Abstract
NS1, the 83-kDa major nonstructural protein of minute virus of mice (MVM), is a multifunctional nuclear phosphoprotein which is required in a variety of steps during progeny virus production, early as well as late during infection. NS1 is the initiator protein for viral DNA replication. It binds specifically to target DNA motifs; has site-specific single-strand nickase, intrinsic ATPase, and helicase activities; trans regulates viral and cellular promoters; and exerts cytotoxic stress on the host cell. To investigate whether these multiple activities of NS1 depend on posttranslational modifications, in particular phosphorylation, we expressed His-tagged NS1 in HeLa cells by using recombinant vaccinia viruses, dephosphorylated it at serine and threonine residues with calf intestine alkaline phosphatase, and compared the biochemical activities of the purified un(der)phosphorylated (NS1O) and the native (NS1P) polypeptides. Biochemical analyses of replicative functions of NS1O revealed a severe reduction of intrinsic helicase activity and, to a minor extent, of ATPase and nickase activities, whereas its affinity for the target DNA sequence [ACCA]2–3 was enhanced compared to that of NS1P. In the presence of endogenous protein kinases found in replication extracts, NS1O showed all functions necessary for resolution and replication of the 3′ dimer bridge, indicating reactivation of NS1O by rephosphorylation. Partial reactivation of the helicase activity was found as well when NS1O was incubated with protein kinase C.
Replication of the single-stranded, linear DNA genomes of parvoviruses involves the formation of a series of monomeric and concatemeric duplex DNA intermediates produced by a unidirectional, single-strand copy mechanism (19, 66). This mode of replication resembles the rolling-circle DNA replication mechanism described for bacteriophages, single-strand plasmids, and geminiviruses (3, 35). The first step of parvovirus DNA replication, the conversion of the single-stranded genome to a monomeric duplex, is executed by cellular components only and is primed directly from the 3′ hydroxyl group of the nucleotide that is base paired through the folding back of a terminal palindromic structure (4, 18). For later stages in the infectious cycle, replication initiates at site-specific, single-strand nicks introduced by a virally encoded initiator protein into origin sequences which are located at either end of the genome (17, 23, 64). The minimal origin sequence at the 3′ end of parvovirus minute virus of mice (MVM) DNA has been determined (16). The DNA motif responsible for the specific interaction with the initiator protein (25) as well as its target nick site and the covalent attachment have been mapped (16).
The viral initiator protein involved in MVM DNA replication is a pleiotropic 83-kDa nuclear phosphoprotein called NS1 (for nonstructural protein 1) (18, 24). A number of studies carried out in vivo (22, 39, 48) or in vitro (4, 17, 21, 56) with wild-type NS1 or derivatives obtained by site-directed mutagenesis have clearly demonstrated the key role of this protein during distinct steps of parvovirus DNA replication. Indeed, NS1 proved to be the only viral protein necessary for viral DNA replication in all cell types. In particular, NS1 is required for the hairpin transfer of the right-end telomere of monomeric replicative forms (4) and for the resolution of concatemeric replication intermediates (17, 21) as determined with recombinant NS1 proteins produced by vaccinia virus (53) or baculoviruses (1). One-step partial purification of NS1 has been achieved by column chromatography based on immunoaffinity (70), conventional methods (13), or, alternatively, Ni2+-NTA–agarose columns, taking advantage of a [His]6 tag engineered to the N terminus of the viral product (56), and has allowed a variety of biochemical activities, related to its replicative functions, to be assigned to the NS1 protein. In addition, some of these activities could be mapped to distinct domains of the multifunctional protein by the use of site-directed NS1 mutants (see Fig. 1A). Thus, NS1 forms oligomers (54, 60); exhibits intrinsic ATP-binding, ATPase, and helicase activities (13, 70); binds site specifically to an [ACCA]2–3 element that is present at multiple positions in the viral genome (11, 25); mediates the site-specific single-strand nicking of replication origins located in the left-hand (12, 56) and right-hand (27) terminal sequences; and becomes covalently attached to the 5′ end of replicated viral DNA (4, 16, 20, 23). Besides its multiple functions during viral DNA replication, NS1 possesses a C-terminal acidic transcription-activating domain (37) that is able to trans regulate the parvovirus promoters as well as various heterologous cellular and viral promoters (29, 38, 61, 67, 68). Furthermore, NS1 can induce cytotoxic and/or cytostatic stress in sensitive host cells (6, 7, 52), for which the N- and C-terminal parts of the polypeptide appear to be important (38).
FIG. 1.
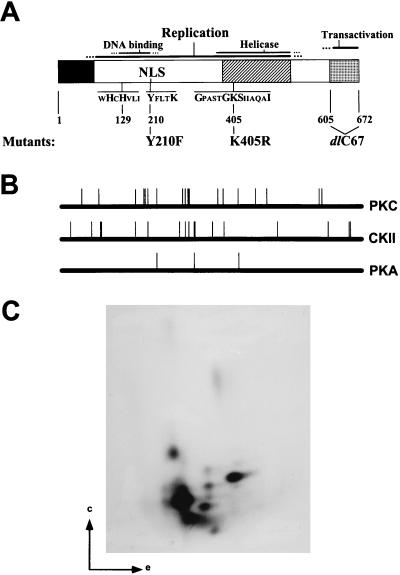
NS1 phosphorylation. (A) Schematic representation of NS1 and its functional domains as determined by mutational analyses (for references, see the text). The black box corresponds to the common N terminus of NS1 and NS2. The hatched box indicates the region of homology between NS1 and SV40 LT. The stippled box denotes the transactivation domain which has been deleted in the mutant NS1dlC67. NLS, nuclear localization signal (194KKx18KKK216). The sequences corresponding to the metal coordination site (uHuHuuu) and linking tyrosine (YxxxK), which are both essential for the nickase function of NS1, as well as the nucleotide binding site (Gx4GKSx5I) are indicated. The substitution mutants Y210F and K405R as well as the C-terminal deletion mutant dlC67 (lacking the last 67 amino acids), which are used in this study, are outlined at the bottom of the scheme. (B) NS1 consensus sites of phosphorylation by PKC, CKII, and PKA as determined by computer alignments with HUSAR. (C) Tryptic phosphopeptide map of in vivo-labeled NS1 isolated from A9 cells incubated with [32P]orthophosphate after MVMp infection. NS1 was immunoprecipitated with antiserum αNSN, digested with trypsin, and analyzed by two-dimensional electrophoresis (e) and chromatography (c).
The multitude of NS1 activities in the course of a viral infection lead one to speculate that the functions of this polypeptide might be regulated at least in part through posttranslational modifications. Indeed, NS1 was found to be phosphorylated in vivo (1, 14, 24, 50). Precedents for regulatory pathways involving posttranslational modifications, in particular phosphorylation, can be found in the control of the cell cycle (33) and neoplastic transformations (44). The dependency of parvoviruses on host cell entry into S phase (18), as well as their preferential replication in and toxicity to neoplastic cells (15, 52), raises the possibility that NS1 is also regulated through phosphorylation. It is worth noting that there are striking functional, structural, and even sequence homologies between NS1 and the simian virus 40 large T antigen (SV40 LT) (2), which both initiate viral DNA replication, trans regulate homologous and heterologous promoters, and disturb host cells. Interestingly, it has been reported that the replicative functions of SV40 LT are modulated through phosphorylation (31, 59) to ensure that viral DNA replication occurs during S phase of the cell cycle. Analysis of distinct replicative functions of LT revealed that this coordination is achieved specifically through the origin-unwinding function of the protein (8, 46, 49). Little is known about the pattern or timing of phosphorylation of the parvovirus NS1 protein. Yet, there is evidence indicating that MVM NS1 phosphorylation occurs early in infection and persists throughout the infection cycle, while the phosphorylation pattern changes in the course of a viral infection (14, 18, 24). Phosphorylated amino acids in porcine parvovirus NS1 were found to be for the most part serine and, to a lesser extent, threonine residues (14, 50). No tyrosine phosphorylation of NS1 has been reported so far (1, 14, 50). The implications of phosphorylation for NS1 functions, however, have remained elusive to date.
To investigate a possible impact of phosphorylation on NS1 activities in vitro, NS1 was purified from recombinant vaccinia virus-infected HeLa cells (53, 56) and tested for a variety of biochemical activities, either in its native form or after dephosphorylation with calf intestine alkaline phosphatase. The native protein and its dephosphorylated derivative, incubated with endogenous protein kinases present in fully competent replication extracts, were both able to carry out all functions necessary for resolution and replication of the 3′ dimer bridge plasmid containing the MVM left-end replication origin (17). In contrast, in the absence of any added protein kinases, the ATPase and, especially, the intrinsic helicase activities of the dephosphorylated polypeptide were markedly reduced compared to those of native NS1, while specific binding to the 3′ origin was enhanced. Rephosphorylation, achieved with a commercially available protein kinase C (PKC) preparation, led to a partial reactivation of the helicase activity of dephosphorylated NS1.
MATERIALS AND METHODS
Viruses and cells.
Recombinant vaccinia viruses were propagated in monolayer cultures of BSC-40 or HeLa cells and purified over a sucrose cushion as previously described (41), except for the release of virus from infected cells by three cycles of freezing and thawing instead of sonification. MVMp was propagated in A9 cells. HeLa, BSC-40, and A9 cells were grown as monolayers in Dulbecco’s modified Eagle medium (DMEM) containing 5% fetal calf serum. HeLa-S3 cells were grown in suspension in Joklik’s medium containing 5% fetal calf serum, using spinner bottles.
Production and purification of native and dephosphorylated NS1 polypeptides.
