Abstract
The RNA elements that are required for replication of defective interfering (DI) RNA of the JHM strain of mouse hepatitis virus (MHV) consist of three discontinuous genomic regions: about 0.46 to 0.47 kb from both terminal sequences and an internal 58-nucleotide (nt)-long sequence (58-nt region) present at about 0.9 kb from the 5′ end of the DI genome. The internal region is important for positive-strand DI RNA synthesis (Y. N. Kim and S. Makino, J. Virol. 69:4963–4971, 1995). We further characterized the 58-nt region in the present study and obtained the following results. (i) The positive-strand RNA structure in solution was comparable with that predicted by computer modeling. (ii) Positive-strand RNA secondary structure, but not negative-strand RNA structure, was important for the biological function of the region. (iii) The biological function had a sequence-specific requirement. We discuss possible mechanisms by which the internal cis-acting signal drives MHV positive-strand DI RNA synthesis.
Infectious cDNA clones and cloned defective interfering (DI) RNAs of many RNA viruses have been used to study viral RNA elements that are necessary for viral RNA replication (cis-acting replication signals). Generally, cis-acting replication signals of RNA viruses include at least one of the genomic termini. Some viral cis-acting replication signals, like the DI RNAs of the murine coronavirus, mouse hepatitis virus (MHV) strain JHM (MHV-JHM), have an additional region(s) from the genome’s interior (10, 11, 16).
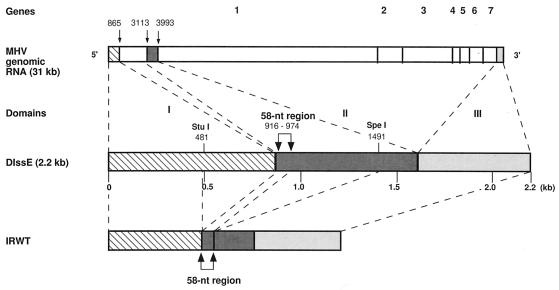
MHV, a prototypic coronavirus, contains a single-stranded, positive-sense RNA genome approximately 31 kb in length (3, 13, 14, 24). The smallest (2.2-kb) naturally-occurring MHV DI RNA, DIssE (18), consists of three noncontiguous regions of the parental MHV-JHM genome (21): the most 5′ region (domain I) corresponds to the most 5′ 0.86 kb of genomic RNA; the second, 0.75-kb region (domain II) corresponds to a 3.1- to 3.9-kb region from the 5′ end of the genome; and the third region (domain III) corresponds to the most 3′ 0.6 kb of genomic RNA (Fig. 1). Among these regions, cis-acting replication signals were initially identified as an approximately 470-nucleotide (nt)-long region of the most 5′ terminus of domain I, about 460 nt of the most 3′ terminus of domain III, and an internal sequence of 134 nt (the 0.13-kb region) that belongs to domain II (11). Further deletion analysis of the 0.13-kb region showed that the minimum length of the region for DI RNA replication was 58 nt (the 58-nt region); we previously referred to the 58-nt region as the 57-nt sequence (11). Characterization of DIssF, which is another MHV-JHM DI RNA, showed that its cis-acting replication signal also includes the 58-nt region (16).
FIG. 1.
Diagram of the structure of IRWT compared with those of MHV-JHM genomic RNA and DIssE RNA. The three domains (I through III) of DIssE RNA are indicated above the diagram of DIssE. The 58-nt region represents the internal cis-acting replication signal.
To look at the role of the internal cis-acting replication signal in DI RNA replication, we compared the sequences of the 0.13-kb regions derived from various MHV strains (11). Overall, the sequences of the 0.13-kb regions from various MHV strains are similar to each other. Computer-based secondary-structural analysis of the 0.13-kb regions revealed that most of the MHV strains form the same or similar main stem-loop structures in the positive strand, yet MHV-A59 forms a smaller main stem-loop structure (11). The RNA secondary structures in the negative strands are much less uniform among the MHV strains. Most DI RNAs that contain MHV-JHM-derived 5′- and 3′-terminal cis-acting replication signals plus internal 0.13-kb regions derived from various MHV strains replicate in MHV-infected cells at 37°C, except for MRP-A59, which contains an MHV-A59-derived 0.13-kb region (11). Interestingly, MRP-A59 replicates at 39.5°C but not at 37°C (11). Negative-strand RNA synthesis of MRP-A59 occurs at 37°C, whereas positive-strand MRP-A59 synthesis from accumulated negative-strand DI RNA does not occur after shifting the incubation temperature from 39.5 to 37°C, demonstrating that the internal cis-acting replication signal functions in positive-strand DI RNA synthesis but not in negative-strand RNA synthesis (11). We speculate that MRP-A59 forms an RNA structure that is suitable for RNA replication at 39.5°C but not at 37°C, while DI RNAs containing the 0.13-kb region derived from other MHV strains form the biologically active structure at 37°C. Because the secondary structure of the positive strand in the MHV-A59-derived 0.13-kb region differs from those of other MHVs, we hypothesize that the secondary structure of the internal cis-acting replication signal in the positive-strand RNA may be important for positive-strand DI RNA synthesis (11).
