Abstract
The adenovirus type 5 (Ad5) early 1B 55-kDa protein (E1B-55kDa) is a multifunctional phosphoprotein that regulates viral DNA replication and nucleocytoplasmic RNA transport in lytically infected cells. In addition, E1B-55kDa provides functions required for complete oncogenic transformation of rodent cells in cooperation with the E1A proteins. Using the far-Western technique, we have isolated human genes encoding E1B-55kDa-associated proteins (E1B-APs). The E1B-AP5 gene encodes a novel nuclear RNA-binding protein of the heterogeneous nuclear ribonucleoprotein (hnRNP) family that is highly related to hnRNP-U/SAF-A. Immunoprecipitation experiments indicate that two distinct segments in the 55-kDa polypeptide which partly overlap regions responsible for p53 binding are required for complex formation with E1B-AP5 in Ad-infected cells and that this protein interaction is modulated by the adenovirus E4orf6 protein. Expression of E1B-AP5 efficiently interferes with Ad5 E1A/E1B-mediated transformation of primary rat cells. Furthermore, stable expression of E1B-AP5 in Ad-infected cells overcomes the E1B-dependent inhibition of cytoplasmic host mRNA accumulation. These data suggest that E1B-AP5 might play a role in RNA transport and that this function is modulated by E1B-55kDa in Ad-infected cells.
The replication cycle of adenoviruses (Ad) is divided by convention into two stages which are separated by the onset of viral DNA replication (reviewed in reference 66). During the late phase of infection, cellular protein synthesis is shut off, due to a translational block of host cell mRNAs (reviewed in reference 80). Further, most cellular mRNAs fail to accumulate in the cytoplasm despite continued nuclear synthesis and processing (4). In contrast, late viral mRNAs are selectively exported to the cytoplasm and are efficiently translated late after infection (1, 5). This severe inhibition of cellular gene expression appears to be mediated by viral proteins that operate at the level of translation and nucleocytoplasmic mRNA transport (2, 54, 79).
The selective accumulation of viral mRNAs during the late phase of infection is mediated by a protein complex that includes the Ad early 1B 55-kDa (E1B-55kDa) and E4orf6 proteins (8, 29, 63). The E1B-E4 protein complex appears to modulate viral and cellular mRNA transport after transcription and processing but before translocation of mRNAs through the nuclear pores (41). Immunofluorescence and immunoelectron microscope studies showed that both proteins are localized within and about the periphery of nuclear viral inclusion bodies (52) believed to be the sites of viral transcription and/or replication (34, 55). This observation is consistent with the idea that the E1B-E4 protein complex regulates RNA metabolism at an intranuclear step, possibly by facilitating the movement of mature viral mRNA to the nuclear pore complex (11, 40, 50). The selective transport is not dependent on the identity of individual mRNAs. Cellular mRNAs transcribed from recombinant viral chromosomes are transported to the cytoplasm late after infection, even at a time when the endogenous cellular transcript is restricted to the nucleus (20, 30).
Ornelles and Shenk (52) have proposed a model by which the E1B-E4 protein complex facilitates the transport and accumulation of viral mRNAs while simultaneously blocking the same process for most host mRNAs. According to their proposal, the E1B-E4 complex relocalizes a cellular factor required for nucleocytoplasmic transport of mRNAs from the sites of host cell transcription and processing to the viral replication/transcription centers. This model is consistent with the observation that cellular splicing factors and heterogeneous nuclear ribonucleoprotein particle (hnRNP) proteins are redistributed to the sites of viral RNA transcription and DNA accumulation during the late phase of infection (34). In addition, subcellular fractionation of Ad12-transformed cells demonstrated that E1B-54kDa exists in a high-molecular-weight complex in the nucleus, indicating that the 54-kDa protein associates with one or more cellular components (28). The observation that the Ad5 E1B-55kDa-mediated accumulation of viral mRNAs is dependent on residual splicing sites in different viral mRNAs suggests that nuclear proteins which are involved in heterogeneous nuclear RNA processing may be targets for the E1B-55kDa protein (11, 40). A similar function, which is independent of E1B-55kDa, has been reported for the E4orf6 and E4orf3 proteins (51). Both proteins seem to encode redundant functions required for efficient tripartite leader splicing during a lytic virus infection (50, 51). These observations suggests that E1B-55kDa and two proteins from the E4 region modulate general pathways in mRNA formation. The demonstration that Ad5 E1B-55kDa but not E4orf6 interferes with mRNA export in Saccharomyces cerevisiae (42) suggests that the late functions required for selective transport of viral mRNA are encoded predominantly in the 55-kDa polypeptide. However, the molecular mechanism by which the E1B-E4 protein complex modulates mRNA transport and the identity of the putative transport factor are still unknown.
We have identified a novel protein referred to as E1B-associated protein (E1B-AP5) that binds specifically to E1B-55kDa in vitro and in vivo. E1B-AP5 is a nuclear RNA-binding protein of the hnRNP protein family that is highly related to hnRNP-U/SAF-A. The E1B-55kDa/E1B-AP5 protein interaction is mediated by two segments in the 55-kDa polypeptide which partly overlap regions responsible for p53 binding. Substantially more E1B-55kDa can bind to E1B-AP5 in the absence of the E4orf6 protein, suggesting that the E4 protein modulates complex formation. In Ad5 E1A/E1B-mediated transformation assays, expression of E1B-AP5 causes a marked reduction in the number of transformed cells. Furthermore, we present evidence that stable expression of E1B-AP5 overcomes the E1B-55kDa-dependent shutoff of host cell mRNA export in Ad-infected cells. Our data indicate that E1B-AP5 might play an important role in nucleocytoplasmic mRNA transport and is at least one of the cellular proteins that is targeted by E1B-55kDa in the selective accumulation of mRNAs in late-Ad-infected cells.
MATERIALS AND METHODS
Generation of 32P-labeled GST fusion proteins and library screening.
The wild-type Ad5 E1B-55kDa-expressing pGEX construct was made by replacing the 5′-untranslated region (Ad5 nucleotides [nt] 1974 to 2018) of p11/55.X (62) with an oligonucleotide with the sequence 5′-GATCCATGGAGCGAAGAAACCCATCTGAGCGGGGGGTAC-3′ and 5′-CCCCCGCTCAGATGGGTTTCTTCGCTCCATG-3′, bounded by sequences recognized by BamHI and KpnI. The DNA sequence of this construct was confirmed. After addition of BamHI linkers to the blunted XbaI site in p11/55.BX, the 2,088-bp BamHI fragment was inserted into the BamHI site of the pGEX-2Tk polylinker (35).
Preparation and purification of glutathione S-transferase (GST) and GST fusion proteins was as described previously (62). To generate the radiolabeled GSTE1B-55kDa fusion protein, we used a modified version of a procedure described by Kaelin et al. (35). Briefly, GSTE1B-55kDa bound to glutathione-Sepharose beads was equilibrated in 1× HMK buffer (20 mM Tris-chloride [pH 7.5], 100 mM NaCl, 12 mM MgCl2). The fusion protein was labeled in 1× HMK buffer for 30 min at 30°C with 250 U of the catalytic subunit of cyclic AMP-dependent protein kinase (Sigma) and 250 μCi of [γ-32P]ATP (5,000 Ci/mmol; Amersham). The reaction was terminated by the addition of HMK stop buffer (10 mM sodium phosphate [pH 8.0], 10 mM sodium pyrophosphate, 10 mM EDTA, 1 mg of bovine serum albumin per ml). The supernatant was removed by aspiration, and the Sepharose was washed five times with 5 bead volumes of NETN (20 mM Tris-chloride [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 [NP-40]). The labeled GST fusion protein was eluted for 30 min in 3 bead volumes of 50 mM reduced glutathione–100 mM Tris-chloride (pH 8.0)–120 mM NaCl under constant agitation.
The expression library screen was performed as described previously (35). A total of 2 × 106 recombinant phages of a HeLa cDNA library (Clontech) were screened. Three rounds of screening were carried out to obtain pure phage populations. DNA was prepared from the clones of interest and digested with EcoRI to obtain the cDNA inserts, which were subsequently subcloned into pBKRSV (Stratagene) for DNA sequencing.
To isolate overlapping cDNA clones of E1B-AP5, the HeLa cDNA library was rescreened with 32P-labeled DNA fragments from E1B-AP5/2 and a 3′ restriction fragment from clone E1B-AP5/3 as described previously (14). Positive clones were plaque purified and analyzed by subcloning into pBKRSV and DNA sequencing.
Homology searches.
