Abstract
Proteolytic processing of the picornaviral polyprotein mediated by the differential action of virus-encoded proteinase(s) is pivotal to both RNA genome replication and capsid formation. Possibly to enlarge the array of viral proteins, picornaviral polyprotein processing results in intermediate and mature products which apparently have distinct functions within the viral life cycle. For hepatitis A virus (HAV), we report here on the autoproteolysis of precursor polypeptides comprising the only viral proteinase, 3Cpro, and on their role in viral particle formation. Following transient expression of a nested set of 3Cpro-containing proteins (P3, 3ABC, 3BCD, 3CD, 3BC, and 3C) in eukaryotic cells, the extent of processing was determined by analyzing the cleavage products. The 3C/3D site was more efficiently cleaved than those at the 3A/3B and 3B/3C sites, leading to the accumulation of the intermediate product 3ABC. In the absence of 3A from the precursor, cleavage at the 3B/3C site was further reduced and a switch to an alternative 3C/3D site was observed. Coexpression of various parts of P3 with the precursor of the viral structural proteins P1-2A showed that all 3C-containing intermediates cleaved P1-2A with almost equal efficiency; however, viral particles carrying the neutralizing epitope form much more readily in the presence of the complete P3 domain than with parts of it. These data support the notion that efficient liberation of structural proteins from P1-2A is necessary but not sufficient for productive HAV capsid formation and suggest that the polypeptides flanking 3Cpro promote the assembly of viral particles.
The picornaviral genome is a single-stranded RNA of approximately 7.5 kb in length, with an open reading frame for a polyprotein whose molecular mass is about 250 kDa. Proteolytic cleavage of the viral polyprotein P1-P2-P3 is central in the viral life cycle and leads to the liberation of the capsid proteins (VPO, VP3, VP1, or VP1-2A) from the P1 or P1-2A domain and of the nonstructural proteins from the P2 and P3 domains. Common to all picornaviruses is the major proteinase 3Cpro, which excises itself from the P3 domain of the polyprotein and catalyzes almost all cleavages within the polyprotein. An additional proteinase, 2Apro, or an unusual nonenzymatic step specifically catalyzes the liberation of the structural proteins’ precursor (23). Hepatitis A virus (HAV) is exceptional, as protein 2A is proteolytically inactive and found attached to VP1 and its precursors (13, 14). For HAV, it has been proposed that P1-2A is the functional precursor of the structural proteins and is liberated from the primary translation product by proteinase 3Cpro (1, 16, 21, 26, 29). Furthermore, unlike most other picornaviruses, HAV replicates very slowly in infected cells, and although the viral structural proteins accumulate and are hence detectable in cell cultures, neither the nonstructural proteins of the P2 and P3 domains nor their precursors were found, possibly due to the low metabolic activity of HAV or to low protein stability (9, 32). To bypass the limitations caused by the virus’ retarded replication in infected cells, various recombinant expression systems were employed to study HAV polyprotein processing. With bacterial and eukaryotic in vitro and in vivo expression, it was shown that 3Cpro is able to liberate all structural and nonstructural proteins from the primary translation product (12, 20, 21, 29–31).
The major P3-processing intermediates of poliovirus, the picornaviral prototype, are proteins 3AB and 3CD, which were shown to have distinct functions in protein processing and genome replication. 3CDpro, the precursor of proteinase 3Cpro and polymerase 3Dpro, cleaves P1 much better than 3Cpro (35) and plays a crucial role in RNA synthesis due to its specific binding to viral RNA structures (2). The multiple functions of 3AB, the precursor of the genome-linked protein VPg (3B), have been studied extensively (34). Although several stable HAV P3-processing intermediates (e.g., 3ABC and 3CD) have been detected, their distinct proteolytic activity and roles within the viral life cycle have not yet been directly assessed (12, 14, 31).
In order to further our understanding of the role of P3 intermediates during the viral life cycle, processing of the complete P3 domain and its proteolytic intermediates was assessed in detail by expressing a nested set of HAV 3Cpro precursor polypeptides in a transient eukaryotic system. The data suggest that 3A, as part of the polypeptide, affects P3 cleavage efficiency and allowed us to propose a processing scheme which argues for an alternative cleavage site C-terminal to the known 3C-3D junction. Although the proteolytic capacities within P1-2A of 3Cpro and its precursors are almost the same, evidence is provided that other P3 proteins in addition to 3C are required for efficient assembly of viral particles, as was demonstrated in experiments in which either P1-2A was coexpressed with different P3-derived constructs or the HAV genome deleted from 3AB or 3D was expressed.
MATERIALS AND METHODS
Construction of plasmids.
