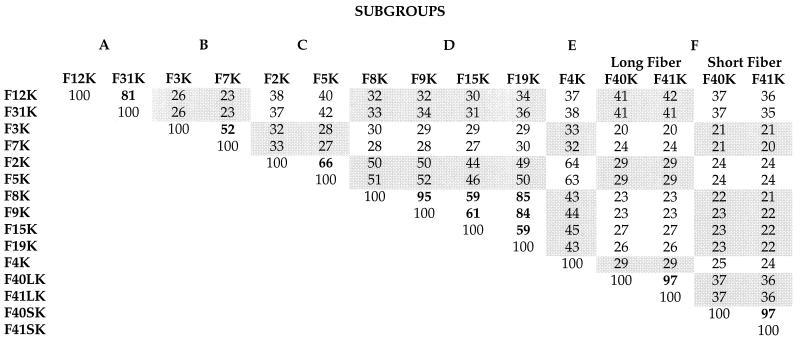
Previous studies have shown that Ad9 (subgroup D) and Ad12 (subgroup A) are blocked from binding to cells by preincubation of cells with fiber protein of the subgroup C viruses, which suggested that they are able to recognize the CAR protein as a cellular fiber receptor (2, 38). These observations prompted us to evaluate the fiber receptor specificity from the different Ad subgroups. As a first step, we aligned the amino acid sequences of 13 fiber knobs by using the Jotun-Hein algorithm. Two major clusters on the phylogenetic tree were distinguished (Fig. 1): the top cluster included Ad serotypes in subgroups A, C, D, and E, and the lower branch included those in subgroups B and F. Knobs within the same subgroup were most similar; Ad2 and Ad5, 66%; Ad9 and Ad15, 59%; Ad12 and Ad31, 81%; and Ad3 and Ad7, 52%. Knob sequences between subgroups were less similar (Table 1). For example, F5K was 63% similar to F4K, 52% similar to F9K, 40% similar to F12K, and only 29% similar to F41LK.
FIG. 1.
Phylogenetic tree of 13 fiber knob amino acid sequences. The sources of the sequences are identified in Materials and Methods. The sequences were aligned as described in the footnote to Table 1.
TABLE 1.
Alignment of 15 fiber knob amino acid sequencesa
Similarity table of fifteen knob amino acid sequences. The sources of the sequences are identified in Materials and Methods. The sequences were aligned from the TLWT hinge sequences or comparable sequence (8, 47) to the translational stop codon by using the program Megalign with the Jotun-Hein Algorithm with a PAM 250 weight table. Similarity scores between serotypes within a subgroup are printed in boldface type.
On the basis of these comparisons, we selected eight knobs for constructing His-tagged baculovirus expression vectors of F12K (subgroup A), F3K (subgroup B), F2K and F5K (subgroup C), F9K (subgroup D), and F4K (subgroup E). Since Ad41 (subgroup F) has both a long fiber and a short fiber (57), both F41LK and F41SK were produced in insect cells. After purification, the knob proteins were used in competition binding assays with 3H-labeled parental viruses on A549 cells (Table 2). Fiber knobs of Ad2, Ad4, Ad9, and Ad12 and the long fiber of Ad41 inhibited attachment by all viruses tested, with the exception of Ad3, which was inhibited only by its homologous fiber knob. This suggested that with the exception of Ad3, all serotypes tested could use CAR as a cellular fiber receptor. Serotype Ad9 was only partially inhibited by F2K, F4K, F5K, F9K, F12K, and F41LK. Inhibition of Ad9 binding increased to 90% after the cells were treated with 5 mM EDTA (data not shown). This confirmed an earlier observation that cellular binding by Ad9 depends on both fiber-receptor and penton base–αv-integrin interactions, the latter of which is divalent-cation dependent and inhibited by chelators such as EDTA and EGTA. The F41SK protein inhibited none of the serotypes, including the parental virus Ad41, whereas the F41LK protein did. Therefore, this suggests that the Ad41 long fiber can function as the cellular attachment protein for Ad41.
TABLE 2.