Replication of the cloned naturally occurring MHV-A59-derived DI RNA, MIDI (4, 17, 27), and a synthetic DI RNA transcript, B36, which consists mostly of MHV-A59-derived sequences (22), does not require the 58-nt-long region of MHV-A59 sequence that corresponds to the MHV-JHM 58-nt region. Deletion analyses of MIDI showed that the minimum sequence required for DI RNA replication was the 5′-end 466 nt and the 3′-end 461 nt, excluding poly(A) (17). However, whether RNA transcripts that consist of only the 5′-end 466 nt and the 3′-end 461 nt of MHV-A59 genomic termini can replicate is not known. In our hands, synthetic DI RNA transcripts that consisted of 0.5 kb from both termini of MHV-A59 and those that consisted of 0.5 kb from both termini of MHV-A59 plus the 0.13-kb region of MHV-A59 failed to replicate in MHV-A59-infected cells (unpublished data), whereas another MHV-A59-derived RNA, which had the MHV-JHM 0.13-kb internal sequence inserted between MHV-A59-derived termini, replicated in MHV-infected cells at 37°C (unpublished data). Therefore, we speculated that replication of MHV-A59-derived DI RNAs probably requires an internal region that is equivalent to the 58-nt region of MHV-JHM DI RNA (11). A functionally similar RNA element in MHV-A59 DI RNA may be located quite differently from the MHV-JHM 58-nt region. We further speculated that parental MHV genomic RNA may contain multiple genomic regions which have the same biological function as the internal cis-acting replication signal of MHV-JHM DI RNA. Some of them may be necessary for efficient MHV genomic RNA replication, whereas MHV DI RNA contains only one or two of these functional regions (11).
Knowing how the internal cis-acting replication signal drives MHV DI RNA positive-strand RNA synthesis will shed light on the mechanism of coronavirus positive-strand RNA synthesis, which is still poorly characterized. MHV DI RNAs replicate extremely efficiently in DI RNA-transfected cells (20); their replication can start from the introduced positive-strand DI RNA transcripts (20) as well as negative-strand transcripts (8), and a unique cold-sensitive variant in DI RNA replication is available (11). These properties of MHV DI RNAs provide an excellent model system to study the coronavirus RNA replication mechanism.
We tested our hypothesis that the secondary structure of the internal cis-acting replication signal in positive-strand RNA is important for positive-strand DI RNA synthesis (11), and our data indeed support this hypothesis.
MATERIALS AND METHODS
Viruses and cells.
Plaque-cloned MHV-A59 was used as a helper virus (12). Mouse DBT cells (6) were used for the preparation of seed viruses, and mouse L2 cells were used for RNA transfection experiments.
Plasmid construction.
The names of all oligonucleotides as well as their sequences, unique restriction sites, and binding sites are shown in Table 1. For all plasmid construction involving PCR, we used the same PCR conditions: plasmid DNA and oligonucleotide primers were incubated at 93°C for 30 s, 37°C for 45 s, and 72°C for 90 s in PCR buffer (0.05 M KCl, 0.01 M Tris-HCl [pH 8.0], 0.0025 M MgCl2, 0.01% gelatin, 0.17 mM [each] deoxynucleoside triphosphate, and 5 U of Taq polymerase [Promega]) for 25 cycles. A complete cDNA of DIssE, DE5-w4, was used as a parental clone for DI cDNA construction (20).
TABLE 1.
Synthetic oligonucleotides used in this study
| Oligonucleotide | Sequencea | Restriction site | Binding siteb |
|---|---|---|---|
| 52 | 5′-AAGCTTAATACGACTCACTATAGTATAAGAGTGATTGGCGTCCGTAC-3′ | 1–24 (23 nt T7 promoter) | |
| 101 | 5′-CGCTCTTAACTAGTTTG-3′ | SpeI | 1488–1504 of DIssE |
| 130 | 5′-TTCCAATTGGCCATGATCAA-3′ | 21236–1255 of IRWT | |
| 1942 | 5′-GTAGAGTACTATCAAATCTCTTTAGACAACGCCAGTT-3′ | ScaI | 643–678 of IRWT |
| 2167 | 5′-TATGACTCGGCCGAACAGATGTTGT-3′ | EagI | 852–878 of DIssE |
| 10052 | 5′-ATGCTGACAGGCCTGTAG-3′ | StuI | 905–922 of DIssE |
| 10078 | 5′-TCCGAATCAGACTAGTCAACCTGCTC-3′ | SpeI | 528–552 of INT-1 |
| 10082 | 5′-GTGGAGGCCTGTAGTTCTTG-3′ | StuI | 475–494 of IRWT |
| 10083 | 5′-GTTGGAGGCCTGTAGTCTTTG-3′ | StuI | 475–494 of IRWT |
| 10084 | 5′-GTGGAGGCCTGTAGTCCTTGTCGCTGAT-3′ | StuI | 475–502 of IRWT |
| 10115 | 5′-GTGGAGGCCTGTAGTCCTTGTCACCG-3′ | StuI | 475–500 of IRWT |
| 10120 | 5′-CTTTAGACAACGCCAGTT-3′ | 643–664 of IRWT | |
| 10127 | 5′-CTCTTAACTAGTCAACCTGCTCCTTGGCAACGCTGTCCTC-3′ | SpeI | 512–551 of IRWT |
| 10155 | 5′-CAGCTCGCCCGGGGTCC-3′ | SmaI | 4–21 of pT7-4 |
| 10167 | 5′-CTCTTAACTAGTCAACCTGCTCCTTGGCAACACCGTCCTC-3′ | SpeI | 512–551 of IRWT |
| 10245 | 5′-AACTAGTCAACCTGCTCCTTGGCAACTCCGTCCTCTTCTTGGGTATCGGAGACAAG-3′ | SpeI | 491–546 of IRWT |
| 10246 | 5′-AACTAGTCAACCTGCTCCTTGGCAACTCCGTCCTCTTCTTGGGTATCGGGGACAAG-3′ | SpeI | 491–546 of IRWT |
| 10252 | 5′-AACTAGTCAACCTGCTCCTTGGCAACTCCGTCCTCTTCTTGGGTATCTGCGAC-3′ | SpeI | 494–546 of IRWT |
| 10253 | 5′-AACTAGTCAACCTGCTCCTTGGCAACTCCGTCCTCTTCTTGGGTATCCGCGAC-3′ | SpeI | 494–546 of IRWT |
| 10308 | 5′-AACTAGTCAACCCGCTCCTTG-3′ | SpeI | 525–546 of IRWT |
| 10309 | 5′-AACTAGTCAACCTGTTCCTTGGCAAC-3′ | SpeI | 520–546 of IRWT |
| 10310 | 5′-AACTAGTCAACCTGCTCCCTGGCAACGCC-3′ | SpeI | 517–546 of IRWT |
| 10311 | 5′-AACTAGTCAACCTGCTCCTCGGCAACGCCG-3′ | SpeI | 516–546 of IRWT |
