Abstract
Aim
To explore the diagnostic value of serum-derived exosomal miRNAs and predict the roles of their target genes in Alzheimer’s disease (AD) based on the expression of miRNAs in AD patients.
Methods
We determined the relative concentration of exosomal miRNAs by High-throughput Second-generation Sequencing and real-time quantitative real-time PCR.
Results
71 AD patients and 71 ND subjects were collected. The study demonstrated that hsa-miR-125b-1-3p, hsa-miR-193a-5p, hsa-miR-378a-3p, hsa-miR-378i and hsa-miR-451a are differentially expressed in the serum-derived exosomes of AD patients compared with healthy subjects. According to ROC analysis, hsa-miR-125b-1-3p has an AUC of 0.765 in the AD group compared to the healthy group with a sensitivity and specificity of 82.1–67.7%, respectively. Enrichment analysis of its target genes showed that they were related to neuroactive ligand-receptor interactions, the PI3K-Akt signaling pathway, the Hippo signaling pathway and nervous system-related pathways. And, hsa-miR-451a had an AUC of 0.728 that differentiated the AD group from the healthy group with a sensitivity and specificity of 67.9% and 72.6%, respectively. Enrichment analysis of its target genes showed a relationship with cytokine-cytokine receptor interactions and the PI3K-Akt signaling pathway.
Conclusion
The dysregulation of serum exosomal microRNAs in patients with AD may promote the diagnosis of AD. The target genes of miRNAs may be involved in the occurrence and development of AD through various pathways.
Keywords: Alzheimer’s disease, early-onset Alzheimer’s disease, late-onset Alzheimer’s disease, serum, exosome, microRNA
Introduction
Alzheimer’s disease, which is mainly characterized by cognitive impairment and abnormal intellectual and behavioral characteristics, is the most common neurodegenerative disorder. Early-onset AD (EOAD) occurs when the age at onset of AD is under 65 years, while late-onset AD (LOAD) is defined when the age of onset is over 65 years old.1 EOAD patients were more likely to show language deficits, distraction and visuospatial function impairment than LOAD patients and exhibited more memory loss.2 The imaging of EOAD patients showed obvious atrophy in the frontal lobe, parietal lobe and lateral temporal lobe rather than the classic mesial temporal lobe.3 Moreover, EOAD patients presented with more severe gray matter atrophy and more abundant senile plaques, neurofibrillary tangles, and synaptic loss than LOAD patients.2 Recent research results show that there are 15.07 million elderly dementia patients aged 60 and above in China, of which 9.83 million suffer from AD, 3.92 million suffer from vascular dementia, and 1.32 million suffer from other forms of dementia.4 The existing biomarkers for AD diagnosis are invasive and expensive, and cannot be widely used in clinical practice. Recently, Professor Shen Lu searched for diagnostic peripheral biomarkers in the serum of AD patients and ultimately identified seven serum autoantibodies (MAPT, DNAJC8, KDM4D, SERF, CDKN1A, AGER, and ASXL1) as candidate peripheral biomarkers for AD diagnosis.5 Currently, we have no drugs or other treatments to prevent the progression of AD or biomarkers to identify AD in the preclinical phases. Alzheimer’s disease cheap and reliable peripheral biomarkers are important for early diagnosis in routine clinical practice and research. Novel peripheral biomarkers to diagnose AD are urgently needed.
Extracellular vesicles first form as intraluminal vesicles in multivesicular compartments and are secreted upon fusion of these compartments with the plasma membrane as exosomes.6 Exosomes appear as round structures delimited by a lipid bilayer (30 to 150 nm in diameter) and can contain proteins, lipids and nucleic acids, including DNA, mRNA, miRNA and other noncoding RNAs, protecting their contents from degradation.7 Exosomes can be released from neurons, astrocytes, oligodendrocytes and microglia in the brain and are involved in the regulation of synaptic communication, nerve regeneration and degeneration.8 MicroRNAs are small noncoding RNAs (20–22 nucleotides) partially packaged in lipid microvesicles, such as exosomes and apoptotic bodies.9 Mature microRNA is incorporated into a protein complex-RNA induced silencing complex, which binds the target mRNA transcript to repress translation.10 Individual miRNAs can target several mRNAs, and a single mRNA has the potential to be modulated by multiple miRNAs in a complex regulatory network.11 Previous research reports that an up-regulated miRNA-125b could target the 3’untranslated region of mRNA encoding a 15-lipoxygenase (utilized in the conversion of docosahexaenoic acid into neuroprotectin D1) and the vitamin D3 receptor of the nuclear hormone receptor superfamily. They are key neuromolecular factors essential in lipid-mediated signaling, neurotrophic support, defense against reactive oxygen and nitrogen species, and neuroprotection in the CNS. In AD hippocampal CA1 cells, miRNA-125 damaged neurons by downregulating 15-lipoxygenase and the vitamin D3 receptor.12 The study revealed that overexpression of miR-451a downregulated the expression of activating transcription factor 2(ATF2).13 ATF2 mediated the normal physiological metabolism of nerve cells under physiological conditions and is an essential component. When ATF2 expression is deficient, the development of nerve cells cannot occur normally, which can trigger a large number of cell apoptosis and neuronal loss, leading to the occurrence of AD.14 Increasing evidence indicates that these molecules may play important roles in the biological pathways that regulate the formation of Aβ42 and hyperphosphorylation of Tau, as well as in AD patients. We identified 5 exosomal miRNAs by qPCR between AD patients and nondemented (ND) controls to develop noninvasive diagnostic biomarkers in the diagnosis of AD and predicted the roles of their target genes to explore the probable pathogenic mechanism of AD.