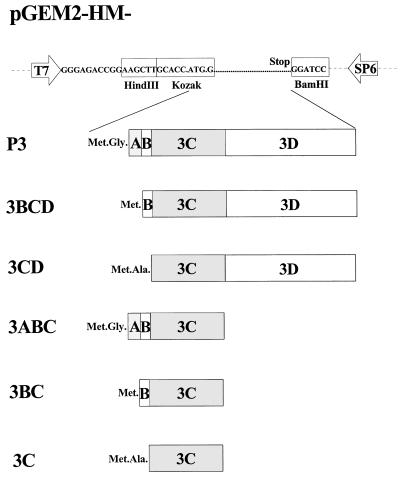
For expression in the vaccinia virus T7 system, various regions of HAV P3 were inserted into pGEM2. For cloning of the exact P3-, 3ABC-, 3BC-, 3C-, 3BCD-, and 3CD-coding regions, we relied on the published 2C/3A, 3A/3B, 3B/3C, and 3C/3D cleavage sites (8, 12, 31). The following primers were used for PCR amplification of subgenomic fragments of the attenuated strain HAV HM175: primer 1, 5′-ATGGAAGCTTGCACCATGGGAATTTCAGATGATGATAATGATAG-3′ (sense 3A); primer 2, 5′-ACCAAGCTTGCACCATGGGGGTATATTATGGTGTAACTA-3′ (sense 3B); primer 3, 5′-GATCAAGCTTGCACCATGGCAACTTTGGAAATAGCAGGACTGG-3′ (sense 3C); primer 4, 5′-TTGGATCCTTACTGACTTTCAATTTTCTTATCAA-3′ (antisense 3C); and primer 5, 5′-TAGGATCCTCATGAAAGGTCACAAATGAAACACT-3′ (antisense 3D). The HindIII and BamHI restriction sites are underlined. With primers 1 and 4, and pT7-HAV1 as the template (11), the 3ABC-coding region (nucleotide positions 4996 to 5943) was amplified by PCR. All nucleotide positions given refer to the nucleotide sequences of the HM175 attenuated strain (5). With primers 1 and 5, primers 2 and 4, primers 2 and 5, primers 3 and 4, and primers 3 and 5, the P3 (nucleotides [nt] 4996 to 7410), 3BC (nt 5218 to 5943), 3BCD (nt 5281 to 7410), 3C (nt 5286 to 5943) and 3CD (nt 5286 to 7410), genomic regions were amplified by PCR. The HindIII-BamHI-restricted PCR fragments were inserted into the respective sites of pGEM2 in such a way that transcription of the HAV-coding sequence would be driven by the T7 promoter. The conservation of the appropriate reading frame was confirmed by DNA sequencing, which identified neutral T-to-C and C-to-T exchanges at nucleotide positions 6211 and 7027 of pT7-HAV1, respectively, that produced a dam-sensitive XbaI restriction site at nucleotide position 6211. A schematic representation of the constructs is given in Fig. 1. The expression plasmid pEXT7-HM/HM-P1-2A(E/S) contains the cleavage site E/S at VP1-2A amino acid position 273/274, which is favored by 3Cpro as described before (26). pT7-HAV1-Δ3AB, coding for P1-P2-3CD and corresponding to the HAV open reading frame with an in-frame deletion of 3AB, was constructed by religating the 10-kb EcoRI-Bsh1365I-restricted and blunt-ended (Klenow polymerase) fragment of pT7-HAV1 encoding P1-P2-P3 (11). The seven C-terminal amino acids of 2C were replaced by a leucine followed by the six C-terminal amino acids of 3B, thus generating a 2C/3C cleavage site derived from the former 3B-3C junction. After religation of the XhoI-linearized and blunt-ended pT7-HAV1, pT7-HAV1-3DΔ was obtained, which codes for the HAV polyprotein P1-P2-3ABCDΔ with a 28% truncation from the C terminus of 3D.
FIG. 1.
Schematic representation of HAV cDNA constructs used for transient coexpression in the vaccinia virus T7 system (7). Transcription of the HAV-coding sequence is driven by the T7 promoter. Amino acid residues at the N terminus of the open reading frames are derived from vector-encoded sequences downstream of the translation initiation codon. To ensure the same efficiency of translation initiation, all P3-derived sequences are preceded by a translation initiation codon flanked by a conserved sequence (15). Termination of translation is ensured by a stop codon.
Expression in COS7 cells.
For transient mammalian expression, we used the vaccinia virus expression system (6, 7). COS7 cells (3 × 105) grown overnight to approximately 70% confluency in 35-mm-diameter wells were transfected with 1 μg of purified cDNA and 9 μl of LipofectAmine according to the manufacturer’s directions (Gibco/BRL-Life Technologies). Three hours after transfection, infection with vTF7-3 (multiplicity of infection, 1) was done for 30 min at 37°C. After the medium was replaced by 2 ml of Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, incubation was continued for 18 h before the cells were scraped off the plate in 250 μl of phosphate-buffered saline containing 0.05% (vol/vol) Tween 20. Seventy microliters of the crude cellular extract was separated on a discontinuous 12% polyacrylamide gel containing 0.1% sodium dodecyl sulfate and blotted onto a nitrocellulose membrane. Efficiency of protein transfer was confirmed by Ponceau-S staining of the membrane. For immunodetection, polyclonal antisera anti-3D (31), anti-3B, anti-3C (30), anti-VPO (9), anti-VP1 (9) and immunoglobulins conjugated to alkaline phosphatase were used.
Characterization of HAV antigenicity and particles.
The crude cell extracts were clarified by centrifugation with a table top centrifuge at 13,000 × g. The supernatants were analyzed in appropriate dilutions by a particle-specific enzyme-linked immunosorbent assay (ELISA), with the neutralizing monoclonal antibody K2-4F2 as capture and as detection antibody (19). Each transfection assay was performed twice and analyzed in duplicate in one or two separate experiments. As negative controls, the individual P3 intermediates and P1-2A were expressed separately. The specific antigenicity was estimated as the mean ELISA signal of the different sets of coexpressions normalized by the mean values of the negative controls, which were set to zero. To characterize the HAV particles produced in vivo, the crude cell extracts were subjected to a continuous sucrose gradient (5 to 30% [wt/wt] in 10 mM Tris-HCl, pH 7.3) with the SW65 rotor (75 min, 48,000 rpm, 5°C). Individual fractions were analyzed by the particle-specific ELISA, and their sucrose concentrations were determined with a refractometer.
