Abstract
Deoxyribonucleoside triphosphate (dNTP) pool imbalances are associated with an increase in the rate of misincorporation and hypermutation during in vitro reverse transcription reactions. However, the effects of in vivo dNTP pool imbalances on the accuracy of reverse transcription are unknown. We sought to determine the effects of in vivo dNTP pool imbalances on retroviral mutation rates and to test our hypothesis that 3′-azido-3′-deoxythymidine (AZT) increases the retroviral mutation rates through induction of dNTP pool imbalances. D17 cells were treated with thymidine, hydroxyurea (HU), or AZT, and the effects on in vivo dNTP pools were measured. Thymidine and HU treatments induced significant dNTP pool imbalances. In contrast, AZT treatment had very little effect on the dNTP pools. The effects of in vivo dNTP pool imbalances induced by thymidine and HU treatments on the retroviral mutation rates were also determined. Spleen necrosis virus (SNV)-based and murine leukemia virus (MLV)-based retroviral vectors that expressed the lacZ mutant reporter gene were used. The frequencies of inactivating mutations introduced in the lacZ gene in a single replication cycle provided a measure of the retroviral mutation rates. Treatment of D17 target cells with 500 μM thymidine increased the SNV and MLV mutant frequencies 4.7- and 4-fold, respectively. Treatment of D17 target cells with 2 mM HU increased the SNV and MLV mutant frequencies 2.1- and 2.7-fold, respectively. These results demonstrate that dNTP pool imbalances are associated with an increase in the in vivo retroviral mutation rates, but AZT treatment results in an increase in the retroviral mutation rates by a mechanism not involving alterations in dNTP pools.
High levels of genetic variation found in retrovirus populations arise through mutation, recombination, and selection (20, 33, 34, 41–44, 46). The clinically significant consequences of the high levels of retrovirus variation include the emergence of virus variants that are resistant to antiretroviral drugs (7, 8, 17, 29, 30, 45, 52, 54, 57), alteration of cellular tropism (38), and modulation of viral pathogenesis (19, 22, 61). High error rates during reverse transcription by reverse transcriptase (RT) are an important source of mutations in retroviral genomes (33, 34, 41–44, 46). RTs lack proofreading activity, and it has been hypothesized that they are evolutionarily selected for low affinity to the template (56). Therefore, the accuracy of reverse transcription depends solely on discrimination between nucleotides prior to their incorporation into the nascent DNA. The host cell DNA polymerases also replicate the provirus through each cell cycle; however, the error rates of mammalian DNA polymerases are much lower than the error rates of RTs, and they do not significantly contribute to the generation of variation in retroviral populations (14). RNA polymerase II transcribes the provirus to generate the genomic RNA of the next generation and could significantly contribute to retroviral variation. The error rate of RNA polymerase II has not been measured; however, our recent results have suggested that at least one-third of the mutations in retroviral genomes occur during the DNA-dependent DNA synthesis stage of reverse transcription (27). A high rate of retroviral recombination rapidly assorts these mutations to further increase variation in retroviral populations. Thus, the processes of mutation and recombination play important roles in generating variation in retroviral populations (20, 33, 34, 41–44, 46).
The error rates of many RTs have been measured in vivo and range from 0.3 × 10−5 to 3 × 10−5 mutations/bp/replication cycle (33, 34, 41–44, 58). Previous studies have shown that the mutation rates of retroviruses can be further increased by treatment of the target cells with nucleoside analogs 5-azacytidine [4-amino-1-β-ribofuranosyl-5-triazine-2(1H)one] and 3′-azido-3′-deoxythymidine (AZT) (25, 44). We have postulated that these nucleotide analogs increase the retroviral mutation rates by altering the levels or the relative concentrations of intracellular deoxyribonucleoside triphosphates (dNTPs) (25, 44). Intracellular dNTP levels and relative concentrations may affect both the RT mutation rates and the spectrum of mutations that arise during reverse transcription. Alterations in dNTP pools have been shown to affect RT error rates in vitro (1, 23, 48) and have been suggested as a possible mechanism for retroviral G-to-A hypermutations in vitro (35, 37, 59). However, the effects of dNTP pool imbalances on the in vivo retroviral mutation rates and the spectrum of mutations generated are unknown.
The effects of dNTP pool imbalances on the replication fidelity and mutagenesis of eukaryotic genomes have been extensively studied (28, 31, 36, 47). Nucleotide pool imbalances are mutagenic to cells and may be induced by disturbing the cellular pathways that regulate dNTP pools. These pathways include the cellular ribonucleotide reductase, the enzyme responsible for synthesizing dNDPs and for regulating the dNTP pools (5, 31, 47). Inhibition of various other enzymatic reactions involved in the synthesis of nucleotides (for example, thymidine kinase) may also induce dNTP pool imbalances (31, 47). Additionally, decreased host cell repair of genomic DNA and increased frequency of chromosome breakage may also occur as a result of dNTP pool imbalances (31, 47).
It has been well documented that intracellular dNTP pools can be altered by treatment of mammalian cells with thymidine or hydroxyurea (HU) (10, 12, 47, 53, 62). In addition, HU treatment in combination with other antiviral nucleoside analogs is currently being used in an effort to control HIV-1 replication (2, 13, 32). Micromolar to millimolar amounts of thymidine have been shown to increase the frequencies of mutants that are resistant to 2,6-diaminopurine and to 6-thioguanine three- and fourfold, respectively (62). The increase in the mutant frequency results from dNTP pool imbalances in the treated cells, which ultimately arise from modulation of the feedback regulation of ribonucleotide reductase (5). HU treatment alters dNTP pools by inhibiting ribonucleotide reductase, resulting in depletion of all dNTPs, with the most significant reductions in the dATP pool. HU may also affect DNA repair by inducing an overall decrease in dNTP pools, which in turn results in an increased sensitivity to UV irradiation and other mutagens (53, 62). The dNTP pool imbalances induced by HU have been shown to inhibit retroviral replication (10, 32). Additionally, HU potentiates the antiviral effects of several dideoxynucleoside analogs (12). In other studies, dNTP pools have been demonstrated to affect retroviral replication (15, 37, 55). Modulation of dNTP pools by using thymidine and cytidine can either restrict or enhance retroviral replication (37). However, the effects of dNTP pool imbalances on in vivo retroviral mutagenesis have not been characterized.
In an effort to determine the effect of dNTP pool imbalances on in vivo retroviral mutation rates, we examined the effects of HU, thymidine, or AZT treatments of D17 cells on in vivo dNTP pools. We also examined the effects of thymidine and HU treatments of D17 target cells on spleen necrosis virus (SNV) and murine leukemia virus (MLV) mutation rates. The results indicate that both thymidine and HU treatments induced dNTP pool imbalances, which are associated with increased retroviral mutation rates.
MATERIALS AND METHODS
Retroviral vectors.
The construction of SNV-based retroviral vector pLW-1 and MLV-based retroviral vector pGA-1 by using standard techniques was previously described (25, 50). In this report, pLW-1 and pGA-1 refer to plasmids, while LW-1 and GA-1 refer to the viruses derived from these plasmids. The SNV-based vector pLW-1 expresses the bacterial β-galactosidase gene (lacZ) and expresses the hygromycin B phosphotransferase gene (hygro) from an internal ribosomal entry site (IRES) (6, 16, 21). The MLV-based vector pGA-1 expresses lacZ and a neomycin phosphotransferase gene (neo) from IRES (24).
Cells, transfections, and infections.