Materials and Methods
Subject and Sample Collection
All patients with AD (n=71) were enrolled between March 2017 and August 2018 at Changsha, Hunan Province, China, and diagnosed with possible or probable AD according to the 2011 version of the diagnostic guidelines for Alzheimer’s disease recommended by the National Institute on Aging-Alzheimer’s Association workgroups.10 The ND group was recruited in May 2018 from subjects who underwent a medical examination at Xiang Ya Hospital Central South University. Baseline data, including age, sex, level of education, and peripheral blood, were collected in each group. All participants were evaluated with the Mini-mental State Examination (MMSE), Montreal Cognitive Assessment scale (MOCA), Activities of Daily Life (ADL), Clinical Dementia Rating (CDR), Hachinski Ischemic Score (HIS) and Hamilton Depression Scale (HAMD). AD patients (n=71) were divided into two groups: early-onset AD (n=12) and late-onset AD (n=59). ND subjects (n=71) were separated into two groups according to age<65 (n=9) years old and age≥65 (n=62) years old.
The study was approved by the Ethics Committee of Xiangya Hospital Central South University Changsha, China (IRB 201606289), and complied with the Declaration of Helsinki. Written informed consent was obtained from all participants or their guardians.
Blood Preparation and Exosome Isolation
Peripheral blood was drawn into coagulation tubes and centrifuged at 3000 × g for 15 min. Serum was aspirated and stored at −80°C. Exosomal preparations were performed using the ExoQuick™ Exosome Isolation Kit(SBI, USA) following the manufacturer’s instructions.Briefly, the serum was centrifuged at 1500 x g for 30 min at 4°C to remove cells and debris. Following this, 1mL clarified serum and 250ul of Exosome Isolation reagent was added. Mix the serum/reagent mixture thoroughly, and then incubate the sample upright at 4°C for 30 minutes. After incubation, the sample was centrifuged at 1500 x g for 30 minutes at 4°C. The supernatant was discarded and the pellet, containing the exosomes at the bottom of the tube, was resuspended in phosphate-buffered saline.
Exosome Identification
The size and morphological features of exosomes were examined by transmission electron microscopy (TEM, JEM1230). Nanoparticle tracking analysis (NTA, ZetaView) was used to observe the diameter distribution and concentration of exosomes. Western blotting was performed according to the manufacturer’s instructions. The blotting membrane was incubated with CD63 antibody (1:500 dilution, Santa Cruz SC-365604)/TSG101 (1:500 dilution, Santa Cruz SC-7964) overnight. After washes with T-TBS, goat anti-rabbit IgG (H+L) secondary antibody was added for 1 h (1:5000 dilution, Thermo Pierce 31,210). The proteins were finally detected using chemiluminescence (Thermo Pierce 34,075).
Deep Sequencing on an Illumina HiSeqTM 2500
We selected 4 AD patients with typical clinical features and 4 healthy individuals with age, gender, and education matching, and performed high-throughput second-generation sequencing of serum exosomes miRNA on Illumina HiSeqTM 2500 of Guangzhou Ruibo Biological Company.
RNA Isolation and MicroRNA qRT-PCR
We identified 775 microRNAs from the serum exosomes of 4 AD patients and 4 ND subjects through sequencing. Validation of 5 serum exosome-enriched miRNAs (hsa-miR-125b-1-3p, hsa-miR-193a-5p, hsa-miR-378a-3p, hsa-miR-378i and hsa-miR-451a) was carried out by qPCR in a cohort of 71 patients with AD and 71 ND subjects.
Total RNA was isolated from exosomes using TRIzol (Ambion) mainly according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using a poly(A) reaction system and a reverse reaction system. Hsa-miR-125b-1-3p, hsa-miR-193a-5p, hsa-miR-378a-3p, hsa-miR-378i and hsa-miR-451a were determined by qRT-PCR (Applied Biosystems, USA) using U6 RNA as an endogenous control.
Statistical Analysis
The age, sex, level of education, and various scales (MMSE, MOCA, ADL, CDR, HIS and HAMD) of all subjects are shown as the mean ± standard deviation (x±S). The differences in the levels of miRNAs (hsa-miR-125b-1-3p, hsa-miR-193a-5p, hsa-miR-378a-3p, hsa-miR-378i and hsa-miR-451a) between the control subjects aged <65 years and those aged ≥65 years were analyzed using the Mann–Whitney U-test. The microRNA expression levels in the early-onset AD group, late-onset AD group and ND group were compared by the Kruskal–Wallis test. Then, we compared the expression of microRNAs in the AD group and the control group by the Mann–Whitney U-test. A receiver operating characteristic (ROC) curve was used to calculate the area under the curve and analyze the sensitivity and specificity of miRNAs in the diagnosis of AD.
All statistical analyses were performed using SPSS version 23.0. A p value of < 0.05 was considered statistically significant.
Prediction of MiRNA Target Genes and Their Functions
First, TargetScan, miRDB, miRTarBase and miRWalk software were applied to predict the target genes of miRNAs. Gene Ontology (GO) provided annotation with three categories, biological process (BP), cellular compartment (CC) and molecular function (MF), for candidate target genes. GO analysis identified significant annotations from all gene annotations by the hypergeometric distribution method in the background of certain species. The Kyoto Encyclopedia of Genes and Genomes pathways also supplied significant annotation of signal transduction and disease pathways for candidate genes by Fisher’s test.