Cross-competition of Ad serotypes by purified knob proteinsa
| Sero- type | Inhibitionb by:
|
||||||
|---|---|---|---|---|---|---|---|
| A (F12K) | B (F3K) | C (F2K) | D (F9K) | E (F4K) | F
|
||
| F41LK | F41SK | ||||||
| Ad12 | +++ | − | +++ | +++ | +++ | +++ | − |
| Ad3 | − | +++ | − | − | − | − | − |
| Ad2 | +++ | − | +++ | +++ | +++ | +++ | − |
| Ad9 | ++ | − | ++ | ++ | ++ | ++ | − |
| Ad4 | +++ | − | +++ | +++ | +++ | +++ | − |
| Ad41 | +++ | − | +++ | +++ | +++ | +++ | − |
Approximately 106 A549 cells (human lung carcinoma cells) were preincubated for 1 h in 250 μl of PBS plus 2 μg of purified fiber knob protein per ml at 37°C. The cells were chilled for 10 min, and 3H-labeled serotypes were added and incubated for 1 h at 4°C. The cells were washed twice. The pellet was taken up in 100 μl of PBS, added to scintillation cocktail, and counted directly.
+++, >95% inhibition; ++, 40 to 60% inhibition; −, no inhibition.
Soluble CAR protein binds to capsids and fiber knob proteins.
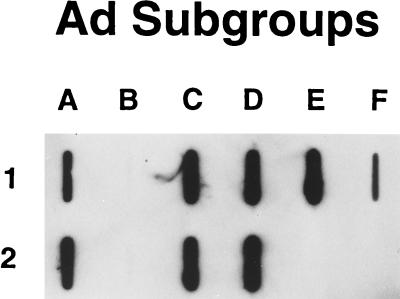
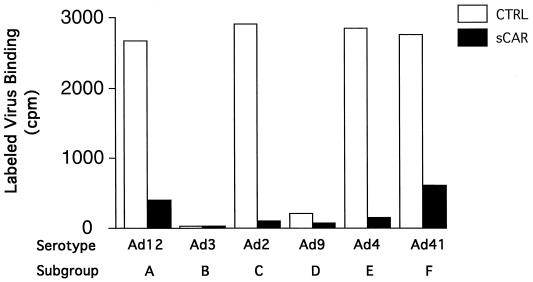
Since Ad2 and Ad5 fibers have been shown to bind to CAR, our cross-competition results strongly suggested that subgroups A, D, E, and F also bind to CAR. A baculovirus vector was constructed that expressed a secreted version of sCAR, which consisted of the extracellular domain of the CAR protein with its native signal sequence (4), a linker sequence, and a FLAG-tag sequence, DYKDDDDK, to facilitate detection and purification. In addition to the serotypes already available to us, viral stocks of three more serotypes, Ad31 (subgroup A), Ad15 (subgroup D), and Ad19 (subgroup D), were produced in 293/ORF6 cells (5). Purified Ads from each of the serotypes were immobilized on nitrocellulose with a slot-blot apparatus and tested for their ability to bind sCAR (Fig. 2). All the serotypes tested, except those from subgroup B, bound sCAR protein. They included Ad12 and Ad31 (subgroup A), Ad2 and Ad5 (subgroup C), Ad9 and Ad15 (subgroup D), Ad4 (subgroup E), and Ad41 (subgroup F). In an additional experiment (data not shown), the subgroup D virus Ad19 also bound to sCAR. The signal for Ad41 appeared weaker than the other signals, perhaps because the short Ad41 fiber does not bind CAR. Not surprisingly, the subgroup B viruses Ad3 and Ad7 did not bind sCAR protein (Fig. 2).
FIG. 2.
sCAR-virus slot blot. Approximately 2 × 1010 particles of each serotype were blotted onto nitrocellulose. The blot was blocked 5% milk in PBS and then incubated overnight with soluble CAR protein. Detection was performed as described in Materials and Methods. The lettering refers to the subgroup classification. The letter-and-number combination refers to the following serotypes: A1 and A2, Ad12 and Ad31; B1 and B2, Ad3 and Ad7; C1 and C2, Ad2 and Ad5; D1 and D2, Ad9 and Ad15; E1, Ad4; F1, Ad41.
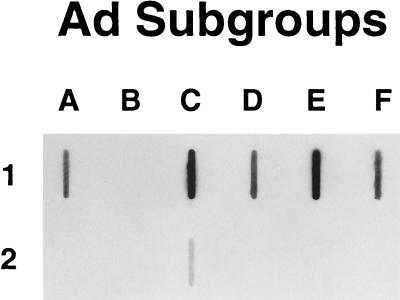
These results were confirmed by blotting purified fiber knob proteins onto a nitrocellulose membrane and incubating the blot with sCAR protein. The results in Fig. 3 demonstrate that with the exception of F3K and F41SK, all the fiber knob proteins tested were capable of binding sCAR protein. Although each of the tested knobs was purified, differences in both purity and measured protein concentrations may explain the differences in observed signal strength. In a reverse experiment in which sCAR protein was first immobilized onto plastic and then labeled viruses were bound, levels of binding were measured that fluctuated no more than two- to threefold among all serotypes tested (data not shown). This suggests that the affinity of each serotype for the CAR protein is by and large comparable. Clearly, further study is needed to address this point.