D17 and C3A2 cells (obtained from the American Type Culture Collection) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; ICN) supplemented with 6% bovine calf serum (HyClone Laboratories), penicillin (50 U/ml; Gibco), and streptomycin (50 μg/ml; Gibco). D17 is a dog osteosarcoma cell line that can be infected with SNV. C3A2 is a D17-derived reticuloendotheliosis virus-based helper line that can be used to package SNV (60). Hygromycin B (Calbiochem) was present in the media at a final concentration of 120 μg/ml (0.23 mM) for C3A2 and D17 cells. C3A2-derived helper cell clones infected with LW-1 were propagated in the presence of polyclonal anti-SNV antibodies. These antibodies have been used previously to suppress SNV reinfection (25, 27, 42–44). PG13 and PA317 cells (American Type Culture Collection) are MLV-based helper cell lines (39, 40). PG13 and PA317 cells were maintained in DMEM supplemented with 10% bovine calf serum, penicillin (50 U/ml), and streptomycin (50 μg/ml). G418 was present at final concentrations of 600 μg/ml (0.79 mM) and 400 μg/ml (0.53 mM) in the media for PG13 and PA317 cells, respectively.
Helper cell clones producing LW-1 and GA-1 virus were derived by infecting C3A2 and PG13 cells, respectively, at a low multiplicity of infection (<0.00005). Therefore, the probability of obtaining cell clones with more than one provirus was less than 0.0005.
Cells were transfected by using the previously described dimethyl sulfoxide-Polybrene method (26). D17 cells were plated at densities of 2 × 105 cells on 60-mm-diameter plates. Twenty-four hours later, the cells were infected with 1.0 ml of virus in the presence of Polybrene (50 μg/ml [final concentration]) as previously described (20). Infections using LW-1 or GA-1 virus were performed for 1 h with 1 ml of virus. Twenty-four hours later, the transfected or infected cells were maintained on medium containing G418 or hygromycin.
Staining of LW-1- or GA-1-infected cells for β-galactosidase activity.
Cells infected with LW-1 or GA-1 were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) 14 days after selection was initiated by using previously described protocols (3). Briefly, cells infected with LW-1 or GA-1 were selected for drug resistance and fixed in 1 ml of 0.05% glutaraldehyde (Sigma) in phosphate-buffered saline for 10 min at room temperature. The cells were rinsed three times with 4-ml aliquots of phosphate-buffered saline: once for 1 min, once for 10 min, and once for 1 min. After the final rinse was removed, 1.25 ml of a solution containing 20 mM K3Fe(CN)6 (Sigma), 20 mM K4Fe(CN)6 (Sigma), 1.5 mM MgCl2 (Fisher), and 1 mg of X-Gal (American Bioinorganics, Inc.) per ml was added to the plates. The plates were then sealed with Parafilm and incubated at 37°C for 24 h. The numbers of blue and white colonies were determined by viewing the cells under a light microscope at a magnification of ×40.
Extraction of dNTPs from D17 cells.
Pools of dNTPs were extracted from D17 cells plated at a density of 2 × 105 cells per 60-mm-diameter dish (approximately 2 × 106 in total) for each measurement. Twenty-four hours later, the medium was replaced with culture medium (DMEM containing 6% calf serum, penicillin, and streptomycin) containing 2 mM HU (Sigma), 500 μM thymidine (Sigma), or 0.1 μM AZT (Sigma). After incubation for 4, 10, or 24 h, the cells were harvested, counted, and extracted as previously described (51). Briefly, the cells were resuspended in 150 μl of 0.4 N perchloric acid (Aldrich), incubated on ice for 20 min, and centrifuged at 4°C for 2 min. The supernatants were neutralized with 1 volume of 0.5 N trioctylamine (Sigma) in Freon (Aldrich) for 4 min and centrifuged at 4°C for 3 min. The upper aqueous phase of the resulting three phases was transferred to another tube, quickly frozen in a dry ice-ethanol bath, and stored at −80°C. To ensure efficient recovery of dNTPs, 50 μl of Tris-EDTA (pH 7.5) was added to the remaining two phases, which were mixed and again centrifuged at 4°C for 3 min. The upper aqueous phase of the reextracted samples was transferred to another tube and quickly frozen in a dry ice-ethanol bath. The two extractions for each sample were pooled, and the amounts of dNTPs were determined by a previously described enzymatic assay (50).
RESULTS
Retroviral vectors and experimental protocol.
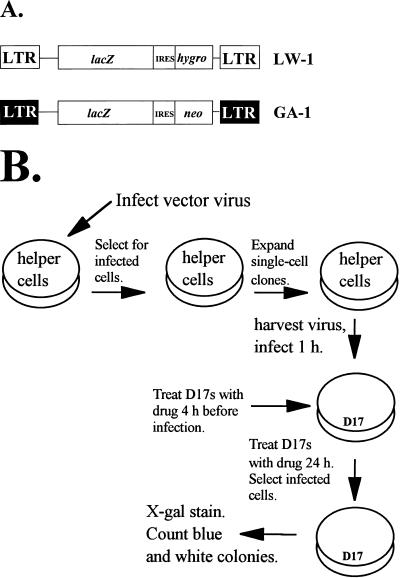
A previously described in vivo assay was used to determine the effects of HU and thymidine on retroviral mutant frequencies (25). The assay used an SNV-based vector (LW-1) and an MLV-based vector (GA-1) (Fig. 1A), both of which expressed the lacZ gene from the viral long terminal repeat (LTR) promoter. The lacZ gene served as a reporter of mutations. The LW-1 vector expressed hygro, and the GA-1 vector expressed neo (16, 24).
FIG. 1.
Rapid in vivo assay to determine the effects of HU and thymidine on retrovirus mutation rates. (A) SNV-based retroviral vector LW-1 and MLV-based retroviral vector GA-1. LW-1 contains the LTRs and cis-acting elements from SNV (shown as white boxes). LW-1 transcribes E. coli lacZ and hygro from the promoter in the LTR. The hygro gene is expressed from the IRES of encephalomyocarditis virus. GA-1 contains the LTRs and cis-acting elements from MLV (shown as black boxes). GA-1 transcribes lacZ and neo from the promoter in the LTR. The neo is expressed from the IRES. (B) Experimental protocol. Helper cell clones producing LW-1 or GA-1 (C3A2 for LW-1 and PG13 for GA-1) were generated by transfecting LW-1 or GA-1 plasmid DNA into the helper cells, harvesting virus, and infecting fresh helper cells (C3A2 for LW-1 and PG13 for GA-1). Following drug selection of cells infected with LW-1 or GA-1, individual colonies were isolated and expanded. D17 target cells for infection were treated with HU or thymidine for 4 h and then infected with virus for 1 h. D17 target cells were maintained in HU- or thymidine-supplemented media for 24 h following infection. After hygromycin or G418 selection of infected cells, the drug-resistant colonies were stained with X-Gal and analyzed for β-galactosidase expression. The numbers of blue (wild-type) and white (mutant) colonies were determined, and the forward mutant frequency was calculated as the ratio of white to total (blue plus white) colonies.
Helper cell clones producing LW-1 were established by first transfecting C3A2 helper cells with pLW-1. Virus was harvested from these helper cells and used to infect fresh C3A2 helper cells (Fig. 1B). Even though superinfection interference reduced the virus titer by as much as 100-fold during infection of C3A2 cells, it was possible to obtain cell clones resistant to hygromycin. After hygromycin selection, individual drug-resistant cell clones were isolated and expanded in the presence of anti-SNV antibodies to suppress reinfection of the virus-producing cells. The LW-1 vector was introduced into the helper cells by infection to avoid any mutations that may have occurred during transfection. In addition, helper cell clones were stained with X-Gal to verify that the lacZ gene was functionally active (data not shown).