RESULTS AND DISCUSSION
Detailed analysis of HAV P3 processing.
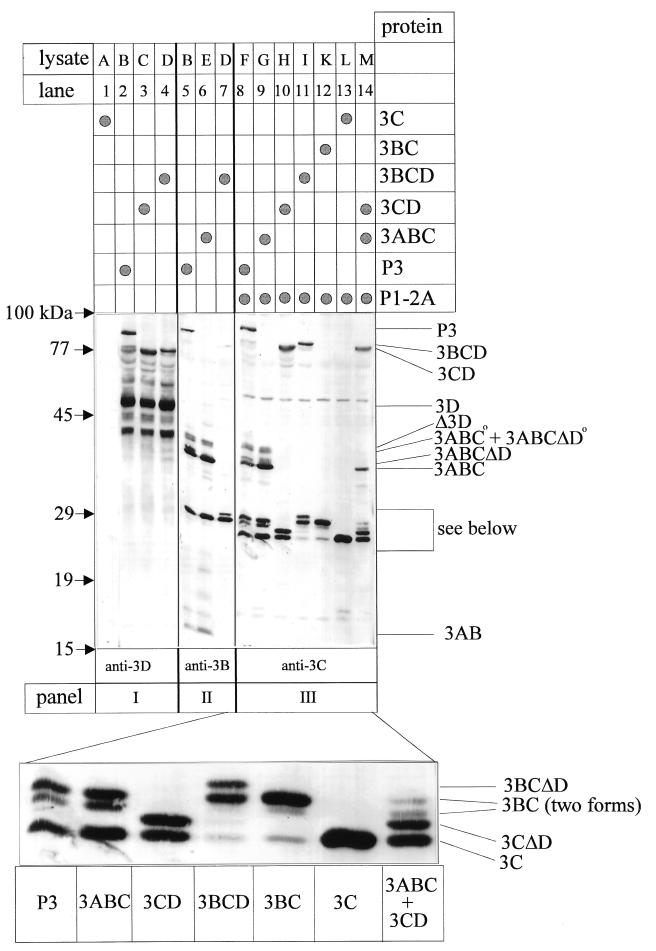
Both 3ABC and 3CD have been simultaneously detected as intermediates of HAV P3 processing, yet which factors regulate the generation of these partially overlapping cleavage products is still unknown (31). To unravel the possible proteolytic pathways within HAV P3, we expressed 3Cpro and its precursors as a nested set of proteins with the vaccinia virus T7 system. As depicted in Fig. 2, the products of cells expressing 3C, 3BC, 3BCD, 3CD, 3ABC, and P3 alone or in the presence of P1-2A were analyzed by immunoblotting with monospecific polyclonal antisera raised against 3D (panel I), 3B (panel II), and 3C (panel III). The patterns of anti-3D reactive proteins derived from P3 (lane 2), 3CD (lane 3), and 3BCD (lane 4) included the uncleaved precursor proteins P3, 3CD, and 3BCD and several processing products. Small amounts of mature 3CD were cleaved off P3 (lane 2) and 3BCD (lane 4), which comigrated with the precursor polypeptide of 3CD-expressing cells (lane 3). The most prominent cleavage product was 3D, which was liberated in almost equal quantities from P3 and 3BCD. Besides various larger anti-3D reactive polypeptides, substantial amounts of an approximately 40-kDa protein were found which might be products of degradation or of alternative translation initiation. The extract of 3C-expressing cells was not reactive with anti-3D, proving the specificity of the antiserum (lane 1). In extracts of cells expressing P3, 3ABC, and 3BCD, polypeptides with the mobility of 3ABC (lanes 5 and 6), 3BC (lanes 5 to 7), and 3AB (lanes 5 and 6) were the major processing products detectable with anti-3B (panel II; for additional bands, see below). With anti-3C (panel III), all uncleaved precursors P3, 3ABC, 3CD, 3BCD, and 3BC could be identified in addition to processing intermediates and the mature proteinase (lanes 8 to 12, respectively), clear evidence that P3 as well as P3-derived intermediates (3BCD, 3CD, 3ABC, and 3BC) were proteolytically active. Although almost all intermediate and end products were detected as processing products, their relative proportions were markedly different among the various precursors tested, suggesting that the efficiency of cleavage at the different sites within P3 varied significantly and was dependent on 3C-flanking domains. Among the P3-derived and anti-3C reactive polypeptides, 3ABC, 3BC, and 3C were major cleavage products, whereas 3BCD and 3CD were found in minute amounts (lane 8). Additional anti-3C reactive proteins derived from P3 were identified by their comigration with processing products derived from other 3C-containing precursors and will be discussed below. The overall product pattern derived from 3ABC was similar to that of P3 and led to the production of equal amounts of 3C and 3BC (lane 9). In contrast, only small amounts of 3C were generated from precursors 3BCD (lane 11) and 3BC (lane 12), implying that cleavage at the 3B-3C junction is inefficient when 3A is absent from the precursor.
FIG. 2.
Immunoblot analysis after transient expression of HAV P3 and its proteolytically active derivatives. Extracts of cells transinfected with the cDNAs mentioned (see Fig. 1, lysates A to M) and vTF7-3 were analyzed for their P3-processing pattern. After electrophoretic separation on one gel and transfer to a nitrocellulose membrane, immunological detection of recombinant proteins was performed, with antisera indicated at the bottom of the figure. The magnified panel is derived from the lower half of panel III. Immunoreactive products are marked on the right side; the positions of molecular size standards are shown on the left.