FIG. 3.
sCAR-fiber knob protein slot blot. Approximately 2 μg of each purified fiber knob protein was blotted onto a nitrocellulose. The blot was blocked and incubated overnight with sCAR protein. Detection was performed as described in Materials and Methods. The lettering refers to the subgroup classification. The letter-and-number combination refers to the following fiber knob proteins: A1, F12K; B1, F3K; C1 and C2, F2K and F5K; D1, F9K; E1, F4K; F1 and F2, F41LK and F41S-K.
The ability of the sCAR protein to block fiber-mediated attachment to Ramos cells was next evaluated. These cells were chosen because they express high levels of CAR protein but lack αv-integrins. This allows for a direct evaluation of fiber-mediated binding while eliminating any secondary binding mechanism that involves αv-integrins, as has been found for Ad9 (38). First we determined that a final concentration of sCAR protein of 10 μg/ml was needed to completely inhibit Ad2 binding to Ramos cells (data not shown). Then 3H-labeled Ad12, Ad3, Ad2, Ad9, Ad4, and Ad41 (equal number of input counts; the particle numbers as determined by UV spectroscopy were approximately the same) were incubated with sCAR protein for 30 min at room temperature and added to Ramos cells. After a 1-h incubation, the cells were washed and bound virus was determined by scintillation counting (Fig. 4). Our results indicated that all viruses that had previously been shown to bind sCAR bound at comparable levels to Ramos cells except for serotype Ad9, which bound at a much lower level than the other viruses. Although it has been shown that Ad9 is able to use the CAR protein as a cellular fiber receptor, it preferentially uses αv-integrins to bind to cells. Since Ramos cells lack these vitronectin receptors, we hypothesize that other cell surface receptors may interfere with the ability of the short-shafted Ad9 fiber to reach its cell surface receptor CAR, resulting in the observed lower level of Ad9 binding to these cells (38). No binding for Ad3 was found, indicating that Ramos cells do not express the receptor for Ad3 fiber. Preincubation of the viruses with sCAR protein resulted in a 95% drop in the amount of virus bound to the cells. This demonstrated that the sCAR protein blocked the viral fiber proteins by binding to their receptor recognizing knobs and thus prevented them from binding to their native receptor present on Ramos cells. The results also demonstrated for the first time that the sCAR protein can interact directly with the fiber protein without cofactors and, since its intracellular tail is lacking, without intracellular signaling.
FIG. 4.
sCAR protein inhibits binding to Ramos cells. Equal specific activity input doses (9,000 cpm) of radiolabeled Ad serotypes were incubated with sCAR protein at 10 μg/ml for 30 min at RT. Next, the virus was added to 2 × 106 Ramos cells and incubated in suspension for 1 h at RT. The cells were washed, the pellet was resuspended in 100 μl of PBS, and bound virus was determined by scintillation counting. The bars represent cpm bound (mean of three assays ± 5%). CTRL is control binding without sCAR preincubation.
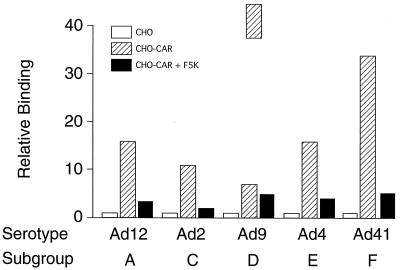
To confirm that the serotypes chosen attached to cells through a fiber-CAR interaction, a binding experiment was set up with wild-type CHO cells, which do not express the CAR protein, and a CHO cell line that had been transformed with the cDNA for human CAR (4). In the control experiment, none of the tested viruses (Ad12, Ad2, Ad9, Ad4, and Ad41) bound above background to wild-type CHO cells (Fig. 5). All viruses, however, bound at high levels to the CHO-CAR cells. Upon preincubation of these cells with saturating amounts of purified F5K protein (final concentration, 2 μg/ml), binding of the viruses dropped to background levels. Preincubation of Ad5 with saturating amounts of sCAR protein also inhibited the binding of this virus by as much as 95% (data not shown). Similar results were obtained when purified sCAR protein was immobilized in polystyrene wells, followed by a binding assay with 3H-labeled Ad virions. After preincubation of wells with F5K protein, virtually no virus binding was found (data not shown). These results confirmed that the tested viruses all used the CAR protein as a cellular fiber receptor.