| 10316 | 5′-GTGGAGGCCTGTAGTCCCTGTCGC-3′ | SpeI | 475–498 of IRWT |
| 10317 | 5′-GTGGAGGCCTGTAGTCCTCGTCGCC-3′ | SpeI | 475–499 of IRWT |
| 10321 | 5′-GAGGCCTGTAGTCCATG-3′ | SpeI | 478–494 of IRWT |
| 10322 | 5′-ACTAGTCAACCTGCTCCATG-3′ | SpeI | 526–545 of IRWT |
| 10323 | 5′-GAGGCCTGTAGTCCTAGTC-3′ | SpeI | 478–496 of IRWT |
| 10324 | 5′-ACTAGTCAACCTGCTCCTAG-3′ | SpeI | 526–545 of IRWT |
| 10325 | 5′-GAGGCCTGTAGTCCGTG-3′ | SpeI | 478–494 of IRWT |
| 10326 | 5′-ACTAGTCAACCTGCTCCGTG-3′ | SpeI | 526–545 of IRWT |
| 10327 | 5′-GAGGCCTGTAGTCCTGGTC-3′ | SpeI | 478–496 of IRWT |
| 10328 | 5′-ACTAGTCAACCTGCTCCTGG-3′ | SpeI | 526–545 of IRWT |
PCR products were obtained with DE5-w4 and oligonucleotides 2167 and 101. The EagI-FspI fragment of the PCR products was then inserted into the PstI site of pT7-4 to yield JHM134+. PCR products were obtained by incubating DE5-w4 with oligonucleotide 10052 and oligonucleotide 101. Insertion of the StuI-SpeI fragment of the PCR products into the DE5-w4 large StuI-SpeI fragment resulted in plasmid INT-1. The SnaBI-SpeI fragment of the PCR products that was obtained by incubating INT-1 with oligonucleotide 10078 and oligonucleotide 52, which consists of T7 promoter sequence and the 5′ end of MHV genomic RNA sequence, was inserted into the large INT-1 SnaBI-SpeI fragment to produce IRWT. PCR products that were obtained by incubating IRWT with oligonucleotide 10082 and oligonucleotide 130 were restricted by StuI and NruI and then inserted into the large IRWT StuI-NruI fragment to produce IRP-1. IRP-2, IRP-3, and IRN-1 were constructed by a similar procedure, except that oligonucleotides 10083, 10084, and 10115, respectively, were used in place of oligonucleotide 10082. IRN-2 was constructed by inserting the SnaBI-SpeI fragment of the PCR products that were obtained by incubating IRWT with oligonucleotide 10127 and oligonucleotide 52 into the large IRWT SnaBI-SpeI fragment. Clones IRP-4, IRP-5, IRP-6, IRP-7, IRPN-2, IRPN-3, IRPN-4, IRPN-5, and IRPN-6 were constructed in a manner similar to clone IRN-2, except that oligonucleotides 10308, 10309, 10310, 10311, 10167, 10245, 10246, 10252, and 10253, respectively, were used in place of oligonucleotide 10127. The construction of IRPN-1 was identical to that of IRP-3, except that IRN-2 was used as a template for PCR. IRPN-7 was constructed in a manner similar to IRP-1, except that oligonucleotide 10316 was used in place of oligonucleotide 10082 and IRP-4 was used as a template for PCR. The construction of IRPN-8 was essentially the same as that of IRPN-7, except that oligonucleotide 10317 and IRP-5, respectively, were used in place of oligonucleotide 10082 and IRP-4. Insertion of the StuI-SpeI fragment of PCR products that were obtained by incubating IRWT with oligonucleotide pairs 10321 and 10322, 10323 and 10324, 10325 and 10326, and 10327 and 10328 into the large IRWT StuI-SpeI fragment resulted in the construction of IRPN-9, IRPN-10, IRPN-11, and IRPN-12, respectively. For each mutant, we sequenced the entire region of the insertion obtained by PCR.
RNA transcription and transfection.
Plasmid DNAs were linearized by XbaI digestion and transcribed with T7 RNA polymerase as previously described (20). We used a DEAE-dextran-mediated procedure for RNA transfection, as previously described (20).
Characterization of RNA secondary structure in solution.
RNA secondary structure analysis in solution was performed by digesting in vitro-transcribed RNA transcripts with various RNases followed by primer extension analysis. We followed the methods of Jacobson et al. (7) and Stern et al. (26) with modifications. Briefly, 2 μg of in vitro RNA transcripts corresponding to the MHV-JHM 0.13-kb region was suspended in 30 μl of a reaction buffer consisting of 30 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 270 mM KCl, 18 mM β-mercaptoethanol, and 100 μg of tRNA/ml. The sample was heated to 68°C for 5 min, slowly cooled to 37°C, and treated with RNases. Preliminary experiments were done to determine optimum enzyme dilutions and incubation temperatures which gave consistent partial digestions of the RNA transcripts. A 3-μl quantity of RNase A (0.25 μg/ml), RNase T1 (0.2 U/ml), or RNase V1 (700 U/ml) in cold enzyme buffer (80 mM HEPES [pH 7.8], 20 mM MgCl2, 300 mM KCl, and 6 mM β-mercaptoethanol) was added separately to the RNA samples and incubated for 17 min at room temperature, for 20 min at room temperature, and for 19 min at 37°C, respectively. The reaction was terminated by the addition of 150 μl of chilled stop buffer (300 mM sodium acetate and 10 mM EDTA) followed by placement on ice. For the partial digestion of RNA transcripts with RNase U2, 1 μl of RNase U2 (200 U/ml) was mixed with 2 μg of RNA in 5 μl of RNase U2 buffer (8 mM sodium citrate [pH 3.5], 0.8 mM EDTA, 0.5 μg of tRNA/ml) and incubated for 12 min at 50°C. The reaction was terminated by the addition of 194 μl of chilled stop buffer. The 5′-end 32P-labeled oligonucleotide 10155, which binds to the plasmid-derived sequence downstream of the 0.13-kb region, was added to the RNA, and primer extension reactions were performed according to the methods of Stern et al. (26). Reaction products were analyzed on a 6% polyacrylamide gel containing 7 M urea.