NS1 was produced from recombinant vaccinia viruses in suspension cultures of HeLa-S3 cells. Cells were collected by low-speed centrifugation and resuspended in serum-free DMEM containing 15 PFU per cell each of vTF7-3 (32) and the appropriate recombinant vaccinia viruses with the NS1 gene under the control of the bacteriophage T7 promoter (51, 53). The cell-virus suspension (5 ml; 107 cells) was transferred into a 150-mm2 tissue culture dish, incubated for 2 h at 37°C, and supplemented (to 20 ml) with DMEM containing 10% fetal calf serum and 190 mM NaCl to favor cap-independent translation from the encephalomyocarditis virus leader sequence present in the pTM-1-based constructs (51, 53, 54, 56). Cultures (2 × 108 cells each) were harvested 18 h postinfection, nuclear extracts were prepared (21), and His-NS1 was purified by using Ni2+-NTA–agarose (Qiagen) columns (56). For dephosphorylation, nuclear extracts containing His-NS1 were adjusted to the appropriate buffer conditions (100 mM Tris-HCl, pH 8.5; 1 mM ZnCl); supplemented with the protease inhibitors phenylmethylsulfonyl fluoride (PMSF; 174 μg/ml), leupeptin (1 μg/ml), aprotinin (1 μg/ml), and pepstatin (1 μg/ml); and incubated for 15 min at 37°C in the presence of 150 μg of calf intestine alkaline phosphatase (Boehringer Mannheim). Dephosphorylated NS1 was immediately purified from other proteins by affinity chromatography, using Ni2+-NTA–agarose columns. NS1 preparations were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were detected by Coomassie blue staining. All mutant NS1 proteins used as controls were tested for their described biochemical properties.
Production and purification of bacterially expressed GST-NS1.
pBacGST-EK-NS1 was constructed by inserting the BamHI fragment of the cloning intermediate pT-GST-NS1 into pGEX-5X-2 (Pharmacia). pT-GST-NS1 was obtained by PCR amplification of a fragment encompassing the glutathione S-transferase (GST) part of pGEX-5X-2 with the primer pair 5′-CAGTATCCATGGCCCCTATAC-3′ and 5′-TCACGCCATGGCCGCTCGA-3′, digested with the restriction endonuclease NcoI, and inserted into pTHis-NS1 (56). For expression of the GST-NS1 fusion protein, Sure bacteria were transformed with pBacGST-EK-NS1 and grown overnight at 37°C. Cultures were diluted 1:4 and further amplified for 3 h at 32°C before production of GST-NS1 was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2.5 h at 32°C. Bacteria were then collected by centrifugation, suspended in sonication buffer (20 mM HEPES-KOH, pH 8.0; 300 mM KCl; 0.05% Nonidet P-40 [NP-40]; 0.1 mM dithiothreitol [DTT]), and digested with 1 mg of lysozyme per ml for 10 min at 37°C. The suspension was supplemented with 25 mM EDTA together with the protease inhibitors PMSF, leupeptin, and aprotinin, and the bacteria were disrupted by sonication. The soluble proteins were cleared from insoluble material by centrifugation at 100,000 × g for 1 h, and soluble NS1 was purified by addition of 300 μl of glutathione-Sepharose (Pharmacia) per liter of culture. After being allowed to attach for 2 h at 4°C and then rinsed extensively with sonication buffer, bound NS1 was eluted with 10 mM glutathione in sonication buffer at pH 7.5. Since GST is able to self-associate, GST-NS1 is present as a dimer, as determined by fast-performance liquid chromatography on Superose 6 columns (57). To avoid this artifact, the 27.5-kDa GST polypeptide was cleaved off by treatment with 0.25 mg of enterokinase (Boehringer Mannheim) per mg of NS1 for 15 min at 30°C, and GST-free NS1 was further purified on a 5′-AMP column (Pharmacia).
In vivo 32P labeling and tryptic phosphopeptide analysis.
After infection with MVMp (10 PFU/cell) in serum-free DMEM for 30 min, A9 cultures were incubated for 18 h in medium containing 5% fetal calf serum. Cultures (107 cells each) were then labeled with [32P]orthophosphate (ICN) (10−10 Ci/cell) in complete medium without phosphate (Gibco/BRL) for an additional 4-h period and harvested directly in 1 ml of RIPA buffer (20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 0.1% SDS; 1% sodium deoxycholate; 1% Triton X-100) containing the protease inhibitors PMSF, leupeptin, and pepstatin as well as phosphatase inhibitors (20 mM NaF, 5 mM β-glycophosphate, 5 mM p-nitrophenyl phosphate, 5 mM sodium molybdate, 1 mM sodium orthovanadate, and 5 mM sodium phosphate). Immunoprecipitations were carried out with 5 μl of αNSN, an antiserum raised against the N-terminal 91 amino acids of MVM NS proteins (26), for 2 h at room temperature. Immune complexes were precipitated with protein A-Sepharose (Pharmacia), washed three times with RIPA buffer, and further purified by 10% SDS-PAGE. 32P-labeled proteins were revealed by autoradiography after being blotted on polyvinylidene difluoride membranes (Millipore) (42), and the band corresponding to NS1 was excised. Digestion of membrane-bound NS1 was performed with 50 U of trypsin (Promega) for 18 h at 37°C. Tryptic peptides contained in the supernatant were recovered by lyophilization and analyzed on thin-layer cellulose plates (Merck) in two dimensions, by electrophoresis (using pH 1.9 buffer) and by chromatography (in phosphochromatography buffer) (5).
Site-specific binding of NS1 to the MVM 3′ origin of replication.
Site-specific binding assays using NS1 and the MVM 3′ origin were performed as described previously (25). Briefly, plasmid pL1-2TC, which contains the minimal active 3′ replication origin (16), was digested with restriction enzymes Sau3A and NarI, and the DNA fragments were labeled at the 3′ end by filling in with Sequenase (Amersham), [α-32P]dGTP, and unlabeled dATP, dCTP, and dTTP. Binding assays were carried out in 100 μl of a solution containing 20 mM Tris-HCl (pH 8.0), 10% glycerol, 1% NP-40, 5 mM DTT, and 100 mM NaCl, supplemented with labeled pL1-2TC DNA fragments, 500 ng of oligo-d[I,C], 0.5 mM adenosine 5′-O-(3-thiotriphosphate) (γ-S-ATP), and 50 ng of purified NS1. After interactions were allowed to take place for 30 min on ice, 2 μl of antiserum αNSN was added and the incubation was continued for another hour. Immune complexes were precipitated with protein A-Sepharose, deproteinized, and analyzed by nondenaturing 7% PAGE in the presence of 0.1% SDS.
Site-specific nicking of the 3′ origin of replication.
NS1-mediated site-specific nicking and the consequent covalent attachment of NS1 to the 5′ end of the nicked product were analyzed according to the method of Nüesch et al. (56). The substrate containing the 3′ origin of replication was obtained as a 95-bp EcoRI fragment of pL1-2TC. This fragment was end labeled by a fill-in reaction using Sequenase, [32P]dATP, and unlabeled dTTP. Approximately 1 ng of substrate was incubated with 20 ng of NS1 in the presence of 3 mM ATP for 1 h at 37°C. The reaction was stopped by adding 0.1% SDS and 2.5 mM EDTA, and immunoprecipitations were performed with αNSN. The immune complexes were freed from proteins by proteinase K digestion and phenol-chloroform extraction and then analyzed by electrophoresis on 8% sequencing gels.
Helicase assay.
Helicase assays were carried out as described previously (56). M13-VAR, used as the substrate, was prepared by annealing the reverse primer (Amersham) to M13 single-stranded DNA (Amersham) followed by extension for 5 min at room temperature in the presence of Sequenase and deoxynucleoside triphosphates (dNTPs), including [α-32P]dATP. 32P-labeled fragments of various lengths were obtained by addition of dideoxy-GTP and further incubation for 20 min. Purified NS1 (2 to 200 ng) was incubated with 20 ng of substrate for 30 min to 1 h in the presence of 3 mM ATP and, when indicated, an ATP regeneration system that consisted of 25 mM phosphocreatine and 25 ng of phosphocreatine kinase. The reactions were stopped by addition of SDS and EDTA, and the products were analyzed by nondenaturing 7% PAGE in the presence of 0.1% SDS.
ATPase assay.
The NS1 used for ATPase assays was further purified by centrifugation through a 1.5-ml 15 to 40% glycerol gradient at 50,000 rpm, using a TLS55 rotor (Beckman), for 18 h at 4°C. Twenty fractions (75 μl each) were collected from the top, with peak amounts of NS1 being present around fraction 9. The ATPase activities of individual fractions which had been matched for their respective NS1 contents, as determined by Coomassie blue staining after SDS-PAGE, were measured. ATPase assays were performed according to the method of Wilson et al. (70), using 2 to 20 ng of NS1 protein in a solution containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 5 mM DTT, 0.01% NP-40, and 30 μM ATP, supplemented with 0.5 μCi of [γ-32P]ATP (Amersham) (3,000 Ci/mmol) and 0.1 μg of single-stranded DNA, for 20 min at room temperature. The reaction was terminated by addition of 100 μl of 7.5% (wt/vol) acid-washed charcoal in 50 mM HCl–5 mM H3PO4, and free phosphate was separated from unreacted charcoal-bound ATP by centrifugation. A 50-μl sample of the 32Pi-containing supernatant was analyzed by scintillation counting.
In vitro resolution and replication reactions with the left-end bridge fragment.
Resolution of the 3′ dimer bridge of MVM DNA was investigated in vitro as previously described, using pLEB711 as a substrate (17). Approximately 100 ng of purified NS1 expressed from HeLa cells was supplied in either native or dephosphorylated form to HeLa replication extracts. The reaction mixture was incubated for 2 h at 37°C in the presence of dNTPs (including [32P]dATP), ATP, and an ATP regeneration system. The NS1-attached labeled products were recovered by immunoprecipitation with αNSN and digested with the restriction endonuclease ScaI, and the resulting fragments were further separated into NS1-bound and NS1-free fractions by centrifugation. These products were analyzed independently by 1% agarose gel electrophoresis.