Based on the hydrophobic properties of 3A, it has been suggested for both polio and hepatitis A virus that 3A as part of protein 3AB might function as a membrane anchor for the replication primer 3B and thus for the replication complex (25, 34). To assess whether larger 3A-containing polypeptides might also differ from those lacking 3A in their hydrophobicity and hence in their possible cytoplasmic localization, the solubility of P3 and its cleavage products of lysate F was determined. 3C and 3BC were found to be soluble in phosphate-buffered saline, whereas P3 and 3ABC could be solubilized only in the presence of detergent (e.g., 1% Triton X-100). Based on these observations and on the comparison of the various cleavage patterns, it is tempting to speculate that 3A-containing proteins might be either folded differently or localized to distinct cytoplasmic sites and therefore subjected to different processing pathways. Since precursors lacking 3A do not give rise to the mature proteinase, it can be concluded that mature 3Cpro is liberated mainly from 3ABC, which is derived from P3.
Besides the cleavage products observed earlier in other recombinant systems (12, 13, 21, 29–31), additional protein bands migrating more slowly than 3ABC, 3BC, and 3C were now found and identified by their immunoreactivity and electrophoretic mobility as proteins 3ABCΔD, 3CΔD, and 3BCΔD (Fig. 2, lanes 7, 8, 10, 11, 14). Since these polypeptides were detected only among the products of precursors containing 3D and the proteolytically active proteinase (data not shown for the inactive precursors), 3ABCΔD, 3CΔD, and 3BCΔD may be formed by 3C-mediated processing at an alternative 3C/3D cleavage site (V/S) located 15 amino acid residues C-terminal to the Q/R-scissile bond described earlier (31). Note that the Δ3D content of this polypeptide is not detectable by anti-3D, which was raised against the C-terminal part of 3D (31). It is striking that the alternative 3C/3D site was cleaved as efficiently as the Q/R site in precursors 3CD and 3BCD, suggesting that in these precursors which lack 3A, both cleavage sites are accessible to the active site of the proteinase (lanes 10 and 11). HAV 3CΔD might be similar to 3C′, a C-terminally extended form of 3C which has been reported for enteroviruses, human rhinoviruses, and aphthoviruses (28). In poliovirus, protein 3CD is a prominent and stable intermediate product, and the alternative cleavage of 3CD is mediated by proteinase 2A liberating 3C′ and 3D′ (18). Taken together, these results show, for the first time, that proteolytically active precursors spanning the 3C/3D site and lacking 3A can be cleaved at an alternative site. Future experiments should address the potential role of the additional HAV P3-derived polypeptides by testing the in vivo and in vitro viability of HAV mutants carrying mutated alternative cleavage sites.
As an additional new product derived from P3 and 3ABC, 3BC was detected in two forms which migrated slightly apart and which were identified by their immunoreaction with anti-3C (lanes 8 and 9). Only minute amounts of the faster-migrating 3BC were detected among the anti-3C reactive products of 3BCD and 3BC (lanes 11 and 12) and among the anti-3B reactive products of P3 and 3ABC (lanes 5 and 6). The reduced immunoreactivity with anti-3B (compare lanes 5 and 8 or lanes 6 and 9) suggests that the faster-migrating form of 3BC might be truncated at its N terminus. Moreover, a polypeptide of approximately 40 kDa (marked ° in the figure) was detected by the anti-3B and anti-3C sera (lanes 5, 6, 8, and 9). Since this polypeptide was found among the products derived from P3 as well as from 3ABC in its proteolytically active and inactive form (latter not shown), 3ABC° is likely the product of posttranslational modification of 3ABC. Our results on the detailed analysis of P3 processing obtained by recombinant expression in eukaryotic cells indicate that next to the major products 3ABC, 3BC, and 3C, additional polypeptides are formed either by modification or cleavage at an alternative 3C/3D site. The data clearly show that 3C cleavage efficiency within P3 depends on the presence of 3C-flanking regions, particularly 3A.