FIG. 5.
Qualitative analysis of Ad binding to CHO-CAR cells. Approximately 106 CHO-CAR cells, or CHO-CAR cells preincubated with F5K protein at 5 μg/ml, were incubated for 1 h at 4°C with radiolabeled Ad serotypes. The cells were washed, the pellet was resuspended in 100 μl of PBS, and bound virus was determined by scintillation counting. Open bars represent binding of the viruses to CHO cells standardized to 1 (mean of 3 assays ± 6%). Hatched bars represent binding to CHO-CAR cells (fold increase over binding to CHO). Solid bars represent binding to CHO-CAR cells after preincubation with F5K protein (fold increase over binding to CHO). The counts measured (in cpm) were as follows: Ad12, 323, 5,021, 865; Ad2, 944, 10,299, 1,500; Ad9, 50, 320, 210; Ad4, 293, 4,772, 1,135; Ad41, 255, 8,567, 1,158.
DISCUSSION
The CAR protein has recently been identified as the cellular fiber receptor for the subgroup C viruses Ad2 and Ad5 and the coxsackie B viruses (4, 49). In this study, we have demonstrated that at least seven more Ad serotypes can use CAR as a cellular fiber receptor: Ad12 and Ad31 (subgroup A); Ad9, Ad15, and Ad19 (subgroup D); Ad4 (subgroup E); and Ad41 (subgroup F). Our results confirm the earlier observation that Ad2 fiber can block the binding of Ad12 to A549 and HeLa cells (2) and that Ad12 fiber knob can block Ad2. The fact that soluble CAR bound to both Ad12 and Ad31 was not surprising, since the fiber knobs of these serotypes are 81% similar at the amino acid level (Table 1).
From a previous study, it appeared that Ad9 (subgroup D) used the same cellular fiber receptor as Ad2 (38). A comparison of the fiber knob sequences of Ad8, Ad9, Ad15, and Ad19 revealed high similarity scores ranging from 59% between F15K and F19K to 85% between F9K and F19K to 95% between F8K and F9K (Table 1). Therefore, it is not surprising that sCAR bound to both the blotted Ad15 and Ad19 virions (Fig. 1 and data not shown). Given the high similarity between Ad8 and Ad9 fiber knob sequences, we would expect that Ad8 will bind to CAR as well.
It has been hypothesized that the only serotype in subgroup E, Ad4, is an Ad of mixed genotype in that the E1, E2, and E3 regions of the viral genome are homologous to sequences typically found in subgroup B viruses whereas the fiber gene and E4 are probably derived from subgroup C virus (3, 15). The Ad2 and Ad4 fiber knob sequences are 64% similar (Table 1), and it is not surprising that both recognize the same cellular fiber receptor.
The subgroup F serotype Ad41 has been shown to contain two distinct fiber genes; one encodes a fiber shaft consisting of 22 β-repeats of 15 amino acids each, and the other encodes a shaft with 12 β-repeats of 15 amino acids each (48, 57); these are designated the long-shafted (41L) and short-shafted (41S) fibers, respectively. We have demonstrated that fiber 41L recognizes CAR as a cellular fiber receptor protein. Competition experiments with purified fiber 41S knob protein failed to inhibit the binding of 3H-labeled Ad41 to A549 cells, suggesting that F41L functions as the attachment protein for this serotype (Table 2). Subgroup F has one more serotype, Ad40, that has also been shown to contain both a long-shafted protein and a short-shafted fiber protein (24). The knobs of the long fibers of Ad40 and Ad41 show 97% similarity at the amino acid level (26, 48). Therefore, it is highly probable that the long-shafted Ad40 fiber will also serve as the attachment protein.
It is interesting that an overall sequence similarity of only 29% for F2K and F41LK still results in recognition and binding to CAR as a cellular fiber receptor (Table 1). At this similarity level, however, it is still possible that conserved amino acids will play a role in the fiber-receptor interaction, possibly augmented by interactions based on structural secondary and tertiary features conserved between the different fiber knobs, such as loops, β-sheets (56), and clefts or canyons. As has been found for numerous protein-protein recognition sites (23), conserved amino acids and structures may cojointly provide multiple contacts between the fiber and the CAR proteins, resulting in attachment of the virus to the cell. Such contact points may vary between the numerous fiber-CAR pairs, as may the relative affinity of the different fibers for the CAR protein.