In vitro phosphorylation of NS1.
Purified dephosphorylated NS1 (100 ng) was treated with one of the following protein kinases for 30 min at 37°C in the presence of 10 μCi [γ-32P]ATP in a solution containing 20 mM HEPES-KOH (pH 7.5), 5 mM MgCl2, 5 mM KCl, and 0.1 mM DTT: PKC (Sigma; 0.1 U), casein kinase II (CKII; Boehringer Mannheim; 5 mU), the catalytic subunit of cAMP/GMP-dependent kinase (PKA; Sigma; 20 μg), or cdc2 complex (UBI; 10 ng). In some experiments, instead of defined protein kinases, 500 ng of total protein from HeLa cell extracts was used to phosphorylate NS1. The reaction was terminated by addition of 0.1% SDS and 2.5 mM EDTA and incubation for 30 min at 70°C. 32P-labeled NS1 was purified by immunoprecipitation with αNSN and analyzed by SDS-PAGE.
RESULTS
Production of native and dephosphorylated NS1 polypeptides.
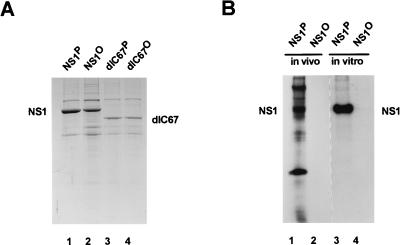
To evaluate the complexity of NS1 phosphorylation, potential phosphorylation sites were predicted by computer analyses and in vivo phosphorylation experiments were carried out with MVM-infected cells (Fig. 1B and C). Computer searches limited to three main protein kinases (PKC, CKII, and PKA) revealed the existence of over 35 consensus phosphorylation sequences within the NS1 polypeptide (Fig. 1B). This complexity was confirmed by tryptic peptide mapping of in vivo-labeled NS1 (Fig. 1C). Depending on the cell line tested and the time of analysis during infection, 8 to 12 distinct phosphorylated peptides could be detected, as illustrated in Fig. 1C for NS1 produced in A9 cells 18 h postinfection. At least four of these phosphopeptides were consistently present in NS1 preparations from all MVM-infected cell lines tested and therefore might contain candidate target phosphorylation sites necessary for NS1 functions (14). To investigate the relevance of NS1 phosphorylation for MVM propagation, the present study was undertaken to test whether phosphorylation indeed has an impact on NS1 replication functions, at least in vitro. To this end, NS1 was produced by recombinant vaccinia viruses in HeLa cells (53), dephosphorylated (or not) on serine and threonine residues by using calf intestine alkaline phosphatase, and purified on Ni2+-NTA–agarose through an N-terminal [His]6 tag (56). The efficiency of the dephosphorylation procedure was determined by addition of vaccinia virus-produced NS1 that was 32P labeled either in vivo, using orthophosphate, or in vitro, by treatment with PKC in the presence of [γ-32P]ATP. As shown in Fig. 2, the dephosphorylation procedure efficiently removed the 32P label from NS1 (Fig. 2B) under conditions which caused relatively little NS1 degradation (Fig. 2A), as determined after SDS-PAGE by autoradiography and Coomassie blue staining, respectively. Besides using full-length NS1, we performed many of the subsequent biochemical analyses in parallel with the mutant polypeptide NS1dlC67 (54), which lacks the C-terminal 67 amino acids corresponding to the transcription-activating domain (38). This NS1 derivative, which mimics the underphosphorylated 65-kDa NS1* species detected in MVM-infected cells (25), was included to determine whether a phosphorylation site(s) essential for viral DNA replication is located within this C-terminal domain of NS1.
FIG. 2.
Production, dephosphorylation, and purification of NS1. (A) Full-length (wild-type) and C-terminally truncated (dlC67) NS1 were produced by recombinant vaccinia viruses in HeLa cells. Nuclear extracts containing NS1 were treated (dephosphorylated samples, with superscripted “O”) or not treated (native samples, with superscripted “P”) with calf intestine alkaline phosphatase, and the NS1 polypeptides were purified by affinity chromatography by means of their N-terminal [His]6 tags. The purified samples were analyzed by SDS-PAGE and Coomassie blue staining. Lanes: 1 and 2, wild-type NS1; 3 and 4, NS1dlC67; 1 and 3, native NS1; 2 and 4, dephosphorylated NS1. (B) Wild-type NS1 produced by recombinant vaccinia viruses was metabolically 32P labeled in vivo, using 106 HeLa cells, and extracted by repeated freezing and thawing. Alternatively, dephosphorylated NS1 was 32P labeled in vitro, using PKC. The 32P-labeled proteins were then mixed with the same amount of NS1-containing nuclear extract as used in panel A. Phosphatase-treated or untreated samples were then purified on Ni2+-NTA–agarose columns and analyzed by SDS-PAGE and autoradiography. Lanes: 1 and 2, NS1 labeled in vivo in HeLa cells; 3 and 4, NS1 labeled in vitro, using recombinant PKC; 1 and 3, mock treatment; 2 and 4, treatment with calf intestine alkaline phosphatase. The low degree of purification of in vivo 32P-labeled NS1 in lane 1 was probably due to the formation of aggregates caused by the freezing and thawing procedure used to release the 32P-labeled proteins from the cells.
Biochemical characterization of dephosphorylated versus native NS1.
Purification of functionally active NS1, development of a variety of distinct biochemical assays, and generation of specific NS1 derivatives by site-directed mutagenesis allowed a number of activities, such as site-specific interaction with a consensus DNA motif (25), single-strand site-specific nicking and covalent attachment to the left-end origin (13, 56), intrinsic helicase function, and ATPase activity (70), to be assigned to the NS1 protein. These developments prompted us to determine whether phosphorylation had any impact on these properties of NS1 by comparing native and dephosphorylated NS1 polypeptides purified from recombinant vaccinia virus-infected HeLa cells.
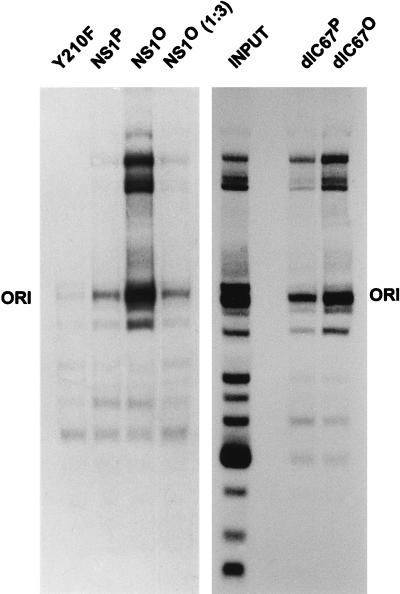
To measure the site-specific binding of NS1 to the [ACCA]2–3 element located within the 3′ origin of replication, plasmid pL1-2TC was used as a substrate after digestion with Sau3A and NarI and end labeling of the restriction fragments by a fill-in reaction with Sequenase (25). The 32P-labeled plasmid fragments were incubated with purified NS1 in the presence of nonspecific competitor DNA [oligo-d(I,C)] and nonhydrolyzable γ-S-ATP. Specific NS1-DNA complexes were immunoprecipitated with αNSN, an antiserum raised against the N-terminal 91 amino acids of MVM NS proteins (26); digested with proteinase K; and analyzed by nondenaturing PAGE in the presence of SDS. Mutant NS1 Y210, which contains a substitution of the linkage tyrosine for covalent attachment to replicated viral DNA (56) and which has been shown to be severely impaired for site-specific binding to the origin, served as a negative control in these assays. As illustrated in Fig. 3, full-length NS1 and C-terminally deleted NS1dlC67 were both able to immunoprecipitate the plasmid fragment containing the 3′ origin when supplied in either the native or the dephosphorylated form, indicating that NS1 phosphorylation is not required for the specific interaction with the [ACCA]2–3 element. On the contrary, both dephosphorylated NS1 proteins (the wild type and the C-terminal deletion mutant) consistently exhibited a more than threefold-higher affinity for the origin-containing fragment than did their native counterparts. The immunoprecipitation of large plasmid fragments can be assigned to the known nonspecific DNA-binding activity of NS1, with the un(der)phosphorylated (NS1O) and native (NS1P) NS1 forms showing the same ratio of specific to nonspecific binding [Fig. 3; compare NS1P with NS1O (1:3)].
FIG. 3.
Site-specific binding of native versus dephosphorylated wild-type NS1 and NS1dlC67. Purified NS1 was incubated with 32P-labeled, Sau3A- and NarI-digested pL1-2TC plasmids containing the MVM active 3′ origin of replication, in the presence of γ-S-ATP. NS1-DNA complexes were immunoprecipitated with αNSN, and their DNA constituents were revealed by autoradiography after nondenaturing 7% PAGE in the presence of 0.1% SDS. The fragment containing the MVM origin is denoted ORI. The NS1 mutant Y210F served as a control.