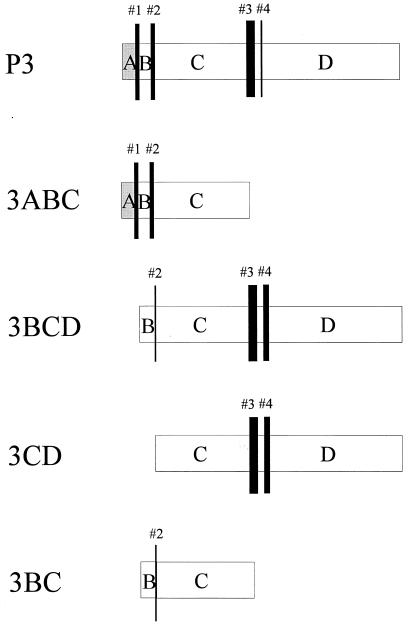
By comparing the quantity of processing products derived from P3 with those derived from its potential 3C-containing intermediates, the relative cleavability of the P3 sites was estimated. The prevalence of cleavages (numbered 1 to 4) within the context of all precursors tested is depicted in Fig. 3, with the thickness of the bars being proportional to the cleavage efficiency. As judged from the intensity of product bands, particularly from those shown in Fig. 2III, it was obvious that 3C-mediated cleavage at sites within P3 is affected by the presence of 3C-flanking regions. In 3BC, where 3C is only flanked by 3B, the N terminus of 3C (site 2) was relatively inefficiently cleaved, whereas processing at sites 3 and 4 in 3CD gave rise to substantial amounts of both mature 3C and 3CΔD. In polypeptide 3BCD, the extent of cleavage at either end of 3C (sites 2, 3, and 4) was similar to what was observed when cleavage could occur at only one terminus of 3C (see 3CD and 3BC). Autoproteolysis at the termini of 3Cpro was discussed after the crystal structure of HAV 3Cpro was resolved (3). Based on the flexibility of the terminal regions of 3C, it was suggested that processing at the 3B/3C site, but not at the 3C/3D site, is likely to be intramolecular. Since the expression system used here does not allow direct differentiation between intra- and intermolecular cleavage reactions, our data neither support nor disprove this hypothesis. A shift in the relative cleavability of the P3 sites was observed in 3C precursors containing the 3A moiety. Although cleavage at site 3 is still the most prominent in P3, subsequent processing of junctions 1 and 2 liberating 3AB, 3C, and 3BC is equally probable. Cleavages at sites 3C/3D (no. 3) followed by 3A/3B (no. 1) and 3B/3C (no. 2) presumably represent the major processing pathway allowing the production of the stable P3 intermediates 3BC and 3ABC. The stability of these processing intermediates might imply that they play a role in the viral life cycle in addition to being the source of the mature proteinase. In fact, we have shown that 3ABC strongly binds HAV RNA, and its accumulation might be a prerequisite for its function in RNA replication (17). Polypeptides containing N-terminal sequences of 3D (e.g., 3BCΔD and 3CΔD) were found predominantly from precursors lacking 3A (3BCD and 3CD). Since these polypeptides were also detected among the processing products of P3, they can be regarded as products of a minor processing pathway occuring indeed at a low but significant level. Unfortunately, due to the low HAV expression level in infected cells (10), the role of the minor processing intermediates in HAV replication will be difficult to elucidate. In an initial attempt to directly assess the function of P3 in one step of the viral life cycle, we now describe experiments which indicate that P3 intermediates might affect HAV particle formation.
FIG. 3.
Hierarchical organization of cleavages within P3 and its proteolytic intermediates. Based on the intensity of product bands shown in Fig. 2, the extent of cleavage within various 3C-containing precursor polypeptides was estimated for the 3A/3B (no. 1), 3B/3C (no. 2), and regular (no. 3) and alternative 3C/3D (no. 4) sites and is symbolized by the thickness of the bars. A thick bar represents efficient cleavage, whereas a thin bar symbolizes inefficient processing. Cleavages of sites 1, 2, and 3 are part of the major processing pathway. Cleavage at the alternative 3C/3D site (no. 4) was initially deduced from the product pattern of 3BCD and 3CD, which includes substantial amounts of 3CΔD. 3CΔD was also found among the P3-derived products, and thus cleavage at site 4 in combination with cleavages at sites 1 and 2 might present a minor processing pathway within HAV P3.
Effect of HAV P3 intermediate polypeptides on the formation of viral particles.
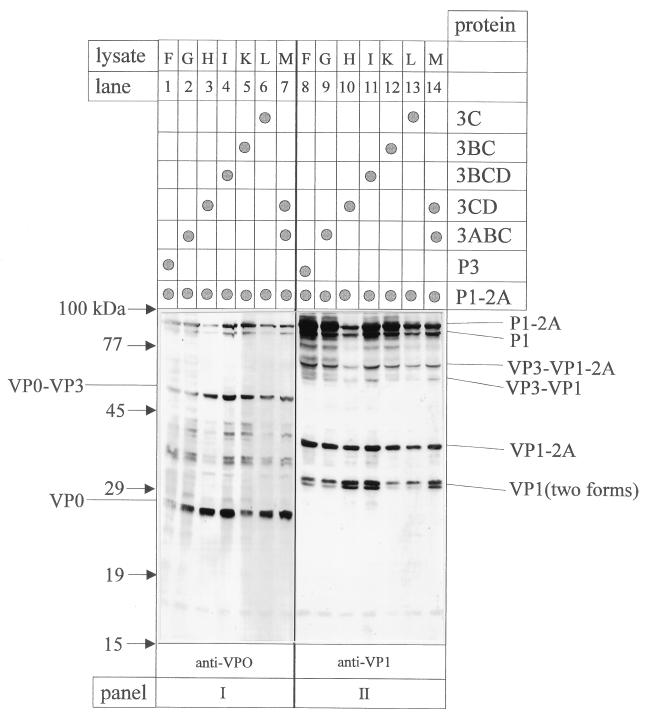
As reported elsewhere, P1-2A is the precursor polypeptide required for efficient assembly of empty HAV particles which was used as substrate for P3 and its 3C-containing products in cis and in trans (1, 4, 27). After coexpression of P1-2A with the various precursors of 3C, both the liberation of viral structural proteins as well as the assembly of particulated viral antigen were analyzed. Here we show that, irrespective of the P3 derivatives used for coexpression with P1-2A (in trans), the overall pattern of structural proteins was essentially the same as that demonstrated by immunoblot analysis with anti-VP0 and anti-VP1 (Fig. 4). P1-2A, P1, VP3-VP1-2A, VP3-VP1, VPO-VP3, VP1-2A, VP1, and VPO were immunodetected in all extracts. Somewhat higher proportions of products were found when 3CD and 3BCD (lysates H and I) were used as source of viral proteinase, an observation which cannot be explained at present. As discussed earlier, the two immunologically reactive forms of VP1 are products of 3Cpro-mediated cleavage at alternative VP1/2A cleavage sites (26). When anti-VP4 was used to differentiate between VP0 and VP2, no cleavage of VP0 into VP2 and VP4 was observed (data not shown).