Homology searches in the GenBank and SwissProt data bases and alignments were carried out by using the Genetics Computer Group FASTA program (version 8.1) on an INDY workstation (Silicon Graphics) and the BLAST homology finder program in the National Center for Biotechnology Information database, remotely. Protein motifs and patterns were analyzed by using the FIND PATTERN program in the Prosite dictionary of protein sites and patterns.
RNA and protein analysis.
The methods for isolation of total and cytoplasmic RNA and Northern blot analysis have been described previously (13, 46). Total RNA isolated from different cell lines was a generous gift from M. Rehli (Universität Regensburg). The human β-actin gene segment (46) was labeled with [α-32P]dCTP and used as cellular probe DNA. The viral L3 and L5/E4 cDNAs were used as viral probe DNAs (29).
To produce an E1B-AP5-specific antigen for immunization, a 2,570-bp fragment corresponding to nt 174 and 2744 in the E1B-AP5 cDNA was generated by PCR with primers fw (5′-GCGCGCAGGATCCGGATGGATGTGCGCCGTCTGAAGGTGAACG-3′) and rev (5′-GCGCGCGTCGACCTACTGTGTACTTGTGCCACCCTGTG-3′), which introduce a BamHI site at the 5′ end and a SalI site at the 3′ end, respectively. This fragment was inserted between the BamHI and SalI sites of pGEX-5X1 (Pharmacia) to generate pGEXE1B-AP5. The GST fusion protein was expressed in Escherichia coli DH5α, affinity purified, and used to immunize female New Zealand White rabbits (Charles River). Sera were collected 14 days after the fourth boost, yielding a polyclonal antibody to E1B-AP5. Monoclonal antibodies were also used in our studies: 2A6 (65) and 9C10 (Oncogene Science) are specific for E1B-55kDa, RSA3 (44) is specific for the E4orf6 protein, B6 (58) is specific for the E2A-72kDa protein; and 12CA5 (Boehringer) is specific for the HA epitope.
For the analysis of proteins by Western blotting or immunoprecipitation (62), total-cell extracts were prepared in lysis buffer (50 mM Tris-chloride [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.15% NP-40, 0.05 mM phenylmethylsulfonyl fluoride) per 90-mm-diameter dish. After 1 h on ice, the lysate was sonicated and the insoluble debris was pelleted at 10,000 × g at 4°C. If necessary, cells were labeled for 1 h with 50 μCi of [35S]methionine (Amersham) per 90-mm plate before extract preparation. After electrophoresis, proteins were detected by autoradiography or enhanced chemiluminescence (Amersham). The proteins were quantitated from a TIFF file by using the Analyze Particles program (NIH Image 1.52).
Indirect immunofluorescence.
Ad5- and mock-infected cells grown on glass coverslips were washed twice in phosphate-buffered saline (PBS) and subsequently fixed by incubation with 4% paraformaldehyde in CSK buffer {100 mM KCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA [ethylene glycol-bis(β-aminoethylether)-N,N,N′,N′-tetraacetic acid], 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.8], 1.5 mM phenylmethylsulfonyl fluoride} for 10 min at room temperature. The cells were permeabilized with 0.5% Triton X-100 in CSK buffer for 15 min at room temperature. The cells were washed three times in PBS containing 0.1% Tween 20 and incubated in blocking buffer (0.5% blocking reagent, 10 mM Tris-chloride [pH 7.5], 150 mM NaCl) for 30 min at room temperature. Samples were incubated with the primary antibodies for 1 h at room temperature, washed three times with PBS–0.1% Tween 20, and then incubated with 10 μg of fluorescein- or Cy3-conjugated goat antibodies specific for mouse or rabbit immunoglobulin G (Dako) for 1 h. After being washed five times in PBS, the samples were mounted in PBS-glycerol containing 0.5 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml and viewed with a Leitz Aristoplan Photomicroscope by using epifluorescence illumination.
In vitro binding assays.
In vitro-radiolabeled proteins were made in a coupled transcription-translation system (TnT; Promega), as specified by the manufacturer, with 1 μg of DNA, 40 μCi of [35S]methionine (1,000 Ci/mmol; Amersham) and T7 or T3 RNA polymerase. For in vitro binding, 5 μl of reticulolysate was added to 200 μl of binding buffer (50 mM Tris-chloride [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.15% NP-40, 1 mM dithiothreitol, 0.05 mM phenylmethylsulfonyl fluoride) containing 20 μl of purified GST fusion proteins and rotated for 1 h at 4°C. Matrices were washed five times with 1 ml of binding buffer before being subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
RNA- and ssDNA-binding analyses.
Binding of in vitro-translated E1B-AP5 to ribonucleotide homopolymer and single-stranded DNA (ssDNA) was carried out essentially as described previously (37). Briefly, radiolabeled E1B-AP5 protein was generated by in vitro translation and incubated with 3 μg of the corresponding ribonucleotide homopolymer (Pharmacia) and ssDNA-agarose (GIBCO) in 250 μl of DA250 buffer (10 mM HEPES/NaOH [pH 7.9], 250 mM NaCl, 1.5 mM MgCl2) for 1 h at 4°C. E1B-AP5 protein bound to agarose beads was washed five times in DA buffer containing 0.25, 0.5, or 1 M NaCl and analyzed by SDS-PAGE followed by autoradiography.
Cells and viruses.
All cell lines were grown as monolayer cultures in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. To generate stable cell lines expressing epitope-tagged E1B-AP5, 8 μg of pSVfluE1B-AP5 was cotransfected with 1.5 μg of pBabe-puro (47) into H1299 cells (45) by calcium phosphate coprecipitation and stable transfectants (H12-AP5 cells) were isolated in medium containing 0.5 μg of puromycin (Sigma) per ml. pSVfluE1B-AP5 expressing epitope-tagged E1B-AP5 under the control of the simian virus 40 promoter was generated by inserting the BamHI-SalI fragment from pGEXE1B-AP5 into pAS2 (kindly provided by S. Elledge). The E1B-AP5 cDNA fused to the HA tag from pAS2 was then inserted between the EcoRI and SalI sites of pSVK3 (Pharmacia).
H5wt300 served as the wild-type Ad5 parent in these studies. The following mutant viruses were used: H5dl338 carries a 524-bp deletion in the E1B coding region located between nt 2805 and 3329 (54). The linker insertion mutations H17, A143, H180, H224, A262, R309, H326, H354, S380, R443, and F484 in the E1B-55kDa gene have been described previously (76). The mutants containing these mutations are recombinants of Ad2 and Ad5 and are named first by a letter representing the restriction site at the insertion and then by a number representing the amino acid residue at or just preceding the 4-residue insertion. Mutant H5in3328(+) produces an E1B protein containing an 11-amino-acid (aa) insert between Ile-438 and Trp-439 (72). In the E1B mutant H5pm490A/491A, the codons for Ser-490 and Ser-491 have been changed to those for alanine (72). H5dl355 contains a 14-bp deletion in the E4orf6 gene between nt 2331 and 2346 (29) and does not express the E4orf6 protein (29); H5dl341 contains a 1-bp deletion in the E4orf3 gene and does express the E4orf3 protein (64). The Ad5 wild-type virus was propagated on A549 cells (21) or H1299 cells (45). E1B and E4 mutant viruses were propagated on 293 cells (25) or W162 cells (73), respectively. Virions purified by cesium chloride equilibrium density centrifugation were used for all infections.
Ad DNA replication was determined by PCR exactly as described previously (62).
Transformation assays.
Primary cultures of baby rat kidney (BRK) cells were prepared from kidneys of 6-day-old Sprague-Dawley rats as described previously (48) and grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. For transformation assays, subconfluent cells were transfected 2 days postplating by the calcium phosphate procedure (26) with salmon sperm carrier and plasmid DNAs exactly as described previously (48). Three weeks after transfection, foci were stained with 1% crystal violet in 25% methanol.
The following plasmids were used in this study: pXC15 (pAd5 XhoI-C), which contains the left end (1 to 15.5 map units) of the Ad5 genome and expresses the E1A and E1B proteins (43), and plasmid pC53-SN3, which encodes human wild-type p53 from the pCMV/neo vector.
Nucleotide sequence accession number. The EMBL and GenBank accession no. of E1B-AP5 is AJ007509.
RESULTS
To isolate cDNAs encoding cellular proteins capable of interacting specifically with E1B-55kDa, we screened 2 × 106 recombinant phages of a HeLa cell λgt11 expression library with the radiolabeled GSTE1B-55kDa protein. A total of 12 clones encoding fusion proteins which bound specifically to 32P-GSTE1B-55kDa with high affinity were identified. These fell into five classes, based on sequence analysis, and were referred to as E1B-associated proteins E1B-AP1 to E1B-AP5. In this study, we have focused on the E1B-AP5 family of clones. The characterization of the remaining E1B-associated proteins will be described elsewhere.