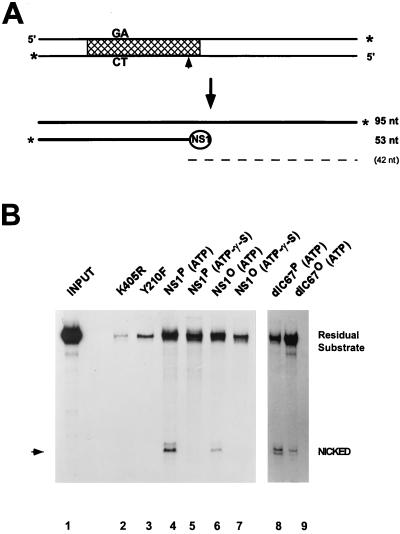
The NS1-mediated nicking of the 3′ origin and the consequent covalent attachment of the polypeptide to the 5′ end of the nicked strand are essential to initiate resolution and replication of MVM DNA replication intermediates (17, 21, 22). As depicted in Fig. 4A, this site- and strand-specific endonuclease activity can be measured in vitro by using purified NS1 and a cloned 3′ origin (a 3′-end-labeled 95-bp EcoRI fragment of pL1-2TC) under low-salt conditions without any additional cellular components (56). Under these conditions, the nicking reaction is rather inefficient, given that site-specific binding to the NS1 recognition motif [ACCA]2–3 is not essential. In contrast, nicking is dependent on the supply of hydrolyzable ATP and of NS1 that contains intact ATP-binding, metal coordination, and linking tyrosine consensus domains (56). These requirements were confirmed by the present study showing that significant nicking was achieved by native full-length and dlC67 NS1 in the presence of ATP, but not by the NTP-binding site (substitution of arginine for lysine at position 405 [K405R]) and active-site tyrosine (Y210F) mutants or in the presence of γ-S-ATP instead of hydrolyzable ATP (Fig. 4B). When dephosphorylated NS1 was tested in this assay, nicking was also found to occur in an ATP-dependent way, albeit to a 5 to 10 times lower extent than with native NS1. Considering the efficiency of dephosphorylation (Fig. 2B), these results suggest that phosphorylation is not a prerequisite for NS1 nicking activity. Yet the significantly lower capacity of dephosphorylated NS1 for 3′ origin nicking, versus that of native NS1, evident in the present assay points to this NS1 function as a potential target for phosphorylation-mediated up-modulation. Current investigations, in which 3′ origin nicking reactions are being performed in the presence of a purified cellular cofactor, the parvovirus initiation factor (PIF), support these findings (10).
FIG. 4.
Site-specific nicking of native versus dephosphorylated wild-type NS1 and NS1dlC67. (A) Diagram of substrate and denatured products of the nicking reaction. Labeled 3′ ends are marked by asterisks, the cross-hatched area delineates the minimal active left-end origin (16), and the nick site is indicated by an arrowhead. The circled NS1 depicts the covalently linked NS1 at the 5′ end of the nicked strand. The dashed line indicates the predicted unlabeled 42-nucleotide (nt) single-strand product DNA. (B) For each assay, 3′-end-labeled substrate was incubated with purified NS1 in the presence of ATP or γ-S-ATP as indicated. DNA which became covalently attached to NS1 was then immunoprecipitated with αNSN, deproteinized, and analyzed by 8% sequencing gel electrophoresis. The migration of the nicked product is indicated by an arrowhead. NS1 mutants K405R and Y210F served as negative controls. Lanes: 1, input substrate (1:20 dilution); 2 and 3, negative controls, using mutant NS1; 4 to 7, wild-type NS1; 8 and 9, NS1dlC67; 2 to 4, 6, 8, and 9, reactions in the presence of 2 mM ATP; 5 and 7, reactions in the presence of ATP-γ-S. Residual substrate consists of the unnicked positive strand coimmunoprecipitated with the NS1-attached nicked strand, as well as contaminating substrate DNA which was not removed by the washing procedure.
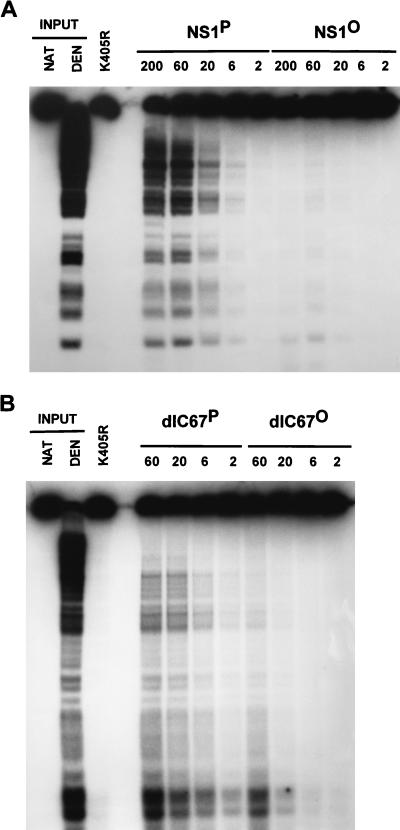
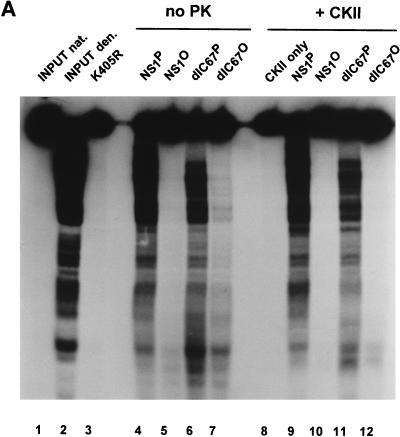
NS1 is thought to facilitate strand displacement synthesis during MVM DNA replication through its intrinsic helicase activity. In the presence of hydrolyzable ATP, NS1 proved able to unwind DNA fragments of a size up to 600 nucleotides from circular M13 DNA templates (56). Figure 5 presents a titration experiment in which the unwinding activities of HeLa cell-derived native and dephosphorylated NS1 were compared, using the M13-VAR substrate that consists of 32P-labeled fragments of various lengths annealed to circular M13 DNA (56). The NS1 mutant K405R, which is helicase deficient due to an amino acid substitution at the conserved lysine 405 position in the nucleoside triphosphate (NTP)-binding domain (53, 56), served as a negative control to ascertain the NS1 dependence of the unwinding reactions measured. Wild-type and C-terminally deleted NS1dlC67 both exhibited helicase activity, the extent of which was reduced more than 30-fold as a result of dephosphorylation. Therefore, the NS1 helicase function appears to be under a tight control mediated by phosphorylation.
FIG. 5.
Helicase activity of purified native and dephosphorylated wild-type NS1 (A) and NS1dlC67 (B). Helicase assays were carried out by incubating 20 ng of M13-VAR (32P-labeled fragments of various lengths annealed to circular M13 DNA) with the indicated amounts (in nanograms) of NS1 in the presence of ATP. The reaction products were analyzed by nondenaturing 7% PAGE in the presence of 0.1% SDS. NS1 mutant K405R served as a negative control. Native (NAT) and heat-denatured (DEN) input DNA are shown on the left as references.
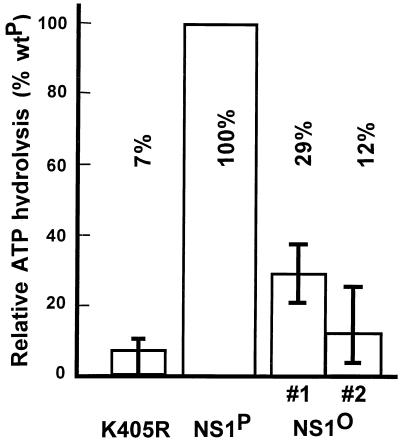
Furthermore, we investigated a possible effect of NS1 phosphorylation on the ATPase activity that is involved in a variety of biochemical activities described for NS1. Purified NS1 was supplied with [γ-32P]ATP in the presence of single-stranded DNA as a cofactor (13), and the release of labeled phosphate was measured by scintillation counting. To minimize contamination with endogenous ATPases present in HeLa cell extracts, and due to fluctuations inherent in the assay, NS1 preparations were subjected to a further purification step involving centrifugation through a 15 to 40% glycerol gradient. Fractions 7 to 9 (the last fraction containing the peak amount of NS1) were analyzed individually, matched for their NS1 content as determined by Coomassie blue staining after SDS-PAGE. The average values from multiple experiments, performed with titrated amounts of NS1, were calculated, with their standard deviations, for two independent dephosphorylated NS1 preparations (no. 1 and 2) and expressed as relative ATPase activities versus those of native NS1 samples. As presented in Fig. 6, dephosphorylation was associated with a marked impairment (four- to eightfold) of the ATPase activity of native NS1 protein, although a significant residual activity could still be detected. To confirm that the measured ATPase activity was derived from NS1, the mutant K405R was used as a negative control. Similar mutations within the NTP-binding domains of ADV NS1 (13) and other ATPases (65) have been shown to completely abolish the ATPase activity of the respective polypeptides. It is worth noting that these measurements were made at NS1P and NS1O concentrations within the linear part of the dose response, i.e., under conditions in which ATP was not limiting, allowing the comparison of actual ATPase activities rather than the affinities of the respective polypeptides for ATP.
FIG. 6.
Effect of dephosphorylation on ATPase activity of NS1. Released 32Pi was determined by scintillation counting after incubation of [γ-32P]ATP with native or dephosphorylated NS1. Average values from multiple assays using different sucrose gradient fractions are shown with their standard deviation bars for two independent NS1O preparations (#1 and #2). The NS1 mutant K405R served as a negative control. Data are expressed relative to the ATPase activity of native wild-type NS1 (wtP).
NS1 replication activity.
All of the above-presented analyses of native and dephosphorylated NS1 polypeptides have taken advantage of assays developed to study the distinct replicative functions of this protein in the absence of additional accessory proteins (36, 56, 63). To investigate whether dephosphorylated NS1, which is deficient for nickase and helicase functions, could regain these activities when properly rephosphorylated, we compared the abilities of native and dephosphorylated NS1 polypeptides to resolve and replicate a plasmid containing the 3′ head-to-head dimer bridge in crude cellular replication extracts that could be assumed to contain the appropriate protein kinases (see Fig. 8).