Results
Baseline Clinical Characteristics
The clinical characteristics of the AD patients (n=71) and the ND subjects (n=71) are shown in Table 1. The mean age below 65 years old was not significantly different between two groups (p = 0.89). However, the mean age of the AD patients over 65 years old was different from that of the control subjects over 65 years old (p<0.05). The number of males in both groups was the same as that of females (p = 1.00). The education levels of the AD and control groups were not significantly different (p=0.34). As expected, the relevant scale assessments, the MMSE, MOCA, CDR, HAMD, and HIS, were significantly different between the two groups (p<0.05, all).
Table 1.
Clinical Characteristics of the AD Patients and ND Subjects
| AD (n=71) | Control (n=71) | P | |
|---|---|---|---|
| Age (years, mean ± SD) | |||
| <65 | 57.27±4.22 | 57.56±4.67 | 0.89 |
| ≥65 | 78.75±7.04 | 74.32±5.83 | <0.05 |
| Sex (male/female, N) | 39/32 | 39/32 | 1.00 |
| Education (years, mean ± SD) | 9.46±4.28 | 9.37±6.18 | 0.34 |
| MMSE (mean ± SD) | 12.96±7.29 | 27.31±1.44 | <0.05 |
| MOCA (mean ± SD) | 5.23±6.62 | 25.07±3.42 | <0.05 |
| CDR (mean ± SD) | 1.79±0.74 | 0.04±0.13 | <0.05 |
| HAMD (mean ± SD) | 3.32±2.63 | 0.97±1.27 | <0.05 |
| HIS (mean ± SD) | 2.24±0.96 | 1.70±0.80 | <0.05 |
Identification of Circulating Exosomes
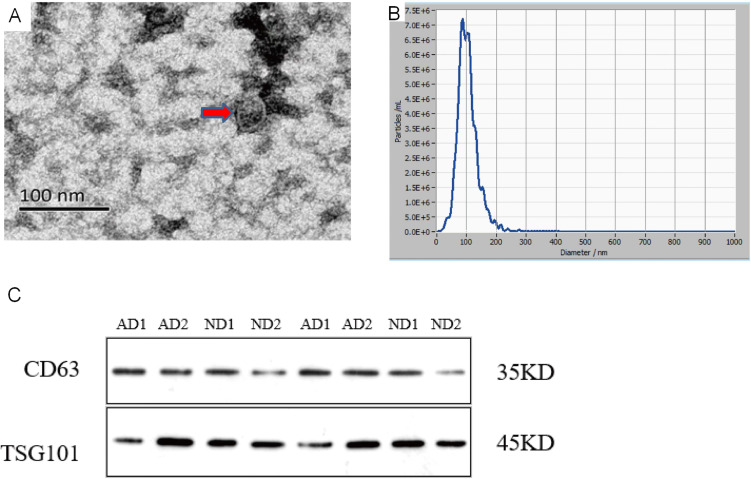
The vesicles appear as round structures enclosed by double-leaflet membranes, and their size ranges from 30 to 150 nm in diameter by TEM (Figure 1A). The processing of extracellular vesicles for observation by conventional TEM usually causes their shrinking, leading to an artifactual cup-shaped morphology.7 The shape difference in vesicles is caused by different methods of preparing samples.15 The yielded concentration of vesicles was 9.8×107 particles/mL with a 2000 dilution factor based on an original concentration of 2.0×1011 particles/mL, and their size distribution was within 96.5±33.1 nm, as measured with NTA (Figure 1B). Western blotting confirmed the expression of CD63 and TSG101 in serum extracts from the AD patients and the control participants (Figure 1C). CD63 is the most commonly used marker protein for exosome identification. TSG101 is involved in various sorting machineries and is important for exosome biogenesis.7
Figure 1.
Exosome Identification (A). The identification of serum exosomes by transmission electron microscope.The exosome appear as round structures enclosed by double-leaflet membranes, with a diameter of approximately 30–150nm (B). The identification of serum exosomes by Nanoparticle Tracking Analysis.The particle size of exosome is 96.5±33.1nm, with an original concentration of 2.0 × 1011Particles/mL (C). The identification of exosomes specific proteins by Western blot.CD63 is a transmembrane protein with a molecular weight of around 35kDa. TSG101 belongs to the ESCRT related protein with a molecular weight of around 45kDa.
High-Throughput Second-Generation Sequencing
MiRNAs showing differential expression shown in Table 2.
Table 2.