FIG. 4.
Immunoblot analysis after transient coexpression of HAV P1-2A with P3 and its proteolytically active derivatives. The extracts of cells transfected with cDNAs coding for the proteins listed at the right (lysates F to M) were analyzed for their P1-2A-processing pattern (see also Fig. 2III). After electrophoretic separation on the same gel and transfer to a nitrocellulose membrane, immunological detection of recombinant proteins was performed with the antisera indicated at the bottom. Immunoreactive products are marked on the sides; the positions of molecular size standards are shown on the left.
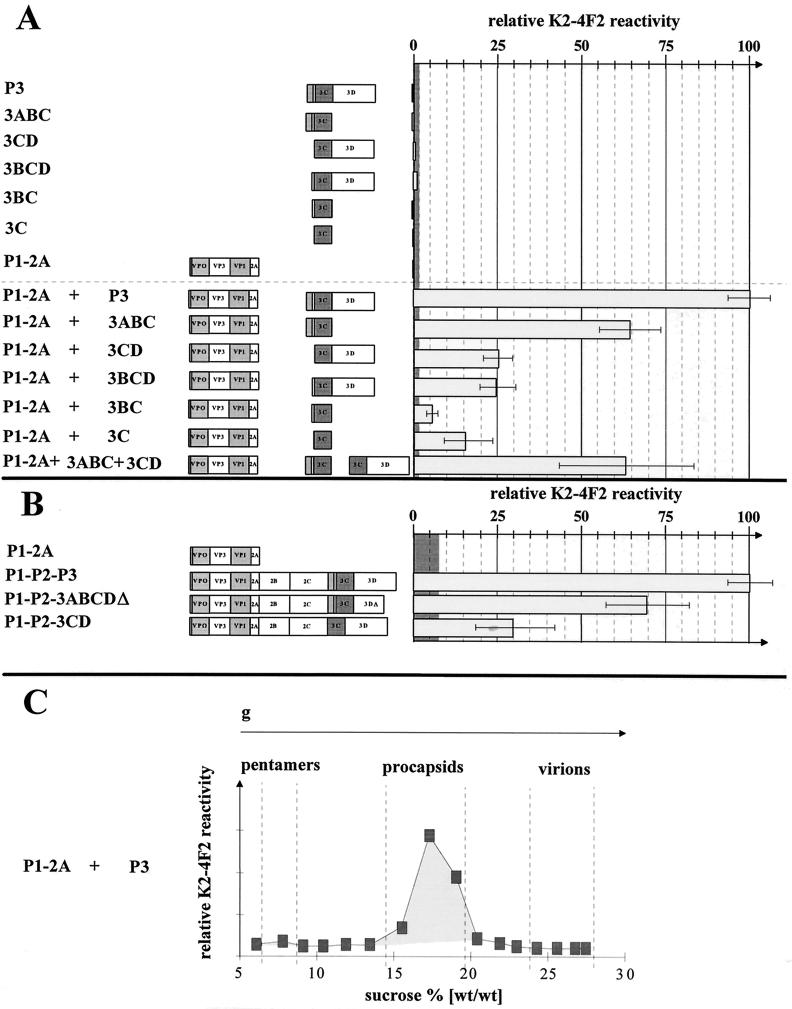
To determine the formation of viral particles in the same extracts, we performed a particle-specific ELISA, using the monoclonal antibody K2-4F2 that specifically detects the neutralizing epitope present on 14-S-pentamers and higher-ordered structures (19, 24). Surprisingly, the extent of HAV particle formation contrasted markedly with the expression rate and processing pattern of P1-2A. Although similar proportions of the mature structural proteins were liberated by the action of P3 and its intermediates, the efficiency of particle formation was dependent on the presence of 3C-flanking polypeptides. The entire P3 protein was most effective in the formation of empty viral particles (sedimenting with approximately 70 S) as shown by the particle-specific ELISA (Fig. 5A) and also after sedimentation through a 5 to 30% (wt/wt) sucrose gradient showing a sedimentation profile similar to those reported earlier (Fig. 5C) (33, 36). Deletion of 3D from P3 led to a one-third reduction of K2-4F2-reactive antigenicity, whereas the absence of 3A or 3AB diminished the antigenicity to about one-fourth of the entire P3-coding region (100%). With 3C or 3BC, a further remarkable reduction in particle formation relative to 3CD or 3BCD was observed. To test whether the functions of 3ABC and 3CD can be complemented in trans, both proteins were coexpressed with P1-2A. The efficiency of particle formation was not increased relative to that of 3ABC alone, suggesting that either some of the alternative intermediates of P3 processing or all P3 proteins are required in cis for efficient assembly. Neither the extract of cells expressing P3 and its proteolytically active derivatives nor P1-2A alone was reactive with the monoclonal antibody, showing its specificity for processed and assembled viral particles. As the extent of 3C-mediated processing of P1-2A did not correlate with particle formation, 3C-flanking proteins play an important role in virion assembly.
FIG. 5.