Our results further show that the F41S knob protein has no effect on the cell attachment of its parental virus Ad41 or any of the other tested viruses to the CAR protein. Several hypotheses can be offered as to the possible function of the short-shafted fiber. It has been shown for Ad40 that the virion penton base proteins are complexed with either the short- or the long-shafted fiber and that both combinations may occur on individual virions (24). Both the Ad40 and Ad41 penton base proteins lack the RGD sequence (31, 48), which promotes internalization of Ad particles by αv-integrins (55). The short-shafted fiber may promote internalization by interacting with receptors other than the αv-integrins. Experiments to evaluate this hypothesis are in progress.
The short-shafted Ad40 and Ad41 fibers are nearly as unrelated to the long-shafted fibers as they are to fibers of different subgroups (24, 34, 48). The subgroup F enteric viruses comprise only two serotypes, although at least 176 strains have been identified based on DNA restriction patterns (24, 26, 48). Studies of the heterogeneity of the fiber DNA and protein sequence have led to the hypothesis that the long-shafted fibers of Ad40 and Ad41 are similar enough for genetic recombination to occur with some frequency (26, 31, 48). The 41S fiber, which has been hypothesized to have evolved from the Ad5 fiber (24), may have lost its ability to recognize CAR as a cellular fiber receptor. Therefore, the 41S fiber may be a natural, if redundant, fiber mutant.
Our finding that the fibers of so many Ad serotypes are capable of using the CAR protein as a cellular receptor does not exclude the possibility that receptors other than CAR are recognized or used by these viruses. In this respect, a mimotope resembling the MHC class I α2 domain has been identified (20) as a potential cellular fiber binding site. A synthetic MHC-I α2 icosamer, although completely dissimilar to the sequence of the extracellular domain of the human CAR protein (4, 49), has been reported to show a net neutralization effect on Ad5 binding to HeLa cells (20).
In the past, Ads have been classified on the basis of their hemagglutination (HA) properties with erythrocytes from 13 different species (18, 40). Subgroup D has been further divided into group DI viruses (Ad9 and Ad19), which strongly hemagglutinate human and rat erythrocytes, group DII viruses (Ad15 and Ad22), which strongly hemagglutinate rat erythrocytes, and group DIII viruses, which do not hemagglutinate any erythrocytes (12, 18, 40). The subgroup C and E viruses Ad2, Ad5, and Ad4 show weak HA with rat erythrocytes, whereas the subgroup A viruses show virtually no HA at all (18). The Ad serotypes we have tested show widely differential HA activity but still recognize CAR as a cellular fiber receptor. Incubation of human erythrocytes, which express the CAR protein, with an anti-CAR antibody inhibits hemagglutination by the subgroup DI viruses Ad8 and Ad9 (5a). Taken together, these observations suggest that recognition of CAR and the ability to hemagglutinate erythrocytes are properties of fiber proteins that are distinct but may overlap.
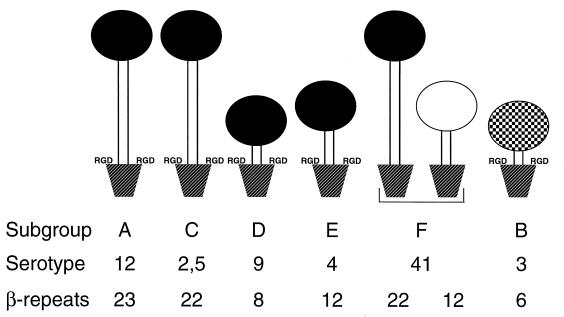
Viruses belonging to subgroups A, C, D, E, and F all bind the same cellular fiber receptor, CAR, yet they cause different patterns of disease. It is therefore unlikely that the fiber-receptor interaction is the sole determinant of viral tissue tropism (33, 50, 51). Observed differences in subgroup tropism may be significantly influenced by several attachment-related factors like the length of the fiber shaft and the receptor specificity of the penton base (Fig. 6). The shaft length of the fiber proteins varies from 23 to 6 β-repeats (8). Differences in fiber shaft length influence the role of penton base in binding, as previously shown for Ad9 (38), as do differences in the receptor specificities of penton base (53). When these differences are accounted for, the subgroups can be grouped as those with long CAR-recognizing fibers with RGD-containing pentons (subgroups A and C) or RGD-minus pentons (subgroup F), intermediate-length CAR-recognizing fibers with RGD-containing pentons (subgroup E), short CAR-recognizing fibers with RGD-containing pentons (subgroup D), and short non-CAR-recognizing fibers with RGD-containing pentons (subgroup B) (Fig. 6). We propose, therefore, that fiber shaft length is a prime determinant of the Ad attachment strategy. As long as the number of β-repeats in the fiber shaft remains above a critical number, the virus will use its fiber exclusively for attachment, followed by internalization by the αv-integrins (55). In this respect, it is of interest that serotype Ad4 (subgroup E), whose fiber has 12 β-repeats, binds to A549 cells exclusively through the fiber protein (data not shown). Any number of fiber shaft β-repeats below the critical number will result in a viral binding strategy that consists of binding, to various degrees, to CAR in combination with direct binding to the αv-integrins. This binding strategy may be a contributing factor to tropism for short-shafted viruses such as those in subgroups D and B.