FIG. 8.
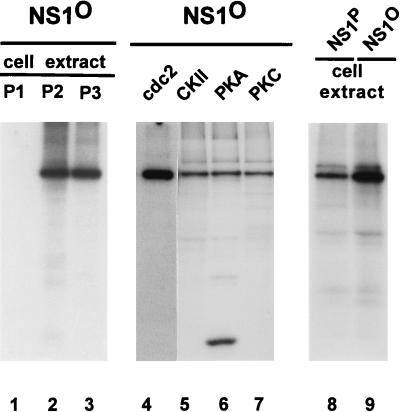
In vitro phosphorylation of NS1. Phosphatase-treated or native wild-type NS1 was incubated with fractionated cell extract or commercially available protein kinases (PK) in the presence of [γ-32P]ATP, immunoprecipitated with αNSN, and revealed by autoradiography after SDS-PAGE. HeLa S100 cell extract was fractionated by chromatography on a phosphocellulose P11 column (Whatman) into P1 (150 mM NaCl flowthrough), P2 (150 to 400 mM NaCl elution), and P3 (400 to 1,000 mM NaCl elution) fractions. cdc2, cdc2 complex. Lanes: 1 to 7 and 9, dephosphorylated NS1 used as a substrate; 8, native NS1 used as a substrate; 1, protein kinases from fraction P1; 2, protein kinases from fraction P2; 3, protein kinases from fraction P3; 4 to 7, commercially available protein kinases (cdc2 complex, CKII, PKA, and PKC, respectively); 8 and 9, protein kinases from HeLa cell replication extracts. The heavily labeled 32-kDa protein in lane 6 probably corresponds to the autophosphorylated catalytic subunit of PKA present in large amounts in the reaction.
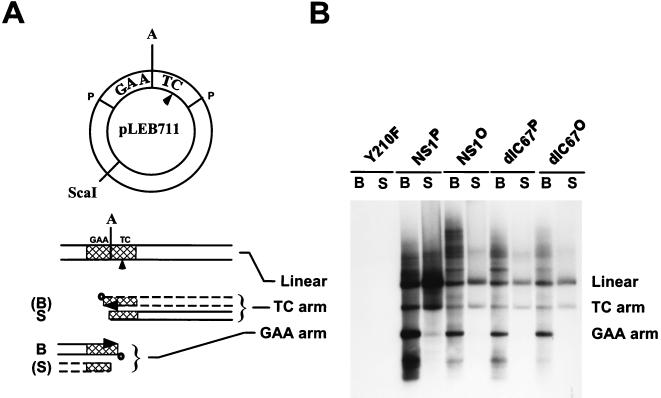
NS1 activity was tested in vitro by using HeLa cell extracts supplemented with pLEB711 as a substrate, in the presence of dNTPs (including [32P]dATP), ATP, and an ATP-regenerating system. The 32P-labeled products were analyzed by agarose gel electrophoresis after immunoprecipitation and ScaI restriction digestion (17). As depicted in Fig. 7, in the presence of replication-competent NS1, the dimer bridge containing pLEB711 was nicked asymmetrically, leading predominantly to the extension of the GAA arm by NS1-mediated strand displacement synthesis and to the production of a covalently closed turnaround form of the TC arm. This asymmetric resolution pattern of the palindrome structure (with the arms named after the unpaired sequence making up a “bubble” in the 3′-terminal hairpin of the viral DNA) (17) has been recently interpreted in detail (19). As shown in Fig. 7B, native NS1P gave rise to the expected pattern of left-end bridge resolution products. No replication was detected with the mutant NS1:Y210F, which contains a mutation in the active-site tyrosine. Dephosphorylated NS1 also proved competent for dimer bridge resolution and replication when incubated with cell extracts, although the efficiency was somewhat lower than with the native NS1 (Fig. 7B). This result clearly demonstrates that the phosphatase treatment did not cause an irreversible inactivation of NS1. The significant replication capacity of the dephosphorylated NS1 in cell extracts contrasts with the almost complete inactivity of this protein in the helicase assay (Fig. 5). This difference might be ascribed either to NS1 helicase function being dispensable for replication or to the presence of protein kinases in HeLa replication extracts which are able to rephosphorylate the phosphatase-treated NS1. The failure so far to isolate NS1 helicase mutants that are still active in replication assays (36, 56, 57) rather argues against the former possibility. Furthermore, the replication extracts used for in vitro replication assays were subsequently found to be able to rephosphorylate phosphatase-treated NS1 (see below and Fig. 8). Similar results were obtained when A9 cell extract was used in replication experiments instead of HeLa cells (data not shown). Like wild-type protein, the dephosphorylated NS1dlC67 mutant (lacking the C-terminal 67 amino acids) exhibited resolution and replication activity similar to those of its native counterpart in this assay (Fig. 7B). The lower yield of pLEB711 resolution and replication achieved by NS1dlC67P compared with wild-type NS1P in the present experiment can be ascribed to the fact that the mutant protein was supplied in lesser amounts than was the wild type. These results confirm that the C-terminal domain of NS1 is not essential for replication activity, as previously indicated (38), and suggest that the presumed up-modulation of NS1 resolution and replication activity by phosphorylation can take place to a significant extent in the absence of this domain.
FIG. 7.
In vitro resolution of the 3′ dimer bridge by native and dephosphorylated wild-type NS1 and NS1dlC67 in fully competent HeLa cell replication extracts. (A) Diagram of the plasmid substrate pLEB711 (17, 22) and the resolution products. The MVM dimer bridge, derived from a dimer replication intermediate as a PstI fragment (P), is marked GAA and TC across the axis of symmetry A, according to the sequence corresponding to the unpaired bubble in the left-end palindrome. The dimer bridge is cross-hatched in the reaction products. The nick site, as determined by Cotmore and Tattersall (16), is marked with an arrowhead, and the ScaI restriction site used to linearize the replication products is indicated. The resolution of the dimer bridge is asymmetric and gives rise to major (solid lines) and minor (dashed lines) products (for details, see references 17, 19, and 22). B, NS1 (boldfaced “donut”)-bound extended forms derived from nicking and strand displacement synthesis across the axis of symmetry; S, NS1-free covalently closed turnaround forms after resolution. (B) The 3′ dimer bridge containing plasmid pLEB711 was subjected to resolution and replication in fully competent HeLa cell replication extracts in the presence of dNTPs (including [α-32P]dATP), ATP, and an ATP regeneration system, using native or dephosphorylated wild-type NS1 and NS1dlC67. The reactions were stopped by adding 0.2% SDS and heating at 70°C for 30 min to disrupt noncovalent binding of NS1 to the replicated DNA. The newly synthesized 32P-labeled DNA was then immunoprecipitated with αNSN and digested with ScaI, allowing the resolved NS1-attached (B) products and NS1-free (S) products to be isolated and analyzed separately by 1% agarose gel electrophoresis and autoradiography. The linearized unresolved plasmid, labeled either by nonspecific nick translation (S) or by rolling-circle replication and incomplete resolution (B) of the circular plasmid, as well as the complete resolution products (TC and GAA arms) are indicated. As reported previously, in vitro resolution of pLEB711 by NS1 is asymmetric, producing predominantly the extended form of the GAA arm (NS1 bound) and the turnaround form of the TC arm (NS1 free).
Reactivation of NS1 helicase activity by in vitro phosphorylation.
To test the assumption that phosphatase-treated NS1 can be reactivated by in vitro phosphorylation, we analyzed for an NS1 function that could be measured in the absence of cellular components (i.e., without any endogenous protein kinases). The helicase activity of NS1 was studied in this regard, given its above-mentioned dramatic suppression as a result of NS1 dephosphorylation. First, we determined whether phosphatase-treated NS1 could indeed be rephosphorylated in vitro by incubation with [γ-32P]ATP in the presence of either crude cell extracts, as a supplier of protein kinases, or commercially available PKA, PKC, CKII, or cdc2 complex. As shown in Fig. 8, dephosphorylated NS1 was indeed a target for rephosphorylation by these various kinases under in vitro conditions. As expected from previous studies showing that a variety of phosphoproteins can be phosphorylated in vitro at sites which are apparently not targets in vivo, native NS1 could also be further phosphorylated in vitro, but to a lesser extent than phosphatase-treated NS1 (Fig. 8, lane 8).
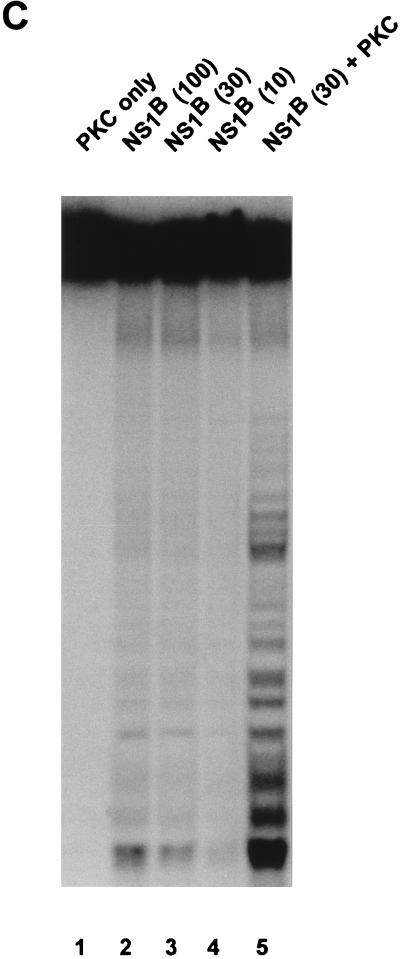
It was then investigated whether the helicase activity lost after phosphatase treatment could be recovered, at least in part, by rephosphorylation of the viral protein in vitro. To this end, phosphatase-treated NS1 was incubated with protein kinases that either were commercially available in a sufficiently purified state (i.e., did not present significant helicase and/or DNase activity [CKII and PKC] or were present in crude extracts which could be used at dilutions exhibiting no measurable endogenous helicase activity. CKII failed to reactivate the helicase function of phosphatase-treated wild-type NS1 or NS1dlC67 and did not affect the helicase activity of the native protein (Fig. 9A). In contrast, PKC proved able to restore substantially the helicase activity of dephosphorylated wild-type NS1 as well as NS1dlC67 (Fig. 9B). Since the extents of NS1 phosphorylation achieved by the two protein kinases were similar (Fig. 8), this result suggests that the up-regulation of NS1 depends on its phosphorylation at a specific site(s). Furthermore, the helicase activity of phosphatase-treated NS1 could also be rescued by incubation of the viral product with phosphocellulose fraction P2 (150 to 400 mM NaCl elution) derived from HeLa replication extracts (Fig. 9B), but not with fraction P3 (400 to 1,000 mM NaCl elution) (data not shown). In this regard, it is worth noting that PKC and CKII segregate into fractions P2 and P3, respectively (57, 71).