HAV particle formation after recombinant expression of P1-2A in trans or cis with P3 and its deleted forms. Extracts of cells transfected with cDNA constructs as depicted on the left and infected with vTF7-3 were analyzed for their K2-4F2 reactivity. The monoclonal antibody K2-4F2 specifically detects the HAV neutralization epitope, which is only exposed on viral particles. The relative immunoreactivity is shown for recombinant trans (A) and cis (B) expression. In panel A, the reactivity of cells coexpressing P1-2A and P3 was 100%; in panel B, the reactivity of P1-P2-P3-expressing cells was 100%. The extracts used in panel A correspond to those shown in Fig. 2 and 4. As negative controls, extracts of cells expressing P1-2A, P3, and its derivatives were used; their mean value was set to zero. The grey zone represents the standard deviation of the negative controls. In panel C, particles formed after coexpression of P1-2A and P3 were separated on a sucrose gradient (5 to 30% sucrose [wt/wt]) and detected in individual fractions by their reactivity with the monoclonal antibody K2-4F2. Sedimentation standards were obtained from a parallel gradient on which HAV particles from infected cells were separated.
In order to test the role of 3C-flanking regions in the context of the complete open reading frame, we compared HAV antigen production and processing of the polyprotein derived from the complete genome with those derived from constructs deleted in 3AB and 3D. For this, P1-P2-P3, P1-P2-3CD, and P1-P2-3ABCDΔ were expressed with the aid of vTF7-3. Compared to P3 alone, the overall production of P3 proteins was lower when the complete HAV genome was expressed. Neither the deletion of 3AB nor that of parts of 3D affected overall polyprotein processing. Instead of 3ABC and 3BC, 2C-3C was found to be a dominant processing product of P1-P2-3CD, and a truncated 3D was produced from P1-P2-3ABCDΔ (data not shown). The relative amount of K2-4F2-reactive antigenicity was about 70% for P1-P2-3ABCDΔ and 30% for P1-P2-3CD compared to that for P1-P2-P3 (Fig. 5B). Probably due to differences in promoter activity, the total amount of antigen production was lower in the cis (Fig. 5B) than in the trans expression experiments (Fig. 5A). These results are in accordance with the data obtained after expression of P1-2A and P3-derived fragments in trans and directly confirm our conclusion that 3A and 3D seem to be required for formation of a processing-assembly complex comprising P3 and P1-2A proteins. It is interesting that various nonstructural proteins (e.g., 2C and 3D) were recently found to be attached to purified particles of foot-and-mouth disease virus and poliovirus (22). As mentioned above, 3A- and 3D-containing polypeptides reside in the noncytosolic cell fraction where P1-2A was also found (data not shown). Thus, 3A and/or 3D within the context of P3 might be necessary to colocalize the proteolytic activity with its substrate. It is also possible that parts of P3 promote 13-S pentamer assembly in a chaperone-like manner before they are further processed into 14-S structures comprising five molecules each of VP0, VP3, and VP1-2A (4). The results presented here clearly show that coexpression of P1-2A and P3 is sufficient and best suited for the efficient production of HAV empty particles for vaccine and diagnostic applications.
ACKNOWLEDGMENTS
We are grateful to B. Moss for vaccinia virus vTF7-3, H. Andres for monoclonal antibody K2-4F2, and Wellcome Inc. for the 3B antiserum.
C.P. was a recipient of a fellowship from the Studienstiftung des Deutschen Volkes. M.J. was supported by a grant of the state of Schleswig-Holstein. The work was supported by the Deutsche Forschungsgemeinschaft (DFG, SFB 367, project B7).
REFERENCES
- 1.Anderson D A, Ross B C. Morphogenesis of hepatitis A virus: isolation and characterization of subviral particles. J Virol. 1990;64:5284–5289. doi: 10.1128/jvi.64.11.5284-5289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of the viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann E M, Mosimann S C, Chernaia M M, Malcolm B A, James M N G. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borovec S V, Anderson A D. Synthesis and assembly of hepatitis A virus-specific proteins in BS-C-1 cells. J Virol. 1993;67:3095–3102. doi: 10.1128/jvi.67.6.3095-3102.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J I, Ticehurst J R, Feinstone S M, Rosenblum B, Purcell R. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987;61:3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elroy-Stein O, Fuerst T R, Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci USA. 1989;86:6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic expression system based on a recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauss-Müller V, Jürgensen D, Deutzmann R. Autoproteolytic cleavage of recombinant 3C proteinase of hepatitis A virus. Virology. 1991;182:861–864. doi: 10.1016/0042-6822(91)90630-t. [DOI] [PubMed] [Google Scholar]
- 9.Gauss-Müller V, Lottspeich F, Deinhardt F. Characterisation of hepatitis A virus structural proteins. Virology. 1986;155:732–736. doi: 10.1016/0042-6822(86)90234-5. [DOI] [PubMed] [Google Scholar]
- 10.Gauss-Müller, V. Unpublished observations.