Preparation of virus-specific intracellular RNA and Northern blotting.
Viral-specific intracellular RNAs were extracted 7 h postinfection (p.i.) as previously described (18). Poly(A)-containing RNAs were selected by oligo(dT)-cellulose column chromatography. The RNAs were denatured and electrophoresed through a 1% agarose gel containing formaldehyde (19) and transferred onto nylon filters (ICN Pharmaceutical, Inc.). Northern blot analysis was performed with a 32P-labeled random-primed probe corresponding to 85 to 474 nt from the 5′ end of DE5-w4.
Direct sequencing analysis of DI-specific RT-PCR products.
DI RNA-specific cDNA was synthesized by incubating intracellular RNA with oligonucleotide 1942, which binds a region spanning nt 643 to 678 from the 5′ end of IRWT; this binding site represents a junction site between the 3′-end cis-acting signal and the inserted fragment containing the 58-nt region. After cDNA synthesis, avian myeloma virus reverse transcriptase (RT) (Promega) was inactivated by heating the sample to 100°C for 10 min. The DI-specific RT-PCR products were obtained by incubating the cDNA with oligonucleotides 1942 and 52 and were separated by agarose gel electrophoresis. Direct PCR sequencing was performed as previously described (9, 28), using oligonucleotide 10120 as a primer.
RESULTS
RNA secondary structure of the 58-nt region in solution.
To test our hypothesis that the positive-strand RNA secondary structure of the internal cis-acting replication signal is important for positive-strand DI RNA synthesis (11), we used enzymatic probing methods to determine whether the RNA secondary structure of the 58-nt region predicted by computer modeling (31) was comparable to the actual RNA secondary structure in solution. Although 58 nt defines the minimum size needed for biological function, we used in vitro transcripts containing the entire 0.13-kb region for this analysis; we hoped that MHV-derived sequences surrounding the 58-nt region might stabilize the structure formed by the 58 nt.
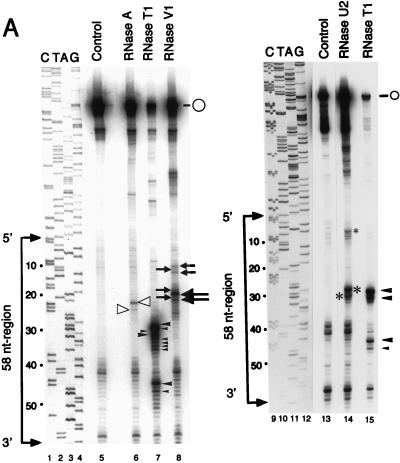
The RNA transcript corresponding to the internal region of MHV-JHM RNA was transcribed in vitro from plasmid JHM134+ and treated with various RNases. We chose RNase T1, RNase A, and RNase U2, which react with single-stranded regions, and RNase V1, which reacts with double-stranded regions, so that when analyzed by primer extension, specific stops would be generated that reflect the accessibility of the region to RNases. Figure 2A represent two of these primer extension analyses. We conducted nine independent experiments for RNase A and RNase T1 treatments and four independent experiments for RNase U2 and RNase V1 treatments. We found that there were slight variations in the results of separate experiments, yet some sites were consistently cut by RNases. Figure 2B shows the locations of consistent RNase cleavage sites. RNase A showed consistently strong cleavage at the residues in positions 21 (21 U) and 23; cleavage of 23 U by RNase A was more evident in other experiments (data not shown). RNase T1 cleaved efficiently at 28 G, 31 G, and 43 G in repeated experiments. RNase T1 frequently showed weak cleavage activity at two residues 3′ to 28 G, four residues 3′ to 31 G, and 45 C. The minor RNase T1 cleavage at 46 A to 49 G shown in lane 7 of Fig. 2A was not consistently observed, as shown in lane 15 of Fig. 2A. RNase U2 cleaved efficiently at 27 A and 29 A and less efficiently at 6 A in repeated experiments. RNase V1 consistently cleaved at 18 C and 20 A. In most experiments, weak cleavage was observed at 9 C, 10 C, 11 U, and 19 G (Fig. 2A). The RNase V1 cleavage that is shown in Fig. 2A as occurring several nucleotides downstream of 20 A was not obvious in other experiments.
FIG. 2.