FIG. 9.
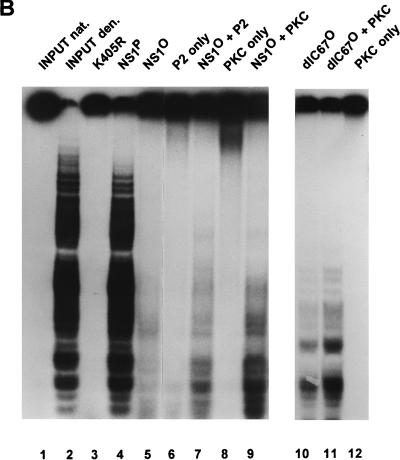
In vitro reactivation of NS1 helicase function with protein kinases. Helicase assays were carried out as described in the legend to Fig. 5, using 10 ng of native or 30 ng of phosphatase-treated wild-type NS1 or NS1dlC67 from recombinant vaccinia virus-infected HeLa cells or the indicated amounts (in nanograms) of bacterially produced NS1. The NS1 mutant K405R served as a negative control. Native (nat.) and denatured (den.) input DNAs are shown as references. (A) Vaccinia virus-produced NS1 was incubated with 5 mU of CKII (+ CKII) or without protein kinase (no PK). Lanes: 1, native input substrate; 2, heat-denatured input substrate; 3, mutant NS1 used as negative control; 4, 5, 9, and 10, wild-type NS1; 6, 7, 11, and 12, NS1dlC67; 4, 6, 9, and 11, native NS1; 5, 7, 10, and 12, dephosphorylated NS1; 4 to 7, no protein kinase added; 8 to 12, CKII added to the reactions. (B) Dephosphorylated vaccinia virus-produced NS1 was incubated with the P2 fraction derived from HeLa cells (as in Fig. 8), or PKC (0.1 U). Lanes: 1, native input substrate; 2, heat-denatured input substrate; 3, mutant NS1 used as a negative control; 4, native wild-type NS1, used as positive control; 4, 5, 7, and 9, wild-type NS1; 10 and 11, NS1dlC67; 5, 7, and 9 to 11, dephosphorylated NS1; 6 and 7, fraction P2 added to the reaction; 8, 9, 11, and 12, PKC added to the reaction. (C) Bacterially produced NS1 (NS1B) was tested either in the absence (lanes 2 to 4) or in the presence (lane 5) of 0.1 U of PKC. The amount of NS1 used in each assay is indicated in parentheses (in nanograms). Lane 1, helicase assay performed with 0.1 U of PKC in the absence of NS1.
To ascertain that the above-mentioned modulations of helicase activity were due to the phosphorylation state of NS1 and not to side effects caused by the treatments used, NS1 was also produced in bacteria. NS1 was expressed as a GST-NS1 fusion protein, purified on glutathione-Sepharose, and further analyzed after cleavage of the 27.5-kDa GST N terminus with enterokinase. Despite possible protein phosphorylation in bacteria (43), only a little helicase activity could be detected with up to 100 ng of bacterially produced NS1 per reaction in the absence of added protein kinase (Fig. 9). However, when the reaction was supplied with PKC, a striking increase in the helicase activity of the bacterially produced NS1 preparation was observed (more than 10% of the level determined for an equivalent amount of native, HeLa cell-derived NS1). Therefore, these results substantiated the dependence of NS1 helicase function on a phosphorylation pattern that cannot be achieved effectively in a bacterial context but is provided to a significant extent by incubation with PKC.
DISCUSSION
The NS1 protein of MVM, a parvovirus, exhibits a number of functional analogies to the LT of SV40, a papovavirus. NS1 is able to form oligomers and to bind and hydrolyze NTPs, and it shows DNA helicase activity, site-specific interaction with target DNA motifs, and transcriptional regulation, in addition to serving as the essential initiator protein for viral DNA replication, all properties shared with SV40 LT (for reviews, see references 31 and 59). Furthermore, NS1 shows striking amino acid homology to the SV40 LT within the putative helicase domain (2). Due to these similarities between the two viral proteins, it is intriguing to use the well-studied SV40 LT as a paradigm for the control of MVM NS1 functions. The replicative activity of SV40 LT is regulated positively and negatively by phosphorylation. The cyclin-dependent kinase cdc2 activates LT by phosphorylation of threonine 124 (46, 49), leading to the formation of double hexamers competent for bidirectional unwinding of the SV40 origin in vitro. Interestingly, mutation of T124 to alanine, while blocking this activation of the unwinding function, has no effect on the intrinsic DNA helicase activity of the LT molecule (49). Recently, it has been shown that phosphorylation of T124 activates the LT minimal domain required for specific binding to the central pentanucleotide repeats in the SV40 core origin, even in the absence of the LT amino acid sequences involved in hexamer formation (47). This suggests that although T124 is located outside the minimal DNA-binding domain, phosphorylation of this residue may induce conformational changes within the DNA-binding domain itself. Negative regulation of LT occurs predominantly through phosphorylation of serine 120 and/or 123, which is catalyzed in vitro by a novel form of CKI in a reaction which appears to be highly dependent on the tertiary structure of LT (8, 9). The exact step of the interaction between LT and the SV40 origin at which this control takes place has yet to be determined. Presumably these opposing regulatory phosphorylations, in addition to others, operate in the infected cell to modulate the different activities of LT in an optimal temporal scheme.
The present in vitro study comparing the replicative properties of NS1O and NS1P, both produced in HeLa cells, showed that distinct biochemical activities are modulated by the phosphorylation state of the polypeptide. By analogy with SV40 LT, this modulation suggests that NS1 functions may be regulated by phosphorylation in infected cells, although this remains to be shown. Un(der)phosphorylated NS1 is still able to bind to its cognate origin sequence, and it does so with substantially higher affinity than the native, phosphorylated polypeptide. This gain of function clearly demonstrates that the dephosphorylation procedure used here does not affect the integrity of the polypeptide. In contrast, dephosphorylation was found to be associated with a reduction of nickase, helicase, and ATPase activities. These opposite effects of phosphorylation on distinct NS1 activities (enhanced DNA binding but reduced enzymatic activities of NS1O versus those of NS1P) are consistent with a putative regulation of NS1 by protein kinases. One effect of such phosphorylation might be to shift the bulk of accumulated NS1 from one functional state to another as the requirements for transcriptional regulation and viral DNA synthesis change during the infection process. However, this remains somewhat speculative at present, since it is not known whether the pattern of residues modified in the predominant, phosphorylated fraction of NS1 does, in fact, change with time during the viral growth cycle. Extrapolated in vivo, our data could indeed indicate that newly synthesized NS1 has to become phosphorylated first before it becomes competent for viral DNA replication, while other functions requiring site-specific binding but no DNA unwinding activity might be favored in the absence of phosphorylation. In this regard, it is worth noting that dephosphorylated wild-type NS1 and NS1dlC67 derived from HeLa cells, as well as un(der)phosphorylated NS1 produced in bacteria, became activated for helicase function upon phosphorylation by PKC preparations. The fact that the wild-type NS1 and NS1dlC67 proteins are modulated in the same way through their phosphorylation state indicates that the C-terminal transactivation domain (37) is unlikely to serve as a major (positive or negative) regulatory component for NS1 replicative functions. This argues against the possibility that the hypophosphorylated NS1dlC67, like NS1* present in infected cells (24), constitutes a product escaping the requirements for DNA replication of the full-length protein.