E1B-AP5 is a new member of the hnRNP family.
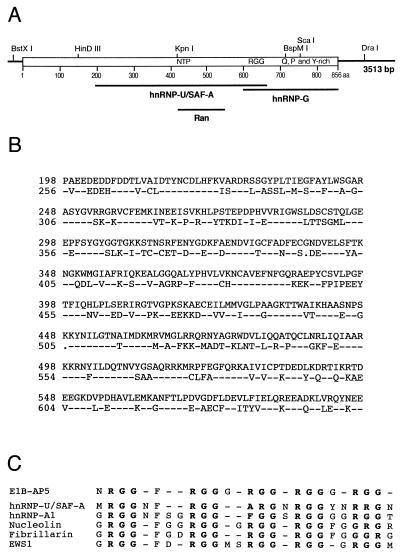
Several E1B-AP5 cDNA fragments were assembled into a contiguous stretch of 3,513 bp (Fig. 1A). Analysis of the E1B-AP5 cDNA sequence revealed a large open reading frame from nt 174 to 2742 (Fig. 1B). The proposed start codon lies in a favorable initiation context (38), and the predicted protein contains 856 aa residues with a molecular mass of 95,805 Da and a pI of 6.5.
FIG. 1.
E1B-AP5 cDNA maps, nucleotide sequence, and predicted amino acid sequence. (A) cDNA clones E1B-AP5/1 and E1B-AP5/2 were isolated with the [γ-32P]ATP-labeled GSTE1B-55kDa protein probe from a λgt11 HeLa cDNA expression library. cDNA clones E1B-AP5/3 to E1B-AP5/7 were isolated from the same library by rescreening with E1B-AP5/2 and later with a fragment from the 3′ end of E1B-AP5/3. The thick black bar on top represents the 2,568-bp open reading frame of E1B-AP5. Thin bars denote the 5′ and 3′ untranslated sequences, respectively. The locations of some unique restriction enzymes are indicated above the bars. (B) The complete E1B-AP5 cDNA sequence was generated by assembling restriction fragments from E1B-AP5/6 and E1B-AP5/7 in pBKRSV. Sequence was determined from each cDNA clone twice on both strands by sequence-derived oligonucleotide primers. The predicted amino acid sequence is shown in the single-letter code.
A comparison of the predicted amino acid sequence of the open reading frame of E1B-AP5 to the protein databases by using the FASTA and BLAST homology finder revealed three regions of E1B-AP5 that are highly related to previously reported protein sequences (Fig. 2A). The most striking similarity in the sequence is to the human 120-kDa hnRNP-U/SAF-A protein (37). This protein exists in two isoforms (18), both of which have been reported to bind homopolymeric RNA (37) and vertebrate scaffold attachment region DNA elements (17). The predicted protein sequence of isoform 1 is 56% identical over a 400-aa stretch to a region of E1B-AP5 between aa 198 and 598 (Fig. 2B). E1B-AP5 is not related to the second isoform of hnRNP-U/SAF-A. The central region of E1B-AP5 contains a Gxxxx-GKS/T phosphate-binding loop (residues 428 to 435) and the DxxG Mg2+-binding motif (residues 547 to 550), both of which are required for guanine nucleotide binding (7). Significantly, these motifs are arrayed in a configuration in E1B-AP5 in the same order and with comparable spacing to sequences in the small GTP-binding protein Ran from several species (22% identity in a 122-aa overlap). A second region in the carboxy-terminal part of E1B-AP5 (residues 612 to 666) contains closely spaced RGG repeats (Fig. 2C). The RGG box motif is involved in RNA binding (37) and is present in a number of viral and cellular proteins with proposed roles in RNA metabolism (reviewed in reference 9). Finally, toward the carboxy-terminal end of the predicted E1B-AP5 sequence lies a third region of similarity (23% identity in a 280-aa overlap) with the amino terminus of the RNA-binding protein hnRNP-G (69) (Fig. 2A). This protein is a component of ribonucleosomes and binds to RNA presumably through one RNP consensus RNA-binding domain. The carboxy-terminal 255-aa domain of E1B-AP5 is rich in proline, glutamine, and tyrosine residues. Based on the significant sequence similarity to hnRNP-U/SAF-A, hnRNP-G, and other hnRNP proteins, e.g., A1, K, and L (data not shown), we propose that E1B-AP5 is a member of the hnRNP protein family.
FIG. 2.
Sequence homologies to hnRNP-U/SAF-A, hnRNP-G, and Ran. (A) The regions that have been identified within the predicted amino acid sequence of E1B-AP5 are shown below in their relative positions along the E1B-AP5 polypeptide (open box). The locations of the NTP-binding consensus sequence (NTP), RGG boxes (RGG), and glutamine-, proline-, and tyrosine-rich region (Q, P and Y-rich) at the carboxy terminus are indicated. (B) Direct sequence comparison of the related sequences of E1B-AP5 and hnRNP-U/SAF-A. The dashes in the hnRNP-U/SAF-A sequence are identical amino acids in E1B-AP5, and dots indicate gaps in the hnRNP-U/SAF-A sequence alignment. (C) Alignment of E1B-AP5 with RGG box domains from several proteins. The consensus sequences of hnRNP-U/SAF-A, hnRNP-A1, nucleolin, fibrillarin, and EWS1 were derived from the sequences listed in reference 9.
Analysis of E1B-AP5 mRNA and protein expression.
To determine the size and tissue distribution of the E1B-AP5 mRNA, Northern blot analysis was performed on a variety of human tissues and cell lines with a 1.6-kb fragment from the E1B-AP5 coding sequence as a probe (Fig. 3A). Two bands of approximately 3.2 and 3.8 kb were detected in all RNA preparations examined. Probing total RNA from HeLa cells with a 279-bp DraI restriction fragment from the most 3′ end of the untranslated region resulted in the detection of only the 3.8-kb mRNA (data not shown), indicating that the two mRNAs differ in the length of their 3′ untranslated region.
FIG. 3.
Analysis of E1B-AP5 mRNA and protein expression. (A) Northern blot analysis. A 20-μg portion of total RNA isolated from the indicated human tissues and cell lines were subjected to electrophoresis through a 1.2% agarose gel containing formaldehyde, transferred to a nitrocellulose filter, and hybridized to a [α-32P]dCTP-labeled E1B-AP5 coding sequence probe from E1B-AP5/5. (Monoc, monocytes; Macrop, macrophages; Lmypho, lymphocytes; Fibrob, fibroblasts; HepG2, hepatocytes; HaCAT, keratinocytes; MelIm, melanoma). The locations of the 18S and 28S rRNAs are indicated. RNA loading was determined by staining the RNA with ethidium bromide prior to transfer to nitrocellulose filters. (B) Immunoprecipitation of E1B-AP5 and hnRNP-U/SAF-A proteins. Radiolabeled E1B-AP5 (lanes 1 to 3) and hnRNP-U/SAF-A (lanes 4 to 6) proteins were generated by in vitro translation and subjected to immunoprecipitation with the anti-E1B-AP5 antiserum (α-E1B-AP5; lanes 2 and 5) and a matched preimmune serum (pre; lanes 3 and 6). The precipitates were analyzed by SDS-PAGE and autoradiography. Lanes designated “input” received the same amount of in vitro-translated proteins added to each immunoprecipitation reaction mixture. The positions of markers are indicated. (C) Western blot analysis of E1B-AP5 protein expression. Total-cell extracts were prepared and subjected to SDS-PAGE, transferred to nitrocellulose filters, and probed with the rabbit anti-E1B-AP5 antiserum. The positions of markers are indicated on the right. (D) Indirect immunofluorescence analysis. A549 cells were probed with the rabbit anti-E1B-AP5 antiserum (α-E1B-AP5) followed by fluorescein-conjugated sheep anti-rabbit antibodies. Magnification, ×100.
To identify the protein encoded by the E1B-AP5 gene in vivo, a polyclonal rabbit anti-E1B-AP5 antiserum was made against the GSTE1B-AP5 fusion protein. To exclude the possibility that the anti-E1B-AP5 antiserum cross-reacts with the highly related 120-kDa hnRNP-U/SAF-A protein, we carried out immunoprecipitation experiments with in vitro-translated E1B-AP5 and hnRNP-U/SAF-A proteins by using the anti-E1B-AP5 antiserum (Fig. 3B). Only in vitro-translated E1B-AP5 protein was efficiently precipitated with the antiserum, while no reactivity was detectable with radiolabeled hnRNP-U/SAF-A or a matched preimmune serum.