miRNAs Showing Differential Expression
| miRNA | HC Mean | AD mean | AD/HClog2 (Foldchange) | P value | Up/Down |
|---|---|---|---|---|---|
| hsa-miR-148a-3p | 17,112.6625 | 35,300.595 | 1.0446 | 0.0457 | Up |
| hsa-miR-16-5p | 776.1175 | 2850.38 | 1.8768 | 0.0086 | Up |
| hsa-miR-19b-3p | 626.865 | 2269.56 | 1.8562 | 0.0094 | Up |
| hsa-miR-483-5p | 583.1125 | 151.8575 | −1.9411 | 0.0159 | Down |
| hsa-miR-4488 | 115.9425 | 26.97 | −2.104 | 0.0343 | Down |
| hsa-miR-223-3p | 50.825 | 149.49 | 1.5564 | 0.0357 | Up |
| hsa-miR-125b-1-3p | 33.8425 | 9.7575 | −1.7943 | 0.0468 | Down |
| hsa-miR-4492 | 22.3275 | 4.34 | −2.3631 | 0.0144 | Down |
| hsa-miR-9-5p | 15.14 | 41.9275 | 1.4695 | 0.0352 | Up |
| hsa-miR-4466 | 12.6075 | 1.31 | −3.2666 | 0.0145 | Down |
| hsa-miR-93-3p | 8.735 | 21.8625 | 1.3236 | 0.0352 | Up |
| hsa-miR-324-5p | 7.3075 | 29.68 | 2.022 | 0.0126 | Up |
| hsa-miR-490-3p | 5.6475 | 23.205 | 2.0388 | 0.0414 | Up |
| hsa-miR-24-1-5p | 4.85 | 0.8625 | −2.4914 | 0.0267 | Down |
| hsa-miR-374b-5p | 4.035 | 11.2525 | 1.4796 | 0.0450 | Up |
| hsa-miR-6727-5p | 3.3875 | 0 | −8.4041 | 0.0095 | Down |
| hsa-miR-6724-5p | 3.27 | 0.1775 | −4.2034 | 0.0341 | Down |
| hsa-miR-29c-5p | 3.15 | 14.8075 | 2.2329 | 0.0162 | Up |
| hsa-miR-4787-5p | 3.0575 | 0.29 | −3.3982 | 0.0319 | Down |
| hsa-miR-3940-3p | 2.8025 | 8.7175 | 1.6372 | 0.0316 | Up |
| hsa-miR-4707-3p | 2.405 | 0 | −7.9099 | 0.0155 | Down |
| hsa-miR-205-5p | 1.405 | 7.8075 | 2.4743 | 0.0253 | Up |
| hsa-miR-4467 | 1.315 | 7.2125 | 2.4554 | 0.0269 | Up |
| hsa-miR-3138 | 0.7475 | 5.05 | 2.7561 | 0.0213 | Up |
| hsa-miR-3140-3p | 0.49 | 3.6425 | 2.8941 | 0.0308 | Up |
| hsa-miR-3188 | 0.3525 | 2.615 | 2.8911 | 0.0360 | Up |
| hsa-miR-4504 | 0.27 | 3.085 | 3.5142 | 0.0277 | Up |
| hsa-miR-4683 | 0.27 | 2.5225 | 3.2238 | 0.0307 | Up |
| hsa-miR-6829-5p | 0.205 | 3.2675 | 3.9945 | 0.0453 | Up |
| hsa-miR-2277-3p | 0.1025 | 2.005 | 4.2899 | 0.0448 | Up |
| hsa-miR-4659a-3p | 0 | 3.9375 | 8.6211 | 0.0003 | Up |
| hsa-miR-3911 | 0 | 2.1425 | 7.7432 | 0.0073 | Up |
| hsa-miR-103a-2-5p | 0 | 4.3125 | 8.7524 | 0.0079 | Up |
| hsa-miR-4747-5p | 0 | 1.8525 | 7.5333 | 0.0118 | Up |
| hsa-miR-541-3p | 0 | 1.7375 | 7.4409 | 0.0166 | Up |
| hsa-miR-450a-2-3p | 0 | 1.305 | 7.0279 | 0.0173 | Up |
| hsa-miR-6891-5p | 0 | 2.2325 | 7.8025 | 0.0324 | Up |
| hsa-miR-3148 | 0 | 2.0425 | 7.6742 | 0.0336 | Up |
| hsa-miR-3173-3p | 0 | 1.6425 | 7.3597 | 0.0338 | Up |
| hsa-miR-154-5p | 0 | 1.635 | 7.3531 | 0.0390 | Up |
| hsa-miR-3680-3p | 0 | 1.8175 | 7.5058 | 0.0399 | Up |
| hsa-miR-4437 | 0 | 1.9725 | 7.6239 | 0.0431 | Up |
| hsa-miR-3146 | 0 | 1.3975 | 7.1267 | 0.0442 | Up |
| hsa-miR-6759-5p | 0 | 1.6425 | 7.3597 | 0.0454 | Up |
Notes: High-throughput second-generation sequencing of exosomal miRNAs detected a total of 775 miRNAs. Compared with the ND group, 424 miRNAs were upregulated and 351 miRNAs were downregulated in the AD group. There were 44 miRNAs with significant differences in expression between the two groups (P<0.05), and the main differences in miRNAs expression. Among them, 34 were upregulated and 10 were downregulated.