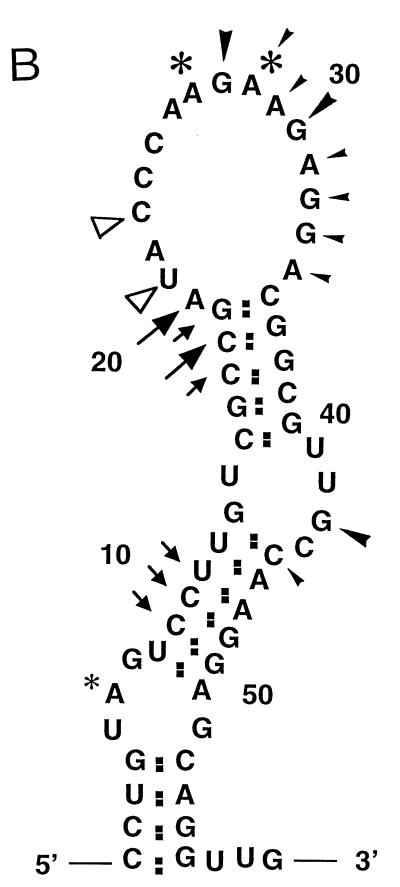
Enzymatic probing of positive-strand RNA of the 58-nt region. (A) RNA transcripts including the 0.13-kb region were synthesized in vitro and treated with no RNase (lanes 5 and 13), RNase A (lane 6), RNase T1 (lanes 7 and 15), RNase V1 (lane 8), or RNase U2 (lane 14). Primer extension was performed with a 32P-labeled oligonucleotide that hybridizes downstream of the 0.13-kb region. The primer extension products were applied to a 6% sequencing gel. Dideoxysequencing of the corresponding sequence is shown in lanes 1 to 4 and 9 to 12. RNase cleavage sites that were consistently observed in the repeated experiments are marked; large and small symbols represent strong and weak cleavage sites, respectively. Open arrowheads, RNase A cuts; solid arrowheads, RNase T1 cuts; asterisks, RNase U2 cuts; arrows, RNase V1 cuts. The open circles show the positions of full-size primer extension products. (B) Schematic representation of the results obtained by enzymatic probing. The positions of RNase cleavage sites with respect to the computer-predicted RNA secondary structure of the positive strand of the 58-nt region are shown. The free energy of the end loop was −6.6 Kcal/mol. The symbols are the same as for panel A.
Although some unexpected consistent RNase cleavage occurred, e.g., an RNase V1 cut at 20 A, a weak RNase T1 cut at 45 C, and RNase T1 cleavage at several non-G sites, we concluded that the RNA secondary structure deduced from this primer extension analysis and that of the computer-predicted structure (11) were similar; we discuss the relative importance of the unexpected RNase cuts in Discussion below. Based on these experiments, we are confident that the computer modeling of RNA secondary structure is reliable enough to serve as a foundation for our subsequent experiments.
Asymmetric mutational analysis of the 58-nt region.
To test whether the positive-strand RNA structure of the 58-nt region was important for biological function, we examined whether DI RNA replication was affected by altering the 58-nt region’s RNA secondary structure on each strand. To create these mutants, we used a property particular to G-U base pairs that selectively disrupts the structure in either the positive or negative strands. G-U pairs can replace A-U pairs or G-C pairs in one strand, but on the opposite strand the A-C base pairs cannot form and the stem structure will be disrupted. Thus, by changing a G-C pair to a G-U pair, an A-U pair to a G-U pair, a G-U pair to a G-C pair, or a G-U pair to an A-U pair in the DI RNA and assaying the DI RNA replication, we could selectively evaluate the need for a specific structure in the two strands.
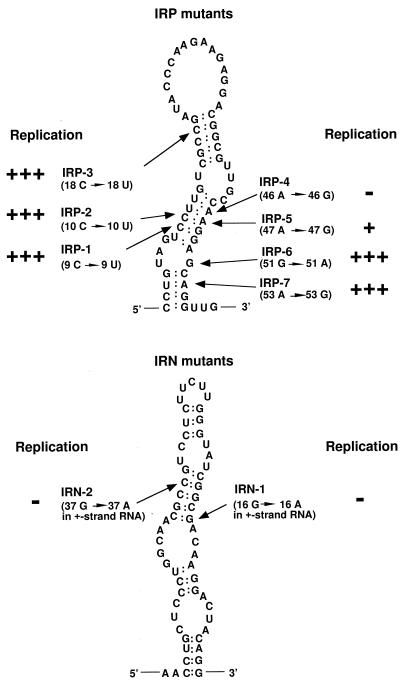
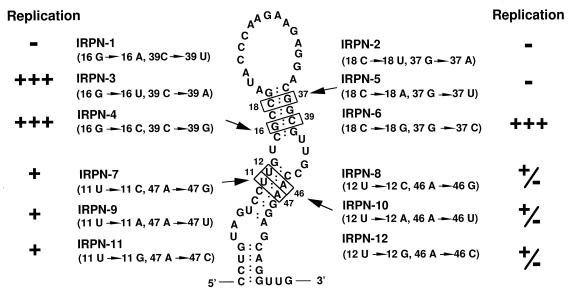
IRWT, which contained the MHV-JHM-derived 58-nt region, was used as a wild-type (wt) DI RNA. For convenience in DNA construction, IRWT contained 0.27-kb-long extra sequence that is not necessary for DI RNA replication (10, 11) between the 58-nt region and the 3′ cis-acting replication signal (Fig. 1). We constructed two series of IRWT-derived mutants, one of which maintained the positive-strand RNA secondary structure of the 58-nt region but not the negative-strand RNA secondary structure (Fig. 3); these were IRP-1, IRP-2, IRP-3, IRP-4, IRP-5, IRP-6, and IRP-7. Computer modeling showed that the 58-nt region of the negative-strand RNA of each of these mutant DI RNAs had an RNA secondary structure extensively different from those of the others, and none of the negative-strand RNA secondary structures of the mutant DI RNAs’ 58-nt regions were similar to that of the wt 58-nt region (data not shown). A second series of mutants (IRN-1 and IRN-2) contained the wt negative-strand RNA secondary structure of the 58-nt region and had altered positive-strand RNA secondary structures (Fig. 3). Using computer modeling, we examined all possible single-nucleotide substitutions from a G-C pair to a G-U pair, from an A-U pair to a G-U pair, from a G-U pair to a G-C pair, or from a G-U pair to an A-U pair in both the positive and negative strands of the 58-nt region and found that nucleotide substitution at only these nine sites caused strand-specific alterations in RNA secondary structure; i.e., they did not alter RNA secondary structure in one strand but did alter it in the opposite strand.
FIG. 3.
Positions of nucleotide substitutions in IRP and IRN mutants. The position from the 5′ end of the 58-nt region and the nucleotide substitution of each mutant are in parentheses below the mutant name. The diagram of IRP mutants represents the wt secondary structure of the positive-strand RNA of the 58-nt region. The diagram of IRN mutants represents the wt secondary structure of the negative-strand RNA of the 58-nt region. The replication efficiency of each DI RNA is summarized from the data shown in Fig. 4. DI RNAs that replicated at an efficiency similar to that of IRWT are indicated by “+++”, and those that replicated at approximately 5% of IRWT efficiency are indicated by “+”. DI RNAs that failed to replicate are indicated by “−”.