FIG. 6.
A model for Ad tropism based on fiber shaft length, fiber receptor recognition, and penton base receptor recognition. The serotypes from the five Ad subgroups that recognize CAR have different fiber shaft lengths and are complexed with the pentameric penton base that has RGD sequences, although penton base can occur without RGD sequences (Ad40 and Ad41). Fibers from subgroups A and C to F are able to recognize the CAR protein as a cellular fiber receptor. The number of β-repeats in the fiber shaft of those serotypes varies from 23 (Ad12) to 8 (Ad9). Once the number of β-repeats drops to 8, attachment is enhanced by direct interaction of the penton base RGD loops with the cellular αv-integrins. The cellular fiber receptors for fiber 41 short and fiber 3 remain unidentified.
Serotypes Ad40 and Ad41 (subgroup F) are viruses with long-shafted CAR-recognizing fibers, which lack an RGD sequence in their penton base protein (31, 48). Internalization of these viruses may be facilitated by cellular molecules other than the αv-integrins, by using binding sites that have not been identified yet. Several in vitro studies of these enteric Ads have demonstrated that in addition to attachment and internalization, Ad tropism is governed by many factors that work at the cellular, transcriptional, and translational level (21, 22, 48, 50). This, however, may be vastly different in vivo (31). The elucidation of the mechanisms that underlie both the extracellular and the intracellular aspects of Ad tropism will have great impact on the molecular biology of Ads as well as on the application of Ad-based vectors in human gene therapy.
ACKNOWLEDGMENTS
We thank Duncan McVey, Joe Bruder, and Lou Cantolupo for critically reading the manuscript in its various states of development. We are indebted to Miguel Carrión and Marilyn Menger for help with purification of the His-tagged fiber knob proteins.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1989. [Google Scholar]
- 2.Bai M, Campisi L, Freimuth P. Vitronectin receptor antibodies inhibit infection of HeLa and A549 cells by adenovirus type 12 but not by adenovirus type 2. J Virol. 1994;68:5925–5932. doi: 10.1128/jvi.68.9.5925-5932.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey A, Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology. 1994;205:438–452. doi: 10.1006/viro.1994.1664. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M A, Crowell R L, Finberg R W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Brough D E, Lizonova A, Hsu C, Kulesa V A, Kovesdi I. A gene transfer vector-cell line system for complete functional complementation of adenovirus early region E1 and E4. J Virol. 1996;70:6497–6501. doi: 10.1128/jvi.70.9.6497-6501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Celi, L., and R. W. Finberg. Unpublished data.
- 6.Chroboczek J, Bieber F, Jacrot B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology. 1992;186:280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- 7.Chroboczek J, Jacrot B. The sequence of adenovirus fiber: similarities and differences between serotypes 2 and 5. Virology. 1987;161:549–554. doi: 10.1016/0042-6822(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 8.Chroboczek J, Ruigrok R W H, Cusack S. Adenovirus fiber. In: Doerfler W, Böhm P, editors. The molecular repertoire of adenoviruses. I. New York, N.Y: Springer-Verlag; 1995. pp. 163–200. [Google Scholar]
- 9.Defer C, Belin M T, Caillet-Boudin M L, Boulanger P. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Guilmi A M, Barge A, Kitts P, Gout E, Chroboczek J. Human adenovirus serotype 3 (Ad3) and the Ad3 fiber protein bind to a 130-kDa membrane protein on HeLa cells. Virus Res. 1995;38:71–81. doi: 10.1016/0168-1702(95)00043-p. [DOI] [PubMed] [Google Scholar]
- 11.Douglas J T, Rogers B E, Rosenfeld M E, Michael S I, Feng M, Curiel D T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 12.Eiz B, Pring-Åkerblom P. Molecular characterization of the type-specific γ-determinant located on the adenovirus fiber. J Virol. 1997;71:6576–6581. doi: 10.1128/jvi.71.9.6576-6581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fender P, Kidd A H, Brebant R, Öberg M, Drouet E, Chroboczek J. Antigenic sites on the receptor-binding domain of human adenovirus type 2 fiber. Virology. 1995;214:110–117. doi: 10.1006/viro.1995.9949. [DOI] [PubMed] [Google Scholar]