The Expression of Serum Exosomal miRNAs
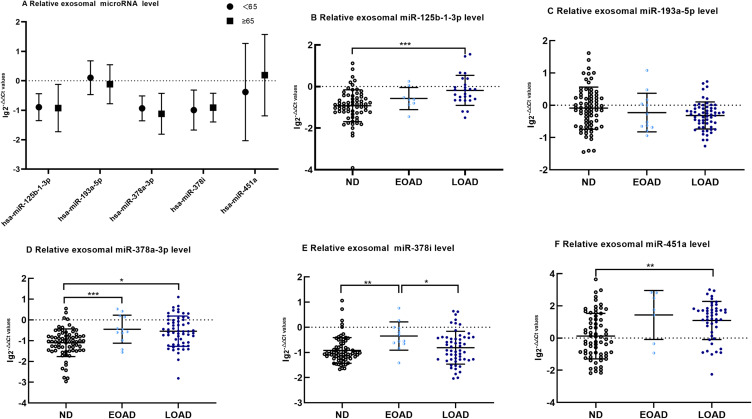
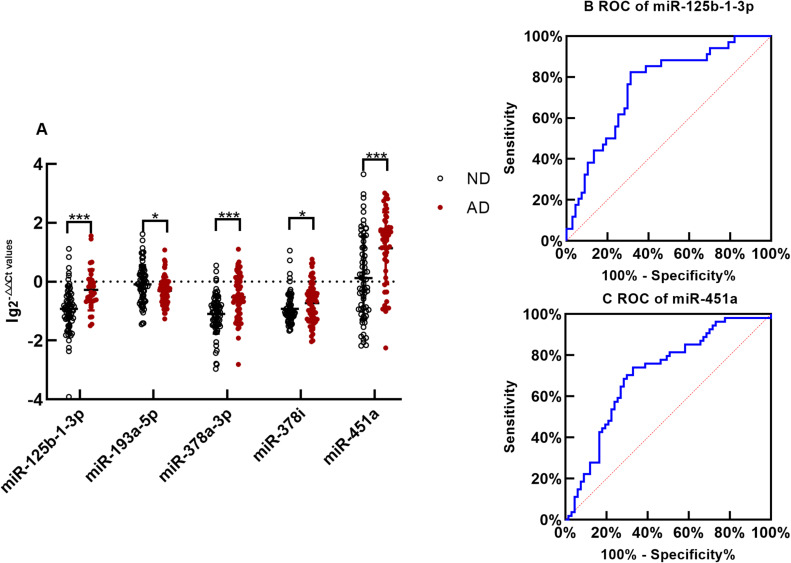
After removing the miRNAs with 0 in sequencing samples, the top 5 miRNAs with the highest expression were screened for qRT-PCR validation. There was no difference in the expression of exosomal miR-125b-1-3p, miR-193a-5p, miR-378a-3p, miR-378i and miR-451a between the ND control subjects aged <65 years old and those aged ≥65 years old (Figure 2A, p>0.05, all). Compared with those of the control group, the levels of exosomal miR-125b-1-3p, miR-378a-3p, miR-378i and miR-451a were significantly increased in the AD group (Figure 3A, p<0.001, 0.001, 0.05 and 0.001, respectively). The AD patients had lower levels of exosomal miR-193a-5p than the controls (Figure 3A, p<0.05). The results for miR-125b-1-3p showed modest improvement in distinguishing the AD patients from the healthy controls (Figure 3B, AUC=0.765, sensitivity=82.1% and specificity=67.7%). MiR-451a showed a strong ability to distinguish the AD patients from the controls (Figure 3C, AUC=0.728, sensitivity=67.9% and specificity=72.6%). However, serum exosomal miR-193a-5p, miR-378a-3p and miR-378i had a weak ability to distinguish the AD patients from the healthy subjects (AUC<0.7, all).
Figure 2.
The difference of exosomal microRNA level different age group of ND with t-tests or among EOAD, LOAD and ND are compared with nonparametric tests (Kruskal–Wallis test). (A) There are no significant difference in age<65 years old group and age ≥65 years old group of exosomal microRNA (B). Comparing with ND, the expression of miR-125b-1-3p in serum exosomes of LOAD is upregulated significantly(p < 0.001). (C) There are no significant difference of miR-193a-5p among EOAD, LOAD and ND. (D) Comparing with ND, the expression of miR-378a-3p of EOAD and LOAD are upregulated significantly(p < 0.001 and p = 0.013, respectively).(E)Comparing with EOAD, the expression of miR-378i of LOAD and ND are downregulated significantly(p = 0.004 and p = 0.039, respectively) (F). Comparing with ND, the expression of miR-451a of LOAD is upregulated significantly(p = 0.001). The y-axis indicate microRNA expression levels by log10 change. *p < 0.05; **p < 0.01;***p < 0.001.
Figure 3.
Exosomal microRNAs qRT-PCR (A) The difference of exosomal microRNA level between AD and ND is compared with nonparametric tests Mann–Whitney U-test. The y-axis indicates microRNA expression levels by log10 change. *p < 0.05,***p < 0.001. The expression of miR-125b-1-3p in serum exosomes of AD is upregulated significantly(p < 0.001);The exosomal miR-193a-5p of AD is downregulated significantly comparing with ND(p=0.020); The expression of miR-378a-3p in serum exosomes of AD is upregulated significantly(p < 0.001); Comparing with ND, the expression of exosomal miR-378i of AD is downregulated significantly(p = 0.042);The expression of miR-451a in exosomes of AD is upregulated significantly (p< 0.001) (B). The receiver operating characteristic(ROC) curve of serum exosomal miR-125b-1-3p. Area Under the Curve(AUC) of miR-125b-1-3p is 0.765 to distinguish AD from normal with a sensitivity and specificity of 82.1% and 67.7% (C). The ROC curve of serum exosomal miR-451a. AUC of miR-451a is 0.728 to distinguish AD from ND with a sensitivity and specificity of 67.9% and 72.6%,respectively.