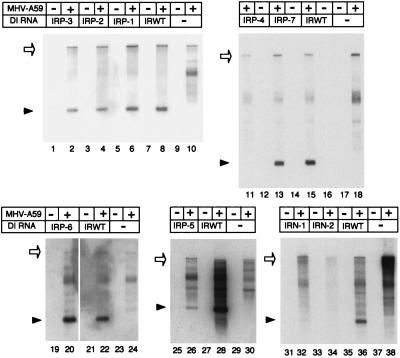
Equal amounts of in vitro-synthesized DI RNA transcripts were transfected into L2 cells infected with MHV-A59 helper virus 1 h prior to transfection and into mock-infected cells. Virus-specific intracellular RNAs were extracted 7 h p.i., and poly(A)-containing RNAs were selected by oligo(dT) column chromatography and separated on 1% agarose-formaldehyde gels. Northern blot analysis with a probe that specifically bound to DI RNAs and MHV mRNA 1 showed in repeated experiments that IRWT, IRP-1, IRP-2, IRP-3, IRP-5, IRP-6, and IRP-7 replicated and that IRP-4, IRN-1, and IRN-2 did not replicate (Fig. 4). IRP-5 replicated at a significantly lower efficiency than either IRWT or any other IRP mutant. Sequence analysis of DI-specific RT-PCR products from DI RNA-transfected cells showed that replicating DI RNAs maintained the sequences of their input 58-nt regions (data not shown). IRP-1, IRP-2, IRP-3, IRP-5, IRP-6, and IRP-7, all of which maintained the positive-strand RNA secondary structure of the 58-nt region but not the negative-strand RNA secondary structure, replicated. These data not only demonstrated that the negative-strand RNA structure was not important for the biological function of the 58-nt region but also indicated the importance of positive-strand RNA secondary structure in the function of the 58-nt region.
FIG. 4.
Replication of IRP and IRN mutants in DI RNA-transfected, MHV-A59-infected cells. Equal amounts of in vitro-synthesized DI RNA were transfected into L2 cell monolayers that had been infected with MHV-A59 helper virus 1 h previously. Cytoplasmic RNA was harvested at 7 h p.i. Poly(A)-containing RNAs were selected by oligo(dT) column chromatography and electrophoresed through a 1% agarose gel containing formaldehyde. Following transfer to nitrocellulose membranes, samples were hybridized to a 32P-labeled probe corresponding to nt 85 to 473 of MHV genomic RNA. +, MHV infection; −, mock infection and mock transfection. MHV-A59 genomic RNA and DI RNA are indicated by open arrows and arrowheads, respectively.
Sequence requirement of the 58-nt region.
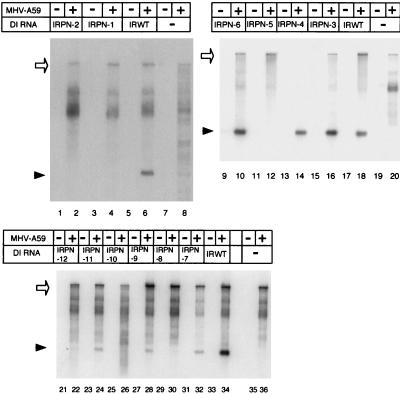
We next investigated why IRN-1, IRN-2, and IRP-4 did not replicate. The failure to replicate IRN-1 and IRN-2 could be due to the destruction of the positive-strand RNA secondary structure of the 58-nt region or to the primary sequence alteration in these mutants. To distinguish between these possibilities, we constructed IRPN-1 and IRPN-2, each of which contained the original mutation of IRN-1 and IRN-2, respectively, with an additional 39 C → 39 U substitution in IRN-1 and an additional 18 C → 18 U substitution in IRN-2 (Fig. 5). These changes had the effect of restoring the overall secondary structure of both the positive and negative strands to that of the wt while keeping in place the primary sequence alteration previously introduced in IRN-1 and in IRN-2. Northern blot analysis of DI RNA replication in DI RNA-transfected cells showed that IRPN-1 and IRPN-2 did not replicate at all (Fig. 6).
FIG. 5.
Positions of nucleotide substitutions in IRPN mutants. The position and nucleotide substitution of each mutant is indicated on the computer-predicted secondary structure of the wt positive-strand RNA of the 58-nt region. The nucleotide base pairs altered in these mutants are boxed and numbered from the 5′ end of the 58-nt region. The replication efficiency of each DI RNA is summarized from the data shown in Fig. 6. DI RNAs that replicated at an efficiency similar to that of IRWT are indicated by “+++”, and those that replicated at approximately 5% of IRWT efficiency are indicated by “+”; those that replicated at less than 1% of IRWT efficiency are indicated by “+/−”. DI RNAs that failed to replicate are indicated by “−”.
FIG. 6.
Replication of IRPN mutants in DI RNA-transfected, MHV-A59-infected cells. RNA transfection and Northern blot analyses were performed as described in the legend to Fig. 4. +, MHV infection; −, mock infection and mock transfection. MHV-A59 genomic RNA and DI RNA are indicated by open arrows and arrowheads, respectively.