- 14.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber W C, Russell D J, Tibbetts C. Fiber gene and genomic origin of human adenovirus type 4. Virology. 1993;196:603–611. doi: 10.1006/viro.1993.1516. [DOI] [PubMed] [Google Scholar]
- 16.Henry L J, Xia D, Wilke M E, Deisenhofer J, Gerard R D. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol. 1994;68:5239–5246. doi: 10.1128/jvi.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herissé J, Galibert F. Nucleotide sequence of the EcoRI fragment of the adenovirus 2 genome. Nucleic Acids Res. 1981;9:1229–1240. doi: 10.1093/nar/9.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hierholzer J C. Further subgrouping of the human adenoviruses by differential hemagglutination. J Infect Dis. 1973;128:541–550. doi: 10.1093/infdis/128.4.541. [DOI] [PubMed] [Google Scholar]
- 19.Hong J S, Mullis K G, Engler J A. Characterization of the early region 3 and fiber genes of Ad7. Virology. 1988;167:758–767. [Google Scholar]
- 20.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC class I α2 domain at the surface of human epithelial and B-lymphoblastoid cells. EMBO J. 1997;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwitz M. The Adenoviridae and their replication. In: Fields B N, Knipe D M, editors. Virology. Vol. 2. New York, N.Y: Raven Press; 1990. pp. 1679–1721. [Google Scholar]
- 22.Horwitz M. Adenoviruses. In: Fields B N, Knipe D M, editors. Virology. Vol. 2. New York, N.Y: Raven Press; 1990. pp. 1723–1740. [Google Scholar]
- 23.Janin J, Clothia C. The structure of protein-protein recognition sites. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- 24.Kidd A H, Chroboczek J, Cusack J, Ruigrok R W H. Adenovirus type 40 virions contain two distinct fibers. Virology. 1993;192:73–84. doi: 10.1006/viro.1993.1009. [DOI] [PubMed] [Google Scholar]
- 25.Kidd A H, Erasmus M J. Sequence characterization of the adenovirus 40 fiber gene. Virology. 1989;172:134–144. doi: 10.1016/0042-6822(89)90115-3. [DOI] [PubMed] [Google Scholar]
- 26.Kidd A H, Erasmus M J, Tiemessen C T. Fiber sequence heterogeneity in subgroup F adenoviruses. Virology. 1990;179:139–150. doi: 10.1016/0042-6822(90)90283-w. [DOI] [PubMed] [Google Scholar]
- 27.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Louis N, Fender P, Barge A, Kitts P, Chroboczek J. Cell binding domain of adenovirus serotype 2 fiber. J Virol. 1994;68:4104–4106. doi: 10.1128/jvi.68.6.4104-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathias P, Wickham T, Moore M, Nemerow G. Multiple adenovirus serotypes use av integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mautner V, Steinthorsdottir V, Bailey A. Enteric adenoviruses. In: Doerfler W, Böhm P, editors. The molecular repertoire of adenoviruses. Vol. 2. Berlin, Germany: Springer-Verlag KG; 1995. pp. 229–282. [DOI] [PubMed] [Google Scholar]
- 32.Mei Y-F, Wadell G. Epitopes and hemagglutination binding domain on subgenus B:2 adenovirus fibers. J Virol. 1996;70:3688–3697. doi: 10.1128/jvi.70.6.3688-3697.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei Y-F, Wadell G. Molecular determinants of adenovirus tropism. In: Doerfler W, Böhm P, editors. The molecular repertoire of adenoviruses. Vol. 3. Berlin, Germany: Springer-Verlag KG; 1995. pp. 213–228. [DOI] [PubMed] [Google Scholar]
- 34.Pieniazek N J, Slemenda S B, Pieniazek D, Velarde J, Jr, Luftig R B. Human enteric adenovirus type 41 (Tak) contains a second fiber protein gene. Nucleic Acids Res. 1990;18:1901. doi: 10.1093/nar/18.7.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pieniazek N J, Slemenda S B, Pieniazek D, Velarde J, Jr, Luftig R B. Sequence of human enteric adenovirus type 41 Tak fiber protein gene. Nucleic Acids Res. 1989;17:9474. doi: 10.1093/nar/17.22.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Pring-Akerblom, P. 1993. Unpublished data.