We validated our results from the discovery phase in the EOAD, LOAD and control cohorts using PCR. The expression of exosomal miR-125b-1-3p levels showed the following trend: control<EOAD<LOAD. MiR-125b-1-3p in LOAD was upregulated significantly compared with that in the control group (Figure 2B, p<0.001). However, there were no significant differences in miR-193a-5p among the three groups, which presented the following trend of expression levels: LOAD<EOAD<control (Figure 2C). Finally, exosomal miR-378a-3p, miR-378i and miR-451a levels showed a consistent trend (control<LOAD<EOAD) and showed an increase in miR-378a-3p in both EOAD and LOAD (Figure 2D p<0.001, 0.05), an increase in miR-378i in EOAD (Figure 2E, p<0.01) and a robust increase in miR-451a in the LOAD group (Figure 2F, p<0.05) versus the control group. MiR-378i expression in the EOAD group was significantly higher than that in the LOAD group (Figure 2E, p<0.05).
Prediction of MiRNA Target Genes and Their Functions
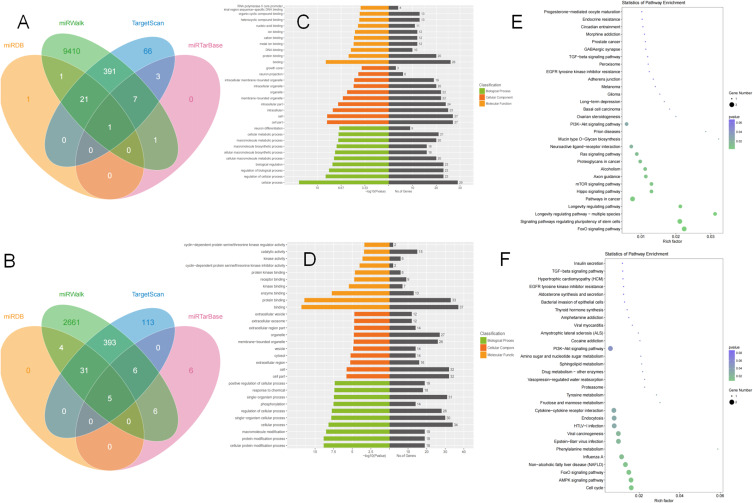
In vivo, miRNA usually involve the expression of target genes in protein translation. So, we focused on the abnormal miRNA and its target genes in AD pathogenesis. We predicted the target genes of miR-125b-1-3p and miR-451a via TargetScan, miRDB, miRTarBase and miRWalk software (Figures 4A and B). The putative target gene of miR-125b-1-3p showing an overlap in four kinds of software was S1PR1 (Gene ID: MIMAT0004592), which was involved in preventing cell apoptosis, promoting cell growth and reproduction, and regulating immune and inflammatory responses. However, 5 target genes, CAB39, OSR1, RAB5A, CUX2 and MIF, for miR-451a were shared between combinations of all four software programs.
Figure 4.
Prediction of MiRNA Target Genes (A and B).The venn of target genes of miR-125b-1-3p and miR-451a (C and D) Gene Ontology of miR-125b-1-3p’s and miR-451a’s candidate genes (E and F) Kyoto Encyclopedia of Genes and Genomes pathway of miR-125b-1-3p’s and miR-451a’s candidate genes.
The top ten terms from the GO results of the predicted miR-125b-1-3p and miR-451a target genes in molecular function (MF), cellular component (CC), and biological process (BP) are shown in Figures 4C and D. In the three categories of miR-125b-1-3p target genes, MF analysis mostly included RNA polymerase II core promoter, metal ion binding, DNA binding, and protein binding; CC analysis mainly included neuron projection and intracellular membrane-bounded organelle. BP analysis mainly involved neuron differentiation, cellular metabolic processes, biological regulation and other processes. Regarding the GO analysis of miR-451a putative genes, MF categories mainly included receptor binding, enzyme binding and protein binding; CC categories mostly covered extracellular exosomes, membrane-bound organelles and extracellular regions; BP categories included positive regulation of cellular processes, phosphorylation and protein modification processes.
KEGG pathways of miR-125b-1-3p’s target genes are involved in axon guidance, neuroactive ligand−receptor interaction, the Hippo signaling pathway, the PI3K-Akt signaling pathway and nervous system-related pathways (Figure 4E). However, hsa-miR-451a had an AUC of 0.728 to differentiate the AD group from the ND group, with a sensitivity and specificity of 67.9% and 72.6%, respectively. Enrichment analysis of its target genes showed a relationship with cytokine−cytokine receptor interactions, the PI3K-Akt signaling pathway, endocytosis, the AMPK signaling pathway and so on (Figure 4F).
Discussion
In 2018, the National Institute on Aging and Alzheimer’s Association referred to β-amyloid deposition, pathologic tau and the neurodegeneration [AT(N)] research framework to define AD, which was undeveloped in general medical practice.16 Less-invasive/less-expensive blood-based biomarker tests along with genetic and clinical analyses are urgently needed to screen for AD among different disorders that can lead to dementia. The current study revealed that miR-125b-1-3p and miR-451a had altered expression in exosomes derived from AD patients, and they had adequate specificity and sensitivity to diagnose AD. Not only was there a difference in EOAD and LOAD based on an age cut off of 65 years, but more importantly, there was a difference shown in clinical characteristics, neuroimaging and neuropathology.2 In the LOAD cohort, differential expression of miR-125b-1-3p, miR-378a-3p and miR-451a was observed. In a cohort of EOAD patients, the expression of miR-378a-3p and miR-378i from serum exosomes was increased compared with that in the ND. Only miR-378i appeared to be significantly different in the EOAD and LOAD cohorts, which suggested that altered exosomal expression of miR-378i may differentiate EOAD from LOAD. However, the EOAD sample was too small, which may lead to bias.