- 11.Harmon S A, Richards O C, Summers D F, Ehrenfeld E. The 5′-terminal nucleotides of hepatitis A virus RNA, but not poliovirus RNA, are required for infectivity. J Virol. 1991;65:2757–2760. doi: 10.1128/jvi.65.5.2757-2760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmon S A, Updike W, Jia X-Y, Summers D F, Ehrenfeld E. Polyprotein processing in cis and in trans by hepatitis A virus 3C protease cloned and expressed in Escherichia coli. J Virol. 1992;66:5242–5247. doi: 10.1128/jvi.66.9.5242-5247.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia X Y, Ehrenfeld E, Summers D F. Proteolytic activity of hepatitis A virus 3C protein. J Virol. 1991;65:2595–2600. doi: 10.1128/jvi.65.5.2595-2600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jürgensen D, Kusov Y Y, Fäcke M, Kräusslich H G, Gauss-Müller V. Cell free translation and proteolytic processing of the hepatitis A virus polyprotein. J Gen Virol. 1993;74:677–683. doi: 10.1099/0022-1317-74-4-677. [DOI] [PubMed] [Google Scholar]
- 15.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:225–229. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusov Y A, Sommergruber W, Schreiber M, Gauss-Müller V. Intermolecular cleavage of hepatitis A virus (HAV) precursor protein P1-P2 by recombinant HAV proteinase 3C. J Virol. 1992;66:6794–6796. doi: 10.1128/jvi.66.11.6794-6796.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusov Y Y, Morace G, Probst C, Gauss-Müller V. Interaction of hepatitis A virus (HAV) precursor proteins 3AB and 3ABC with the 5′ and 3′ termini of the HAV RNA. Virus Res. 1997;51:151–157. doi: 10.1016/s0168-1702(97)00089-0. [DOI] [PubMed] [Google Scholar]
- 18.Lee C K, Wimmer E. Proteolytic processing of poliovirus polyproteins: elimination of 2Apro-mediated, alternative cleavage of polypeptide 3CD by in vitro mutagenesis. Virology. 1988;166:405–414. doi: 10.1016/0042-6822(88)90511-9. [DOI] [PubMed] [Google Scholar]
- 19.MacGregor A W, Kornitschuk M, Hurrell J G R, Lehmann N L. Monoclonal antibodies against hepatitis A virus. J Clin Microbiol. 1983;18:1237–1243. doi: 10.1128/jcm.18.5.1237-1243.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malcolm B A, Chin S M, Jewell D A, Stratton-Thomas J R, Thudium K B, Ralston R, Rosenberg S. Expression and characterisation of recombinant hepatitis A virus 3C proteinase. Biochemistry. 1992;31:3358–3363. doi: 10.1021/bi00128a008. [DOI] [PubMed] [Google Scholar]
- 21.Martin A, Escriou N, Chao S F, Girgard M, Lemon S M, Wychowski C. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the hepatitis A virus polyprotein: functional impact on the infectivity of HAV RNA transcripts. Virology. 1995;213:213–222. doi: 10.1006/viro.1995.1561. [DOI] [PubMed] [Google Scholar]
- 22.Newman J F E, Brown F. Foot-and-mouth disease virus and poliovirus particles contain proteins of the replication complex. J Virol. 1997;71:7657–7661. doi: 10.1128/jvi.71.10.7657-7662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 24.Ping L-H, Lemon S M. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J Virol. 1992;66:2208–2216. doi: 10.1128/jvi.66.4.2208-2216.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisani G, Beneduce F, Gauss-Müller V, Morace G. Recombinant expression of hepatitis A virus protein 3A: interaction with membranes. Biochem Biophys Res Commun. 1995;211:627–638. doi: 10.1006/bbrc.1995.1859. [DOI] [PubMed] [Google Scholar]
- 26.Probst C, Jecht M, Gauss-Müller V. Proteinase 3C-mediated processing of VP1-2A of two hepatitis A virus strains: in vivo evidence for cleavage at amino acid position 273/274 of VP1. J Virol. 1997;71:3288–3292. doi: 10.1128/jvi.71.4.3288-3292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Probst, C. Unpublished observations.
- 28.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 29.Schultheiß T, Kusov Y Y, Gauss-Müller V. Proteinase 3C of hepatitis A virus (HAV) cleaves the HAV polyprotein P2-P3 at all sites including VP1/2A and 2A/2B. Virology. 1994;198:275–281. doi: 10.1006/viro.1994.1030. [DOI] [PubMed] [Google Scholar]
- 30.Schultheiß T, Sommergruber W, Kusov Y Y, Gauss-Müller V. Cleavage specificity of purified recombinant hepatitis A virus 3C proteinase on natural substrates. J Virol. 1995;69:1727–1733. doi: 10.1128/jvi.69.3.1727-1733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tesar M, Pak I, Jia X Y, Richards O C, Summers D F, Ehrenfeld E. Expression of hepatitis A virus precursor protein P3 in vivo and in vitro: polyprotein processing of the 3CD cleavage site. Virology. 1994;198:524–533. doi: 10.1006/viro.1994.1063. [DOI] [PubMed] [Google Scholar]
- 32.Updike W S, Tesar M, Ehrenfeld E. Detection of hepatitis A virus proteins in infected BS-C-1 cells. Virology. 1991;185:411–418. doi: 10.1016/0042-6822(91)90789-e. [DOI] [PubMed] [Google Scholar]
- 33.Winokur P L, McLinden J H, Stapelton J T. The hepatitis A virus polyprotein expressed by a recombinant vaccinia virus undergoes proteolytic processing and assembly into virus-like particles. J Virol. 1991;65:5029–5036. doi: 10.1128/jvi.65.9.5029-5036.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang W, Cuconati A, Paul A P, Cao W, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 35.Ypma-Wong M F, Dewalt P G, Johnson V H, Lamb J G, Semler B L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- 36.Zhu N L, Li G D, Wang Y. Physiochemical and immunological characterization of hepatitis A virus nucleocapsids expressed in a vaccinia virus/T7/EMCV system. Arch Virol. 1994;135:443–449. doi: 10.1007/BF01310029. [DOI] [PubMed] [Google Scholar]