IRPN-1 had a 16 A-39 U base pair in place of the wt 16 G-39 C pair in the positive strand. This change may weaken the RNA secondary structure and cause formation of an altered RNA secondary structure which is no longer functional. Alternatively, the 58-nt region may require the presence of 16 G for its function. To examine these possibilities, we constructed IRPN-3 and IRPN-4. Both clones maintained both positive- and negative-strand RNA secondary structures and had two nucleotide substitutions; IRPN-3 had 16 U and 39 A, and IRPN-4 had 16 C and 39 G. Note that IRPN-3 contained a U-A base pair at positions 16 and 39. Northern blot analysis of intracellular RNAs from DI RNA-transfected cells showed that both IRPN-3 and IRPN-4 replicated (Fig. 6). Sequence analysis of DI-specific RT-PCR products showed that replicating IRPN-3 and IRPN-4 maintained the input sequences of their 58-nt regions. Because IRPN-3, which forms a weak 16 U-39 A base pair, replicated, the presence of a G-C (or C-G) base pair at nucleotides 16 and 39 was not a requirement for the biological function of the 58-nt region. These data demonstrated that nt 16 may be G, U, or C, but not A, and still maintain the function of the 58-nt region.
Nucleotide substitutions introduced in IRPN-2 changed the wt G-C base pair to a weak U-A base pair at positions 18 and 37. The failure of IRPN-2 to replicate may be due either to changing the G-C base pair to a U-A base pair or to substituting 37 A for 37 G. The 18 C → 18 U change was an unlikely reason for the loss of the biological function of the 58-nt region in IRPN-2, because IRP-3, which contained 18 U, replicated well (Fig. 4). To clarify why IRPN-2 did not replicate, we constructed IRPN-5 and IRPN-6; IRPN-5 had an 18 A-37 U base pair, and IRPN-6 had an 18 G-37 C base pair, while each contained wt RNA secondary structure in both strands. Characterization of DI RNA replication in DI RNA-transfected cells showed that IRPN-6, but not IRPN-5, replicated (Fig. 6). Sequence analysis of IRPN-6-specific RT-PCR products demonstrated that there was no sequence change in the 58-nt region of the replicating IRPN-6 (data not shown). DI RNAs containing 18 C-37 G (IRWT), 18 U-37 G (IRP-3), or 18 G-37 C (IRPN-6) replicated, whereas those containing 18 A-37 U (IRPN-5) or 18 U-37 A (IRPN-2) did not replicate, indicating that nt 18 should not be A and/or nt 37 should not be U or A in order to maintain the function of the 58-nt region.
To discover why IRP-4 did not replicate, we constructed three additional mutant DI RNAs: IRPN-8, IRPN-10, and IRPN-12. These DI RNAs had nucleotide substitutions at nt 12 and 46; IRPN-8 had a 12 C-46 G base pair, IRPN-10 had a 12 A-46 U base pair, and IRPN-12 had a 12 G-46 C base pair (Fig. 5). All of these mutants maintained the wt RNA secondary structure of the 58-nt region in both strands (Fig. 5). Characterization of DI RNA replication in DI RNA-transfected cells showed that these DI RNAs replicated very poorly (less than 1% as efficiently as IRWT) (Fig. 6), demonstrating that any sequence substitutions in this base pair are lethal for DI RNA replication. Such a primary sequence requirement may indicate that the nucleotides in these positions have roles in addition to maintaining the integrity of the secondary structure of the positive-strand RNA of the 58-nt region.
We next examined why IRP-5, which maintained the positive-strand RNA secondary structure of the 58-nt region (Fig. 3), replicated at a low efficiency by characterizing three additional DI RNAs, IRPN-7, IRPN-9, and IRPN-11. These DI RNAs had wt RNA secondary structure at both strands and had nucleotide substitutions at nt 11 and 47; IRPN-7 had an 11 C-47 G base pair, IRPN-9 had an 11 U-47 A base pair, and IRPN-11 had an 11 G-47 C base pair (Fig. 5). Characterization of DI RNA replication in DI RNA-transfected cells showed that these three mutants replicated with an efficiency similar to that of IRP-5 (Fig. 6). Sequence analysis of DI-specific RT-PCR products from DI RNA-transfected cells showed that IRPN-7, IRPN-9, and IRPN-11 maintained the sequences of their input 58-nt regions (data not shown). These data demonstrated that the 11 U-47 A base pair was essential for efficient DI RNA replication. Because mutant DI RNAs that contained nucleotide substitutions at the nt 11-47 base pair replicated better than those contained nucleotide substitutions at the nt 12-46 base pair, the sequence requirement for DI RNA replication at the nt 11-47 base pair was less stringent than that at the nt 12-46 base pair.
These studies indicated that, in addition to maintaining the integrity of the positive strand of the 58-nt region, specific primary sequences at specific sites of the region were also important for its biological function.
DISCUSSION
The present study tested a hypothesis that the RNA secondary structure made by the 58-nt region is important for positive-strand DI RNA synthesis. We showed that the RNA secondary structure of the 58-nt region in solution, deduced by enzymatic-probing methods, and that predicted by computer-based secondary structure analysis were similar. The RNA secondary structure of the 58-nt region in the positive strand, but not that in the negative strand, was important for biological function. The function of the 58-nt region tolerated many sequence substitutions, yet there was a sequence requirement. It was less likely that the differences in the 58-nt regions in DI RNAs affected the stability of the RNAs and thus determined DI RNA replication, because we did not see any noticeable differences in the amounts of undegraded DI RNAs among replication-competent and replication-incompetent DI RNAs that were obtained 2, 4, or 6 h after their transfection into non-MHV-infected cells (data not shown).