The expression level of miRNAs may be affected by common diseases such as hypertension, coronary heart disease and diabetes, long-term use of antidepressant drugs such as fluoxetine and escitalopram, and various malignancies.11,17–19 We used rigorous exclusion criteria to prevent the effect of these factors on miRNA expression. We further analyzed the relationship between the expression level of miRNAs and age. The ND participants under 65 and over 65 years old showed no difference in the expression level of miRNAs. Age also did not have a substantial influence on the miRNA profiles in multiple sclerosis and lung cancer.20,21 Coincidentally, exosomal miR-378i in EOAD was obviously higher than that in LOAD, regardless of the lack of a significant difference between the LOAD and ND groups.
However, exosomal hsa-miR-125b-1-3p, hsa-miR-193a-5p, hsa-miR-378a-3p, hsa-miR-378i and hsa-miR-451a demonstrated differential levels between the AD patients and the ND subjects in our results. Only hsa-miR-125b-1-3p and hsa-miR-451a showed satisfactory sensitivity and specificity to distinguish AD patients from ND participants. McKeever also revealed that cerebrospinal fluid exosomal miR-125b-5p was increased in both young-onset and late-onset AD patients compared with ND participants.2 However, Tan found an obvious decrease in miRNA125b in AD serum with a sensitivity and specificity of 80.8% and 68.3%, respectively, by comparing 105 AD patients and 150 ND participants.21
Some works suggested that miRNAs derived from the brain might be transported by exosomes crossing the blood-brain barrier and then excreted in the cerebrospinal fluid and peripheral blood.22–24 Exosomes can protect miRNAs from ribonuclease in the circulatory system.20,25 Thus, miRNAs in exosomes could more precisely diagnose and monitor disease than miRNAs in serum or plasma. The hsa-miR-451a level in blood could be an endogenous reference.23 However, we rigidly adhered to the procedures of sample preparation, sample extraction and cryopreservation to avoid hemolysis leading to the increase in hsa-miR-451a. Whether circulating miRNAs in peripheral blood susceptible to hemolysis can be diagnostic biomarkers remains controversial.Previous research reports that an up-regulated miRNA-125b could target the 3’untranslated region of mRNA encoding a 15-lipoxygenase (utilized in the conversion of docosahexaenoic acid into neuroprotectin D1) and the vitamin D3 receptor of the nuclear hormone receptor superfamily. They are key neuromolecular factors essential in lipid-mediated signaling, neurotrophic support, defense against reactive oxygen and nitrogen species, and neuroprotection in the CNS. In AD hippocampal CA1 cells, miRNA-125 damaged neurons by downregulating 15-lipoxygenase and the vitamin D3 receptor.12 The study revealed that overexpression of miR-451a downregulated the expression of activating transcription factor 2(ATF2).13 ATF2 mediated the normal physiological metabolism of nerve cells under physiological conditions and is an essential component. When ATF2 expression is deficient, the development of nerve cells cannot occur normally, which can trigger a large number of cell apoptosis and neuronal loss, leading to the occurrence of AD.14 The GO enrichment analysis of hsa-miR-125b-1-3p’s target genes is related to neuron differentiation, cellular metabolic process and metal ion binding, which may be involved in iron deposition causing neuronal loss. In the KEGG pathway, hsa-miR-125b-1-3p’s target genes were involved in neuroactive ligand-receptor interaction, the PI3K-Akt signaling pathway, the Hippo signaling pathway and nervous system-related pathways. Furthermore, there was a positive correlation between the expression of miR-125b and cognitive function in APP/PS1 transgenic mice.26 In vivo, injecting miR-125b into the hippocampus of mice impaired associative learning and was accompanied by downregulation of Bcl-W, DUSP and PPP1CA, resulting in an increase in tau phosphorylation.27 Another report showed that hsa-miR-451 may play a role in AD pathogenesis by downregulating Alzheimer’s disease-related disintegrin and metalloprotease 10.28 Interestingly, both hsa-miR-125b-1-3p and hsa-miR-451a’s target genes are involved in cytokine−cytokine receptor interactions and the PI3K-Akt signaling pathway. Further work is required to demonstrate their biological functions and their target genes in the pathogenesis of AD.
Overall, our study revealed that the differential expression of exosomal miRNAs was altered in AD. The expression level of two exosomal miRNAs effectively discriminates AD patients from ND with a certain sensitivity and specificity. However, additional work will be needed to recapitulate the relationship between these miRNAs and Aβ, pTau or other pathological phenotypes in AD.
Acknowledgments
The authors want to acknowledge the study participants for their dedication to the study.
Funding Statement
This study is funded by the Hunan Provincial Health Commission (B202303106781).
Data Sharing Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to containing information that could compromise the privacy of research participants.
Ethics Statement
Clinical experiment protocols were approved by the Xiangya Hospital of Central South University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflict of interests.