A similar procedure was used to establish GA-1-producing helper cell clones. First, PA317 helper cells were transfected with pGA-1. Virus was harvested from the PA317 helper cells and used to infect PG13 helper cells (Fig. 1B). After selection for G418 resistance, individual cell clones were isolated and expanded. PG13 cells package vector RNA with gibbon ape leukemia virus envelope. Since PG13 cells are derived from mouse fibroblasts and mouse cells do not express the receptor for the gibbon ape leukemia virus envelope, the virus-producing cells cannot reinfect themselves (40).
Next, a single cycle of retroviral replication was carried out by harvesting virus from helper cell clones producing either LW-1 or GA-1 and infecting D17 target cells (Fig. 1B). The effects of HU or thymidine treatment on the retroviral mutation rates were determined by maintaining D17 target cells in medium supplemented with various concentrations of HU or thymidine. The target cells were treated with drug-containing medium for 4 h before infection as well as 20 h after infection. The drug treatments were initiated 4 h before infection to ensure that dNTP pools were altered at the time of infection, when reverse transcription is initiated. We expect most of the reverse transcription to be completed 6 h after infection; thus, maintaining the target cells in drug-containing medium for 20 h after infection ensured that an imbalance of dNTP pools was present throughout viral replication.
D17 cell colonies that formed after drug selection of infected cells were stained with X-Gal. Cell clones containing a phenotypically wild-type lacZ stained blue, whereas cell clones containing inactivated lacZ failed to stain and appeared white. The numbers of blue and white colonies were determined 24 h after staining. The mutant frequency was calculated as the ratio of the number of white colonies to the total number of colonies.
HU treatment increases the SNV and MLV mutant frequencies and decreases virus titers.
To determine the effects of HU on the SNV mutant frequency, LW-1 virus was harvested from C3A2 helper cell clones and used to infect D17 target cells in the absence of HU or in the presence of various concentrations of HU. The range of HU concentrations tested was based on previous studies and on empirical observations of effects of the treatments on virus titer (53). The mutant frequencies were determined by X-Gal staining of hygromycin-resistant cell colonies. The results obtained from infections with virus from three C3A2 helper cell clones are shown in Table 1. Treatment of D17 cells with 0.5 to 2.0 mM HU increased the SNV mutant frequency in a concentration-dependent manner, with a 2.1-fold maximum increase in the mutant frequency. Treatment of the D17 cells with various concentrations of HU also reduced viral titers in a concentration-dependent manner. At the highest HU concentration tested, the viral titers were reduced to 7% relative to the control virus titers. This finding indicated that concentrations of HU that substantially affected SNV virus titers also resulted in statistically significant increases in the mutant frequencies.
TABLE 1.
Effect of HU posttreatment on inactivation of the lacZ gene in LW-1-infected cells
| HU concn (mM) | LW-1 clone | No. of colonies
|
Relative virus titera | Mutant frequencyb | Relative mutant frequencyc | |
|---|---|---|---|---|---|---|
| Total | Mutant | |||||
| 0.00 | P5C1 | 3,451 | 233 | 0.07 | ||
| P4C3 | 612 | 31 | 0.05 | |||
| P3C2 | 855 | 56 | 0.07 | |||
| Total | 4,918 | 320 | 1 | 0.07 | 1 | |
| 0.50 | P5C1 | 1,316 | 138 | 0.10 | ||
| P4C3 | 206 | 14 | 0.07 | |||
| Total | 1,522 | 152 | 0.50 | 0.10 | 1d | |
| 0.75 | P5C1 | 884 | 100 | 0.11 | ||
| P4C3 | 178 | 17 | 0.10 | |||
| Total | 1,062 | 117 | 0.35 | 0.11 | 1.6d | |
| 1.00 | P5C1 | 588 | 84 | 0.14 | ||
| P4C3 | 996 | 86 | 0.09 | |||
| Total | 1,584 | 170 | 0.20 | 0.11 | 1.6d | |
| 2.00 | P5C1 | 938 | 144 | 0.15 | ||
| P4C3 | 406 | 47 | 0.12 | |||
| P3C2 | 444 | 73 | 0.16 | |||
| Total | 1,788 | 264 | 0.07 | 0.15 | 2.1d | |
Virus titers for each experimental group were determined by serial dilution and infection. The relative virus titer represents a ratio of the viral titer of the treatment group to the viral titer of the untreated group. The viral titer of the untreated group was 1.2 × 105 CFU/ml.
Ratio of mutant colonies to total colonies.
Ratio of mutant frequencies of the treatment groups to mutant frequency of the control group.
Mutant frequencies of the untreated and treated groups were compared by using a two-sample t test. The mutant frequency was not significantly different after treatment with 0.5 mM HU (P = 0.11) but was significantly increased after treatments with 0.75 mM HU (P < 0.05), 1.0 mM HU (P < 0.05), and 2 mM HU (P < 0.002).
The effects of HU treatment on the MLV mutant frequencies were also determined in assays using three PG13 helper cell clones (Table 2). Similar to the results obtained with SNV, it was found that treatment of the target cells with various concentrations of HU increased the MLV mutant frequency in a concentration-dependent manner, with a maximum increase of 2.7-fold after treatment with 2.0 mM HU. Treatment with the highest concentrations of HU tested also reduced virus titers to 3% relative to the control virus titers. This finding indicated that concentrations of HU that substantially affected MLV virus titers also increased the MLV mutant frequencies.
TABLE 2.
Effect of HU posttreatment on inactivation of the lacZ gene in GA-1-infected cells
| HU concn (mM) | GA-1 clone | No. of colonies
|
Relative virus titera | Mutant frequencyb | Relative mutant frequencyc | |
|---|---|---|---|---|---|---|
| Total | Mutant | |||||
| 0.00 | 14 | 1,291 | 82 | 0.06 | ||
| 33 | 703 | 39 | 0.06 | |||
| 22 | 560 | 28 | 0.05 | |||
| Total | 2,554 | 149 | 1 | 0.06 | 1 | |
| 0.50 | 14 | 1,385 | 176 | 0.13 | ||
| 33 | 471 | 33 | 0.07 | |||
| Total | 1,856 | 209 | 0.3 | 0.11 | 1d | |
| 0.75 | 14 | 232 | 32 | 0.14 | ||
| 33 | 534 | 53 | 0.10 | |||
| Total | 766 | 85 | 0.3 | 0.11 | 1.8d | |
| 1.00 | 14 | 1,643 | 299 | 0.18 | ||
| 33 | 264 | 34 | 0.13 | |||
| Total | 1,907 | 333 | 0.10 | 0.17 | 2.8d | |
| 2.00 | 14 | 227 | 46 | 0.20 | ||
| 33 | 284 | 41 | 0.14 | |||
| 22 | 322 | 43 | 0.13 | |||
| Total | 833 | 130 | 0.03 | 0.16 | 2.7d | |
Virus titers for each experimental group were determined by serial dilution and infection. The relative virus titer represents a ratio of the viral titer of the treatment groups to the viral titer of the untreated group. The viral titer of the untreated group was 1.6 × 104 CFU/ml.
Ratio of mutant colonies to total colonies.
Ratio of mutant frequencies of the treatment groups to mutant frequency of the control group.
Mutant frequencies of the untreated and treated groups were compared by using a two-sample t test. The mutant frequencies were not significantly different after treatment with 0.50 mM HU (P = 0.07) but were significantly different after treatments with 0.75 mM HU (P < 0.01), 1.0 mM HU (P < 0.007), and 2.0 mM HU (P < 0.005).