Immunoblot analysis with total-cell extracts from various cell lines revealed that E1B-AP5 protein migrates with an apparent molecular mass of 120 kDa in SDS-polyacrylamide gels (Fig. 3C), which differs from the calculated molecular mass of 95.8 kDa. The lower mobility is most probably due to posttranslational modifications; epitope-tagged E1B-AP5 protein expressed from the cDNA clone in human cells also migrates at about 120 kDa relative to marker proteins (see Fig. 7A).
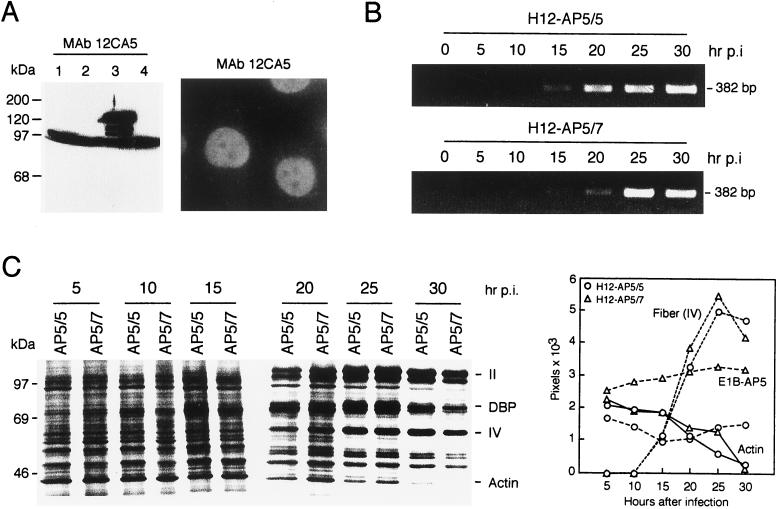
FIG. 7.
Effects of stable expressed fluE1B-AP5 on viral DNA synthesis and host cell shutoff. (A) Expression of fluE1B-AP5 in H12-AP5/7 cells. Total-cell extracts were prepared from H1299 cells (lane 1) and puromycin-resistant cell clones H12-AP/5 (lane 2), H12-AP5/7 (lane 3), and H12-AP5/8 (lane 4), subjected to SDS-PAGE, and analyzed by immunoblotting with monoclonal antibody (MAb) 12CA5. The additional faster-migrating band present in all lanes is due to a cross-reactivity of a cellular protein with monoclonal antibody 12CA5. Indirect immunofluorescence of H12-AP5/7 cells probed with 12CA5 followed by fluorescein-conjugated sheep anti-rabbit antibody. Magnification, ×100. (B) Analysis of viral DNA synthesis. H12-AP5/5 or H12-AP5/7 cells were infected with wt300 virus at an infectivity of 200 PFU per cell. Cells were harvested at the indicated time points postinfection (hr p.i.), and viral DNA synthesis was determined by PCR. The DNA products were subjected to agarose gel electrophoresis and ethidium bromide staining. (C) Analysis of fiber (IV) and actin protein steady-state levels. Cells were infected with wt300 virus and labeled with [35S]methionine for 1 h at 5, 10, 15, 20, 25, and 30 h after infection. A total of 2 × 105 trichloroacetic acid-precipitable counts from each time point was fractionated by SDS-PAGE, and proteins were visualized by autoradiography. Molecular mass markers are indicated on the left. Bands corresponding to cellular actin and several viral polypeptides are listed on the right. The levels of fiber (IV) and actin proteins were quantitated as described and plotted as a function of time. The steady-state level of the E1B-AP5 protein was quantitated after Western blot analysis of the same total-cell extracts (data not shown).
Indirect-immunofluorescence analysis was performed to determine the intracellular localization of endogenous E1B-AP5 protein in H1299 cells by using the E1B-AP5-specific rabbit antiserum (Fig. 3D). The majority of the E1B-AP5 protein is localized to the nucleus but excluded from the nucleolus of these cells. In addition, weak staining in the cytosol indicates that a minor fraction of E1B-AP5 might be localized in the cytoplasm. No specific signal was detected when a matched preimmune serum was used (data not shown).
E1B-AP5 binds to E1B-55kDa in vitro and in vivo.
To confirm further the association of E1B-AP5 with E1B-55kDa, two different protein-binding experiments were performed. First, lysates were prepared from E. coli producing GST fusion proteins with either wild-type E1B-55kDa, human wild-type p53 (33), or GST alone. These fusion proteins were tested in an in vitro binding assay for their ability to capture [35S]methionine-labeled E1B-AP5 protein, prepared by in vitro translation (Fig. 4A). In vitro-translated hnRNP-U/SAF-A and human RCC1 were used as negative controls. Consistent with data from immunoblot analysis (Fig. 3C), in vitro-translated E1B-AP5 migrates with a molecular mass of 120 kDa in an SDS-polyacrylamide gel (Fig. 4A). In vitro binding studies established that only E1B-AP5 bound specifically to GSTE1B-55kDa (Fig. 4A), while no binding was evident with in vitro-translated human RCC1 or hnRNP-U/SAF-A (Fig. 4A).
FIG. 4.
In vitro and in vivo association of E1B-AP5 with E1B-55kDa. (A) E1B-AP5 binds to E1B-55kDa in vitro. In vitro-translated [35S]methionine-labeled RCC1 (lane 1), E1B-AP5 (lane 2), and hnRNP-U/SAF-A (lane 3) proteins were incubated with GSTE1B-55kDa, GST-p53, or GST alone, and proteins bound to washed beads were separated by SDS-PAGE and visualized by autoradiography. Molecular mass markers are indicated on the left in kilodaltons. (B) E1B-55kDa binds to E1B-AP5 in vivo. Subconfluent 293 cells grown on 90-mm-diameter culture dishes were transfected with plasmid pSVfluE1B-AP5 expressing epitope-tagged E1B-AP5 by calcium phosphate coprecipitation. At 36 h after transfection, total-cell extracts were prepared. ATP or GTP was added to a final concentration of 100 μM as indicated, and the extracts were subjected to immunoprecipitation with monoclonal antibody (MAb) 12CA5 followed by immunoblotting. E1B-55kDa was detected with anti-55-kDa rat monoclonal antibody 9C10. Lanes 5 and 6, designated “input,” received 1/20 of the amount of total-cell extract added to each immunoprecipitation reaction mixture.
In the second protein-protein interaction assay, we tested the E1B-55kDa/E1B-AP5 interaction in vivo by using a combined immunoprecipitation-immunoblot assay (Fig. 4B). Human 293 cells, which express high levels of E1B-55kDa protein, were transfected with plasmid pSVfluE1B-AP5 expressing epitope-tagged E1B-AP5. Total-cell extracts were prepared and subjected to immunoprecipitation with monoclonal antibody 12CA5 (49). The presence of a conserved nucleoside triphosphate (NTP)-binding motif in the predicted E1B-AP5 amino acid sequence prompted us to evaluate whether ATP or GTP influence the binding of E1B-AP5 to E1B-55kDa. The immunoprecipitate was then analyzed by immunoblotting with anti-55-kDa rat monoclonal antibody 9C10. The E1B-55kDa protein coprecipitated with E1B-AP5 (Fig. 4B, lane 2), while no binding was detectable in untransfected 293 cells (lane 1). The addition of ATP or GTP (lanes 3 and 4) had no effect on E1B-55kDa binding. Together, these experiments demonstrate that E1B-55kDa specifically associates with E1B-AP5 in vitro and in vivo.
RNA-binding properties of E1B-AP5.
The striking sequence similarity to the RNA-binding proteins hnRNP-U/SAF-A and hnRNP-G, as well as the presence of several highly conserved RGG-boxes in the predicted E1B-AP5 amino acid sequence, prompted us to investigate the nucleic acid-binding properties of the E1B-AP5 protein. To test for RNA and ssDNA binding, radiolabeled E1B-AP5 protein generated by in vitro translation was reacted with ribonucleotide homopolymers and ssDNA-agarose beads at salt concentrations ranging from 0.25 to 1 M NaCl (Fig. 5A). The E1B-AP5 protein exhibited the highest salt-resistant binding to poly(G), intermediate binding to poly(C), and very weak binding to poly(U) and poly(A). Binding to ssDNA was also detected (Fig. 5B). These results are identical to the nucleic acid binding properties of the hnRNP-U/SAF-A protein, which binds preferentially to poly(G) through its RGG box domain (37).
FIG. 5.