References
- 1.Jia L, Fu Y, Shen L, et al. PSEN1, PSEN2, and APP mutations in 404 Chinese pedigrees with familial Alzheimer’s disease. Alzheimers Dement. 2020;16(1):178–191. doi: 10.1002/alz.12005 [DOI] [PubMed] [Google Scholar]
- 2.McKeever PM, Schneider R, Taghdiri F, et al. MicroRNA expression levels are altered in the cerebrospinal fluid of patients with young-onset Alzheimer’s Disease. Mol Neurobiol. 2018;55(12):8826–8841. doi: 10.1007/s12035-018-1032-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mckhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the national institute on aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dementia. 2011;7(3):p. p. 263–269. doi: 10.1002/alz.12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren RJ, Qi JL, Lin SH, et al. The China Alzheimer report 2022. Gen Psych. 2022;35(1):e100751. doi: 10.1136/gpsych-2022-100751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang LJ, Jiao B, Liu XX, et al. Specific serum autoantibodies predict the development and progression of Alzheimer’s disease with high accuracy. Brain Behav Immun. 2023;19(115):543–554. doi: 10.1016/j.bbi.2023.11.018 [DOI] [PubMed] [Google Scholar]
- 6.Cocozza F, Grisard E, Martin-Jaular L, Mathieu M, Thery C. SnapShot: Extracellular Vesicles. Cell. 2020;182(1):262–262 e261. doi: 10.1016/j.cell.2020.04.054 [DOI] [PubMed] [Google Scholar]
- 7.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- 8.Zappulli V, Friis KP, Fitzpatrick Z, Maguire CA, Breakefield XO. Extracellular vesicles and intercellular communication within the nervous system. J Clin Invest. 2016;126(4):1198–1207. doi: 10.1172/JCI81134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maffioletti E, Tardito D, Gennarelli M, Bocchio-Chiavetto L. Micro spies from the brain to the periphery: new clues from studies on microRNAs in neuropsychiatric disorders. Front Cell Neurosci. 2014;8:75. doi: 10.3389/fncel.2014.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagaraj S, Zoltowska KM, Laskowska-Kaszub K, Wojda U. microRNA diagnostic panel for Alzheimer’s disease and epigenetic trade-off between neurodegeneration and cancer. Ageing Res Rev. 2019;49:125–143. doi: 10.1016/j.arr.2018.10.008 [DOI] [PubMed] [Google Scholar]
- 12.Zhao YZ, Bhattacharjee S, Jones BM, Hill J, Dua P, Lukiw WJ. Regulation of neurotropic signaling by the inducible, NF-kB-sensitive miRNA-125b in Alzheimer’s disease (AD) and in primary human neuronal-glial (HNG) cells.. Mol Neurobiol. 2014;50(1):97–106. doi: 10.1007/s12035-013-8595-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao Y, Dong LJ, Takahashi Y, et al. miRNA-451a regulates RPE function through promoting mitochondrial function in proliferative diabetic retinopathy. Am J Physiol Endocinol Met. 2019;316(3):E443–E452. doi: 10.1152/ajpendo.00360.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson AG, Curtis MA, Waldvogel HJ, Faull RLMDrragunow M. Activating transcription factor 2 expression in the adult human brain: association with both neurodegeneration and neurogenesis. Neuroscience. 2005;133(2):437–451. doi: 10.1016/j.neuroscience.2005.02.029 [DOI] [PubMed] [Google Scholar]
- 15.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 16.Jack CR Jr, Bennett DA, Blennow K, et al.; Contributors. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Vijayan M, Bhatti JS, Reddy PH. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog Mol Biol Transl Sci. 2017;146:47–94. [DOI] [PubMed] [Google Scholar]
- 18.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329(5998):1537–1541. doi: 10.1126/science.1193692 [DOI] [PubMed] [Google Scholar]
- 19.Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol. 2013;23(7):602–611. doi: 10.1016/j.euroneuro.2012.06.013 [DOI] [PubMed] [Google Scholar]
- 20.Keller A, Leidinger P, Lange J, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4(10):e7440. doi: 10.1371/journal.pone.0007440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuxia M, Zhennan T, Wei Z. Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J Cancer Res Clin Oncol. 2012;138(12):2045–2050. doi: 10.1007/s00432-012-1285-0 [DOI] [PubMed] [Google Scholar]
- 22.He HN, Liu A, Zhang W, et al. Novel plasma miRNAs as biomarkers and therapeutic targets of Alzheimer’s Disease at the prodromal stage. J Alzh Dis. 2021;83(2):779–790. doi: 10.3233/JAD-210307 [DOI] [PubMed] [Google Scholar]
- 23.Cheng L, Doecke JD, Sharples RA, et al. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry. 2015;20(10):1188–1196. doi: 10.1038/mp.2014.127 [DOI] [PubMed] [Google Scholar]
- 24.Haqqani AS, Delaney CE, Tremblay TL, Sodja C, Sandhu JK, Stanimirovic DB. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS. 2013;10(1):4. doi: 10.1186/2045-8118-10-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheramy M, Hampe CS, Ludvigsson J, Casas R. Characteristics of in-vitro phenotypes of glutamic acid decarboxylase 65 autoantibodies in high-titre individuals. Clin Exp Immunol. 2013;171(3):247–254. doi: 10.1111/cei.12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong H, Li Y, Su B. Identification of circulating miR-125b as a potential biomarker of alzheimer’s disease in APP/PS1 transgenic mouse. J Alzheimers Dis. 2017;59(4):1449–1458. doi: 10.3233/JAD-170156 [DOI] [PubMed] [Google Scholar]
- 27.Banzhaf-Strathmann J, Benito E, May S, et al. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014;33(15):1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng C, Li W, Zhang Z, et al. MicroRNA-144 is regulated by activator protein-1 (AP-1) and decreases expression of Alzheimer disease-related a disintegrin and metalloprotease 10 (ADAM10). J Biol Chem. 2013;288(19):13748–13761. doi: 10.1074/jbc.M112.381392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to containing information that could compromise the privacy of research participants.