It was possible that HU decreased virus titers by killing target cells. To determine if HU treatment was toxic to the target cells, 2 × 105 D17 cells were plated on four small dishes per treatment group. After 24 h, the cell culture medium was replaced with HU-containing medium; the cells were harvested 24 h later, and the numbers of viable cells were determined by trypan blue exclusion. The results indicated that treatment with 0.5, 1.0, and 2.0 mM HU decreased the fractions of viable cells by 11, 39, and 68%, respectively, relative to the untreated controls (data not shown). These observations indicated that the modest cytotoxic effects observed with HU treatment do not account for the severe reductions in virus titers.
Thymidine increases the SNV and MLV mutant frequencies.
The same assay was used to determine the effects of thymidine on the SNV mutant frequencies. The range of thymidine concentrations tested was based on previous studies and on empirical observations of effects of the treatments on virus titer (37). The mutant frequencies were determined by X-Gal staining of hygromycin-resistant colonies. The results obtained from two C3A2 helper cell clones are shown in Table 3. Two independent experiments were performed with the virus harvested from the P3C2 clone. Thymidine treatment of the target cells resulted in a concentration-dependent increase in the SNV mutant frequency, with a maximum increase of 4.7-fold after treatment with 500 μM thymidine. In contrast to the results obtained with HU treatment, thymidine treatment had very little effect on the virus titers; the highest concentration of thymidine tested reduced the virus titer to 30% relative to the control virus titer. This finding indicated that concentrations of thymidine that had little effect on virus titers had a substantial effect on the mutant frequencies.
TABLE 3.
Effect of thymidine on the rate of inactivation of the lacZ gene in LW-1-infected cells
| Thy concn (μM) | LW-1 clone | No. of colonies
|
Relative virus titera | Mutant frequencyb | Relative mutant frequencyc | |
|---|---|---|---|---|---|---|
| Total | Mutant | |||||
| 0.00 | P3C2 | 261 | 16 | 0.06 | ||
| P4C3 | 656 | 40 | 0.06 | |||
| P3C2-2 | 489 | 29 | 0.06 | |||
| Total | 1,406 | 85 | 1 | 0.06 | 1 | |
| 10 | P3C2 | 210 | 21 | 0.8 | 0.10 | 1.6d |
| 50 | P3C2 | 300 | 31 | 0.10 | ||
| P4C3 | 639 | 95 | 0.15 | |||
| P3C2-2 | 133 | 14 | 0.11 | |||
| Total | 1,072 | 140 | 0.9 | 0.13 | 2.2d | |
| 100 | P3C2 | 296 | 40 | 0.14 | ||
| P4C3 | 652 | 119 | 0.18 | |||
| P3C2-2 | 571 | 94 | 0.16 | |||
| Total | 1,519 | 253 | 0.7 | 0.17 | 2.8d | |
| 500 | P3C2 | 146 | 51 | 0.35 | ||
| P4C3 | 170 | 61 | 0.36 | |||
| P3C2-2 | 429 | 98 | 0.23 | |||
| Total | 745 | 210 | 0.3 | 0.28 | 4.7d | |
Virus titers for each experimental group were determined by serial dilution and infection. The relative virus titer represents a ratio of the viral titer of the treatment groups to the viral titer of the untreated group. The viral titer of the untreated group was 1.7 × 105 CFU/ml.
Ratio of the mutant colonies to the total colonies.
Ratio of mutant frequencies of the treatment groups to mutant frequency of the control group.
Mutant frequencies of the untreated and treated groups were compared by using a two-sample t test. The mutant frequencies were significantly different after treatments with 10 μM thymidine (P < 0.03), 50 μM thymidine (P < 0.008), 100 μM thymidine (P < 0.0005), and 500 μM thymidine (P < 0.002).
Thymidine treatment had a similar effect on the MLV mutant frequency. The results obtained with three PG13 cell clones are presented in Table 4. The MLV mutant frequency was increased in a concentration-dependent manner, with a maximum increase of fourfold after treatment with 500 μM thymidine. Thymidine treatment also had very little effect on MLV titers; the highest concentrations of thymidine tested reduced the viral titers only to 34% of the control virus titers. This result indicated that thymidine treatments that had little effect on virus titers had a substantial effect on the MLV mutant frequency.
TABLE 4.
Effect of thymidine on the rate of inactivation of the lacZ gene in GA-1-infected cells
| Thy concn (μM) | GA-1 clone | No. of colonies
|
Relative virus titera | Mutant frequencyb | Relative mutant frequencyc | |
|---|---|---|---|---|---|---|
| Total | Mutant | |||||
| 0.00 | 33 | 417 | 25 | 0.06 | ||
| 14 | 496 | 29 | 0.06 | |||
| 22 | 629 | 29 | 0.05 | |||
| Total | 1,542 | 83 | 1 | 0.05 | 1 | |
| 50 | 33 | 403 | 29 | 0.07 | ||
| 14 | 526 | 37 | 0.07 | |||
| 22 | 443 | 34 | 0.08 | |||
| Total | 1,372 | 100 | 1 | 0.07 | 1.4d | |
| 100 | 33 | 600 | 66 | 0.11 | ||
| 14 | 385 | 46 | 0.12 | |||
| 22 | 552 | 56 | 0.10 | |||
| Total | 1,537 | 168 | 0.85 | 0.11 | 2.2d | |
| 500 | 33 | 223 | 40 | 0.18 | ||
| 14 | 339 | 69 | 0.20 | |||
| 22 | 210 | 43 | 0.20 | |||
| Total | 772 | 152 | 0.34 | 0.20 | 4d | |
Virus titers for each experimental group were determined by serial dilution and infection. The relative virus titer represents a ratio of the viral titer of the treatment groups to the viral titer of the untreated group. The viral titer of the untreated group was 1.4 × 104 CFU/ml.
Ratio of mutant colonies to total colonies.
Ratio of mutant frequencies of the treatment groups to mutant frequency of the control group.
Mutant frequencies of the untreated and treated groups were compared by using a two-sample t test. The mutant frequencies were significantly different after treatments with 50 μM thymidine (P < 0.02), 100 μM thymidine (P < 0.001), and 500 μM thymidine (P < 0.00005).
To determine if thymidine treatment was toxic to the target cells, 2 × 105 D17 cells were plated on four 60-mm-diameter dishes per treatment group. After 24 h, cell culture medium was replaced with thymidine-containing medium; 24 h later, the cells were harvested and the numbers of viable cells were determined by trypan blue exclusion. The results indicated that treatment with 100 and 500 μM thymidine decreased the relative number of viable cells compared to an untreated control by 16 and 40%, respectively (data not shown). Thus, the cytotoxic effect of thymidine treatments may have contributed to the modest threefold reductions in virus titers.
HU and thymidine treatments alter the intracellular dNTP concentrations in D17 cells.
We previously observed that treatment of D17 target cells with 0.1 μM AZT increased the SNV mutation rate sevenfold (25). To test the hypothesis that HU, thymidine, and AZT treatments increased the retroviral mutation rates through induction of dNTP pool imbalances, dNTP pools from D17 cells treated with 2 mM HU, 500 μM thymidine, or 0.1 μM AZT were measured. Approximately 2 × 106 D17 cells were treated with medium either without drug supplementation or with 500 μM thymidine, 2 mM HU, or 0.1 μM AZT. Following incubation for 4, 10, or 24 h in the absence or presence of drugs, cells were harvested by trypsinization and centrifuged, and their dNTPs were extracted as described previously (reference 50 and Materials and Methods). The dNTP pools were measured by using a previously described enzymatic assay (51). The lengths of the incubations were chosen to represent different stages in the infection protocol. The 4-h time point was measured to determine whether dNTP pool imbalances are induced after D17 cells are exposed to drug for 4 h prior to infection. This time point was used to determine whether dNTP pools were altered at the time of virus infection. The 10-h time point corresponds to 6 h after infection; this time point was selected because most of the reverse transcription is expected to be completed at this stage (49, 63). The 24-h time point was chosen to determine whether drug treatments for longer periods further affected retroviral replication, retroviral mutation rates, or cellular dNTP pools.