E1B-AP5 binds to ribonucleotide homopolymers and ssDNA. (A) Quantitation of E1B-AP5 binding to ribonucleotide homopolymers. The percentage of input E1B-AP5 bound to RNA at the indicated NaCl concentrations was determined as described in the text. The mean from three independent experiments is presented. (B) Quantitation of E1B-AP5 binding to ssDNA. The percentage of input E1B-AP5 bound to ssDNA at the indicated NaCl concentrations was determined. The mean from three independent experiments is presented.
Domains in E1B-55kDa required for the interaction with E1B-AP5 in Ad-infected cells.
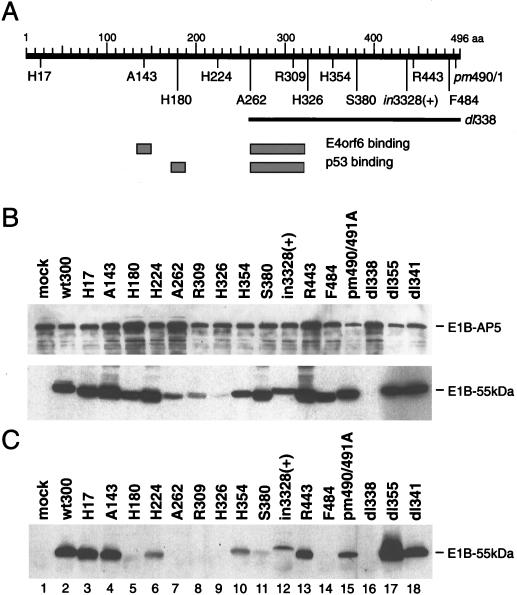
Previous work has demonstrated that the E1B-55kDa polypeptide contains two partially overlapping regions that mediate the interaction with p53 and E4orf6 (36, 62) (Fig. 6A). To determine the domains in the E1B-55kDa protein required to interact with E1B-AP5, we used a series of viruses (Fig. 6A) carrying different mutations in the gene encoding the 55-kDa protein (62, 72, 76). Extracts of Ad-infected MCF-7 cells were prepared, and the expression levels of both proteins were analyzed by immunoblotting (Fig. 6B). The steady-state levels of E1B-AP5 and E1B-55kDa varied for different mutant viruses. Cells infected with mutants A262, R309, and H326 contained very low to nondetectable levels of E1B-55kDa protein 40 h after infection. The same extracts were then subjected to immunoprecipitation and immunoblotting (Fig. 6C), and the amount of protein for each sample was determined as a percentage of that in the wild-type virus by densitometry (data not shown). Several mutations in E1B-55kDa changed its ability to interact with the E1B-AP5 protein. As expected, no E1B-55kDa protein was coprecipitated from dl338-infected extracts. Insertions at aa 224 and 443 and the point mutations at positions 490 and 491 showed decreased binding to E1B-AP5, whereas insertions in the amino-terminal region (aa 17 and 143) had no significant effect on the interaction. The reduction of coprecipitated E1B-55kDa in H354- and in3328(+)-infected cells is most probably due to decreased expression of the Ad protein (Fig. 6B, lanes 10 and 12). In contrast, insertions at aa 180, between aa 262 to 326, and at aa 380 as well as aa 484 strongly interfered with the binding of the E1B-AP5 protein. The 262 to 326 mutations also interfere with the binding of p53 and E4orf6 (36, 62) as well as an apparently distinct function of E1B-55kDa involved in the transcriptional repression of reporter constructs (77). These mutations might disrupt the tertiary structure of the Ad protein, which results in an completely inactivated, nonfunctional polypeptide. Nevertheless, this result suggests that three regions of the 55-kDa polypeptide are required for binding to the E1B-AP5 protein and that p53 and E1B-AP5 share at least one binding domain in the amino-terminal region of E1B-55kDa, because the mutation at position 180 also interferes with the binding of p53 (75).
FIG. 6.
Analysis of the E1B-55kDa/E1B-AP5 protein interaction in mutant and wild-type virus-infected MCF-7 cells. (A) E1B-55kDa mutation sites. The thick black bar at the top represents the 496 residues of the 55-kDa polypeptide. Number 496 denotes the last amino acid. The insertion mutations and the point mutation pm490A/491A (pm490/1) are shown below in their relative positions along the E1B-55kDa polypeptide. The deletion in dl338 is denoted by a thin bar. The p53 and E4orf6 interaction domains (gray boxes) are shown according to their positions along the 55-kDa polypeptide below the E1B protein and were defined by Yew et al. (75) and Rubenwolf et al. (62), respectively. (B) Expression of E1B-55kDa and E1B-AP5 virus-infected cells. Whole-cell extracts used for the coimmunoprecipitation experiment containing 40 μg of protein were subjected to PAGE followed by Western blotting with anti-E1B-55kDa (2A6) hybridoma supernatant and anti-E1B-AP5 antiserum. (C) Coimmunoprecipitation of E1B-55kDa. E1B-55kDa bound to E1B-AP5 protein was coprecipitated with anti-E1B-AP5 rabbit antiserum from the same whole-cell extracts, resolved on SDS–10% polyacrylamide gels, and visualized by Western immunoblot analysis with the 2A6 anti-55-kDa monoclonal antibody.
Also, significantly more E1B-55kDa coprecipitated with E1B-AP5 in the absence of E4orf6 in dl355-infected cells (Fig. 6C, lane 17). This effect was reproduced in three separate experiments and was not observed with the E4orf3 mutant virus dl341. This observation strongly indicates that the E1B-55kDa-associated protein E4orf6 may modulate the E1B-55kDa/E1B-AP5 interaction in productively infected cells.
Stable expression of E1B-AP5 prevents the shutdown of host cell mRNA export.
The results of the experiments presented above, together with our finding that E1B-AP5 is highly related to members of the hnRNP family of proteins, suggested that E1B-AP5 might play some role in nuclear mRNA metabolism. Over the past few years, it has been well established that E1B-55kDa regulates nucleocytoplasmic mRNA transport in complex with the E4orf6 protein (8, 29). Furthermore, it has been proposed that the viral protein complex simultaneously inhibits cellular and activates viral mRNA transport by binding to and relocalizing a nuclear host factor required for mRNA export from the sites of host transcription and processing to the viral replication centers (52, 54). Thus, if E1B-AP5 is one of these host factors that is modulated by E1B-55kDa and if facilitated export of viral mRNAs is due to competition for E1B-AP5 function, overexpression of this protein should interfere, at least in part, with the Ad-induced block to the cytoplasmic accumulation of cellular mRNAs.
To test this prediction, we generated a cell line that expresses an epitope-tagged E1B-AP5 protein. Plasmids pSVfluE1B-AP5 and pBabe-puro were simultaneously introduced into H1299 cells by cotransfection. Drug-resistant clones were isolated, and expression of exogenous fluE1B-AP5 protein was examined by Western blot analysis and indirect immunofluorescence (Fig. 7A). Cell clone H12-AP5/7 expresses high levels of fluE1B-AP5 protein and was used for further analyses. The drug-resistant cell clone H12-AP5/5 does not express fluE1B-AP5 and was used as a control cell line. Indirect immunofluorescence analysis of H12-AP5/7 cells with monoclonal antibody 12CA5 confirmed a predominant nuclear staining for the fluE1B-AP5 protein (Fig. 7A), while no specific signal was obtained with H12-AP5/5 cells and the same antibody (data not shown).
To explore the physiological consequences of fluE1B-AP5 expression on the viral life cycle, we first compared viral DNA replication in H12-AP5/5 cells with that in H12-AP5/7 cells (Fig. 7B). Although the rate of viral DNA synthesis was similar in both cell lines, the onset of replication was delayed by several hours in H12-AP5/7 cells. This delay was reproduced in two separate experiments, but the significance of this effect is unclear. Similar effects have been described for mutant viruses dl355 and dl338, which do not express functional E4orf6 and E1B-55kDa proteins, respectively (29, 54). The ability of wt300 virus to express late viral polypeptides and efficiently shut down host protein synthesis in the absence or presence of fluE1B-AP5 was then evaluated by analyzing the amounts of synthesized viral fiber (IV), cellular actin, and E1B-AP5 proteins (Fig. 7C). Fiber (IV) protein accumulated to abundant levels in both cell lines at 20 to 25 h after infection, while the level of actin continuously decreased from 20 h postinfection in H12-AP5/5 and H12-AP5/7 cells. In contrast, however, the steady-state level of the E1B-AP5 protein remained constant throughout the lytic virus infection in both cell lines (Fig. 7C).