Enzymatic probing of the RNA secondary structure showed a close similarity between the computer-predicted RNA secondary structure and that in the solution; most RNase cleavage sites generally fit well with the computer-predicted RNA secondary structure. However, there were some differences. Because RNase V1 cleaved 20 A, which was predicted to be a single-strand region, there is a possibility that 20 A has an interaction with some other region of the RNA. RNase T1 frequently cleaved at 33 G and 34 G, both of which were predicted to be parts of the end loop (Fig. 2B), yet these sites were not as efficiently cut as those at residues 28 G and 31 G. The weak cleavages by these RNases may indicate that some RNA molecules form a different RNA secondary structure(s) at the end loop in the solution, e.g., some population of RNA transcripts may form 24 C-33 G and 25 C-33 G base pairs; these interactions may produce a smaller end loop structure. The size of the end loop may not be very crucial for the function of the 58-nt region, because the MHV-1 0.13-kb region is biologically functional and computer modeling predicted a small end loop structure (11). RNase T1 frequently cleaved at the non-G residues 29 A, 30 A, 32 A, 35 A, and 45 C. However, such anomalous cleavages were less obvious in some experiments (Fig. 2A, compare lanes 7 and 15). Because RNase T1 digests only 3′ of G residues, but no other nucleotides, at the amount of the enzyme used in this study (0.0006 U), these non-G cleavages could be an artifact of the experiments; i.e., these bands might be the results of premature termination of primer extension products. The appearance of these anomalous cleavages at non-G residues by RNase T1 during the study of RNA secondary structure has also been shown by others (2, 7).
How does the positive-strand RNA secondary structure of the 58-nt region function in positive-strand DI RNA synthesis? One possibility is that the 58-nt region is a part of the viral polymerase recognition site; the MHV RNA replication mechanism may recognize the internal cis-acting replication signal and other regions of cis-acting replication signals to initiate positive-strand RNA synthesis from the 5′ end of DI RNA. There is a precedent for this possibility; single-strand RNA bacteriophage Qβ contains two internal viral polymerase recognition sites for negative-strand RNA synthesis; each internal recognition site is separated from its functional initiation site by about 1.4 and 2.8 kb, respectively, from the 3′ end (1, 23, 25). Another possible mechanism of the positive-strand RNA secondary structure of the 58-nt region in positive-strand DI RNA synthesis is that the 58-nt region may interact with another region of DI RNA to facilitate the stabilization of an RNA secondary structure, which is important for the initiation of positive-strand RNA synthesis. We found a sequence-specific requirement of the biological function of the 58-nt region: sequence alteration at some sites did not affect function, whereas single-nucleotide substitutions at other sites severely affected it. It is possible that the specific sites within the 58-nt region interact with other cis-acting regions through sequence-specific contacts, and sequence substitution at particular sites within the 58-nt region may disrupt such RNA-RNA interaction. Another possibility is that viral or host protein, which is necessary for the biological function of the 58-nt region, specifically binds to the region, and a mutation in the 58-nt region may interfere with this specific protein binding. Several host proteins that bind to various regions of MHV RNA have been described (5, 29, 30) and identified (15), yet whether any proteins specifically interact with the 58-nt region is not known.
ACKNOWLEDGMENTS
We thank Heather King for careful reading of the manuscript.
This work was supported by Public Health Service grants AI29984 and AI32591 from the National Institutes of Health.
REFERENCES
- 1.Barrera I, Schuppli D, Sogo J M, Weber H. Different mechanisms of recognition of bacteriophage Qβ plus and minus strand RNAs by Qβ replicase. J Mol Biol. 1993;232:512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- 2.Baudin F, Bach C, Cusack S, Ruigrok R W H. Structure of influenza virus RNP. 1. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonilla P J, Gorbalenya A E, Weiss S R. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Groot R J, van der Most R G, Spaan W J M. The fitness of defective interfering murine coronavirus DI-a and its derivatives is decreased by nonsense and frame shift mutations. J Virol. 1992;66:5898–5905. doi: 10.1128/jvi.66.10.5898-5905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuya T, Lai M M C. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirano N, Fujiwara K, Hino S, Matsumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson S J, Konings D A, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joo M, Banerjee S, Makino S. Replication of murine coronavirus defective interfering RNA from negative-strand transcripts. J Virol. 1996;70:5769–5776. doi: 10.1128/jvi.70.9.5769-5776.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joo M, Makino S. Mutagenic analysis of the coronavirus intergenic consensus sequence. J Virol. 1992;66:6330–6337. doi: 10.1128/jvi.66.11.6330-6337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y-N, Jeong Y S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y-N, Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai M M C, Brayton P R, Armen R C, Patton C D, Pugh C, Stohlman S A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai M M C, Stohlman S A. RNA of mouse hepatitis virus. J Virol. 1978;26:236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H J, Shieh C-K, Gorbalenya A E, Eugene E V, La Monica N, Tuler J, Bagdzhadzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H P, Zhang X, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y-J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontinuous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luytjes W, Gerritsma H, Spaan W J M. Replication of synthetic defective interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology. 1996;216:174–183. doi: 10.1006/viro.1996.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makino S, Fujioka N, Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985;54:329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino S, Joo M, Makino J K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makino S, Lai M M C. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J Virol. 1989;63:5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makino S, Shieh C-K, Soe L H, Baker S C, Lai M M C. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988;166:550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masters P S, Koetzner C A, Kerr C A, Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer F, Weber H, Weissman C. Interactions of Qβ replicase with Qβ RNA. J Mol Biol. 1981;153:631–660. doi: 10.1016/0022-2836(81)90411-3. [DOI] [PubMed] [Google Scholar]
- 24.Pachuk C J, Bredenbeek P J, Zoltick P W, Spaan W J M, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis virus, strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuppli D, Barrera I, Weber H. Identification of replication elements on bacteriophage Qβ minus strand RNA that are essential for template activity with Qβ replicase. J Mol Biol. 1994;243:811–815. doi: 10.1006/jmbi.1994.1682. [DOI] [PubMed] [Google Scholar]
- 26.Stern S, Moazed D, Noller H F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 27.van der Most R G, Bredenbeek P J, Spaan W J M. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winship P R. An improved method for directly sequencing PCR material using dimethyl sulfoxide. Nucleic Acids Res. 1989;17:1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu W, Leibowitz J. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology. 1995;214:128–138. doi: 10.1006/viro.1995.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu W, Leibowitz J. Specific binding of host cellular proteins to multiple sites within the 3′ end of mouse hepatitis virus genomic RNA. J Virol. 1995;69:2016–2023. doi: 10.1128/jvi.69.4.2016-2023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M, Steigler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]