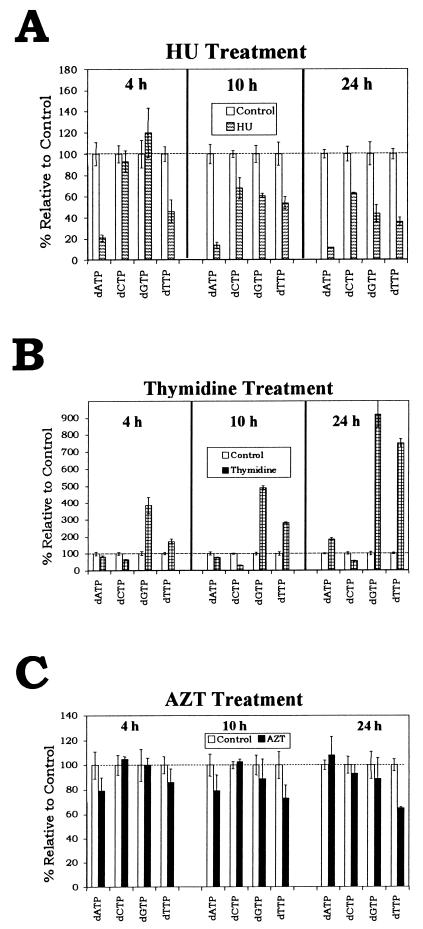
The intracellular dNTP concentrations were measured after D17 cells were maintained in medium supplemented with 2 mM HU for 4, 10, or 24 h. The effect of 2 mM HU treatment on the dNTP concentrations in D17 cells is shown (Fig. 2A). After 4 h of HU treatment, the dATP concentration was reduced to 21% relative to the control (P < 0.02). The dCTP and dGTP concentrations were not changed following HU treatment relative to the controls (P = 0.5 and P = 0.3, respectively). The dTTP concentration was modestly reduced to 46% relative to the control (P < 0.02). After 10 h of HU treatment, the dATP concentration was reduced to 14% relative to the control (P < 0.0004). The dCTP, dGTP, and dTTP concentrations were modestly reduced to 68% (P < 0.03), 61% (P < 0.01), and 54% (P < 0.01), respectively, relative to the controls. After 24 h of treatment, the dATP concentration was reduced to 12% relative to the control (P < 0.03). The dCTP, dGTP, and dTTP concentrations were modestly reduced to 63% (P < 0.04), 44% (P < 0.04), and 36% (P < 0.007), respectively, relative to the controls.
FIG. 2.
Effects of 2 mM HU (A), 500 μM thymidine (B), and 0.1 μM AZT (C) on dNTP pools in D17 cells. dNTP pools were extracted as described in Materials and Methods and determined by using a previously described enzymatic assay. Values for the control were normalized to 100% for each time measurement (dotted line). Two independent experiments were performed for the 4- and 24-h measurements, and three independent experiments were performed for the 10-h measurement. The standard errors of the means are shown by the error bars above and below the value boxes. The absolute levels of dNTPs (picomoles/106 cells) for the untreated controls at the 4-h measurement were as follows: dATP, 19 ± 2; dCTP, 43 ± 3.5; dGTP, 15 ± 2; and dTTP, 134 ± 10. At the 10-h measurement, the values were as follows: dATP, 29 ± 2.7; dCTP, 40 ± 1; dGTP, 18 ± 2; and dTTP, 97 ± 10. At the 24-h measurement, the values were as follows: dATP, 26 ± 1; dCTP, 41 ± 3; dGTP, 18 ± 2; and dTTP, 103 ± 5.
These results indicated that HU affected the intracellular concentrations of dNTPs in the cells after 4 h of treatment, demonstrating that the intracellular concentrations of dNTPs were altered at the time of infection and initiation of reverse transcription. The concentrations of the dNTPs were further altered after 10 h of treatment, and the alterations persisted during the 24-h treatment; thus, HU treatment affected the intracellular concentrations of dNTPs throughout the time period in which reverse transcription occurred.
The intracellular concentrations of dNTPs were also measured after D17 cells were maintained in medium supplemented with 500 μM thymidine for 4, 10, and 24 h. The effect of 500 μM thymidine treatment on dNTP concentrations in D17 cells is shown (Fig. 2B). After 4 h of thymidine treatment, the dGTP and dTTP concentrations were increased to 386% (P < 0.02) and 172% (P < 0.03), respectively, of the level of the controls. The dCTP concentration following thymidine treatment was modestly reduced to 65% (P < 0.03). The dATP concentration was not significantly affected relative to the control (P = 0.23). After 10 h of thymidine treatment, the dGTP and dTTP concentrations were increased to 489% (P < 0.00002) and 283% (P < 0.0001), respectively, relative to the controls. The dATP pool was modestly reduced to 79% relative to the control (P < 0.05), while the dCTP pool was reduced to 33% of the level of the control (P < 0.00004). After 24 h of treatment with 500 μM thymidine, the dTTP and dGTP concentrations were dramatically increased to 922% (P < 0.002) and 751% (P < 0.005), respectively, relative to the controls. After 24 h, the dATP concentration was increased to 185% of the level of the control (P < 0.007), while the dCTP concentration was reduced to 56% of the level of the control (P < 0.05). The results indicated that thymidine induced extensive changes in the levels of the dNTPs, resulting in dramatic increases in the dGTP and dTTP concentrations, a modest increase in the dATP concentrations, and a modest decrease in the dCTP concentrations. The dNTP concentrations were also altered after 4 h of treatment with thymidine, demonstrating that dNTP concentrations were altered at the time of infection and initiation of reverse transcription. The dNTP concentrations continued to change during the 24 h of treatment. These results indicated that treatment of target cells with thymidine resulted in alterations in dNTP concentrations throughout the time period in which reverse transcription occurred.
AZT treatment does not alter intracellular dNTP concentrations in D17 cells.
The effect of 0.1 μM AZT treatment on intracellular dNTP concentrations is shown (Fig. 2C). After 4 and 10 h of treatment, no alterations in dNTP concentrations relative to controls were observed (P > 0.1). Following 24 h of treatment, the dTTP concentration was modestly reduced to 65% of the level of the control (P < 0.01); however, no other alterations in dNTP concentrations were observed (P > 0.5).
Since AZT incorporation into the nascent DNA is expected to cause chain termination, it was conceivable that the presence of AZT triphosphate present in some of the extracted samples affected the measurements of intracellular dNTP concentrations as determined by the enzymatic assay. Since the templates used for the measurement of the dCTP, dGTP, and dATP concentrations did not contain any incorporation sites for thymidine, the presence of AZT was not expected to affect the measured concentrations of these dNTPs. To determine whether AZT affected the dTTP concentration measurements, an assay was performed as previously described to determine if adjustments of the measured dTTP concentrations were necessary (11). First, dTTPs extracted from cells treated with AZT were measured; next, known amounts of dTTPs were added to the samples and again the dTTP concentrations were measured. Since the addition of dTTP to the samples resulted in an additive increase in the dTTP concentrations measured, we concluded that the presence of AZT triphosphates in the samples did not affect the dTTP pool measurements (data not shown). Taken together, the results indicated that treatment with 0.1 μM AZT did not significantly affect the dNTP pools in D17 cells.
HU and thymidine treatments affect the balance of intracellular dNTP pools.