- 36.Pring-Akerblom P, Adrian T. Characterization of adenovirus subgenus D fiber genes. Virology. 1995;206:564–571. doi: 10.1016/s0042-6822(95)80073-5. [DOI] [PubMed] [Google Scholar]
- 37.Pring-Akerblom P, Adrian T. Sequence characterization of the adenovirus 31 fibre and comparison with serotypes of subgenera A to F. Res Virol. 1995;146:343–354. doi: 10.1016/0923-2516(96)80597-8. [DOI] [PubMed] [Google Scholar]
- 38.Roelvink P, Kovesdi I, Wickham T. Comparative analysis of adenovirus fiber-cell interaction: Ad2 and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment. J Virol. 1996;70:7614–7621. doi: 10.1128/jvi.70.11.7614-7621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roelvink P W, Corsaro B G, Granados R R. Characterization of the Helicoverpa armigera and Pseudaletia unipuncta granulovirus enhancin genes. J Gen Virol. 1995;76:2693–2705. doi: 10.1099/0022-1317-76-11-2693. [DOI] [PubMed] [Google Scholar]
- 40.Rosen L. Hemagglutination-inhibition technique for typing adenovirus. Am J Hyg. 1960;71:120–128. doi: 10.1093/oxfordjournals.aje.a120085. [DOI] [PubMed] [Google Scholar]
- 41.Schnurr D, Dondero M E. Two new candidate adenovirus serotypes. Intervirology. 1993;36:79–83. doi: 10.1159/000150325. [DOI] [PubMed] [Google Scholar]
- 42.Signaes C, Akusjarvi G, Pettersson U. Adenovirus 3 fiber polypeptide gene: implications for the structure of the fiber protein. J Virol. 1985;53:672–678. doi: 10.1128/jvi.53.2.672-678.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprengel J, Schmitz B, Heuss-Neitzel D, Zock C, Doerfler W. Nucleotide sequence of human adenovirus type 12 DNA: comparative functional analysis. J Virol. 1994;68:379–389. doi: 10.1128/jvi.68.1.379-389.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevenson S C, Rollence M, Marshall-Neef J, McLelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J Virol. 1997;71:4782–4790. doi: 10.1128/jvi.71.6.4782-4790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevenson S C, Rollence M, White B, Weaver L, McClelland A. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J Virol. 1995;69:2850–2857. doi: 10.1128/jvi.69.5.2850-2857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart P L, Fuller S D, Burnett R M. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stouten P F W, Sander C, Ruigrok R W H, Cusack S. New triple helical model for the shaft of the adenovirus fibre. J Mol Biol. 1992;226:1073–1084. doi: 10.1016/0022-2836(92)91053-r. [DOI] [PubMed] [Google Scholar]
- 48.Tiemessen C T, Kidd A H. The subgroup F adenoviruses. J Gen Virol. 1995;76:481–497. doi: 10.1099/0022-1317-76-3-481. [DOI] [PubMed] [Google Scholar]
- 49.Tomko R P, Xu R, Philipson L. HCAR and MCAR: The human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wadell G. Adenoviruses. In: Zuckerman A J, Banatvala J E, Pattison J R, editors. Principles and practice of clinical virology. 2nd ed. Chichester, United Kingdom: John Wiley & Sons; 1990. pp. 267–287. [Google Scholar]
- 51.Wadell G, Allard A, Johansson M, Svensson L, Uhnoo I. Enteric adenoviruses. Ciba Found Symp. 1987;128:63–91. doi: 10.1002/9780470513460.ch5. [DOI] [PubMed] [Google Scholar]
- 52.Wadell G, Hammarskjöld M-L, Winberg G, Varsanyi T M, Sundell G. Genetic variability of adenoviruses. Ann N Y Acad Sci. 1980;354:16–42. doi: 10.1111/j.1749-6632.1980.tb27955.x. [DOI] [PubMed] [Google Scholar]
- 53.Wickham T J, Carrión M E, Kovesdi I. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific motifs. Gene Ther. 1995;2:750–756. [PubMed] [Google Scholar]
- 54.Wickham T J, Filardo E J, Cheresh D A, Nemerow G R. Integrin avb5 selectively promotes adenovirus mediated cell membrane permeabilization. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins avb3 and avb5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 56.Xia D, Henry L J, Gerard R D, Deisenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure. 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]
- 57.Yeh H-Y, Pieniazek N, Pieniazek D, Gelderblom H, Luftig R B. Human adenovirus type 41 contains two fibers. Virus Res. 1994;33:179–198. doi: 10.1016/0168-1702(94)90054-x. [DOI] [PubMed] [Google Scholar]