Interestingly, all functions of NS1 investigated here are dependent on an intact ATP-binding domain (13, 25, 56) and, thus, are most likely affected by interaction with this cofactor. Consequently, regulation of the NS1 ATPase by phosphorylation might be of central importance for other activities of NS1. ATP-dependent interaction of proteins with their cognate DNA motifs has been described for a variety of processes (34, 40, 45, 58) in addition to NS1-driven initiation of viral DNA replication (25). A constitutive, high-affinity association of polypeptides with target DNA, as is the case for many transcription factors, may not be suitable for pleiotropic proteins, since it would restrict some of their multiple activities. The involvement of ATP as a cofactor promotes a way to modulate the interaction of the protein with the DNA, e.g., by induction of a reversible conformational alteration within the polypeptide on hydrolysis of the trinucleotide and subsequent release of the resulting AMP (34). For NS1, site-specific binding to the origin has been shown to be dependent on the presence of ATP or a nonhydrolyzable ATP analog such as γ-S-ATP, and conditions leading to reduced ATP hydrolysis increase the affinity of NS1 for its cognate element (25), possibly due to the involvement of bound ATP in the formation of higher-order oligomers (54, 69). Therefore, the higher affinity of NS1O for the [ACCA]2–3 element may be assigned, at least in part, to the reduced ATPase activity, i.e., to the greater probability of the dephosphorylated polypeptide being in the ATP-bound form. In this respect, NS1O can be assumed to participate more efficiently in the formation of preinitiation complexes with other replication factors known to be required for MVM DNA replication, such as RP-A, PCNA, DNA polymerase, or the newly described initiation factor PIF (12). In addition, NS1 has been shown to interact with multiple recognition sites, distributed over the entire genome (25), which have been proposed to serve in association with NS1 for assembly of the replicative-form DNA into nucleosome-like structures (30). This kind of high-affinity interaction of NS1 with its cognate element might also involve preferentially un(der)phosphorylated NS1. On the other hand, a constitutive tight association of NS1 with the target DNA motif might interfere with the origin unwinding that allows nicking to occur on the exposed single strand and with the NS1 helicase activity that is necessary for unwinding of the double-stranded template as the replication fork proceeds. This, together with the energy requirements of helicase and nicking reactions, would point to the preferential involvement of phosphorylated NS1P, which has higher ATPase activity than NS1O, in these later steps of viral DNA replication. The reduction of the NS1O ATPase function can be expected to be especially deleterious for processive unwinding of the template DNA during strand displacement synthesis, a process requiring larger amounts of energy than local denaturation of the origin during site-specific nicking. This is in agreement with the observation that compared to that of the native polypeptide, the nicking activity of NS1O is reduced to a lesser degree than helicase activity. It should be stated, however, that DNA unwinding and site-specific nicking are complex reactions that can be controlled at several levels besides ATP turnover; i.e., their modulation may not be merely a reflection of the NS1 ATPase activity. This is exemplified by the NTP binding site mutant K405R, which is deficient in oligomerization (54) and ATP-dependent origin recognition (24), also exhibits a complete loss of helicase and site-specific nickase functions in spite of the residual ATPase activity measured (56).
NS1 is a pleiotropic protein that contributes to aspects of the parvovirus life cycle besides replication, such as transactivation of the P38 promoter, which directs the expression of the capsid genes (61). NS1 is also a cytotoxic effector molecule whose expression can lead to the eventual death of the target cell (6). These NS1-dependent events necessary for virus propagation take place in a temporal order (62). Thus, expression of the structural genes peaks after the burst of viral DNA replication and prior to the appearance of visible cytotoxic effects (18). We have shown that NS1 dephosphorylation suppresses functions associated with initiation of viral DNA replication, while the site-specific affinity of NS1 for the [ACCA]2–3 DNA motif is enhanced. NS1 induces the P38 promoter by binding to the repeated cognate motifs located within the so-called tar element, responsible in cis for transactivation of this promoter. It is therefore tempting to speculate that the enhanced affinity of hypophosphorylated NS1 for this particular binding site leads to an increase in P38 promoter activity, thus tipping the balance between parvovirus DNA replication and structural-gene expression. The modulation of the biochemical properties of NS1 in regard to the phosphorylation state could favor capsid protein production at later stages of infection and thereby promote the assembly of progeny virus particles. It remains to be determined, however, whether preformed NS1 undergoes dephosphorylation and/or newly synthesized NS1 fails to become phosphorylated as the virus cycle progresses.
Though demonstrated here under in vitro conditions for a few distinct NS1 activities, the phosphorylation-dependent activation of NS1 replicative functions may take place in infected cells. This suggestion is supported by the finding that cell extracts can fully reconstitute the replicative activities of dephosphorylated NS1, resulting in resolution and replication of the cloned dimer bridge fragment. Moreover, helicase activity, the NS1 function most affected by dephosphorylation, could be restored by rephosphorylation due to kinases present in the replication extracts or purified recombinant PKC (28). This in vitro specificity has prompted us to search for distinct protein kinases involved in the activation of NS1 replicative functions. During MVM infection, NS1 is phosphorylated at multiple positions, and it is presently unknown which of these phosphorylation sites is (or are) relevant for activation of NS1 replicative functions. It is interesting that the helicase activity of NS1O is rescued specifically by PKC preparations, since this protein kinase family has also been implicated in neoplastic transformation at the cellular level (44). This correlates with the longstanding observation that parvoviruses preferentially replicate in transformed cells, compared to the nontransformed parental cells (15).
Recently we established an in vitro replication system devoid of endogenous protein kinases (28, 55). This system supports initiation of replication by NS1P but not NS1O, providing further evidence that dephosphorylation of NS1 indeed down-modulates initiation of replication and that protein kinases are responsible for restoring NS1 activity. This tool should allow us to screen for distinct protein kinases capable of phosphorylating NS1 and, thus, activating the protein for replicative functions.
ACKNOWLEDGMENTS
We are indebted to Bernard Moss for making available the plasmid pTM-1 and the vTF7-3 virus. We gratefully thank Susan Cotmore for sharing constructs used for our biochemical analyses and the production of antibodies and for stimulating discussions and critical comments.
This work was supported by the Commission of the European Communities, the German-Israeli Foundation for Scientific Research and Development, and Public Health Service grant AI26109 from the National Institutes of Health. R.C. was also supported in part by a fellowship from La Ligue National Contre le Cancer.
REFERENCES
- 1.Astell C R, Liu Q, Harris C E, Brunstein J, Jindal H K, Tam P. Minute virus of mice cis-acting sequences required for genome replication and the role of the trans-acting viral protein, NS1. Prog Nucleic Acid Res Mol Biol. 1996;55:245–285. doi: 10.1016/s0079-6603(08)60196-8. [DOI] [PubMed] [Google Scholar]
- 2.Astell C R, Mol C D, Anderson W F. Structural and functional homology of parvovirus and papovavirus polypeptides. J Gen Virol. 1987;68:885–893. doi: 10.1099/0022-1317-68-3-885. [DOI] [PubMed] [Google Scholar]
- 3.Baas P D, Jansz H S. Single-stranded DNA phage origins. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- 4.Baldauf A Q, Willwand K, Mumtsidu E, Nüesch J P F, Rommelaere J. Specific initiation of replication at the right-end telomere of the closed species of minute virus of mice replicative-form DNA. J Virol. 1997;71:971–980. doi: 10.1128/jvi.71.2.971-980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. In: Hunter B M S T, editor. Protein phosphorylation. 201, part B. San Diego, Calif: Academic Press, Inc.; 1991. pp. 110–152. [DOI] [PubMed] [Google Scholar]
- 6.Brandenburger A, Legendre D, Avalosse B, Rommelaere J. NS1 and NS2 proteins may act synergistically in the cytopathogenicity of parvovirus MVMp. Virology. 1990;174:576–584. doi: 10.1016/0042-6822(90)90110-d. [DOI] [PubMed] [Google Scholar]
- 7.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cegielska A, Virshup D M. Control of simian virus 40 DNA replication by the HeLa cell nuclear kinase casein kinase I. Mol Cell Biol. 1993;13:1202–1211. doi: 10.1128/mcb.13.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cegielska A, Moarefi I, Fanning E, Virshup D M. T-antigen kinase inhibits simian virus 40 DNA replication by phosphorylation of intact T antigen on serines 120 and 123. J Virol. 1994;68:269–275. doi: 10.1128/jvi.68.1.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, J., and J. P. I. Nüesch. Unpublished observations.