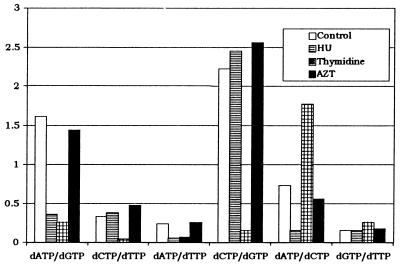
We postulated that in addition to alterations in the levels of dNTPs, imbalances in the ratios of specific pairs of dNTPs might affect the fidelity of RT. To test this hypothesis, we analyzed the dNTP pool imbalances for all six of the possible dNTP pairs (Fig. 3). The ratios of the individual dNTPs were calculated to determine the specific pool imbalances that were induced by 10 h of treatment with HU, thymidine, or AZT.
FIG. 3.
Effects of HU, thymidine, and AZT on dNTP pool imbalances. The ratios of the levels of the four dNTPs are shown following 10 h of treatment with 2 mM HU, 500 μM thymidine, and 0.1 μM AZT. The value of the ratio is shown on the y axis, and the specific dNTP ratio is shown on the x axis.
HU treatment for 10 h induced large imbalances in the dATP/dGTP, dATP/dTTP, and dATP/dCTP pool ratios. The dATP/dGTP pool ratio in the untreated control was 1.61, compared to 0.36 following HU treatment (22% of control). Similarly, the dATP/dTTP pool ratio was 0.24 in the untreated control but was 0.06 following HU treatment (25% of control), and the dATP/dCTP pool ratio was 0.73 in the untreated control, compared to 0.15 following HU treatment (21% of control). Thus, these dNTP pool ratios were dramatically altered four- to fivefold as a result of HU treatment. In contrast, only modest alterations of less than twofold were observed in the dCTP/dTTP, dCTP/dGTP, and dCTP/dTTP ratios. The dCTP/dTTP ratio was increased to 127% of the control, the dCTP/dGTP ratio was increased to 110% of the control, and the dGTP/dTTP ratio was increased to 113% of the control.
The effects of thymidine treatment on the dNTP pool imbalances are also shown in Fig. 3. Thymidine treatment induced drastic imbalances from 2- to 14-fold in the dATP/dGTP, dCTP/dTTP, dATP/dTTP, dCTP/dGTP, and dATP/dCTP pool ratios. The dATP/dGTP pool ratio in the untreated control was 1.61, compared to 0.26 following thymidine treatment (16% of control). The dCTP/dTTP pool ratio was 0.33 in the untreated control but was 0.04 following thymidine treatment (12% of control). The dATP/dTTP pool ratio was 0.24 in the untreated control, compared to 0.07 following thymidine treatment (29% of control). The dCTP/dGTP pool ratio was 2.22 in the untreated control, compared to 0.15 following thymidine treatment (7% of control). Finally, the dATP/dCTP pool ratio was 0.73 in the untreated control and 1.77 following thymidine treatment (242% of control). In contrast, the dGTP/dTTP ratio was modestly (less than twofold) increased to 173% of the control.
The effects of AZT treatment on the dNTP pool imbalances are also shown in Fig. 3. AZT treatment had very little effect on all six dNTP pool ratios. The dATP/dGTP and dATP/dCTP pool ratios were modestly (less than twofold) reduced to 89 and 77% of the control ratios, respectively. In addition, the dCTP/dTTP, dATP/dTTP, dCTP/dGTP, and dGTP/dTTP pool ratios were modestly (less than twofold) increased to 142, 108, 115, and 120%, respectively, relative to control ratios.
DISCUSSION
dNTP pool imbalances in vivo are associated with an increased retroviral mutation rate.
The experiments described here indicate that alterations in the in vivo dNTP pools are associated with an increase in the retroviral mutation rates. HU and thymidine treatments resulted in very different alterations of in vivo dNTP pools, but both treatments were associated with an increased rate of mutations in retroviral genomes.
Thymidine treatment induced dramatic dNTP pool imbalances in D17 cells and increased the SNV and MLV mutation rates as much as 4.7- and 4-fold, respectively. The thymidine-induced dNTP pool imbalances resulted from expansion of the dGTP and dTTP pools and reduction of the dCTP pool. Following 24 h of thymidine treatment, the dATP, dGTP, and dTTP pools continued to expand. These results are consistent with the outcomes expected from regulation of ribonucleotide reductase in the presence of high levels of dTTP (61).
HU treatment induced less extensive pool imbalances and only increased the SNV and MLV mutation rates by 2.1- and 2.7-fold, respectively. Even though the concentrations of all dNTPs were reduced, the dATP concentrations were reduced to the greatest extent. dATP is known to bind to ribonucleotide reductase and stimulate the conversion of NDPs to dNDPs. One possibility is that the depletion of the dATP pool resulted in reduced stimulation of the ribonucleotide reductase, which in turn resulted in reduction of the other dNTP pools.
It should be noted that thymidine and HU treatments had very different effects on the intracellular dNTPs pools. Although these alterations were both qualitatively and quantitatively different, both treatments resulted in increases in the retroviral mutation rates. The observed alterations in the dNTP pool ratios can be used to predict the nature of substitution mutations that will be increased. For example, since thymidine treatment decreased the C-to-T ratio eightfold, it is expected that the rate of C-to-T transitions will be increased. Similar analyses of other dNTP pool imbalances suggest that HU treatment will increase the rates of A-to-G, A-to-T, and A-to-C substitutions. The dNTP pool imbalances induced with thymidine treatment are expected to increase the rates of A-to-G, C-to-T, A-to-T, C-to-G, C-to-A, and T-to-G substitutions. It is also conceivable that overall depletion of dNTP pools may lead to a decrease in the rate of polymerization; the decreased rate of polymerization may promote dissociation of RT from the template, leading to an increase in mutations involving template-switching events (deletions, deletions with insertions, and duplications). Additional studies are needed to determine the nature of mutations induced by these two types of dNTP pool imbalances.
HU and thymidine treatments also resulted in a reduction in viral titers. The reduction in viral titers could not be explained by an increased rate of mutation and inactivation of the selectable marker genes. Based on the size of the selectable marker genes (∼1 kb) and a mutation rate of approximately 2 × 10−5/bp/replication cycle, it is estimated that approximately 2% of the selectable marker genes will be inactivated through mutations (27, 41–43). Even a fivefold increase in the retroviral mutation rate would result in reduction of the viral titer to only 90% relative to the control virus titer. Since we observed reductions of the viral titer to 7% after HU treatment and to 34% after thymidine treatment, we conclude that the treatment inhibits retroviral replication by another mechanism. HU treatment may have had a greater effect on the viral titer because it resulted in severe depletions of all dNTP pools, whereas thymidine treatment resulted in depletion of only the dCTP pools. Therefore, we hypothesize that the depletion of dNTP pools after HU treatment may interfere with efficient reverse transcription, leading to a reduction in viral titer. The reduction of the viral titer to 34% after thymidine treatment may have resulted from cytotoxicity to the target cells observed after the treatment.
AZT treatment increases the retroviral mutation rate by a mechanism not involving dNTP pool alterations.
AZT treatment of D17 cells did not significantly affect the dNTP pools. AZT has been shown to induce dNTP pool imbalances in some cell lines at concentrations much greater (>100-fold) than those used in these studies (9). However, AZT does not affect dNTP pools in peripheral blood mononuclear cells (10), nor does AZT affect dNTP pools in many cell lines (18).