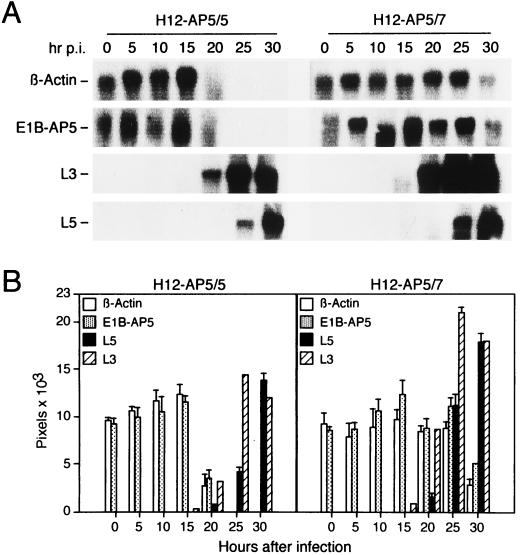
We next tested the effect of overexpressed fluE1B-AP5 on the cytoplasmic accumulation of viral late (L3-hexon and L5-fiber) and cellular β-actin and E1B-AP5 mRNAs in wt300-infected H12-AP5/5 and H12-AP5/7 cells by Northern blot analyses (Fig. 8). The nuclear export of β-actin and E1B-AP5 mRNAs was efficiently blocked in H12-AP5/5 cells at 20 to 25 h after infection, whereas these mRNAs were exported at near normal rates in H12-AP5/7 cells at the same time points. Viral late L3 and L5 mRNAs appeared in the cytoplasm around 20 h and accumulated to high levels at 30 h after infection in both cell lines (Fig. 8A), although L3-hexon and L5-fiber mRNAs accumulated in the cytoplasm at a somewhat higher rate in H12-AP5/7 cells than in the control cell line (Fig. 8B). Thus, high levels of fluE1B-AP5 protein interfere with the virus-induced block of cytoplasmic β-actin and E1B-AP5 mRNA accumulation and simultaneously enhance the export of L3 and L5 mRNA to the cytoplasm. These results, together with those presented above, indicated that overexpressed E1B-AP5 did not inhibit the virus-dependent translational block of β-actin mRNAs in the cytoplasm, since the levels of actin protein were dramatically reduced in infected H12-AP5/7 cells (Fig. 7C). Surprisingly, the levels of the E1B-AP5 protein did not change significantly during the late phase of infection, although the nuclear export of E1B-AP5 mRNAs was efficiently blocked in H12-AP5/5 cells. This observation suggests that the E1B-AP5 protein has a fairly long half-life or perhaps, by analogy to p53, is metabolically stabilized in Ad-infected cells.
FIG. 8.
Effects of stable expressed fluE1B-AP5 on cytoplasmic mRNA accumulation in wt300-infected cells. (A) Northern blot analysis of β-actin, E1B-AP5, L3, and L5 mRNA species in infected H12-AP5/5 and H12-AP5/7 cells. Cytoplasmic RNA was prepared at the indicated time points after infection (hr p.i.). Equal quantities of these RNAs were subjected to electrophoresis, transferred to nitrocellulose membranes, and hybridized with [α-32P]dCTP-labeled β-actin, E1B-AP5, L3, and L5 DNA probes. The bands corresponding to the viral and cellular mRNAs are indicated at the left. (B) The levels of β-actin, E1B-AP5, L3, and L5 mRNAs were quantitated as described and plotted as a function of time. The mean and standard deviation is presented for three independent experiments.
E1B-AP5 interferes with Ad5 E1-mediated transformation of primary rat cells.
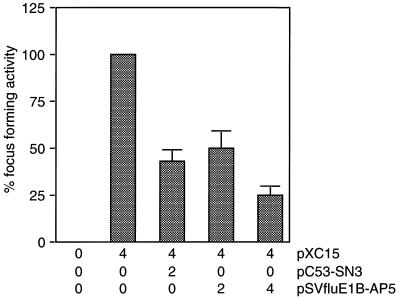
E1B-55kDa transforms primary cells in cooperation with E1A by binding to and blocking p53-mediated transcriptional activation (75). Because p53 and E1B-AP5 seem to interact with the same region in the 55-kDa polypeptide, we tested the effect of E1B-AP5 expression on primary rat cell transformation mediated by Ad5 E1A and E1B proteins. Primary baby rat kidney (BRK) cells were transfected with plasmids expressing E1A and E1B oncogenes in combination with human wild-type p53 or epitope-tagged E1B-AP5 (Fig. 9). Consistent with the previous observations that wild-type p53 inhibits oncogene-mediated focus formation (16, 19), coexpression of p53 with Ad5 E1 proteins reduced the number of transformed cells by almost 60%. Remarkably, inclusion of pSVfluE1B-AP5 in the transformation mixture resulted in a 70 to 80% reduction of dense foci in a concentration-dependent manner. Thus, E1B-AP5, like p53, efficiently interferes with the ability of Ad E1A and E1B proteins to elicit neoplastically transformed foci upon transfection of primary cells in tissue culture. This effect is not due to toxic effects of E1B-AP5 expression, since high levels of E1B-AP5 protein did not reduce the plating efficiency of transiently transfected H1299 cells under puromycin selection (data not shown). According to our data, it seems possible that E1B-AP5 modulates the E1-mediated transformation process by competing with p53 for the interaction with E1B-55kDa and releasing p53 from the Ad protein.
FIG. 9.
E1B-AP5 inhibits Ad5 E1A/E1B-mediated focus formation. Primary BRK cells were transfected with the indicated amounts of plasmids (micrograms of DNA per 3 × 106 cells). Focus-forming activity is presented as a percentage of E1A plus E1B activity. The average number of dense foci for pXC15 was 128 in four independent experiments.
DISCUSSION
In this report, we have described the isolation of a cDNA encoding a protein, E1B-AP5, that physically interacts with the Ad E1B-55kDa oncoprotein. E1B-AP5 is related to members of the hnRNP family. These abundant pre-mRNA binding proteins have been implicated in multiple steps of mRNA processing and seem to play an important role in the localization and transport of RNAs (reviewed in reference 15). The most significant similarity of the predicted protein sequence of E1B-AP5 is to the hnRNP-U/SAF-A protein (Fig. 2), including an acidic amino-terminal region, a putative guanine nucleotide-binding site in the central part of E1B-AP5, and closely spaced RGG repeats toward its carboxyl terminus. However, E1B-AP5 and hnRNP-U/SAF-A differ significantly in their most amino- and carboxy-terminal regions, which might indicate that the two proteins have different functions in RNA metabolism. In addition, the primary structure of E1B-AP5 contains several more limited homologies to other hnRNPs, including A1, G, K, and L. The RGG box domain in the E1B-AP5 protein presumably contributes to its RNA-binding activities (reviewed in reference 9). In hnRNP-U/SAF-A, the RGG box domain is absolutely required for RNA binding (37). Other studies demonstrate that this motif is required for the nuclear targeting of the RNA binding protein Nlp3, an important mediator of mRNA export in S. cerevisiae (39). In light of these sequence similarities, we propose that E1B-AP5 is a new member of the hnRNP family.
Given its role in late viral mRNA transport, we find it intriguing that E1B-55kDa is found associated with a host factor that might be involved in RNA metabolism. Our results suggest that E1B-AP5 provides functions required for nucleocytoplasmic mRNA turnover including mRNA processing and/or mRNA transport. E1B-AP5 is localized predominantly in the nucleus (Fig. 3), it specifically associates with the E1B-55kDa protein (Fig. 4 and 6), and the same regions that mediate the binding of E1B-AP5 to E1B-55kDa (Fig. 6) have been found to be absolutely required for host cell shutoff (76). More significantly, high levels of the E1B-AP5 protein stimulate the export of late viral transcripts and simultaneously prevent the shutdown of host cell mRNA export (Fig. 8). These data are compatible with the hypothesis that the E1B-55kDa protein facilitates the cytoplasmic accumulation of viral transcripts by binding to a nuclear host factor necessary for mRNA export (52). Earlier work demonstrated that E1B-55kDa exerts its late effects after an RNA molecule is spliced and polyadenylated (41, 54). In the absence of the E1B-55kDa polypeptide, late viral transcripts exit the nuclear matrix fraction inefficiently and fail to accumulate in a nuclear downstream compartment (41). Thus, E1B-AP5 may facilitate the release or movement of mature transcripts from the nuclear matrix to the nuclear pores, which seems to be the rate-limiting step for late viral mRNA transport (41). Furthermore, the E4orf6 protein contains a Rev-like nuclear export signal, and it shuttles between the nucleus and cytoplasm (12). Therefore, if E1B-AP5 bridges between viral mRNAs and the E1B-55kDa/E4orf6 complex, it could serve as an adapter to connect newly synthesized mRNAs to an active nucleocytoplasmic shuttling machinery.