- 11.Christensen J, Cotmore S F, Tattersall P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen J, Pederson M, Aasted B, Alexandersen S. Purification and characterization of the major nonstructural protein (NS-1) of Aleutian mink disease parvovirus. J Virol. 1995;69:1802–1809. doi: 10.1128/jvi.69.3.1802-1809.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbau R, Nüesch J P F, Salome N, Rommelaere J. Presented at the VIIth International Parvovirus Workshop, Heidelberg, Germany. 1997. Phosphorylation study of minute virus of mice NS1 protein. [Google Scholar]
- 15.Cornelis J J, Becquart P, Duponchel N, Salomé N, Avalosse B L, Namba M, Rommelaere J. Transformation of human fibroblasts by ionizing radiation, a chemical carcinogen, or simian virus 40 correlates with an increase in susceptibility to the autonomous parvoviruses H-1 virus and minute virus of mice. J Virol. 1988;62:1679–1686. doi: 10.1128/jvi.62.5.1679-1686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotmore S F, Nüesch J P F, Tattersall P. Asymmetric resolution of a parvovirus palindrome in vitro. J Virol. 1993;67:1579–1589. doi: 10.1128/jvi.67.3.1579-1589.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 19.Cotmore S F, Tattersall P. DNA replication in the autonomous parvoviruses. Semin Virol. 1995;6:271–281. [Google Scholar]
- 20.Cotmore S F, Tattersall P. A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. J Virol. 1989;63:3902–3911. doi: 10.1128/jvi.63.9.3902-3911.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotmore S F, Nüesch J P, Tattersall P. In vitro excision and replication of 5′ telomeres of minute virus of mice DNA from cloned palindromic concatemer junctions. Virology. 1992;190:365–377. doi: 10.1016/0042-6822(92)91223-h. [DOI] [PubMed] [Google Scholar]
- 22.Cotmore S F, Tattersall P. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J Virol. 1992;66:420–431. doi: 10.1128/jvi.66.1.420-431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotmore S F, Tattersall P. The NS-1 polypeptide of minute virus of mice is covalently attached to the 5′ termini of duplex replicative-form DNA and progeny single strands. J Virol. 1988;62:851–860. doi: 10.1128/jvi.62.3.851-860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotmore S F, Tattersall P. The NS1 polypeptide of the autonomous parvovirus MVM is a nuclear phosphoprotein. Virus Res. 1986;4:243–250. doi: 10.1016/0168-1702(86)90003-1. [DOI] [PubMed] [Google Scholar]
- 25.Cotmore S F, Christensen J, Nüesch J P F, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2–3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotmore S F, Tattersall P. Organization of nonstructural genes of the autonomous parvovirus minute virus of mice. J Virol. 1986;58:724–732. doi: 10.1128/jvi.58.3.724-732.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cotmore S F, Tattersall P. Presented at the VIIth International Parvovirus Workshop, Heidelberg, Germany. 1997. Resolution of the MVM right-end (5′) hairpin: viral sequences and trans-acting factors required to allow NS1 to nick the origin. [Google Scholar]
- 28.Dettwiler S, Corbau R, Rommelaere J, Nüesch J P F. Presented at the VIIth International Parvovirus Workshop, Heidelberg, Germany. 1997. Replicative functions of MVM NS1 are regulated by members of the protein kinase C family. [Google Scholar]
- 29.Doerig C, Hirt B, Antonietti J-P, Beard P. Nonstructural protein of parvoviruses B19 and minute virus of mice controls transcription. J Virol. 1990;64:387–396. doi: 10.1128/jvi.64.1.387-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doerig C, McMaster G, Sogo J, Bruggmann H, Beard P. Nucleoprotein complexes of minute virus of mice have a distinct structure different from that of chromatin. J Virol. 1986;58:817–824. doi: 10.1128/jvi.58.3.817-824.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fanning E. Simian virus 40 large T antigen: the puzzle, the pieces, and the emerging picture. J Virol. 1992;66:1289–1293. doi: 10.1128/jvi.66.3.1289-1293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuerst T R, Niles E G, Studier F W, Moss B. Eucaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann I. Cyclin-dependent protein kinases and the regulation of the eucaryotic cell cycle. In: Marks F, editor. Protein phosphorylation. Weinheim, Germany: VCH Verlagsgesellschaft mbH; 1996. pp. 179–201. [Google Scholar]
- 34.Hübscher U, Maga G, Podust V N. DNA replication accessory proteins. In: dePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 525–543. [Google Scholar]
- 35.Inamoto S, Yoshioka Y, Ohtsubo E. Site- and strand-specific nicking in vitro at oriT by the traY-traI endonuclease of plasmid R100. J Biol Chem. 1991;266:10086–10092. [PubMed] [Google Scholar]
- 36.Jindal H K, Yong C B, Wilson G M, Tam P, Astell C R. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS1 protein uncouple ATPase and DNA helicase functions. J Biol Chem. 1994;269:3283–3289. [PubMed] [Google Scholar]
- 37.Legendre D, Rommelaere J. Targeting of promoters for trans activation by a carboxy-terminal domain of the NS-1 protein of the parvovirus minute virus of mice. J Virol. 1994;68:7974–7985. doi: 10.1128/jvi.68.12.7974-7985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legendre D, Rommelaere J. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J Virol. 1992;66:5705–5713. doi: 10.1128/jvi.66.10.5705-5713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Rhode S L., III Mutation of lysine 405 to serine in the parvovirus H-1 NS1 abolishes its functions for viral DNA replication, late promoter trans activation, and cytotoxicity. J Virol. 1990;64:4654–4660. doi: 10.1128/jvi.64.10.4654-4660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lusky M, Hurwitz J, Seo Y S. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J Biol Chem. 1993;268:15795–15803. [PubMed] [Google Scholar]
- 41.Mackett M, Smith G L, Moss B. The construction and characterization of vaccinia virus recombinants expressing foreign genes. In: Glover D M, editor. DNA cloning: a practical approach. Oxford, United Kingdom: IRL Press; 1985. [Google Scholar]
- 42.Mansfield M. Protein blotting, a practical approach. Oxford, United Kingdom: IRL Press; 1994. pp. 33–52. [Google Scholar]
- 43.Marks F. The brain of the cell. In: Marks F, editor. Protein phosphorylation. Weinheim, Germany: VCH Verlagsgesellschaft mbH; 1996. pp. 1–35. [Google Scholar]
- 44.Marks F, Gschwendt M. Protein kinase C. In: Marks F, editor. Protein phosphorylation. Weinheim, Germany: VCH Verlagsgesellschaft mbH; 1996. pp. 81–116. [Google Scholar]
- 45.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 46.McVey D, Ray S, Gluzman Y, Berger L, Wildeman A G, Marshak D R, Tegtmeyer P. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J Virol. 1993;67:5206–5215. doi: 10.1128/jvi.67.9.5206-5215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McVey D, Woelker B, Tegtmeyer P. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J Virol. 1996;70:3887–3893. doi: 10.1128/jvi.70.6.3887-3893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merchlinsky M J, Tattersall P J, Leary J J, Cotmore S F, Gardiner E M, Ward D C. Construction of an infectious molecular clone of the autonomous parvovirus minute virus of mice. J Virol. 1983;47:227–232. doi: 10.1128/jvi.47.1.227-232.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moarefi I F, Small D, Gilbert I, Höpfner M, Randall S K, Schneider C, Russo A A, Ramsperger U, Arthur A K, Stahl H, Kelly T J, Fanning E. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J Virol. 1993;67:4992–5002. doi: 10.1128/jvi.67.8.4992-5002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molitor T W, Joo H S, Collett M S. Identification and characterization of a porcine parvovirus nonstructural polypeptide. J Virol. 1985;55:554–559. doi: 10.1128/jvi.55.3.554-559.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moss B, Elroy Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 52.Mousset S, Ouadrhiri Y, Caillet-Fauquet P, Rommelaere J. The cytotoxicity of the autonomous parvovirus minute virus of mice nonstructural proteins in FR3T3 rat cells depends on oncogene expression. J Virol. 1994;68:6446–6453. doi: 10.1128/jvi.68.10.6446-6453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nüesch J P, Cotmore S F, Tattersall P. Expression of functional parvoviral NS1 from recombinant vaccinia virus: effects of mutations in the nucleotide-binding motif. Virology. 1992;191:406–416. doi: 10.1016/0042-6822(92)90202-z. [DOI] [PubMed] [Google Scholar]
- 54.Nüesch J P, Tattersall P. Nuclear targeting of the parvoviral replicator protein molecule NS1: evidence for self-association prior to nuclear transport. Virology. 1993;196:637–651. doi: 10.1006/viro.1993.1520. [DOI] [PubMed] [Google Scholar]
- 55.Nüesch J P, Corbau R, Tattersall P, Rommelaere J. Presented at the 15th Annual Meeting of the American Society for Virology, London, Ontario, Canada. 1996. Replication activities of MVM NS1 are regulated through phosphorylation. [Google Scholar]
- 56.Nüesch J P, Cotmore S F, Tattersall P. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology. 1995;209:122–135. doi: 10.1006/viro.1995.1236. [DOI] [PubMed] [Google Scholar]
- 57.Nüesch, J. P. F. Unpublished observations.
- 58.Parsons R E, Stenger J E, Ray S, Welker R, Anderson M E, Tegtmeyer P. Cooperative assembly of simian virus 40 T-antigen hexamers on functional halves of the replication origin. J Virol. 1991;65:2798–2806. doi: 10.1128/jvi.65.6.2798-2806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prives C. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell. 1990;61:735–738. doi: 10.1016/0092-8674(90)90179-i. [DOI] [PubMed] [Google Scholar]
- 60.Pujol A, Deleu L, Nüesch J P F, Cziepluch C, Jauniaux J-C, Rommelaere J. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J Virol. 1997;71:7393–7403. doi: 10.1128/jvi.71.10.7393-7403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhode S L, III, Richard S M. Characterization of the trans-activation-responsive element of the parvovirus H-1 P38 promoter. J Virol. 1987;61:2807–2815. doi: 10.1128/jvi.61.9.2807-2815.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoborg R V, Pintel D J. Accumulation of MVM gene products is differentially regulated by transcription initiation. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- 63.Skiadopoulos M H, Faust E A. Mutational analysis of conserved tyrosines in the NS1 protein of the parvovirus minute virus of mice. Virology. 1993;194:509–517. doi: 10.1006/viro.1993.1289. [DOI] [PubMed] [Google Scholar]
- 64.Snyder R O, Samulski R J, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 65.Sung P, Higgins D, Prakash L, Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988;7:3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tattersall P, Ward D C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976;263:106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- 67.Vanacker J-M, Rommelaere J. Non-structural proteins of autonomous parvoviruses: from cellular effects to molecular mechanisms. Semin Virol. 1995;6:291–297. [Google Scholar]
- 68.Vanacker J-M, Corbau R, Adelmant G, Perros M, Laudet V, Rommelaere J. Transactivation of a cellular promoter by the NS1 protein of the parvovirus minute virus of mice through a putative hormone-responsive element. J Virol. 1996;70:2369–2377. doi: 10.1128/jvi.70.4.2369-2377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang E H, Prives C. ATP induces the assembly of polyomavirus large tumor antigen into hexamers. Virology. 1991;184:90–98. doi: 10.1016/0042-6822(91)90858-9. [DOI] [PubMed] [Google Scholar]
- 70.Wilson G M, Jindal H K, Yeung D E, Chen W, Astell C R. Expression of minute virus of mice major non-structural protein in insect cells: purification and identification of ATPase and helicase activities. Virology. 1991;185:90–98. doi: 10.1016/0042-6822(91)90757-3. [DOI] [PubMed] [Google Scholar]
- 71.Woodgett J R. Use of synthetic peptides mimicking phosphorylation sites for affinity purification of protein-serine kinases. In: Hunter B M S T, editor. Protein phosphorylation. 200, part A. San Diego, Calif: Academic Press, Inc.; 1991. pp. 169–178. [DOI] [PubMed] [Google Scholar]