Previously, we demonstrated that treatment of D17 target cells with AZT increased the SNV mutant frequency 10-fold and the MLV mutant frequency 2-fold. Based on these results and previous reports indicating that AZT induces dNTP pool imbalances, we previously hypothesized that AZT increases the retroviral mutation rates by inducing a dNTP pool imbalance (25). The results of this study strongly suggest that this hypothesis is incorrect. The results reported here demonstrate that AZT concentrations that increase the SNV mutation rate sevenfold have very little effect on the intracellular dNTP pools. Conversely, HU and thymidine treatments, which have a much greater impact on the intracellular dNTP pools, increase the SNV mutation rate to a lesser extent than the AZT treatment. Finally, HU and thymidine treatments increase the mutation rates of SNV and MLV to similar extents, whereas AZT treatment increases the SNV mutation rate to a much greater extent (10-fold) than the MLV mutation rate (2-fold). Taken together, these results strongly suggest that a mechanism other than alterations of dNTP pools is responsible for increasing the retroviral mutation rates after AZT treatment.
Mutational specificity and retroviral mutation rates may vary between cell types.
To determine whether mutational specificity is correlated with natural dNTP pool imbalances, we compared the dNTP pool imbalances in D17 cells to the specificity of mutations induced during retroviral replication (25, 27, 42–44). Retroviral mutation rates were previously determined in D17 cells using the SNV-based retroviral shuttle vector BK-2 (25, 27). In these experiments G-to-A transitions were the predominant substitution mutations, accounting for 60% (38 of 63) of the substitution mutations (25, 27). Measurement of dNTP pools in D17 cells determined the natural pool asymmetry in D17 cells. The dTTP pool (97 ± 10 pmol/106 cells at the 10-h time point) is larger than the dCTP pool (40 ± 1 pmol/106 cells), and the dATP pool (29 ± 2.7 pmol/106 cells) is larger than the dGTP pool (18 ± 1.5 pmol/106 cells). This natural pool imbalance may influence the mutational specificity of substitution mutations, causing the majority of substitution mutations to be G-to-A transitions. The role of natural pool imbalances as a determinant of replication fidelity and specificity was demonstrated in studies using lacZα as a reporter during phagemid replication (64). These results suggest that the intracellular dNTP concentrations may affect the specificities as well as the rates of transition mutations during retroviral replication.
Based on in vitro and in vivo data, overall sizes of dNTP pools as well as the natural pool imbalances are likely to influence mutation rates and spectra. Thus, mutation rates in different cell lines might vary if the levels or natural pool imbalances differ. Similarly, mutation rates in cell lines may differ from mutation rates in the in vivo target cells depending on the levels and asymmetry of the dNTP pools. Finally, the activation state of the cell has been shown to influence the levels of dNTPs (10); therefore, the mutation rates of retroviruses may differ between proliferating and quiescent cells.
dNTP pool imbalances affect virus titers.
These experiments characterize the effect of dNTP pool imbalances on retrovirus titers in one round of retroviral replication. It was previously shown that HU inhibits HIV-1 replication in culture (12, 32); however, this inhibition was measured after several days of HU treatment, during which many rounds of retroviral replication occurred. Our experiments characterized the inhibition of the virus titer in a single replication cycle, which indicated that 2 mM HU decreased the virus titers of MLV and SNV by 97 and 93%, respectively. Similarly, thymidine treatment has been shown to inhibit retroviral replication in cell cultures (62). Quantitation of the thymidine-induced inhibition demonstrated that treatment of cells with 500 μM thymidine decreased virus titers to 34% of the control for the MLV- and SNV-based vectors.
Intracellular dNTP pools may affect the rate and spectrum of retroviral mutations.
These results suggest that dNTP pools in the target cells for retrovirus infection may be a critical determinant of retrovirus replication fidelity. Based on previous studies, thymidine treatment is more likely to increase the rates of substitution mutations and G-to-A hypermutations than the rates of other mutations (28, 43, 59). The probability of misincorporation of some nucleotides over others is based on the stability of the mispair and the ability of the polymerase to extend the mispair and complete DNA synthesis. The probability of a specific mispair forming is likely to be determined by the relative levels of dNTPs; thus, the spectrum and rates of substitution mutations are expected to be affected by the dNTP pool imbalances in the cells. Frameshift mutations involving primer-template slippage and deletion mutations involving template switching by RT are likely to result from the evolutionarily selected low processivity of RT (56). However, it is conceivable that decreased levels of dNTP substrates promote pausing by RT, which would increase the rate of dissociation of the RT from the template. If so, then the increased rate of template dissociations may result in an increase in mutations involving template-switching events (deletions, deletions with insertions, and duplications [33, 34, 41–44]).
Implications for antiviral therapy.
The results demonstrate that induction of dNTP pool alterations increases the in vivo retroviral mutation rate. These results also confirm previous studies indicating that reductions in the dNTP pools have an inhibitory effect on viral replication (10). The results of this study show that treatment of patients with HU may result in an increase in the rate of mutations in HIV-1 genomes, which in turn may facilitate development of drug resistance and escape from the host immune response. However, it is arguable whether alterations in the retroviral mutation rates will have an effect on the extent of variation present in HIV-1 populations (4). Based on a mathematical model, it has been hypothesized that small changes in the selective growth advantage for the virus will have a greater impact on variation in the population than large changes in the viral mutation rates. Further studies are needed to determine the role of retroviral mutation rates in development of drug resistance and pathogenesis.
ACKNOWLEDGMENTS
We thank Jeffery Anderson, Benjamin Beasley, Que Dang, Krista Delviks, Elias Halvas, Wei-Shau Hu, Carey Hwang, Evguenia Svarovskaia, Yegor Voronin, and Wen-hui Zhang for critical reading of the manuscript. We especially thank Wei-Shau Hu for valuable intellectual input and discussions throughout the project.
This work was supported by Public Health Service grant CA58875 from the National Institutes of Health.