Considerable evidence indicates that cellular DNA replication, transcription, RNA processing, and RNA transport occur in association with intranuclear structures (reviewed in reference 70). Interestingly, it has been reported that the E1B-AP5-related protein hnRNP-U/SAF-A may function in the organization of chromosomal DNA and, along with other hnRNP proteins, may be involved in the formation and maintenance of nuclear structures (17, 22, 61). Considering a similar function of E1B-AP5 during infection with Ad, one might speculate that this protein is a component of nuclear compartments that mediate mRNA transport to the cytoplasm and/or favor viral DNA replication. With the onset of viral DNA replication, the rapidly growing number of transcriptionally active viral chromosomes colonize transport-gated nuclear microenvironments and promote the export of late viral mRNAs by a mechanism dependent on the E1B/E1B-AP5 protein interaction. Such a mechanism would be compatible with the idea that the transport selectivity observed in Ad-infected cells results from the displacement of gated cellular transcription and/or transport units by viral chromosomes in the late phase of infection (74). Accordingly, high levels of E1B-AP5 would efficiently interfere with the proposed competition for these specialized compartments, as we observed (Fig. 8). Moreover, this hypothesis would also account for the finding that mRNA export of activated cellular genes in the late phase of Ad infection is coupled to transcription and is dependent on the expression of a functional E1B-55kDa protein (32, 74). Since E1B-55kDa is present in several intranuclear localizations (52, 68), we suggest that the 55-kDa E1B protein occupies remaining or newly established gated environments by binding to E1B-AP5. Apparently, this interaction could modulate the export of these cellular mRNA molecules.
E1B-AP5 contains two additional regions of similarity to cellular proteins that might link its function to posttranscriptional regulation, signal transduction pathways, and cellular proto-oncogenes. The carboxy-terminal region of E1B-AP5, which exhibits significant sequence similarity to hnRNP-G, contains clustered proline residues that resemble potential binding sites for Src homology 3 (SH3) domains (10). This domain, which includes approximately 60 amino acids, is found in a very large group of proteins, including cytoskeletal elements and signaling proteins (3). We note that the putative RNA-binding protein G3BP, which contains, similarly to E1B-AP5, the SH3 ligand motif PXXP (78) (E1B-AP5 residues 698 to 701 and 708 to 711), has been found to bind to the SH3 domain of the p21ras GTPase-activating protein (53). In addition, hnRNP-K has been shown to bind to the SH3 domains of the cellular proto-oncogenes c-src and p95vav (31, 71), indicating that both proteins might play a role in the regulation of mRNA biogenesis. Moreover, the central region of E1B-AP5 shows substantial homology to the guanine nucleotide-binding motif present in the nuclear G protein Ran, an important regulator of nuclear import and export processes (reviewed in reference 24). This strongly implicates E1B-AP5 as a guanine nucleotide-binding protein, but we do not yet have data to prove that this is the case. Apparently, such an activity could potentially regulate the function of E1B-AP5. Should both activities of E1B-AP5 be substantiated, it is possible that E1B-AP5 function is linked to proteins that mediate protein-protein associations and reversible GTP binding and, along with Src homology 2 (SH2) domains, regulate cytoplasmic and/or nuclear signaling as well as cell cycle progression (59).
Over the past few years, it has been well established that the multifunctional E1B-55kDa protein provides additional important functions in lytically infected cells that are probably unrelated to the inhibition of mRNA transport. It has been shown that E1B-55kDa directly inhibits host protein synthesis (1) and blocks the E1A-induced accumulation of the cellular tumor suppressor protein p53 in combination with E4orf6 (27, 56). Furthermore, p53/E1B-55kDa complexes have been implicated in viral replication (60) and inhibition of p53-mediated G1 growth arrest or apoptosis (57, 67, 75). In addition, it was shown recently that the Ad protein relieves growth constraints of the cell cycle by mechanisms independent of p53 (23). The latter observation strongly suggests that E1B-55kDa abates growth restrictions on viral replication by interacting with other cellular proteins that, like p53, regulate cell cycle progression. If cell cycle-dependent viral replication and E1B-AP5 function are linked, it is tempting to speculate that E1B-55kDa modulates cell cycle regulation, at least in part, by binding to E1B-AP5. In fact, such an activity could also explain the reduction of transformed cells in our transformation assays (Fig. 9). On the other hand, because p53 and E1B-AP5 share at least one binding region on E1B-55kDa (Fig. 6), it is also possible that E1B-AP5 decreases the number of transformants by competing with p53 for the interaction with E1B-55kDa.
An intriguing alternative for the function of E1B-AP5 is that it participates in both cell cycle regulation and mRNA processing, including mRNA transport. Based on the multiple functions of E1B-55kDa in lytic infection and in Ad-induced transformation, it will therefore be important to investigate whether E1B-AP5 links the two regulatory processes. In this context, it will be interesting to analyze the contribution of E1B-AP5 for the use of E1B mutant viruses in tumor therapy, as recently suggested by Bischoff et al. (6).
ACKNOWLEDGMENTS
We are very grateful to those named in the text for donating gifts of reagents, to A. Richter for providing plasmid pSAF-A, and to J. Köstler for performing DNA sequencing.
S.G. was supported by a graduate program from the Deutsche Forschungsgemeinschaft Therapieforschung Onkologie. Work in the laboratory of T.S. was supported by the National Cancer Institute (grant CA41086). T.S. is an American Cancer Society Professor and an Investigator of the Howard Hughes Medical Institute. T.D. was supported by a grant from the Infektionsforschung, AIDS-Stipendienprogramm DKFZ, Heidelberg, Germany.
REFERENCES
- 1.Babich A, Feldman L T, Nevins J R, Darnell J E, Weinberger C. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translation discrimination. Mol Cell Biol. 1983;3:1212–1221. doi: 10.1128/mcb.3.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss L E, Ginsberg H S, Darnell J J. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Sagi D, Rotin D, Batzer A, Mandiyan V, Schlessinger J. SH3 domains direct cellular localization of signaling molecules. Cell. 1993;74:83–91. doi: 10.1016/0092-8674(93)90296-3. [DOI] [PubMed] [Google Scholar]
- 4.Beltz G A, Flint S J. Inhibition of HeLa cell protein synthesis during adenovirus infection. J Mol Biol. 1979;131:353–373. doi: 10.1016/0022-2836(79)90081-0. [DOI] [PubMed] [Google Scholar]
- 5.Binger M H, Flint S J. Accumulation of early and intermediate mRNA species during subgroup C adenovirus productive infections. Virology. 1984;136:387–403. doi: 10.1016/0042-6822(84)90175-2. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 7.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 8.Bridge E, Ketner G. Interaction of adenoviral E4 and E1b products in late gene expression. Virology. 1990;174:345–353. doi: 10.1016/0042-6822(90)90088-9. [DOI] [PubMed] [Google Scholar]
- 9.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 10.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction pathways. Cell. 1995;809:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 11.Dix I, Leppard K N. Regulated splicing of adenovirus type 5 E4 transcripts and regulated cytoplasmic accumulation of E4 mRNA. J Virol. 1993;67:3226–3231. doi: 10.1128/jvi.67.6.3226-3231.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobner T, Wolf I, Emrich T, Lipp M. Differentiation-specific expression of a novel G-protein-coupled receptor from Burkitt’s lymphoma. Eur J Immunol. 1992;22:2795–2799. doi: 10.1002/eji.1830221107. [DOI] [PubMed] [Google Scholar]
- 14.Dobner T, Wolf I, Mai B, Lipp M. A novel divergently transcribed human histone H2A/H2B gene pair. DNA Sequence. 1991;1:409–413. doi: 10.3109/10425179109020799. [DOI] [PubMed] [Google Scholar]
- 15.Dreyfuss G, Hentze M, Lamond A I. From transcript to protein. Cell. 1996;85:963–972. doi: 10.1016/s0092-8674(00)81298-2. [DOI] [PubMed] [Google Scholar]
- 16.Eliyahu D, Michalovitz D, Eliyahu S, Pinhasi-Kimhi O, Oren M. Wild-type p53 can inhibit oncogene-mediated focus formation. Proc Natl Acad Sci USA. 1989;86:8763–8767. doi: 10.1073/pnas.86.22.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fackelmayer F O, Dahm K, Renz A, Ramsperger U, Richter A. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur J Biochem. 1994;221:749–757. doi: 10.1111/j.1432-1033.1994.tb18788.x. [DOI] [PubMed] [Google Scholar]
- 18.Fackelmayer F O, Richter A. Purification of two isoforms of hnRNP-U and characterization of their nucleic acid binding activity. Biochemistry. 1994;33:10416–10422. doi: 10.1021/bi00200a024. [DOI] [PubMed] [Google Scholar]