REFERENCES
- 1.Bebenek K, Abbots J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 2.Birion F, Lucht F, Peyramond D, Fresard A, Vallet T, Nugire F, Grange J, Malley S, Hamedi-Sangsari F, Vila J. Pilot clinical trial of the combination of hydroxyurea and didanosine in HIV-1 infected individuals. Antiviral Res. 1996;29:111–113. doi: 10.1016/0166-3542(95)00931-0. [DOI] [PubMed] [Google Scholar]
- 3.Cepko C. XGAL staining of cultured cells. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1992. pp. 9.11.9–9.11.12. [Google Scholar]
- 4.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 5.Cohen A, Barankiewicz J, Lederman H M, Gelfand E W. Purine and pyrimidine metabolism in human T lymphocytes. Regulation of deoxyribonucleotide metabolism. J Biol Chem. 1983;258:12334–12340. [PubMed] [Google Scholar]
- 6.Davies M V, Kaufman R J. The sequence context of the initiation codon in the encephalomyocarditis virus leader modulates efficiency of internal translation initiation. J Virol. 1992;66:1924–1932. doi: 10.1128/jvi.66.4.1924-1932.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Farrash M A, Kuroda M J, Kitazaki T, Masuda T, Kato K, Hatanaka M, Harada S. Generation and characterization of a human immunodeficiency virus type 1 (HIV-1) mutant resistant to an HIV-1 protease inhibitor. J Virol. 1994;68:233–239. doi: 10.1128/jvi.68.1.233-239.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgibbon J E, Howell R M, Haberzettl C A, Sperber S J, Gocke D J, Dubin D T. Human immunodeficiency virus type 1 pol gene mutations which cause decreased susceptibility to 2′,3′-dideoxycytidine. Antimicrob Agents Chemother. 1992;36:153–157. doi: 10.1128/aac.36.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frick L W, Nelson D J, St. Clair M H, Furman P A, Krenitsky T A. Effects of 3′-azido-3′-deoxythymidine on the deoxynucleotide triphosphate pools of cultured human cells. Biochem Biophys Res Commun. 1988;154:124–129. doi: 10.1016/0006-291x(88)90659-6. [DOI] [PubMed] [Google Scholar]
- 10.Gao W-Y, Cara A, Gallo R C, Lori F. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA. 1993;90:8925–8928. doi: 10.1073/pnas.90.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao W-Y, Johns D G, Mitsuya H. Enzymatic assay for quantification of deoxynucleoside triphosphates in human cells exposed to antiretroviral 2′,3′-dideoxynucleosides. Anal Biochem. 1994;222:116–122. doi: 10.1006/abio.1994.1462. [DOI] [PubMed] [Google Scholar]
- 12.Gao W-Y, Johns D G, Mitsuya H. Anti-human immunodeficiency virus type 1 activity of hydroxyurea in combination with 2′,3′-dideoxynucleosides. Mol Pharmacol. 1994;46:767–772. [PubMed] [Google Scholar]
- 13.Giacca M, Zanussi S, Comar M, Simonelli C, Vaccher E, de Paoli P, Tirelli U. Treatment of human immunodeficiency virus infection with hydroxyurea: virologic and clinical evaluation. J Infect Dis. 1996;174:204–209. doi: 10.1093/infdis/174.1.204. [DOI] [PubMed] [Google Scholar]
- 14.Glickman B W, Saddi V A, Curry J. Spontaneous mutations in mammalian cells. Mutat Res. 1994;304:19–32. doi: 10.1016/0027-5107(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 15.Goulaouic H, Subra F, Mouscadet J F, Carteau S, Auclair C. Exogenous nucleosides promote the completion of MoMLV DNA synthesis in G0-arrested Balb c/3T3 fibroblasts. Virology. 1994;200:87–97. doi: 10.1006/viro.1994.1166. [DOI] [PubMed] [Google Scholar]
- 16.Gritz L, Davies J. Plasmid encoded hygromycin-b resistance: the sequence of hygromycin b phosphotransferase and its expression. Gene. 1979;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 17.Gu Z, Gao Q, Li X, Parniak M, Wainberg M A. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J Virol. 1992;66:7128–7135. doi: 10.1128/jvi.66.12.7128-7135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao Z, Cooney D A, Hartman N R, Perno C F, Fridland A, De Vico A L, Sarngadharan M G, Broder S, Johns D G. Factors determining the activity of 2′,3′dideoxynucleosides in suppressing human immunodeficiency virus in vitro. Mol Pharmacol. 1988;34:431–435. [PubMed] [Google Scholar]
- 19.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 20.Hu W-S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and a high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang S, Krausslich H, Nicklin M J H, Duke G, Palmenberg A, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Japour A J, Welles S, D’Aquilla R T, Johnson V A, Richman D D, Coombs R W, Reichelderfer P S, Kahn J O, Crumpacker C S, Kuritzkes D R. Prevalence and clinical significance of zidovudine resistance mutations in human immunodeficiency virus isolated from patients after long-term zidovudine treatment. J Infect Dis. 1995;171:1172–1179. doi: 10.1093/infdis/171.5.1172. [DOI] [PubMed] [Google Scholar]
- 23.Ji J, Loeb L A. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992;31:954–958. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen R A, Rothstein S J, Reznikoff W J. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177:65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- 25.Julias J G, Kim T, Arnold G, Pathak V K. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J Virol. 1997;71:4254–4263. doi: 10.1128/jvi.71.6.4254-4263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai S, Nishizawa M. New procedures for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim T, Mudry R A, Jr, Rexrode II C A, Pathak V K. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. J Virol. 1996;70:7594–7602. doi: 10.1128/jvi.70.11.7594-7602.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunz B A, Kohalmi S E. Modulation of mutagenesis by deoxyribonucleotide levels. Annu Rev Genet. 1991;25:339–359. doi: 10.1146/annurev.ge.25.120191.002011. [DOI] [PubMed] [Google Scholar]
- 29.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 30.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 31.Loeb L A, Kunkel T A. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 32.Lori F, Malykh A G, Foli A, Maserati R, De Antoni A, Minoli L, Padrini D, Degli Antoni A, Barchi E, Jessen H, Wainberg M A, Gallo R C, Lisziewicz J. Combination of a drug targeting the cell with a drug targeting the virus controls human immunodeficiency virus type 1 resistance. AIDS Res Hum Retroviruses. 1997;13:1403–1409. doi: 10.1089/aid.1997.13.1403. [DOI] [PubMed] [Google Scholar]
- 33.Mansky L M, Temin H M. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J Virol. 1994;68:494–499. doi: 10.1128/jvi.68.1.494-499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez M A, Vartanian J-P, Wain-Hobson S. Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc Natl Acad Sci USA. 1994;91:11787–11791. doi: 10.1073/pnas.91.25.11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res. 1989;181:305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 37.Meyerhans A, Vartanian J-P, Hultgren C, Plikat U, Karlsson A, Wang L, Eriksson S, Wain-Hobson S. Restriction and enhancement of human immunodeficiency type 1 replication by modulation of intracellular deoxynucleoside triphosphate pools. J Virol. 1994;68:535–540. doi: 10.1128/jvi.68.1.535-540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cell lines based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parthasarathi S, Varela-Echavarria A, Ron Y, Preston B D, Dougherty J P. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J Virol. 1995;69:7991–8000. doi: 10.1128/jvi.69.12.7991-8000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci USA. 1990;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pathak V K, Temin H M. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J Virol. 1992;66:3093–3100. doi: 10.1128/jvi.66.5.3093-3100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patick A, Rose R, Greytok J, Bechtold C M, Hermsmeier M A, Chen P T, Barrish J C, Zahler R, Colonno R J, Lin P-F. Characterization of a human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston B D, Poiesz B J, Loeb L A. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 47.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 48.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 49.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Sherman P A, Fyfe J A. Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal Biochem. 1989;180:222–226. doi: 10.1016/0003-2697(89)90420-x. [DOI] [PubMed] [Google Scholar]
- 52.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snyder R D. Consequences of the depletion of cellular deoxynucleoside triphosphate pools on the excision-repair process in cultured human fibroblasts. Mutat Res. 1988;200:193–199. doi: 10.1016/0027-5107(88)90082-6. [DOI] [PubMed] [Google Scholar]
- 54.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in the HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 55.Temin H M. Studies on carcinogenesis by avian sarcoma viruses. V. Requirement for new DNA synthesis and for cell division. J Cell Physiol. 1966;69:53–64. [Google Scholar]
- 56.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varela-Echavarria A, Garvey N, Preston B D, Dougherty J P. Comparison of Moloney murine leukemia virus mutation rate with the fidelity of its reverse transcriptase in vitro. J Biol Chem. 1992;267:24681–24688. [PubMed] [Google Scholar]
- 59.Vartanian J-P, Meyerhans A, Sala M, Wain-Hobson S. G→A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci USA. 1994;91:3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe S, Temin H M. Construction of a helper cell line for reticuloendotheliosis virus cloning vectors. Mol Cell Biol. 1983;3:2241–2249. doi: 10.1128/mcb.3.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Sag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 62.Wilkinson Y A, McKenna P G. The effects of thymidine on deoxyribonucleotide pool levels, cytotoxicity and mutation induction on Friend mouse erythroleukemia cells. Leuk Res. 1989;13:615–620. doi: 10.1016/0145-2126(89)90130-6. [DOI] [PubMed] [Google Scholar]
- 63.Yin P D, Pathak V K, Rowan A E, Teufel II R J, Hu W-S. Utilization of nonhomologous minus-strand DNA transfer to generate recombinant retroviruses. J Virol. 1997;71:2487–2494. doi: 10.1128/jvi.71.3.2487-2494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Mathews C K. Natural DNA precursor pool asymmetry and base sequence context as determinants of replication fidelity. J Biol Chem. 1995;270:8401–8404. doi: 10.1074/jbc.270.15.8401. [DOI] [PubMed] [Google Scholar]