- 19.Finlay C A, Hinds P W, Levine A J. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 20.Gaynor R B, Hillman D, Berk A J. Adenovirus early region 1A protein activates transcription of a novel gene introduced into mammalian cells by infection or transfection. Proc Natl Acad Sci USA. 1984;81:1193–1197. doi: 10.1073/pnas.81.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giard R J, Aaronson S A, Todaro G J, Arnstein P, Kersey J H, Dosik H, Parks W P. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 22.Göhring F, Fackelmayer F O. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry. 1997;36:8276–8283. doi: 10.1021/bi970480f. [DOI] [PubMed] [Google Scholar]
- 23.Goodrum F A, Ornelles D A. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J Virol. 1997;71:548–561. doi: 10.1128/jvi.71.1.548-561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Görlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 25.Graham F L, Smiley J, Russel W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 26.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 27.Grand R J, Grant M L, Gallimore P H. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 28.Grand R J, Mustoe T, Roberts S, Gallimore P H. The quaternary structure of the adenovirus 12 early region 1B 54K protein. Virology. 1995;207:255–259. doi: 10.1006/viro.1995.1074. [DOI] [PubMed] [Google Scholar]
- 29.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hearing P, Shenk T. Sequence-independent autoregulation of the adenovirus type E1A transcription unit. Mol Cell Biol. 1985;5:3214–3221. doi: 10.1128/mcb.5.11.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobert O, Jallal B, Schlessinger J, Ullrich A. Novel signaling pathway suggested by SH3 domain-mediated p95vav/heterogeneous ribonucleoprotein K interaction. J Biol Chem. 1994;269:20225–20228. [PubMed] [Google Scholar]
- 32.Huang W, Flint S J. The tripartite leader sequence of subgroup C adenovirus major late mRNAs can increase the efficiency of mRNA export. J Virol. 1998;72:225–235. doi: 10.1128/jvi.72.1.225-235.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiménez-Garcia L F, Spector D L. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- 35.Kaelin W G, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 36.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 37.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983;47:1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M S, Henry M, Silver P. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 40.Leppard K N. Selective effects on adenovirus late gene expression of deleting the E1b 55K protein. J Gen Virol. 1993;74:575–582. doi: 10.1099/0022-1317-74-4-575. [DOI] [PubMed] [Google Scholar]
- 41.Leppard K N, Shenk T. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 1989;8:2329–2336. doi: 10.1002/j.1460-2075.1989.tb08360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang S, Hitomi M, Tartakoff A M. Adenoviral E1B-55kDa protein inhibits yeast mRNA export and perturbs nuclear structure. Proc Natl Acad Sci USA. 1995;92:7372–7375. doi: 10.1073/pnas.92.16.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Logan J, Pilder S, Shenk T. Functional analysis of adenovirus type 5 early region 1B. Cancer Cells. 1984;2:527–532. [Google Scholar]
- 44.Marton M J, Baim S B, Ornelles D A, Shenk T. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J Virol. 1990;64:2345–2359. doi: 10.1128/jvi.64.5.2345-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitsudomi T, Steinberg S M, Nau M M, Carbone D, D’Amico D, Bodner H K, Oie H K, Linnoila R I, Mulshine J L, Minna J D, Gazdar A F. p53 gene mutations in non-small-lung cell cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171–180. [PubMed] [Google Scholar]
- 46.Moore M, Schaack J, Baim S B, Morimoto R I, Shenk T. Induced heat shock mRNAs escape the nucleocytoplasmic transport block in adenovirus-infected HeLa cells. Mol Cell Biol. 1987;7:4505–4512. doi: 10.1128/mcb.7.12.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niman H L, Houghten R A, Walker L E, Reisfeld R A, Wilson I A, Hogle J M, Lerner R A. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nordqvist K, Öhman K, Akusjärvi G. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol Cell Biol. 1994;14:437–445. doi: 10.1128/mcb.14.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Öhman K, Nordqvist K, Akusjärvi G. Two adenovirus proteins with redundant activities in virus growth facilitates tripartite leader mRNA accumulation. Virology. 1993;194:50–58. doi: 10.1006/viro.1993.1234. [DOI] [PubMed] [Google Scholar]
- 52.Ornelles D A, Shenk T. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J Virol. 1991;65:424–429. doi: 10.1128/jvi.65.1.424-429.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker F, Maurier F, Delumeau I, Duchesne M, Faucher D, Debussche L, Dugue A, Schweighoffer F, Tocque B. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol. 1996;16:2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puvion-Dutilleul F, Puvion E. Sites of transcription of adenovirus type 5 genomes in relation to early viral DNA replication in infected HeLa cells: a high resolution in situ hybridization and autoradiographical study. Biol Cell. 1991;71:135–147. doi: 10.1016/0248-4900(91)90060-z. [DOI] [PubMed] [Google Scholar]
- 56.Querido E, Marcellus R, Lai A, Rachel C, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan M P. Enhanced proliferation, growth factor induction and immortalization by adenovirus E1A 12S in the absence of E1B. Oncogene. 1994;9:2639–2647. [PubMed] [Google Scholar]
- 58.Reich N C, Sarnow P, Duprey E, Levine A J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 59.Ren M, Coutavas E, D’Eustachio P, Rush M G. Effects of mutant Ran/TC4 proteins on cell cycle progression. Mol Cell Biol. 1994;14:4216–4224. doi: 10.1128/mcb.14.6.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridgway P J, Hall A R, Myers C J, Braithwaite A W. p53/E1b58kDa complex regulates adenovirus replication. Virology. 1997;237:404–413. doi: 10.1006/viro.1997.8782. [DOI] [PubMed] [Google Scholar]
- 61.Romig H, Fackelmayer F O, Renz A, Ramsperger U, Richter A. Characterization of SAF-A, a novel nuclear DNA-binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 1992;11:3431–3440. doi: 10.1002/j.1460-2075.1992.tb05422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubenwolf S, Schütt H, Nevels M, Wolf H, Dobner T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol. 1997;71:1115–1123. doi: 10.1128/jvi.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarnow P, Hearing P, Anderson C W, Reich N, Levine A J. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J Mol Biol. 1982;162:565–583. doi: 10.1016/0022-2836(82)90389-8. [DOI] [PubMed] [Google Scholar]
- 65.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 66.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Vol. 2. New York, N.Y: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 67.Shepherd S E, Howe J A, Mymryk J S, Bayley S T. Induction of the cell cycle in baby rat kidney cells by adenovirus type 5 E1A in the absence of E1B and a possible influence of p53. J Virol. 1993;67:2944–2949. doi: 10.1128/jvi.67.5.2944-2949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smiley J K, Young M A, Flint S J. Intranuclear location of the adenovirus type 5 E1B 55-kilodalton protein. J Virol. 1990;64:4558–4564. doi: 10.1128/jvi.64.9.4558-4564.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soulard M, Della Valle V, Siomi M C, Pinol Roma S, Codogno P, Bauvy C, Bellini M, Lacroix J C, Monod G, Dreyfuss G, et al. hnRNP G: sequence and characterization of a glycosylated RNA-binding protein. Nucleic Acids Res. 1993;21:4210–4217. doi: 10.1093/nar/21.18.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strouboulis J, Wolffe A P. Functional compartmentalization of the nucleus. J Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- 71.Taylor S J, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 72.Teodoro J G, Halliday T, Whalen S G, Takayesu D, Graham F L, Branton P E. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J Virol. 1994;68:776–786. doi: 10.1128/jvi.68.2.776-786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinberg D H, Ketner G. A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc Natl Acad Sci USA. 1983;80:5383–5386. doi: 10.1073/pnas.80.17.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang U C, Huang W, Flint S J. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J Virol. 1996;70:4071–4080. doi: 10.1128/jvi.70.6.4071-4080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 76.Yew P R, Kao C C, Berk A J. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology. 1990;179:795–805. doi: 10.1016/0042-6822(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 77.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 78.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Feigenbaum D, Schneider R J. A late adenovirus factor induces eIF-4E dephosphorylation and inhibition of cell protein synthesis. J Virol. 1994;68:7040–7050. doi: 10.1128/jvi.68.11.7040-7050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Schneider R T. Adenovirus inhibition of cellular protein synthesis and the specific translation of late viral mRNAs. Semin Virol. 1993;4:229–236. [Google Scholar]